In their commentary, Golczyk et al. (2014) state that the level of plastid DNA (ptDNA) remains approximately constant during mature, aging, and senescent stages of leaf development, in apparent contrast with our conclusion of ptDNA decline from previous studies. However, the ptDNA decline that we reported was found during proplastid-to-chloroplast development, before the mature-to-senescent leaf stages. We found little change in ptDNA after chloroplast maturation. Furthermore, we argue that their data fail to demonstrate constancy in ptDNA during development: Most of their data are not quantitative and demonstrate neither constancy nor change in ptDNA content. We also show that our inability to detect 4′,6-diamidino-2-phenylindole (DAPI)–stainable DNA in some plastids is not due to “artifact-prone methods,” as suggested by Golczyk et al. In our opinion, the fundamental issues concern whether ptDNA content declines during leaf development, the magnitude of that decline, and by extension, the functional relevance of such a decline. Our conclusions for three plant species with respect to changes in ptDNA content during leaf development are summarized in Figure 1, based on four methods that provide quantitative data on ptDNA mass per plastid and per cell and on molecular structure. The only quantitative data provided by Golczyk et al. (2014) are based on amounts of DNA between two closely spaced quantitative PCR (qPCR) primers and thus depend on the unvalidated assumption that such tiny DNA fragments represent genome-sized ptDNA molecules. We conclude that an accurate assessment of ptDNA must consider not only ptDNA quantity but also molecular quality, a parameter not addressed by Golczyk et al. In our opinion, the manner of data presentation is the principal difference leading to our opposing conclusions.
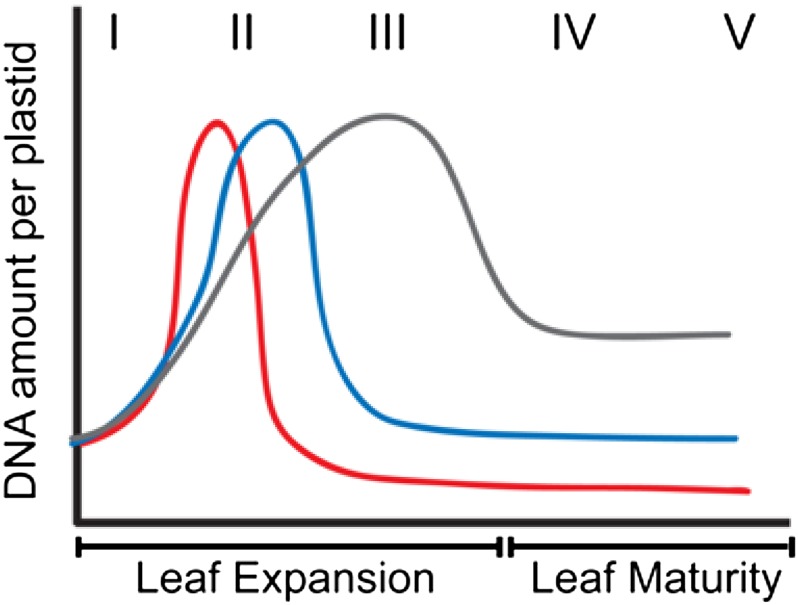
Figure 1.
Schematic Representation of Changes in the Amount of ptDNA per Plastid during Development in Three Plant Species.
Increase in ptDNA amount due to ptDNA replication occurs very early in development in maize (red line), followed by a rapid decline. For Arabidopsis (blue line), the increase in ptDNA occurs slightly later and the decline in ptDNA amount is much later. For tobacco (gray line), ptDNA increases more gradually and the decline is less severe. The Roman numerals indicate stages of leaf development. I to III represent expanding leaves, and IV and V represent expanded leaves. (Reprinted from Rowan and Bendich [2009], Figure 1.)
QUANTITATIVE CONCLUSIONS FROM NONQUANTITATIVE DATA
Although Golczyk et al. (2014) used four methods for assessing the presence of ptDNA during the development of green leaves, three of these methods involved microscopic examination that did not provide quantitative information for either ptDNA per plastid, ptDNA per cell, or ptDNA per nuclear DNA (nucDNA) amount. DAPI-DNA fluorescence revealed DNA within chloroplast nucleoids that was characterized by “visual inspection…indicating unaltered DNA contents per organelle” in young and mature leaves. Furthermore, for none of the four plant species examined (Arabidopsis thaliana, sugar beet [Beta vulgaris], tobacco [Nicotiana tabacum], and maize [Zea mays]) was statistical data provided concerning the number of cells or plastids chosen for analysis or the fraction of cells or plastids that exhibited DAPI-DNA fluorescence at different stages of leaf development. Using electron microscopy, fibrils were identified as DNA-containing areas, but no statistical analysis of the data was provided and no quantitative conclusion was drawn.
