The nucleolus, the most prominent nuclear structure, is involved in ribosome biogenesis and nuclear and nucleolar chromatin organization. Nucleolins are among the most abundant nucleolar proteins. In contrast with mammals, plants encode two nucleolins. Characterization of Arabidopsis thaliana nuc2 mutants shows the importance of these proteins in chromatin silencing and ribosomal DNA organization.
Abstract
In plants as well as in animals, hundreds to thousands of 45S rRNA gene copies localize in Nucleolus Organizer Regions (NORs), and the activation or repression of specific sets of rDNA depends on epigenetic mechanisms. Previously, we reported that the Arabidopsis thaliana nucleolin protein NUC1, an abundant and evolutionarily conserved nucleolar protein in eukaryotic organisms, is required for maintaining DNA methylation levels and for controlling the expression of specific rDNA variants in Arabidopsis. Interestingly, in contrast with animal or yeast cells, plants contain a second nucleolin gene. Here, we report that Arabidopsis NUC1 and NUC2 nucleolin genes are both required for plant growth and survival and that NUC2 disruption represses flowering. However, these genes seem to be functionally antagonistic. In contrast with NUC1, disruption of NUC2 induces CG hypermethylation of rDNA and NOR association with the nucleolus. Moreover, NUC2 loss of function triggers major changes in rDNA spatial organization, expression, and transgenerational stability. Our analyses indicate that silencing of specific rRNA genes is mostly determined by the active or repressed state of the NORs and that nucleolin proteins play a key role in the developmental control of this process.
INTRODUCTION
In eukaryotes, 45S rDNA (encoding 18S, 5.8S, and 25S rRNA) is arranged in tandem arrays, with copy numbers ranging from hundreds to several thousands in plants. However, only a small proportion of rRNA genes are transcribed. The active fraction forms loops that emanate from knobs of heterochromatin to form the nucleolus (Sáez-Vásquez and Gadal, 2010; Layat et al., 2012; Grummt and Längst, 2013). Although the epigenetic pathways to silence rDNA have largely been studied in Arabidopsis thaliana hybrid plants (Lawrence et al., 2004), the mechanisms that activate and/or repress hundreds of rDNA units have not yet been fully elucidated in a nonhybrid plant. Recent reports reveal that proteins involved in histone and DNA modifications might play major roles in the activation and/or repression of specific subsets of rRNA genes in Arabidopsis plants (Earley et al., 2010; Pontvianne et al., 2012).
Nucleolin is one of the most abundant nucleolar proteins and is expected to play a major role in the multiple functions of the nucleolus. Moreover, it has been implicated in DNA replication, recombination, and repair (Tuteja and Tuteja, 1998), nucleolar localization of telomerase (Khurts et al., 2004), and chromatin remodeling (Angelov et al., 2006; Mongelard and Bouvet, 2007). However, in contrast with animals and yeast, the genomes of different plant species encode at least two nucleolin-like proteins (Supplemental Figure 1). This suggests that duplicated nucleolin genes might play redundant and/or specific roles during plant growth and development or in response to environmental conditions.
In Arabidopsis, two genes encoding nucleolin proteins have been described: NUC1 and NUC2 (formerly named NUC-L1 and NUC-L2). The first is highly and ubiquitously expressed in normal growth conditions (Kojima et al., 2007; Petricka and Nelson, 2007; Pontvianne et al., 2007). Knockout mutations of the gene are not lethal but lead to severe defects in plant growth and development (Petricka and Nelson, 2007; Pontvianne et al., 2007). Moreover, the absence of this protein in nuc1 (formally named nuc-L1) mutant plants induces nucleolar disorganization, Nucleolus Organizer Region (NOR) decondensation, pre-rRNA accumulation (Petricka and Nelson, 2007; Pontvianne et al., 2007), and the derepression of specific 45S rDNA variants (Kojima et al., 2007; Pontvianne et al., 2010). By contrast, little is known about NUC2 and its role in rDNA chromatin organization and expression. This led us to investigate more precisely the biological significance of the second nucleolin gene in Arabidopsis. We report here that Arabidopsis plants require the expression of both NUC1 and NUC2 for survival. NUC1 and NUC2 both bind chromatin and control rDNA variant expression, but they act through different and antagonistic molecular mechanisms. In nuc2 loss-of-function plants, rRNA genes are globally hypermethylated, and the copy number of rDNA variants is affected. Finally, the data suggest that delocalization of silent 45S rDNA from inactive to active NORs is able to induce transcriptional activation of normally repressed rRNA genes.
RESULTS
NUC2 Is a Functional Protein-Coding Gene
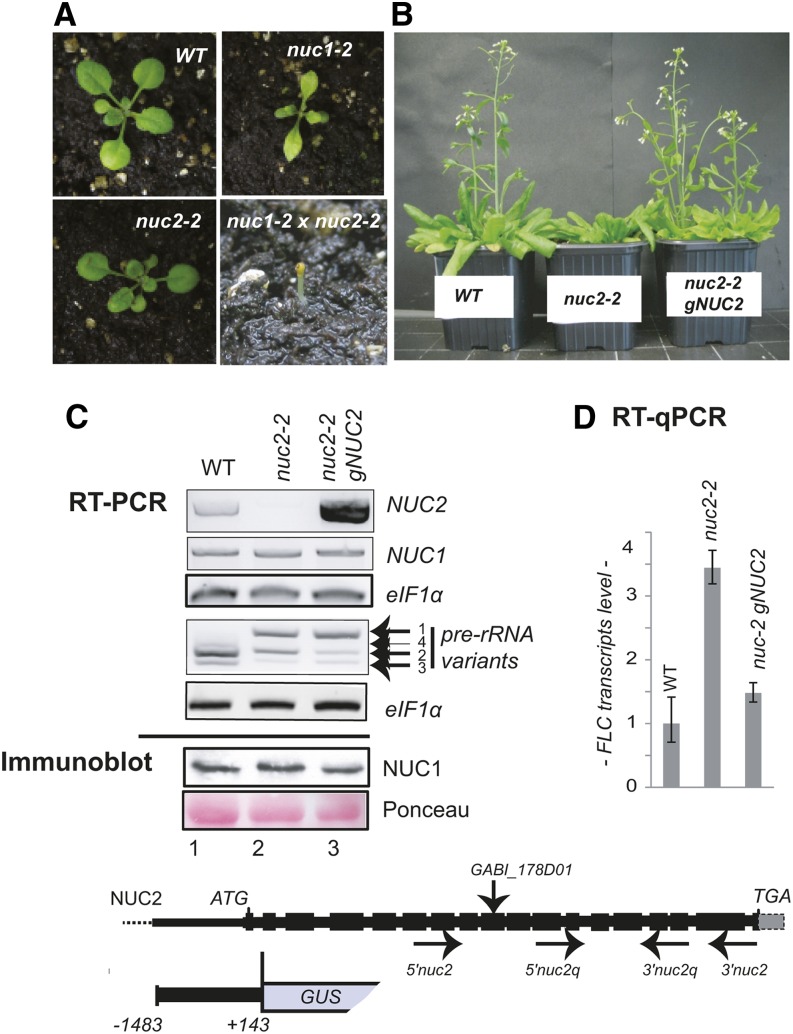
To address the role and biological significance of the second nucleolin gene in plants, we characterized two homozygous nuc2 mutant lines, inducing partial (nuc2-1) or complete (nuc2-2) loss of NUC2 gene expression (Figure 1; Supplemental Figure 2).
Figure 1.
Arabidopsis NUC2 Is a Functional Protein Gene.
(A) Photographs of Arabidopsis wild-type, nuc2-2 (GABI_178D01), nuc1-2 (SALK_002764), and nuc1 nuc2 plants.
(B) Arabidopsis wild-type, nuc2-2, and nuc2-2 gNUC2 plants were grown on soil under a 16-h/8-h light/dark cycle. Late flowering is observed in nuc2-2 mutants compared with wild-type and nuc2-2 gNUC2 plants.
(C) Top panel, RT-PCR using cDNA prepared from 15-d-old wild-type (lane 1), nuc2-2 (lane 2), and nuc2-2 gNUC2 (lane 3) seedlings to detect NUC2, NUC1, and 3′ ETS pre-rRNA transcripts, respectively. Amplification of eIF1α transcripts was performed to verify similar amounts of cDNA in each sample. Bottom panel, immunoblot reaction using α-At-NUC1. The membrane was stained with Ponceau S before hybridization to verify similar amounts of proteins.
(D) RT-qPCR analysis to determine relative levels of FLC transcripts in wild-type, nuc2-2, and nuc2-2 gNUC2 plants. The drawing at bottom shows the Arabidopsis NUC2 gene from the ATG start codon to the TGA stop codon. Black boxes correspond to exons separated by introns. The positions of primers 5′nuc2/3′nuc2 and 5′nuc2q/3′nuc2q used in RT-PCR experiments are shown. A sequence upstream from ATG (nucleotide 1483), the first intron, and the exon from the NUC2 gene fused to GUS are represented. The positions of primers 5′nuc1/3′nuc1 and rRNA 5′3ets/3′3ets are shown in Figure 4 and Supplemental Figure 3. The T-DNA insertion in the nuc2-2 mutant plant GABI_178D01 is indicated.
[See online article for color version of this figure.]
Under long-day conditions (16-h day/8-h night), nuc2-2 mutant seedlings emerge and grow quite similarly to wild-type plants (Figure 1A), while nuc1 plants display growth defects (Kojima et al., 2007; Petricka and Nelson, 2007; Pontvianne et al., 2007, 2010). We further observed that nuc1 nuc2 double mutants are not viable (Figure 1A). Indeed, homozygous nuc1 and nuc2-2 double mutations are seedling lethal, indicating that NUC2 is a functional gene and that both nucleolin genes are required for plant survival. Similar results were obtained when the allele nuc1-3 was introduced into nuc2-1 mutant plants. We also observed that nuc1 mutant plants heterozygous for NUC2 are viable but display stronger growth and developmental defects than homozygous nuc1 or nuc2 single mutant plants (Supplemental Figure 3).
Interestingly, nuc2-2 mutants flower later than wild-type plants (Figure 1B). After 53 d of growth, the wild type displays ∼17 flowers per plant, in contrast with ∼1 flower per plant for nuc2-2. The number of rosette leaves at the time of bolting is ∼10 and ∼20 in wild-type and nuc2-2 plants, respectively. We do not observe any obvious floral and/or silique phenotype in nuc2-2 plants. Late flowering is related to the disruption of the NUC2 gene, since complementation of nuc2-2 plants with the Arabidopsis NUC2 genomic sequence restores wild-type flowering time (Figures 1B and 1C). Moreover, quantitative RT-PCR (RT-qPCR) analyses reveal a higher accumulation of FLOWERING LOCUS C (FLC) transcripts in nuc2-2 than in wild-type plants (Figure 1D), which is restored in nuc2-2 plants carrying the gNUC2 transgene. These observations are further supported by RNA sequencing analysis showing that FLC is upregulated in the nuc2-2 plants. No other genes related to flowering timing or development were detected among the 33 upregulated or 17 downregulated genes in nuc2-2 compared with wild-type plants (Supplemental Figure 4).
The Arabidopsis genome encodes at least four variants of 45S rDNA. While rDNA VAR1 copies are globally transcriptionally inactive, rDNA VAR2, VAR3, and VAR4 units are permissive to transcription in adult plants (Pontvianne et al., 2010; Abou-Ellail et al., 2011). Disruption of the NUC1 gene induces the derepression of rDNA VAR1 copies (Pontvianne et al., 2010). Interestingly, strong accumulation of pre-rRNA VAR1 and a decrease of pre-rRNA VAR2 and VAR3 levels are observed in nuc2-2 mutant plants (Figure 1C). Wild-type profiles of VAR1 and VAR2 pre-rRNA are not restored to the wild-type level in the nuc2-2 plants complemented with the gNUC2 construct. By contrast, we do not detect major differences between the level of NUC1 transcripts or protein in nuc2-2 or nuc2-2 gNUC2 plants compared with wild-type plants.
Thus, the results described above show that NUC2 is a functional gene that influences flowering time and rRNA gene expression, two phenotypes that appear to be uncoupled.
NUC2 Promoter Activity in Specific Plant Tissues and Organs
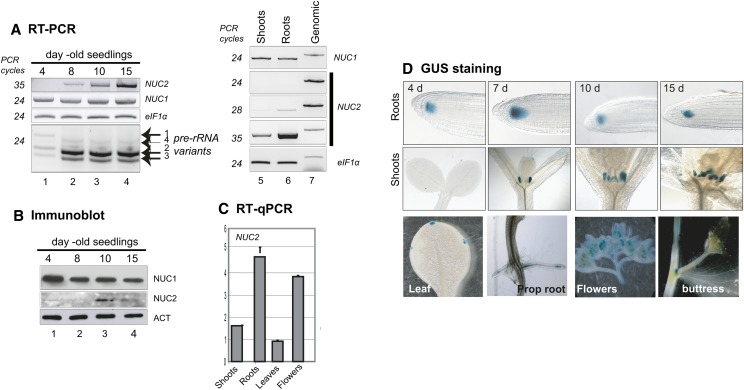
Database analyses using Genevestigator (www.genevestigator.com/gv/) and BAR (www.bar.utoronto.ca/welcome.htm) allow the identification of major fluctuations in the levels of NUC2 transcripts among different plant tissues, organs, developmental stages, and/or environmental conditions (Figure 2; Supplemental Figure 5). RT-PCR analyses confirm that NUC2 transcripts are detectable 8 d after germination and then accumulate at 10 and 15 d. A peak of NUC2 protein accumulation is also detected at 10 d. Interestingly, a concomitant decrease of the pre-rRNA VAR1 level is observed 8 d after germination. By contrast, the levels of NUC1 transcript and protein remain similar from 4 to 15 d after germination and accumulate at similar levels in both roots and shoots (Figures 2A and 2B). In adult plants, we observed that the NUC2 transcript level is higher in roots and flowers than in shoots and leaves (Figures 2A and 2C).
Figure 2.
NUC2 Gene Expression during Seedling Development and in Plant Organs.
(A) RT-PCR to show NUC2 and NUC1 transcripts during seed germination (lanes 1 to 4), in shoots (lane 5), and in roots (lane 6). Absence of genomic contamination in the cDNA samples was verified by amplification of NUC1, NUC2, and eIF1α genomic DNA, which generates higher molecular size bands (lane 7). The number of cycles (24, 28, and 35) of each PCR is indicated.
(B) Immunoblot experiment to determine the levels of NUC1 and NUC2 proteins during seed germination. α-Actin (ACT) was used to verify similar amounts of proteins.
(C) RT-qPCR to determine the relative levels of NUC2 transcripts in shoots, roots, leaves, and flowers.
(D) Analysis of NUC2 promoter activity in Arabidopsis plants transformed with the pAtNUC2:GUS construct. Top and middle rows, Arabidopsis seeds were sown on MS medium and maintained for 48 h at 4°C (day 0) and then transferred to room culture, at 22°C under continuous light conditions, for 4, 7, 10, and 15 d. Bottom row, GUS staining in leaf hydathode cells, roots, flowers, and buttress. The NUC2 gene fused to GUS is represented in Figure 1.
To gain insight into the developmental control of NUC2 gene expression, we further characterized NUC2 promoter activity using plants bearing pNUC2:GUS (for β‑glucuronidase) transcriptional fusions. In the root apical meristem, GUS staining is detected at 4, 7, 10, and 15 d after germination (Figure 2D, top row). By contrast, staining is detected in stipules only after 7 d of germination and then at 10 and 15 d (Figure 2D, middle row), in agreement with our RT-PCR analyses. In adult plants, expression is detected specifically in leaf hydathode cells, lateral roots, flowers, and the buttress (Figure 2D, bottom row). On the other hand, NUC2 promoter activity is induced in response to auxin treatment and during silique development, earlier in developing anthers than in developing pistils (Supplemental Figures 6 and 7).
Accordingly, these results show that NUC2 is developmentally controlled in most plant tissues and organs analyzed. Importantly, NUC2 expression and protein abundance increase during the establishment of rDNA VAR1 repression throughout seedling development.
NUC1 Protein Might Control NUC2 Expression
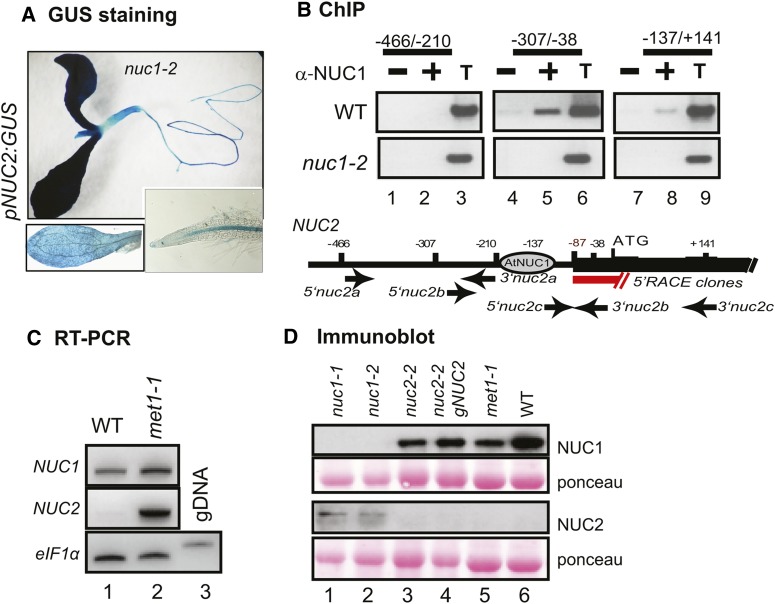
Disruption of NUC1 induces NUC2 overexpression (Pontvianne et al., 2007, 2010). To determine if NUC1 might control NUC2 expression at the transcriptional level, we transformed nuc1 mutant plants with the pNUC2:GUS reporter gene construct. As shown in Figure 3A, the nuc1 mutation considerably strengthens the pNUC2:GUS signal and extends NUC2 promoter activity to the entire seedling (compare with Figure 2D, staining in the stipules and roots). In fact, the pNUC2:GUS signal in the nuc1 background is similar to the pNUC1:GUS signal in wild-type plants (Petricka and Nelson, 2007; Pontvianne et al., 2007). Next, the possibility that NUC1 protein physically interacts with chromatin at the NUC2 locus was tested by chromatin immunoprecipitation (ChIP) using an anti-NUC1 serum (Figure 3B). In wild-type chromatin, NUC2 sequences were detected with primer pairs corresponding to the 5′ region (lanes 5 and 8) but not with primers matching the upstream intergenic region (lane 2). The 5′ end of NUC2 transcripts maps to the position −87 nucleotides upstream from the ATG translation initiation codon; therefore, NUC1 likely binds the NUC2 promoter domain rather than its 5′ untranslated region domain. No significant signals were detected in PCR using chromatin incubated with protein A alone (lanes 1, 4, and 7) or using chromatin extracted from nuc1-2 seedlings and incubated with antibodies against NUC1 (lanes 1, 2, 4, 5, 7, and 8). These in vivo analyses are corroborated by in vitro electrophoretic mobility shift assay (EMSA) showing that NUC1 recombinant protein can bind different regions of NUC2 promoter DNA indiscriminately. These observations suggest that the specificity of NUC1 for region −307 to −38 might be conferred by the NUC2 promoter chromatin structure or by other associated proteins in planta (Supplemental Figure 8A).
Figure 3.
NUC1 Participates in the Regulation of NUC2 Gene Expression.
(A) Analysis of NUC2 promoter activity in nuc1 mutant plants transformed with the pNUC2:GUS construct. The bottom panels show shorter staining to better visualize GUS activity in leaves and roots.
(B) Chromatin from wild-type plants was isolated and incubated either with protein A only (lanes 1, 4, and 7) or with antibodies against NUC1 (lanes 2, 5, and 8). Immunoprecipitated DNA was analyzed by PCR to detect NUC2 sequences from −466 to −210 (lanes 1 to 3), from −307 to −38 (lanes 4 to 6), and from −137 to +141 (lanes 7 to 9). The positions of primers used to amplify NUC2 sequences are indicated. The transcription initiation site is mapped by 5′ rapid amplification of cDNA ends at 87 nucleotides upstream from ATG. An aliquot of total chromatin sample before the immunoprecipitation reaction was used to control PCR amplifications (lanes 3, 6, and 9). Chromatin isolated from the nuc1 mutant was used to control the specificity of At-NUC1 antibodies in the ChIP reaction (nuc1-2 panels).