By contrast, the data presented in our previous articles for ptDNA of Arabidopsis, tobacco, Medicago truncatula, pea (Pisum sativum), and maize were quantitative, revealing an increase followed by a decrease in ptDNA during the transition from proplastid to pale-green chloroplast to fully green chloroplast (as well as the etioplast-to-chloroplast transition upon illumination of etiolated maize seedlings) (Oldenburg and Bendich, 2004; Rowan et al., 2004, 2009; Oldenburg et al., 2006; Shaver et al., 2006, 2008; Zheng et al., 2011). Those data (summarized in Figure 1) show that after the decrease to the level in mature chloroplasts, ptDNA remains essentially constant as leaves reach maturity, in agreement with the conclusion of Golczyk et al. As for gerontoplasts in senescing leaves, although Golczyk et al. concluded that the ptDNA level remains unchanged, their description may, in fact, reflect a decline in ptDNA, as they noted that “DNA fluorescence became more and more diffuse and difficult to visualize.” In our work, we analyzed leaves long before the onset of leaf senescence, except for Arabidopsis where we reported “a slight decline in cpDNA content as mature leaves senesce” (Rowan et al., 2009) and concluded that “the demise of organellar DNA can be independent of its effect on senescence” (Oldenburg and Bendich, 2004).
The only quantitative assay of ptDNA reported by Golczyk et al. (2014) was real-time qPCR. Using standard qPCR procedures, our group (Rowan et al., 2009) and Thomas Börner and colleagues (Zoschke et al., 2007) reported constant values for plastid genome equivalents per nuclear genome equivalent (ptDNA/nucDNA) during Arabidopsis leaf development. The qPCR data presented by Golczyk et al. include sugar beet (a plant we have not investigated), tobacco, and maize. Since maize is the plant we have studied most intensively, and the decline in ptDNA is greater for maize than tobacco (Figure 1), we focus on maize.
Golczyk et al. show that for maize, ptDNA/nucDNA increases from ∼800 at stage I of leaf development to ∼1200 to 1400 for stages III to VI. Unfortunately, they provide neither images of nor a definition of “stage” for maize leaves used in their experiments. Furthermore, the reference given for maize “grown in a greenhouse as previously described (Li et al., 2006)” applies only to tobacco and Arabidopsis, not to maize. Thus, we cannot compare the leaf material used by Golczyk et al. directly with the plant tissue we used in our experiments, where both images and detailed descriptions of seedling development were provided (Oldenburg and Bendich, 2004; Oldenburg et al., 2006; Zheng et al., 2011). Using standard qPCR procedures, the ptDNA/nucDNA values we reported ranged from 1200 to 1600 for the mature first leaf blade of 10- to 12-d-old seedlings (stage IV-V in Figure 1) (Zheng et al., 2011). Thus, using the standard qPCR assay, there appears to be no conflict in ptDNA copy number between our data and those of Golczyk et al. The real conflict appears to be related to assumptions about the quality and functionality of the ptDNA present.
In the standard qPCR procedure, the short segment (∼150 bp) of ptDNA that is amplified is usually taken to represent a genome-sized molecule of ptDNA assumed to exist in vivo, thus permitting the inference that “copy number” of the short segment represents the copy number of the plastid genome. This assumption is likely valid for nucDNA that (presumably) exists as intact chromosomal DNA molecules throughout the development of the maize leaf prior to leaf senescence. The quality of ptDNA molecules declines rapidly as plastids mature (described below), so that the copy number of the short segments amplified by standard qPCR may not represent the copy number of the genome or of DNA segments long enough to encode functional products. This hypothesis can be tested using a qPCR procedure in which copy number of long DNA segments (11 kb, for example) is compared with copy number obtained with the standard 0.15-kb segments. We conclude that in addition to copy number data, the quality of ptDNA must be considered when assessing the contribution of ptDNA to plastid function during leaf development.
In addition to the dubious practice of drawing quantitative inferences from visual inspection of microscopy images of plastid nucleoids, the appearance of DAPI-fluorescent nucleoids reveals little of the quality (molecular integrity and molecular weight) of the ptDNA molecules in those nucleoids. Similarly, our quantitative DAPI-DNA data using plastids isolated from the cell reported DNA mass but not quality. DNA quality currently can be assessed only after the DNA is extracted from the cell. Using pulsed-field gel electrophoresis (PFGE) and/or moving pictures of ethidium-stained ptDNA, we found a decline in the size of DNA molecules that accompanied chloroplast maturation in all five plants examined (Oldenburg and Bendich, 2004; Rowan et al., 2004, 2009; Oldenburg et al., 2006; Shaver et al., 2006). We interpreted these data to indicate that ptDNA was damaged by reactive oxygen species (by-products of photosynthesis) and progressively degraded during leaf development. In some plants, like tobacco, ptDNA repair maintains ptDNA integrity longer during leaf development than in others, like maize, resulting in the species differences shown in Figure 1. Thus, the amount of high-quality ptDNA may decline much faster during leaf development than does the amount of total ptDNA, as we reported (Shaver et al., 2006). In addition, the capacity to encode useful plastid gene products could decline before the microscopic appearance of the nucleoid in situ reveals the ptDNA degradation process. As the ptDNA becomes more highly fragmented, the appearance of the nucleoid would deteriorate until the nucleoid is no longer evident in situ. Inspection of the images (Figure 2 and Supplemental Figures 1 to 4) presented by Golczyk et al. (2014) can be interpreted to indicate this progression. Highly fragmented ptDNA, either still aggregated to form a nucleoid or dissociated from the nucleoid, could also affect the interpretation of results from the qPCR assay.
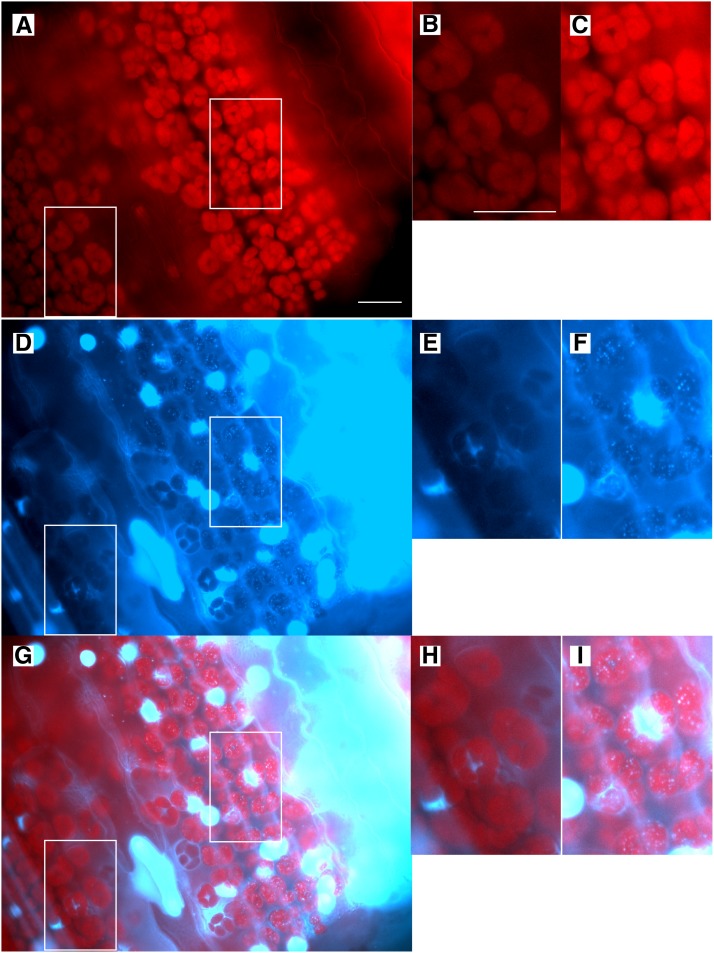
Figure 2.
Fluorescence Microscopy Images of a Tissue Section from the Mature First Leaf Blade of Maize.
The tissue section is from the middle of the leaf blade from 11-d-old seedlings. The top panels show chlorophyll autofluorescence, middle panels show DAPI fluorescence, and bottom panels are merged images. (B), (E), and (H) show plastids in Category 3 (undetectable DAPI-DNA) and (C), (F), and (I) show plastids in Category 1 (discrete DAPI-DNA nucleoids) and are enlargements of boxed regions in (A), (D), and (G). Bar in (A) = 25 µm for (A), (D), and (G), and bar in (B) is 25 µm for (B), (C), (E), (F), (H), and (I). Images were produced as described in Methods and shown in Supplemental Figure 2.