(C) RT-PCR to show NUC2 and NUC1 transcripts in wild-type (lane 1) and met1-1 (lane 2) plants. The amount of cDNA was verified in each reaction by controlling eIF1α transcript levels. Absence of genomic contamination in the cDNA samples was verified by amplification of eIF1α genomic DNA, which generates higher molecular size bands (lane 3).
(D) Immunoblot analysis to detect NUC1 and NUC2 proteins in nuc1 (lanes 1 and 2), nuc2 (lane 3), nuc2-2 gNUC2 (lane 4), met1-1 (lane 5), and wild-type (lane 6) plants. Similar amounts of protein in each membrane were verified by Ponceau S staining.
In standard growth conditions, a methylation island is detected at the 3′ sequence of the NUC2 gene (http://signal.salk.edu/cgi-bin/methylome). However, no changes of CG methylation are observed in this region or in the promoter sequence in nuc1 mutants (Supplemental Figure 8B). Nevertheless, NUC2 is also overexpressed in met1 mutant plants that exhibit a massive loss of DNA methylation at the 3′ sequence (Figure 3C). Therefore, local changes in DNA methylation levels cannot explain NUC2 overexpression in nuc1 plants. In addition, analyses of mutants for the RNA-directed DNA methylation pathway and histone deacetylases/acetylases do not reveal changes in the expression of NUC2 (Supplemental Figure 8C).
To test if NUC1 might also influence NUC2 expression at the posttranscriptional level, we measured NUC2 protein levels in 15-d-old wild-type and mutant plants (Figure 3D). At this stage, NUC2 protein is detected in the two nuc1 mutant lines tested (lanes 1 and 2) but not in wild-type (lane 6), nuc2-2 (lane 3), nuc2-2 gNUC2 (lane 4), or met1 (lane 5) plants. Furthermore, overexpression of the NUC2 gene, under the control of the NUC1 promoter, does not lead to the accumulation of NUC2 protein in wild-type plants (data not shown).
Together, these results indicate that NUC1 is required to inhibit NUC2 gene expression at the transcriptional level and may also influence the accumulation of NUC2 protein.
NUC2 Gene Disruption Affects rDNA Chromatin Organization
Because NUC2 gene disruption increases VAR1 rDNA expression, we further questioned whether (1) RNA pol I transcription and/or (2) rDNA chromatin states are affected in nuc2-2 mutant plants. In nuc1 and histone deacetylase6 (hda6) mutant plants, the expression of pre-rRNA VAR1 is associated with rRNA transcription initiated from an intergenic spacer and with rDNA chromatin decondensation (Earley et al., 2010; Pontvianne et al., 2010).
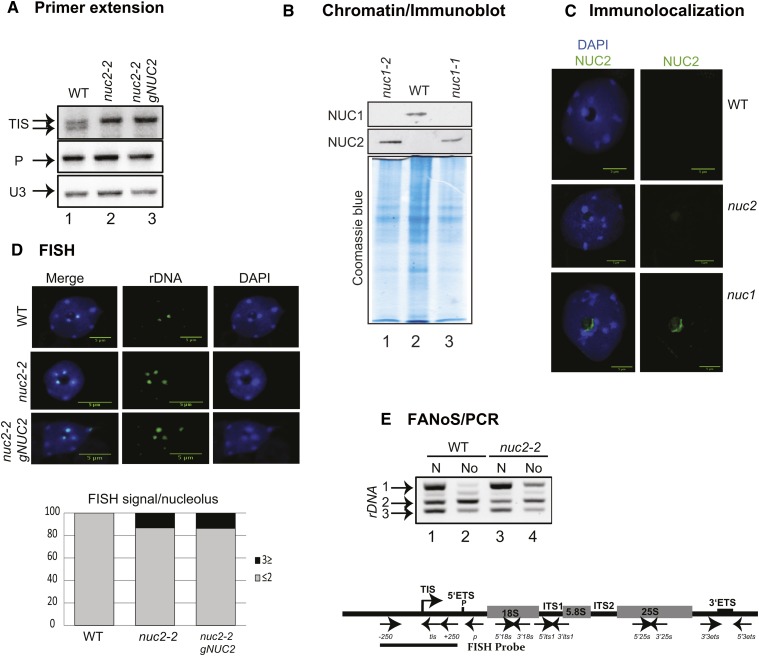
Primer extension analysis (Figure 4A) shows that a single transcription initiation signal (TIS) is detected in nuc2-2 and nuc2-2 gNUC2 samples (lanes 2 and 3), in contrast with the double TIS observed in wild-type plants (lane 1). Nevertheless, quantification of the ratio between the TIS corresponding to primary pre-rRNA and the P signal corresponding to short-lived 5′ end–cleaved pre-rRNA does not reveal significant changes in the amount of primary or processed rRNA precursors in nuc2-2 and nuc2-2 gNUC2 compared with wild-type samples.
Figure 4.
Transcription and rDNA Organization in nuc2-2 Plants.
(A) Primer extension was performed using total RNA extracted from wild-type (lane 1), nuc2-2 (lane 2), and nuc2-2 gNUC2 (lane 3) plants. Primer tis detects transcription from the TIS, and primer p detects pre-rRNA precursors cleaved at the primary cleavage site (P) (Sáez-Vasquez et al., 2004; Pontvianne et al., 2007). Primer extension using primer U3 that accurately maps the 5′ end of U3snoRNA was used to control similar amounts of total RNA in each reaction.
(B) Immunoblot analysis to detect NUC1 and NUC2 protein in nuc1 (lanes 1 and 3) and wild-type (lane 2) chromatin extracts. The Coomassie blue gel staining reveals similar amounts of protein in each chromatin sample.
(C) Immunolocalization of NUC2 in leaves from nuc1 plants. The NUC2 signal (green) colocalizes with chromatin situated in the periphery of the nucleolus and counterstained with DAPI (blue) (nuc1 panels). No NUC2 signal is detected in wild-type or nuc2 plants (used here to control the specificity of the antibodies).
(D) FISH analysis with an rDNA probe containing sequences upstream and downstream from TIS (from −250 to +250) reveals the 45S rDNA loci (green) on nuclei from wild-type, nuc2-2, and nuc2-2 gNUC2 plants. Counterstaining with DAPI (blue) is shown (FISH and DAPI). The bar graphs at bottom show the percentage of rDNA loci associated with the nucleolus in wild-type, nuc2-2, and nuc2-2 gNUC2 plants. Bars = 5 μm.
(E) PCR detection of rRNA gene variants in DNA of purified nuclei (N) or nucleoli (No) of wild-type (lanes 1 and 2) and nuc2-2 (lanes 3 and 4) plants. The positions of 5′3ets and 3′3ets primers used in PCR experiments are shown. The scheme at bottom represents the 45S rDNA unit containing the ETS (5′ ETS and 3′ ETS) and the structural rRNA sequences (18S, 5.8S, and 25S rRNA in gray boxes) separated by internal transcribed spacers (ITS1 and ITS2). Four repeat sequences located in the 3′ ETS are represented (R1 to R4). The TIS and the primary cleavage site (P) in the 5′ ETS are indicated. The positions of primers used to amplify or detect rRNA genes and/or pre-rRNA sequences are shown.
To assess whether NUC2 might influence the expression and subnuclear localization of rDNA, we first tested whether NUC2 associates with a chromatin fraction. Chromatin samples were notably purified from nuc1 mutant plants, in which NUC2 protein overaccumulates, and analyzed by immunoblot. NUC2 protein was detected in nuc1 but not in wild-type chromatin samples (Figure 4B). Similarly, the accumulation of NUC1 protein was detected in wild-type but not in nuc1 mutant extracts. Based on these results, NUC2 subnuclear localization was analyzed in more detail by immunofluorescence on fixed cells. This reveals a discrete localization in the nucleolus near large heterochromatin regions corresponding to heavily 4′,6-diamidino-2-phenylindole (DAPI)-stained DNA in the nuc1 mutant background (Figure 4C; Supplemental Figure 9C). Interestingly, therefore, NUC2 is mainly concentrated in nucleolar regions physically associated with condensed chromatin regions from which VAR1 rDNA has recently been shown to emanate (Pontvianne et al., 2013). Furthermore, the nucleolar localization of NUC1 protein is not affected in nuc2-2 plants (Supplemental Figure 10).
DNA fluorescence in situ hybridization (FISH) analysis of leaf cells shows that up to four rDNA signals are detected in interphase nuclei of wild-type plants (Figure 4D; Supplemental Table 1). These signals correspond to two each of NOR2 and NOR4 (in chromosomes 2 and 4, respectively), as expected in a diploid Arabidopsis cell (Pecinka et al., 2004). Interestingly, ∼18% of nucleoli from nuc2-2 and nuc2-2 gNUC2 cells associate with three or more rDNA signals compared with one or two rDNA signals in wild-type nucleoli. By contrast, no significant differences can be detected in the number or size of the centromeres between the DAPI-labeled wild-type and nuc2-2 and/or nuc2-2 gNUC2 plants.
Finally, to obtain more insight into how disruption of NUC2 induces the transcriptional activation of repressed rDNA VAR1, we purified nucleoli from nuc2-2 mutant plants by fluorescence-assisted nucleolus sorting (FANoS) (Pontvianne et al., 2013). The four rDNA variants are detected in the nuclear fraction from wild-type and nuc2-2 plants (Figure 4E, lanes 1 and 3). By contrast, in the nucleolar fraction, VAR1 rDNA is enriched in nuc2-2 samples (lane 4), while only a faint band is detected in wild-type plants (lane 2). The ratio between rDNA VAR2 and VAR3 signals remains similar in nuc2-2 (lane 4) compared with the wild-type sample (lane 2).
Altogether, these data show that in the nuc2-2 mutant, rDNA subnuclear localization is altered; rDNA VAR1 is localized and expressed in the nucleolus. These results suggest that some modifications and/or rearrangements of rDNA chromatin occur in nuc2-2 mutant plants as NUC2 binds chromatin.
NUC2 Is Required for the Stability of rDNA Variants Copy Number
Derepression of VAR1 rRNA genes in nuc2-2 mutant plants could be explained by the observed reorganization of rDNA from inactive to active NOR chromatin (Jakociunas et al., 2013) but also by possible rDNA copy number variation. This phenomenon has previously been observed in fasciata (fas) mutant plants (Figure 5A), a subunit of the H3/H4 histone chaperone Chromatin Associated Factor1 complex (Mozgová et al., 2010).
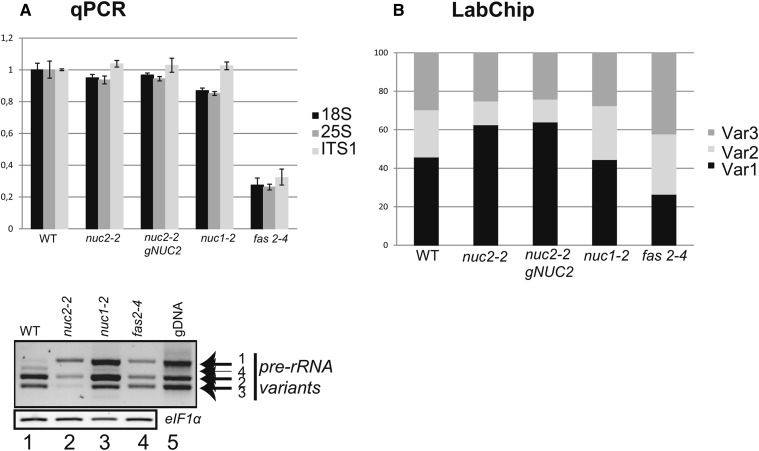
Figure 5.
The Relative Abundance of rDNA Variants Is Affected in Plants with a Disrupted NUC2.
(A) Top, quantitative PCR analysis to amplify 18S, 25S, and ITS1 rDNA sequences from wild-type and mutant (nuc2-2, nuc2-2 gNUC2, nuc1-2, and fas2-4) plants. The bar graphs show relative amounts of 18S (black), 25S (dark gray), and ITS1 (gray) rDNA. Bottom, RT-PCR using cDNA prepared from 15-d-old wild-type (lane 1), nuc2-2 (lane 2), nuc1-2 (lane 3), and fas2-4 (lane 4) seedlings to detect 3′ ETS pre-rRNA transcripts. Amplification of eIF1α was performed to control similar amounts of cDNA in each sample. PCR on genomic DNA from wild-type plants detects the rDNA variants 1 to 4 (lane 5).
(B) PCR amplifications of 3′ ETS sequences using genomic DNA from wild-type, nuc2-2, nuc2-2 gNUC2, nuc1-2, and fas2-4 plants. Relative amounts of each rDNA variant were determined using the LabChip GX system. The bar graphs show the percentage of rDNA VAR1 (black), VAR2 (gray), and VAR3 (dark gray).
We first quantified 18S, 25S, and ITS1 rDNA sequences in wild-type, nuc2-2, and nuc2-2 gNUC2 plants and observed no major variation (Figure 5A, top panel). By contrast, a global decrease of 45S rDNA copy number was detected in fas2-4 mutant plants, as reported previously (Mozgová et al., 2010). Therefore, we further determined the amount of specific rDNA variants. In this analysis, VAR1, VAR2, and VAR3 rDNA copies represent ∼46, 25, and 29%, respectively, of the rRNA genes in wild-type plants. In nuc2-2 mutants, these numbers showed a relative increase of the VAR1 copy number (∼63, 12, and 25%, respectively) (Figure 5B; Supplemental Figure 11). In nuc2-2 gNUC2 plants, the proportions are similar to the ratio observed in nuc2-2 mutant plants, indicating that reintroducing the NUC2 function did not restore normal rDNA variant copy numbers. In addition, analysis of subsequent nuc2-2 and nuc2-2 gNUC2 generations after five selfings in single-seed descent reveals no further changes in the abundance and expression of rDNA variants (data not shown). Finally, no significant changes are observed in nuc1-2, suggesting a specific effect of the loss of NUC2 function on the stability of the VAR1 copy number.
rDNA Methylation State in nuc2-2 Mutants
The expression of rRNA VAR1 was recently shown to be associated with CG hypomethylation of the 5′ external transcribed spacer (ETS) region in nuc1 and hda6 mutant plants (Earley et al., 2010; Pontvianne et al., 2010). The methylation state of 5′ ETS rDNA (from −315 to +243), therefore, was tested by bisulfite sequencing analysis in nuc2-2. This reveals a hypermethylated 5′ ETS region in nuc2-2 mutant lines (∼35%) compared with wild-type plants (∼22%), which was restored to the wild-type level in nuc2-2 gNUC2 samples. Remarkably, the increased methylation level is restricted to the CG context in the 5′ ETS region and is not observed in the upstream promoter domain. CHG and CHH methylation are not affected in nuc2 plants (Figure 6A).
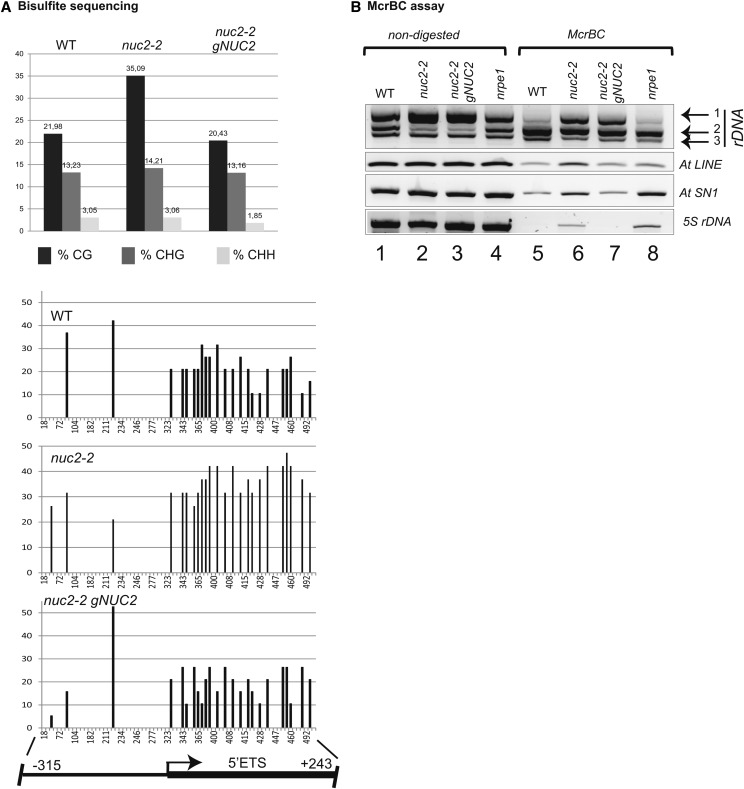
Figure 6.
rDNA Is Hypermethylated in nuc2-2 Mutant Plants.
(A) Bisulfite sequencing analysis. Top, the bar graphs show the percentage of methylated sites in the rRNA gene sequences (from −315 to +243) from wild-type, nuc2-2, and nuc2-2 gNUC2 plants in CG, CHG, and CHH contexts. Bottom, frequencies at which individual cytosines in a CG context are methylated between −315 and +243 relative to TIS. The scheme shows the positions of rDNA sequences upstream (promoter) and downstream (5′ ETS) of TIS. Most of the CG sites localize in the 5′ ETS region.
(B) Genomic DNA from wild-type, nuc2-2, nuc2-2 gNUC2, and nrpe1 plants digested (lanes 5 to 8) or not (lanes 1 to 4) with McrBC. Amplifications of 3′ ETS, At LINE, At SN1, and 5S were performed to detect methylated or unmethylated DNA sequences in nuc2-2, nuc2-2 gNUC2, and control (wild-type and nrpe1) plants.
Because the 5′ ETS sequence is shared by all four rDNA variants, bisulfite sequencing does not allow us to determine which rDNA variants are hypermethylated in nuc2 mutants. Furthermore, bisulfite sequencing of the 3′ ETS could not be used because of the higher number of VAR1 copies (Supplemental Figure 12). Consequently, we decided to perform modified cytosine restriction BC (McrBC) PCR restriction analysis, which consists of specifically cleaving methylated DNA and amplifying specific nonmethylated and noncleaved DNA by PCR (Figure 6B; Supplemental Figure 13). This analysis reveals that VAR1 rDNA is highly amplified in both nuc2-2 and nuc2-2 gNUC2 samples (lanes 6 and 7) as compared with wild-type plants (lane 5). This might either result from a global decrease of DNA methylation level on VAR1 rDNA copies or a higher number of unmethylated VAR1 rDNA copies in these plants. By contrast, similar amounts of nonmethylated rDNA VAR2 and VAR3 are detected (lanes 5 to 8). As observed before, a significant reduction of rDNA VAR2 in nuc2-2 and nuc2-2 gNUC2 plants is detected in undigested DNA samples (lanes 2 and 3) compared with control plants (lanes 1 and 4). Only a faint rDNA VAR1 amplification band is detected in wild-type and nrpe1 plants upon McrBC digestion, revealing that most VAR1 rDNA is methylated in these samples (lanes 5 and 8) (Wierzbicki et al., 2008). Note that At LINE and At SINE repeats, as well as 5S rRNA genes, are hypomethylated in nuc2-2 (lane 6) as in nrpe1 control plants (lane 8).
Altogether, NUC2 disruption results in contrasting variations of rDNA methylation at different loci and on different variants, with a significant decrease of methylated DNA levels on 3′ ETS VAR1-specific regions.
rDNA Organization and Expression in Wild-Type × nuc2 Plants
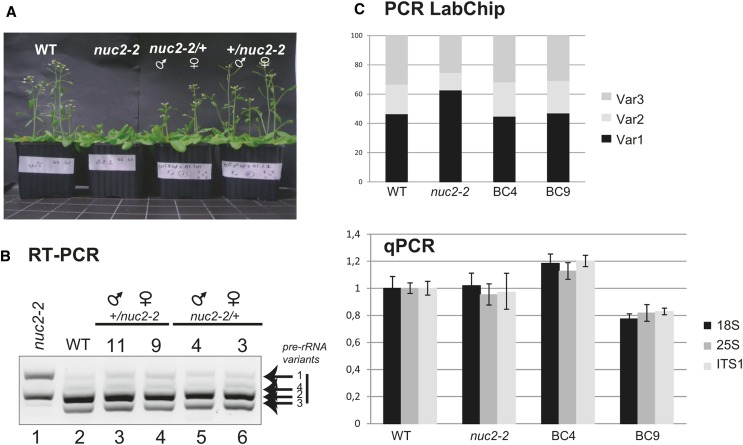
VAR1 rDNA is derepressed in nuc2 mutant plants, a phenomenon associated with a relative increase of VAR1 copy number in the genome of these plants. Therefore, we determined whether or not the introduction of wild-type rDNA copies could restore normal expression patterns. This was achieved by reciprocally crossing wild-type plants with nuc2-2 plants (Figure 7). We first observed that F1 plants from backcrossed nuc2 × wild-type (BC4 and BC3) plants and wild-type × nuc2 (BC11 and BC9) plants display wild-type flowering timing (Figure 7A). Moreover, VAR1 pre-rRNA is repressed, while VAR2 and VAR3 expression is restored to wild-type levels in all the backcrossed lines (Figure 7B). The abundance of rDNA VAR1, VAR2, and VAR3 is basically restored to wild-type levels in the backcrossed nuc2/+ (45, 24, and 31%) and/or +/nuc2 (47, 23, and 30%) plants (Figure 7C, top panel; Supplemental Figure 14). Interestingly, compared with wild-type and nuc2-2 plants, the amount of rDNA is higher in +/nuc2 (BC9) and lower in nuc2/+ (BC4) (Figure 7C, bottom panel).
Figure 7.
Reintegration of Wild-Type rDNA Restores the Abundance of rDNA Variants and the Repression of VAR1 in nuc2-2.
(A) Flowering time analysis of wild-type, nuc2-2, and backcrossed plants using pollen from nuc2-2 (nuc2 × wild type) or from wild-type (wild type × nuc2) plants.
(B) RT-PCR using cDNA prepared from 15-d-old nuc2-2 (lane 1), wild-type (lane 2), +/nuc2 (lanes 3 and 4), and nuc2/+ (lanes 5 and 6) seedlings to detect 3′ ETS pre-rRNA transcripts.
(C) The bar graphs show relative amounts of rDNA VAR1 (black), VAR2 (gray), and VAR3 (dark gray) (top) and 18S (black), 25S (gray), and ITS1 (dark gray) (bottom) rDNA sequences in the wild type, nuc2-2, nuc2/+ (BC4), and +/nuc2 (BC9).
[See online article for color version of this figure.]
These results suggest that the expression of rDNA VAR1 in nuc2-2 and in nuc2-2 gNUC2 plants is intimately linked to changes in the VAR1:VAR2 copies ratio.
NUC1 but Not NUC2 Can Assist Nucleosome Remodeling in Vitro
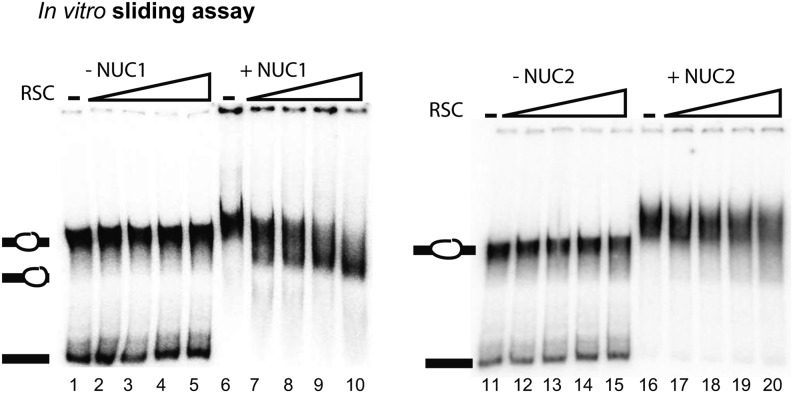
To obtain insight into how the interaction of nucleolin proteins with chromatin might control rDNA organization and/or expression, we studied the nucleosome remodeling of NUC1 and NUC2 in vitro. Mammalian nucleolin has been reported as being able to bind nucleosomes and assist chromatin-remodeling activities in vitro (Angelov et al., 2006; Rickards et al., 2007). To test these potential “activities” of Arabidopsis NUC1 and NUC2, we used an in vitro nucleosome-sliding assay consisting of nucleosomes assembled in vitro and the remodeling activity of chromatin structure remodeling (RSC) (Angelov et al., 2006).
In vitro–reconstituted Xenopus laevis nucleosomes were incubated with increasing amounts of RSC, resulting in an efficient mobilization of the nucleosomes and the formation of end-positioned nucleosomes (Supplemental Figure 9A). To determine mobilization by NUC1 and NUC2, we used suboptimal RSC concentrations at which RSC was unable to remodel nucleosomes alone (Figure 8). Nucleosomes were further incubated with increasing amounts of RSC in the absence or presence of 300 ng of NUC1 (lanes 1 to 10) or NUC2 recombinant proteins (lanes 11 to 20). As expected, no detectable nucleosome sliding was observed in the absence of NUC1 (lanes 2 to 5) or NUC2 (lanes 12 to 15). By contrast, the presence of NUC1 in the remodeling reaction results in an efficient mobilization of central nucleosomes to end-positioned nucleosomes (lanes 7 to 10). No sliding was observed in the presence of NUC2 (lanes 17 to 20). Incubation of NUC1 or NUC2 proteins with nucleosomes without RSC induces a slight shift (lanes 6 and 16). Note that no free probe is detected in fractions containing nucleolin, presumably due to the strong DNA binding activity of NUC1 and NUC2 proteins (Supplemental Figure 9B). Altogether, these analyses indicate that NUC1 and NUC2 can bind nucleosomes but that only NUC1 can assist nucleosome remodeling in vitro.
Figure 8.
NUC1 and NUC2 Binding and Remodeling of Nucleosomes.
Centrally positioned nucleosomes were incubated with increasing (0.2, 0.4, 0.8, and 1.6 μL) amounts of RSC in the absence (lanes 2 to 5 and 12 to 15) or presence (lanes 7 to 10 and 17 to 20) of 300 ng of recombinant His-NUC1 or His-NUC2. Lanes 1 and 11 show nucleosomes alone, and lanes 6 and 16 show nucleosomes with 300 ng of His-NUC1 and His-NUC2, respectively. The migrations of centrally positioned nucleosomes, mobilized nucleosomes, and the 32P radioactively end-labeled DNA are indicated on each EMSA gel.
DISCUSSION
Arabidopsis NUC2 Is a Functional Protein Gene
The genome of Arabidopsis, like most plant genomes, contains two nucleolin protein genes: NUC1 and NUC2 (Supplemental Figure 1). Previously, Arabidopsis NUC1 was demonstrated to be a plant functional homolog of animal and yeast nucleolins (Kojima et al., 2007; Petricka and Nelson, 2007; Pontvianne et al., 2007, 2010). These studies led us to raise the question about the physiological significance of the second nucleolin gene in plants.
Arabidopsis NUC2 knockout plants are viable and do not exhibit a significant growth phenotype, but they have a late flowering phenotype. This suggests that NUC2 is not an essential gene or that NUC1 might rescue the function of NUC2 in nuc2 mutants. Similarly, NUC2 at least partially rescues the loss of NUC1 function in nuc1 plants (Pontvianne et al., 2007). Accordingly, simultaneous disruption of NUC1 and NUC2 is lethal, indicating that NUC2 can fulfill some essential functions of NUC1. Late flowering in nuc2-2 mutant plants associates with the overexpression of FLC, a gene that plays a central role in flowering time regulation in Arabidopsis (Crevillén and Dean, 2011). Remarkably, NUC2 disruption affects the expression of only a small subset of protein-coding and non-protein-coding genes besides FLC. No genes related to 45S pre-rRNA transcription or processing are upregulated or downregulated in nuc2 plants, suggesting that most if not all rDNA phenotypes observed in nuc2 plants are essentially linked to the absence of NUC2 (Figures 1 and 4 to 7; Supplemental Figures 4, 11, 13, and 14).
Nucleolin proteins are histone chaperones (Angelov et al., 2006), and, as expected, NUC1 and NUC2 are able to bind nucleosomes in vitro and are found in chromatin fractions in vivo (Figures 4 and 8). The involvement of histone chaperone activities in flowering time has already been reported in plants with disrupted facilitate chromatin transcription complex (FACT) H2A-H2B histone chaperone activity genes (Lolas et al., 2010; Van Lijsebettens and Grasser, 2010). However, in contrast with the Arabidopsis NUC2 gene, the absence of the SSRP1 and SPT16 FACT subunits leads to reduced expression of FLC and to early flowering. FACT and nucleolin might have different substrate specificities and act at different genomic positions; therefore, it remains to be determined whether their activities act directly on the FLC gene.
NUC2 expression is restricted to some plant tissues and organs and is developmentally controlled during early seedling establishment. NUC2 expression is also significantly lower than NUC1 expression. Interestingly, NUC2 promoter activity is antagonistic to NUC1 promoter activity (Figure 2; Supplemental Figures 5 to 7). The higher expression of NUC2 could be a consequence of the absence or modification of NUC1 protein. Increased expression seems to be at the transcriptional, and possibly also the posttranscriptional, level (Figure 3; Supplemental Figure 8). NUC1 may play a direct role in regulating NUC2 gene expression or, indirectly, by controlling the expression of an unknown repressor. In agreement with this, NUC1 has also been implicated in the expression of RIBOSOMAL PROTEIN genes transcribed by RNA pol II (Kojima et al., 2007). Moreover, transcriptional and posttranscriptional regulation of pol II–transcribed genes by nucleolin proteins has been widely reported in animal cells (Mongelard and Bouvet, 2007; Tajrishi et al., 2011).
Disruption of NUC2 Affects rDNA Genomic Organization and Gene Silencing
The expression of 45S rDNA VAR1 in nuc2-2 mutants is a surprising observation, considering the hypermethylated state of 5′ ETS rDNA in nuc2-2 plants (Figure 6), which contrasts with the hypomethylation and chromatin decondensation of rDNA previously observed in nuc1 plants (Pontvianne et al., 2007, 2010). Nevertheless, how nucleolin affects DNA methylation remains unclear. Moreover, in contrast with nuc1 plants, in which transcription from the TIS is not greatly affected, we observed that most of the detected transcripts initiate from a single nucleotide position in nuc2-2 plants (Figure 4). In wild-type and nuc1 mutant plants, one signal might correspond to the transcription of rDNA VAR2 and another to VAR3. The unique TIS in nuc2-2 could result from decreased levels of VAR3 pre-rRNA, assuming that the TIS of VAR1 pre-rRNA is similar to that of VAR2.
Quantitative analysis of 45S rDNA variants reveals no major changes in the amount of total rDNA in nuc2-2 compared with wild-type plants. However, we observed a significant increase of VAR1 and decrease of VAR2 rDNA copy numbers in these plants (Figure 5). Similarly, plants expressing a low level of NUC2 transcripts (nuc2-1 mutant plants) display changes in the abundance of variants, but in these plants the amount of rRNA genes is increased (Supplemental Figure 2). This is in contrast with fas mutants, which display a reduction of rDNA copies and activation of VAR1, or nuc1 mutants, where the number and abundance of rDNA variants are not affected (Figure 5) (Mozgová et al., 2010; Pontvianne et al., 2013). Disruption of the histone methyltransferases ATXR5/6 also induces the expression of normally silent rDNA VAR1 and an increased or reduced number of silent and active rDNA copies, respectively (Pontvianne et al., 2012). However, in contrast with atxr5/6, we do not detect significant changes in the endoreplication of the rDNA variant copies in the nuc2-2 mutants (data not shown). Remarkably, the relative abundance of rDNA variants is restored to the wild-type level in F1 siblings upon backcrossing the nuc2-2 plants with wild-type plants, with repression of VAR1 and transcription of VAR2 and VAR3 (Figure 7). This shows that reintroduction of rDNA from wild-type plants, rather than the NUC2 gene, is able to reestablish the wild-type functional organization of rDNA in nuc2-2 plants.
In agreement with the observation that NOR2 is transcriptionally silent (Fransz et al., 2002), we have observed that inactive rRNA VAR1 genes localize mainly in NOR2, while the active VAR2 and VAR3 rRNA genes are in NOR4 (J. Sáez-Vásquez, F. Barneche, and C. Pikaard, unpublished data). Although we do not exclude alternative mechanisms, gene conversion (Schuermann et al., 2005) of rRNA could explain the variation in the abundance of rDNA variants in nuc2-2 and nuc2-2 gNUC2 plants. Gene conversion might provoke the translocation of VAR1 copies between different NORs, thereby relocalizing rDNA VAR1 copies in largely hypomethylated regions. VAR1 copies are highly methylated in a wild-type context, and their increased abundance might explain the increased global DNA methylation levels of 5′ ETS rDNA in nuc2-2 plants. However, some rDNA VAR1 copies became hypomethylated, as detected by McrBC analysis (Figure 6; Supplemental Figure 13). In the wild type, VAR1 represents 50% of all rDNA; consequently, the increased number in nuc2-2 plants makes it difficult to determine the contribution of each rDNA variant to global rRNA gene methylation levels (Supplemental Figure 12). It is also possible that NOR2 is not completely silenced during seedling establishment (Figure 9); consequently, some VAR1 units might be transcribed and associated with a single nucleolus in nuc2-2 (Figure 4D). Interestingly, VAR1 rDNA copies (located either in NOR2 or NOR4) associate with the nucleolus in nuc2-2 plants, a relocation that probably correlates with their switch to a transcriptionally active state (Figure 4E).
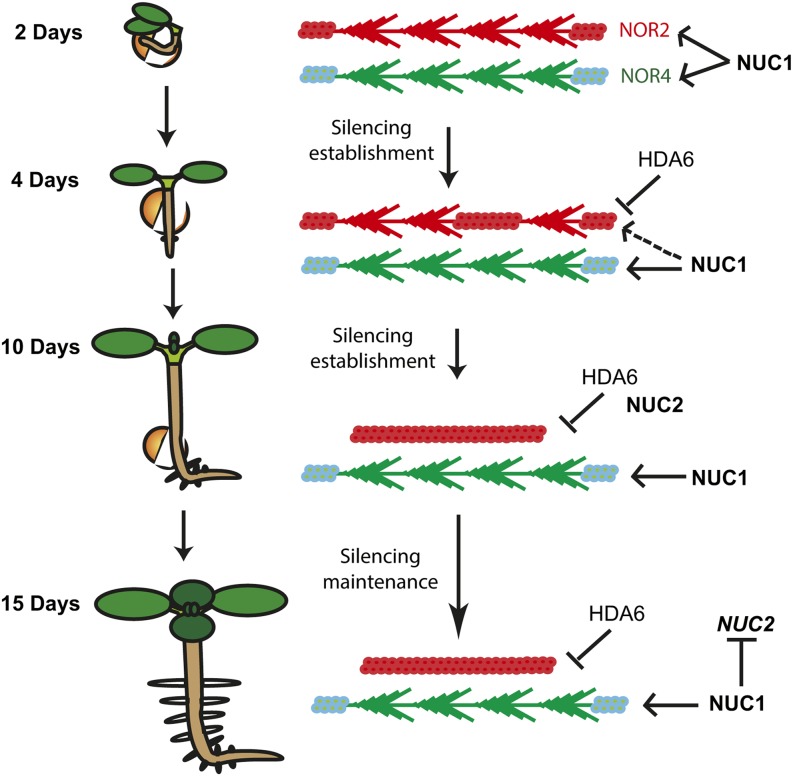
Figure 9.
NUC2 Is Required for Chromatin Dynamics during Developmental Transitions.
In this model, 45S rDNA VAR1 localizes in NOR2 (red) and VAR2 and VAR3 localize in NOR4 (green). Early in seed germination (2 d), both NOR2 and NOR4 are decondensed and transcriptionally active. NUC1 protein is associated with each NOR, as NUC1 binds to active genes (Pontvianne et al., 2010). Then, throughout germination, HDA6 participates in the chromatin silencing of rDNA VAR1 (Earley et al., 2010; Benoit et al., 2013). NOR2 becomes progressively silenced/condensed, and NUC1 dissociates. At 8 to 10 d, NUC2 transcripts and protein accumulate and bind “silenced” chromatin to participate in the heterochromatin establishment of NOR2. At the end of seed germination transition, NUC1 might play a role in repressing NUC2 gene expression at the transcriptional and/or posttranscriptional levels.
So far, we cannot disentangle the respective effects of rDNA variant dosage, delocalization, and activation on the derepression of VAR1 in nuc2 mutant plants. However, it is tempting to speculate that it is due to VAR1 relocation from NOR2 to NOR4. Indeed, it has been demonstrated that rDNA can be integrated and expressed at ectopic locations and form nucleolar structures (Karpen et al., 1988; Oakes et al., 1993; Floutsakou et al., 2013). Therefore, our results fit with a model in which the switch between active and/or repressed rDNA depends on the genomic and chromatin organization of the NOR (Preuss et al., 2008).
A Role of Nucleolin Proteins during Developmental Transition
So far, duplicated genes have been associated with gene dosage and neofunctionalization (Magadum et al., 2013). The results presented in this work, associated with previous studies, indicate that conserved and duplicated nucleolin genes have antagonistic activities in controlling rDNA organization and expression. To our knowledge, only a few duplicated genes with antagonistic roles have been described in Arabidopsis, including the duplicated RecQ helicases involved in homologous recombination and DNA repair (Hartung et al., 2007). How do NUC1 and NUC2 functionally interact in Arabidopsis? NUC2 protein accumulation during seedling development (Figure 2) temporally correlates with NOR condensation and rDNA VAR1 repression (Earley et al., 2010; van Zanten et al., 2011; Benoit et al., 2013). Accordingly, upon heterochromatin establishment of NOR2, the expression of NUC2 could be repressed by NUC1 (Figure 3). Remarkably, both NUC1 and NUC2 proteins associate with chromatin and can bind nucleosomes, although only NUC1 is able to assist remodeling (Figure 8; Supplemental Figure 9). As reported previously, NUC1, as well as human nucleolin, can bind transcriptionally competent rDNA chromatin and might facilitate rDNA transcription (Pontvianne et al., 2010; Cong et al., 2012). Meanwhile, NUC2 might bind nucleosomes to induce and/or maintain a repressive rDNA chromatin state during seedling growth (Figure 9).
In animal and yeast cells, the maintenance of rDNA methylation and gene copy number plays an important role in genome integrity and regulation (Paredes and Maggert, 2009; Ide et al., 2010). In Arabidopsis, it has been demonstrated that rDNA sequences might enhance homologous recombination (Urawa et al., 2001). We also reported high variability of 45S rDNA variants in the genome of Arabidopsis ecotypes (Abou-Ellail et al., 2011). Such a mechanism is subject to evolutionary constraints, as demonstrated by the observation that variations in genome size of Arabidopsis ecotypes are largely due to 45S rDNA copy number (Long et al., 2013). It would be interesting to determine how the variability of rDNA and nucleolin protein sequences, as well as other proteins involved in chromatin-based processes, can influence chromatin organization and the controlled expression of highly repeated sequences such as ribosomal genes.
METHODS
Plant Growth Conditions and Isolation of Mutants
All lines were derived from Arabidopsis thaliana Columbia-0 ecotype. Seeds were first sown on Murashige and Skoog (MS) medium (4.7 g/L MS medium, 0.5 g/L MES, 10 g/L Suc, and 10 g/L plant agar) or on soil and left 2 d at 4°C to synchronize. Plants were then grown either under continuous light (MS plants) or under a 16-h/8-h (light/dark) cycle (soil plants). Seeds corresponding to nuc2-1 (SALK_542511) and nuc2-2 (GABI_178D01) mutant plants were obtained from the Nottingham Arabidopsis Stock Centre (http://nasc.life.nott.ac.uk). The nuc1 and fas2.4 (third generation) mutant lines used in this work have been described (nuc1: Kojima et al., 2007; Pontvianne et al., 2007, 2010; fas2.4: Mozgová et al., 2010; Pontvianne et al., 2013).
Immunoblotting on Total and Chromatin Protein Fractions
Plant material (0.1 g) was homogenized and extracted in 500 μL of 50 mM Tris-HCl, pH 8, 150 mM NaCl, 10 mM EDTA, 50 mM NaF, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride, 10 mM β-mercaptoethanol, and a 1:100 dilution (v/v) of antiprotease cocktail inhibitor (Sigma). The extracts were cleared by centrifugation at 13,000g for 15 min and stored at −80°C.
To obtain protein chromatin fractions from wild-type, nuc1-1, and nuc1-2 plants, chromatin samples were prepared as described previously (Pontvianne et al., 2010) and proteins were extracted in 1× SDS-Laemmli buffer.
Then, SDS-PAGE and immunoblot analysis were performed as described previously (Sáez-Vasquez et al., 2004). The membranes were hybridized with a 1:10,000 dilution of α-At-NUC1 or a 1:7,000 dilution of α-At-NUC2 as described previously (Pontvianne et al., 2007, 2010).
Methods Related to Nucleic Acid
Total RNA was extracted from 15-d-old Arabidopsis plants using Trizol reagent (Amersham Biosciences) and treated with Turbo DNase (Ambion) for 60 min to eliminate contaminant DNA. First-strand cDNA synthesis was performed on 1.5 µg of treated RNA with the oligo(dT) and/or 3′3ets primer (for 3′ ETS rDNA sequences) using the GoScript Reverse Transcription System kit following the manufacturer’s instructions (Promega).
RT-PCR of 24 to 35 cycles was performed using a PTC-200 device (MJ Research). Primer pairs 5′nuc1/3′nuc1, 5′nuc2/3′nuc2, and 5′3ets/3′3ets were used to amplify NUC1, NUC2, and 3′ ETS pre-rRNA transcripts, respectively.
Quantitative PCR was performed using a LightCycler 480 and MESA Blue qPCR Master Mix Plus for SYBR assay (Eurogentec). Primers 5′nuc2q/3′nuc2q and q5′flc/q3′flc were used to detect NUC2 and FLC transcripts, respectively, and primers 5′18S/3′18S, 5′25S/3′25S, and 5′ITS1/3′ITS1 were used to amplify 18S, 25S, and ITS1 rDNA sequences, respectively. The Actin gene was used as an internal standard, and results were analyzed using the comparative cycle threshold method relative to input.
For the relative abundance of each class of rDNA (variant), PCR was performed with primers 5′3ets/3′3ets. PCR products were run on the LabChip GX/GX II (Caliper; Life Sciences) and analyzed with the LabChip GX DNA Assay Analysis software (which calculates the size and concentration of each nucleic acid fragment) according to the manufacturer’s instructions.
Primer extension analysis was performed using 15 µg of total RNA and specific 5′ end–labeled primers as described previously (Sáez-Vasquez et al., 2004). Primers used were tis for the detection of primary pre-rRNA precursors (+104 nucleotides from TIS) and p for the detection of primary pre-rRNA precursors cleaved at the P site (+1274 nucleotides from TIS). Primer 3′U3 was used for the detection of U3snoRNA as a loading control.
Cloning of Arabidopsis NUC2 Promoter Sequences and Detection of GUS Activity
Approximately 1.5 kb of the NUC2 promoter sequence was amplified from genomic DNA and fused to a GUS reporter gene in the vector pGWB433 (Nakagawa et al., 2007). The pAtNUC1:GUS construct in pCAMBIA 1381xa was described previously (Pontvianne et al., 2007).
Histochemical detection of GUS activity was performed using 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc) as a substrate. Seedlings were placed in X-Gluc buffer solution (1 mM X-Gluc, 100 mM Tris-HCl, pH 7, 0.1% Triton X-100, 0.5 mM potassium ferrocyanide, and 0.5 mM potassium ferricyanide) and incubated at 37°C for 12 h. Samples were then incubated in 70% ethanol for 2 h and cleared in Hoyer’s medium. For analysis of siliques, samples were placed in 90% acetone on ice for 1 h, followed by X-Gluc buffer as described above. All microscopic GUS images were observed using a Zeiss Axioskop 2 microscope and recorded using a Leica DC 300 FX digital camera.
ChIP and FISH
Antibodies against At-NUC1 and ChIP reactions were described previously (Pontvianne et al., 2007, 2010). PCR was performed using primers 5′nuc2a/3′nuc2a, 5′nuc2b/3′nuc2b, and 5′nuc2c/3′nuc2c to amplify NUC2 sequences −466/−210, −307/−38, and −137/+141, respectively.
DNA FISH analysis was performed using nuclei from leaves of 4-week-old plants as described previously (Pontvianne et al., 2007). The biotin-labeled probe was detected using digoxigenin (1:200; Roche) followed by sheep anti-digoxigenin antibody conjugated with the fluorochrome Alexa 480 (1:200; Invitrogen). Slides were mounted using Vectashield (Vector Laboratories) mounting medium with 1 µg/mL DAPI and then observed by confocal microscopy (Axio observer.Z1 microscope with the LSM700 scanning module; Zeiss). (A more detailed description of the immuno-FISH experiment is available in Pontvianne et al., 2012).
Immunostaining
Immunofluorescence was performed using 4-week-old leaves (the wild type, nuc1, and nuc2). Treated leaves (Supplemental Methods) were incubated at 4°C overnight with α-At-NUC2 (1:500) and then with anti-rabbit antibody coupled with Alexa 488 (1:1000; Invitrogen) for 3 h at room temperature. Slides were then mounted in Vectashield containing DAPI solution. Observation and imaging were performed using a confocal microscope (LSM 700 from Zeiss).
FANoS
The nuc2-2 plants were crossed with wild-type Columbia-0 plants expressing FIBRILLARIN2 (FIB2) protein fused to yellow fluorescent protein (YFP) and under the control of the 35S cauliflower mosaic virus promoter. Then, F1 and F2 plants were selected to isolate homozygous nuc2-2 plants containing 35S:FIB2:YFP transgene. Three-week-old leaves from the F3 generation were then used to isolate nuclei and nucleoli by FANoS as described previously (Pontvianne et al., 2013). The cell sorter used was an ARIA II special order (BD Biosciences). Total genomic DNA was extracted from nucleoli purified from wild-type and nuc2-2 plants expressing the FIB:YFP construct and amplified by PCR to identify the rDNA associated with the nucleolus (i.e., those that are transcriptionally active).
Bisulfite and McrBC Analyses
Genomic DNA from wild-type, nuc2-2, and nuc2-2 gNUC2 mutant plants was extracted using the DNA Easy Plant Mini kit (Qiagen) following the manufacturer’s instructions.
For bisulfite analysis, 500 ng of DNA was digested overnight using 20 units of BamHI restriction enzyme prior to bisulfite conversion. Bisulfite treatment was performed using the Epitect Bisulfite kit (Qiagen). The primers −315 and +243 were used to amplify rRNA gene sequences (from −315 to +243); the resulting PCR products were cloned in the pGEM-T Easy vector (Promega) and sequenced. Approximately 25 clones per sample were analyzed using the CyMATE method (Hetzl et al., 2007).
For McrBC analysis, 500 ng of DNA was digested using McrBC enzyme (Qiagen), which cuts methylated DNA. Then, PCR was performed on equal amounts of digested and undigested DNA using the HotStart goTag polymerase (Promega). PCR (35 cycles) was performed with primers 5′3ets/3′3ets, 5′5s/3′5s, 5′AtLINE/3′AtLINE, and 5′AtSN1/3′AtSN1 to amplify rDNA sequences (45S and 5S) and transposable elements (At LINE and At SN1). As a control, we used the demethylated nrpe1 mutant.
Nucleosome Reconstitution and Remodeling
Recombinant Xenopus laevis full-length histone proteins were purified from Escherichia coli cells as described earlier (Luger et al., 1999). The 255-bp DNA fragment containing the centrally positioned nucleosome sequence 601 at the middle or at the end of the sequence was amplified by PCR from pGEM3Z-601 (kindly provided by B. Bartholomiew and J. Widom) and end labeled using T4-polynucleotide kinase in the presence of [γ-32P]ATP. Nucleosome reconstitution was performed by the salt dialysis method (Angelov et al., 2006). Carrier DNA (150 to 200 bp, 2 mg) and 50 ng of γ-32P–labeled 601 DNA was mixed with equimolar amounts of histone octamer in nucleosome reconstitution buffer containing 2 M NaCl, 10 mM Tris-HCl, pH 7.4, 1 mM EDTA, and 5 mM β-mercaptoethanol. Nucleosomes (50 ng, 0.2 pmol) were incubated with RSC in remodeling buffer containing 10 mM Tris-HCl, pH 7.4, 5% glycerol, 100 mg/mL BSA, 1 mM DTT, 0.02% Nonidet P-40, 40 mM NaCl, 2.5 mM MgCl2, and 1 mM ATP for 45 min. The reaction was stopped by adding 1 µg of plasmid DNA. Nucleosome position was then analyzed by native PAGE.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At1g48920 (NUC1) and At3g18610 (NUC2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Tree of Nucleolin Proteins from Plants.
Supplemental Figure 2. Characterization of nuc2-1 Mutant Plants.
Supplemental Figure 3. Characterization of Nucleolin Double Mutants.
Supplemental Figure 4. List of Genes Upregulated or Downregulated in nuc2-2 Plants Identified by RNA-seq.
Supplemental Figure 5. Analysis in Silico to Compare Relative Expression Levels of NUC1 and NUC2.
Supplemental Figure 6. NUC1 and NUC2 Expression in Response to Auxin.
Supplemental Figure 7. NUC2 and NUC1 Promoter Activities during Silique Development.
Supplemental Figure 8. Mechanisms Controlling NUC2 Gene Expression.
Supplemental Figure 9. Titration of RSC, EMSA with NUC1 and NUC2, and Immunolocalization of NUC2.
Supplemental Figure 10. Immunolocalization of NUC1 in Wild-Type and nuc2 Mutant Plants.
Supplemental Figure 11. LabChip Data of the Abundance of rDNA Variants in Wild-Type, nuc2, nuc2 gNUC2, nuc1, and fas2 Plants.
Supplemental Figure 12. Bisulfite Analysis of 3′ ETS rDNA Sequences.
Supplemental Figure 13. PCR and LabChip Data with Different PCR Cycles for McrBC Experiments.
Supplemental Figure 14. PCR and LabChip Data with Different PCR Cycles for the Abundance of rDNA Variants in Wild-Type, nuc2, and Wild-Type × nuc2 Backcrossed Plants.
Supplemental Table 1. FISH Data in Wild-Type, nuc2-2, and nuc2-2 gNUC2 Plants.
Supplemental Table 2. Primer List.
Supplemental Methods. Immunostaining, RNA-seq, Bisulfite, EMSA, and Bioinformatics Analyses.
Supplemental Data Set 1. Alignment of Nucleolin Proteins.
Supplementary Material
Acknowledgments
We thank F. Barneche for critical reading of the manuscript and R. Cooke for comments and correction of the English, M. Laudie for bisulfite sequencing, Marie-Christine Carpentier and Christelle Llauro for RNA-seq bioinformatics analysis, Z. Haftek-Terreau for the production of histone recombinant proteins, and B. Bartholomiew and J. Widom for providing pGEM3Z-601. We also thank the flow cytometry facility of Montpellier RIO Imaging. Mutant plants met1-1, fas2-4, and nrpe1 were provided by C. Pikaard, J. Fajkus, and T. Lagrange, respectively. This work was supported by the CNRS (Grants CNRS/JSPS 2008-2009 number PRC449 and ANR SVSE2_SUBCELIF 087217). M.A.-E. was supported by a fellowship from the Egyptian government and N.D. by a fellowship from the Université de Perpignan Via Domitia.
AUTHOR CONTRIBUTIONS
J.S.-V. conceived and directed the project and wrote the article. K.N., P.B., and M.E. designed research. N.D. and M.A.-E. conducted the majority of experiments. F.P. and S.D. performed FANoS and nucleosome assays. S.N. and S.R. performed RNA sequence analysis. H.K., S.U., A.d.B., P.C., and R.M. contributed to the research.
Glossary
- RT-qPCR
quantitative RT-PCR
- ChIP
chromatin immunoprecipitation
- EMSA
electrophoretic mobility shift assay
- TIS
transcription initiation signal
- DAPI
4′,6-diamidino-2-phenylindole
- FISH
fluorescence in situ hybridization
- FANoS
fluorescence-assisted nucleolus sorting
- McrBC
modified cytosine restriction BC
- RSC
chromatin structure remodeling
- FACT
facilitate chromatin transcription complex
- MS
Murashige and Skoog
- X-Gluc
5-bromo-4-chloro-3-indolyl-β-d-glucuronide
- ETS
external transcribed spacer
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abou-Ellail M., Cooke R., Sáez-Vásquez J. (2011). Variations in a team: Major and minor variants of Arabidopsis thaliana rDNA genes. Nucleus 2: 294–299. [DOI] [PubMed] [Google Scholar]
- Angelov D., Bondarenko V.A., Almagro S., Menoni H., Mongélard F., Hans F., Mietton F., Studitsky V.M., Hamiche A., Dimitrov S., Bouvet P. (2006). Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 25: 1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit M., Layat E., Tourmente S., Probst A.V. (2013). Heterochromatin dynamics during developmental transitions in Arabidopsis—A focus on ribosomal DNA loci. Gene 526: 39–45. [DOI] [PubMed] [Google Scholar]
- Cong R., Das S., Ugrinova I., Kumar S., Mongelard F., Wong J., Bouvet P. (2012). Interaction of nucleolin with ribosomal RNA genes and its role in RNA polymerase I transcription. Nucleic Acids Res. 40: 9441–9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevillén P., Dean C. (2011). Regulation of the floral repressor gene FLC: The complexity of transcription in a chromatin context. Curr. Opin. Plant Biol. 14: 38–44. [DOI] [PubMed] [Google Scholar]
- Earley K.W., Pontvianne F., Wierzbicki A.T., Blevins T., Tucker S., Costa-Nunes P., Pontes O., Pikaard C.S. (2010). Mechanisms of HDA6-mediated rRNA gene silencing: Suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev. 24: 1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floutsakou I., Agrawal S., Nguyen T.T., Seoighe C., Ganley A.R., McStay B. (2013). The shared genomic architecture of human nucleolar organizer regions. Genome Res. 23: 2003–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P., De Jong J.H., Lysak M., Castiglione M.R., Schubert I. (2002). Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc. Natl. Acad. Sci. USA 99: 14584–14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Längst G. (2013). Epigenetic control of RNA polymerase I transcription in mammalian cells. Biochim. Biophys. Acta 1829: 393–404. [DOI] [PubMed] [Google Scholar]
- Hartung F., Suer S., Puchta H. (2007). Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 18836–18841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzl J., Foerster A.M., Raidl G., Mittelsten Scheid O. (2007). CyMATE: A new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 51: 526–536. [DOI] [PubMed] [Google Scholar]
- Ide S., Miyazaki T., Maki H., Kobayashi T. (2010). Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327: 693–696. [DOI] [PubMed] [Google Scholar]
- Jakociunas T., Domange Jordö M., Aït Mebarek M., Bünner C.M., Verhein-Hansen J., Oddershede L.B., Thon G. (2013). Subnuclear relocalization and silencing of a chromosomal region by an ectopic ribosomal DNA repeat. Proc. Natl. Acad. Sci. USA 110: E4465–E4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen G.H., Schaefer J.E., Laird C.D. (1988). A Drosophila rRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes Dev. 2: 1745–1763. [DOI] [PubMed] [Google Scholar]
- Khurts S., Masutomi K., Delgermaa L., Arai K., Oishi N., Mizuno H., Hayashi N., Hahn W.C., Murakami S. (2004). Nucleolin interacts with telomerase. J. Biol. Chem. 279: 51508–51515. [DOI] [PubMed] [Google Scholar]
- Kojima H., Suzuki T., Kato T., Enomoto K., Sato S., Kato T., Tabata S., Sáez-Vasquez J., Echeverría M., Nakagawa T., Ishiguro S., Nakamura K. (2007). Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 49: 1053–1063. [DOI] [PubMed] [Google Scholar]
- Lawrence R.J., Earley K., Pontes O., Silva M., Chen Z.J., Neves N., Viegas W., Pikaard C.S. (2004). A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 13: 599–609. [DOI] [PubMed] [Google Scholar]
- Layat E., Sáez-Vásquez J., Tourmente S. (2012). Regulation of Pol I-transcribed 45S rDNA and Pol III-transcribed 5S rDNA in Arabidopsis. Plant Cell Physiol. 53: 267–276. [DOI] [PubMed] [Google Scholar]
- Lolas I.B., Himanen K., Grønlund J.T., Lynggaard C., Houben A., Melzer M., Van Lijsebettens M., Grasser K.D. (2010). The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J. 61: 686–697. [DOI] [PubMed] [Google Scholar]
- Long Q., et al. (2013). Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat. Genet. 45: 884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Rechsteiner T.J., Richmond T.J. (1999). Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 119: 1–16. [DOI] [PubMed] [Google Scholar]
- Magadum S., Banerjee U., Murugan P., Gangapur D., Ravikesavan R. (2013). Gene duplication as a major force in evolution. J. Genet. 92: 155–161. [DOI] [PubMed] [Google Scholar]
- Mongelard F., Bouvet P. (2007). Nucleolin: A multiFACeTed protein. Trends Cell Biol. 17: 80–86. [DOI] [PubMed] [Google Scholar]
- Mozgová I., Mokros P., Fajkus J. (2010). Dysfunction of chromatin assembly factor 1 induces shortening of telomeres and loss of 45S rDNA in Arabidopsis thaliana. Plant Cell 22: 2768–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., et al. (2007). Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100. [DOI] [PubMed] [Google Scholar]
- Oakes M., Nogi Y., Clark M.W., Nomura M. (1993). Structural alterations of the nucleolus in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Mol. Cell. Biol. 13: 2441–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes S., Maggert K.A. (2009). Ribosomal DNA contributes to global chromatin regulation. Proc. Natl. Acad. Sci. USA 106: 17829–17834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka A., Schubert V., Meister A., Kreth G., Klatte M., Lysak M.A., Fuchs J., Schubert I. (2004). Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 113: 258–269. [DOI] [PubMed] [Google Scholar]
- Petricka J.J., Nelson T.M. (2007). Arabidopsis nucleolin affects plant development and patterning. Plant Physiol. 144: 173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., et al. (2010). Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet. 6: e1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., Blevins T., Chandrasekhara C., Feng W., Stroud H., Jacobsen S.E., Michaels S.D., Pikaard C.S. (2012). Histone methyltransferases regulating rRNA gene dose and dosage control in Arabidopsis. Genes Dev. 26: 945–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., Blevins T., Chandrasekhara C., Mozgová I., Hassel C., Pontes O.M., Tucker S., Mokros P., Muchová V., Fajkus J., Pikaard C.S. (2013). Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev. 27: 1545–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F., Matía I., Douet J., Tourmente S., Medina F.J., Echeverria M., Sáez-Vásquez J. (2007). Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol. Biol. Cell 18: 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss S.B., Costa-Nunes P., Tucker S., Pontes O., Lawrence R.J., Mosher R., Kasschau K.D., Carrington J.C., Baulcombe D.C., Viegas W., Pikaard C.S. (2008). Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol. Cell 32: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickards B., Flint S.J., Cole M.D., LeRoy G. (2007). Nucleolin is required for RNA polymerase I transcription in vivo. Mol. Cell. Biol. 27: 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez-Vásquez J., Gadal O. (2010). Genome organization and function: A view from yeast and Arabidopsis. Mol. Plant 3: 678–690. [DOI] [PubMed] [Google Scholar]
- Sáez-Vasquez J., Caparros-Ruiz D., Barneche F., Echeverría M. (2004). A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro. Mol. Cell. Biol. 24: 7284–7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuermann D., Molinier J., Fritsch O., Hohn B. (2005). The dual nature of homologous recombination in plants. Trends Genet. 21: 172–181. [DOI] [PubMed] [Google Scholar]
- Tajrishi M.M., Tuteja R., Tuteja N. (2011). Nucleolin: The most abundant multifunctional phosphoprotein of nucleolus. Commun. Integr. Biol. 4: 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja R., Tuteja N. (1998). Nucleolin: A multifunctional major nucleolar phosphoprotein. Crit. Rev. Biochem. Mol. Biol. 33: 407–436. [DOI] [PubMed] [Google Scholar]
- Urawa H., Hidaka M., Ishiguro S., Okada K., Horiuchi T. (2001). Enhanced homologous recombination caused by the non-transcribed spacer of the rDNA in Arabidopsis. Mol. Genet. Genomics 266: 546–555. [DOI] [PubMed] [Google Scholar]
- Van Lijsebettens M., Grasser K.D. (2010). The role of the transcript elongation factors FACT and HUB1 in leaf growth and the induction of flowering. Plant Signal. Behav. 5: 715–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten M., Koini M.A., Geyer R., Liu Y., Brambilla V., Bartels D., Koornneef M., Fransz P., Soppe W.J. (2011). Seed maturation in Arabidopsis thaliana is characterized by nuclear size reduction and increased chromatin condensation. Proc. Natl. Acad. Sci. USA 108: 20219–20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A.T., Haag J.R., Pikaard C.S. (2008). Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.