ARTIFACTS AND METHODOLOGY
Golczyk et al. (2014) suggest that methodological artifacts could account for our finding of a developmental decline in ptDNA. One example was “possible technical problems with insufficient DAPI dye penetration (Selldén and Leech, 1981; Evans et al., 2010).” However, the data presented in the cited articles are not indicative of poor DAPI penetration of the plastid envelope. Selldén and Leech (1981) wrote that DAPI “did not penetrate either whole cells or isolated protoplasts,” without reference to chloroplasts or whether fixed or unfixed tissue was used. Evans et al. (2010) suggested plastid impermeability as an explanation for why chloroplasts were not convincingly stained in mature tissues, but presented no data on permeability. In our opinion, these two citations mislead the reader as to the possibility that chloroplast impermeability to DAPI is a likely explanation for our inability to detect DNA-containing nucleoids in mature leaf tissue. Furthermore, DAPI-DNA nucleoids are clearly visible in plastids for ∼60% of the cell regions in mature maize leaf blade sections (see below; Table 1, Figure 2; Supplemental Figure 1) and although fainter than in younger leaves, visible in mature leaves of Arabidopsis (Figures 1E and 3H in Rowan et al., 2009), as well as in the images presented by Golczyk et al. (2014).
Table 1. Maize Plastids with and without Discrete DAPI-DNA Fluorescence.
| Category 1: Discretea | Category 2: Diffuse Pattern and Discrete | Category 3: Undetectable | |
|---|---|---|---|
| Cells in tissue sectionsb | 69 | NA | 46 |
| Protoplastsc | 4 | 3 | 8 |
| Isolated plastidsd | 51 | 42 | 38 |
Three categories of DAPI-DNA patterns in plastids were scored by visual examination of fluorescence microscopy images: (1) plastids with many nucleoids as indicated by discrete DAPI-DNA spots, (2) plastids with a diffuse DAPI-DNA signal and a few nucleoids, and (3) plastids with neither a diffuse DAPI-DNA signal nor discrete nucleoids. Examples of Categories 1 and 3 are shown in Figure 2 for tissue sections and for Categories 1 to 3 in Figure 3 for mesophyll protoplasts and Supplemental Figure 3 for isolated plastids.
The number of cells with and without discrete DAPI-DNA nucleoids was determined for both fixed-then-sectioned and sectioned-then-fixed tissues from the first leaf blade of 11- and 10-d-old seedlings, respectively (see Methods and example shown in Supplemental Figure 2H). Since only two categories (discrete and undetectable) were scored, the second category is not applicable (NA) for tissue sections. For fixed-then-sectioned tissue, four fields of view were examined with 45 cells in Category 1 and 28 in Category 3; these values for sectioned-then-fixed were three fields with 24 and 18 cells, respectively.
Fifteen mesophyll protoplasts from the first leaf blade of 10-d-old seedlings were examined.
A total of 131 isolated plastids from the first leaf blade of 9- and 11-d-old seedlings were examined.
At the opposite extreme, Golczyk et al. claim that the membranes of mature chloroplasts are “leaky,” allowing the ptDNA to be degraded by endogenous DNase. Reference was made to Selldén and Leech (1981) showing loss of ptDNA only in damaged plastids when their plastid isolation buffers contained Mg2+. However, our sorbitol and high-salt plastid isolation buffers contain EDTA, so that the leaky membrane/DNase explanation is implausible. Experiments in which exogenous DNase is added to isolated plastid preparations indicate that mature chloroplasts may be more susceptible to membrane damage than proplastids and developing plastids (Selldén and Leech, 1981; Shaver et al., 2006). One procedure for plastid isolation included DNase treatment to remove DNA/chromatin from the outside of plastid membranes (Oldenburg and Bendich, 2004; Rowan et al., 2004). For maize, we found undetectable DAPI-DNA in 85 and 82% of the plastids treated with and without DNase, respectively; for Arabidopsis, the corresponding values were 7 and 7% (Oldenburg and Bendich, 2004; Rowan et al., 2004). In subsequent work, we omitted the DNase and used a high-salt buffer to remove DNA/chromatin and still found plastids with no DAPI-DNA: 9% for pea, 11 to 34% for M. truncatula, 2 to 36% for maize from DAPI-DNA quantification, and 80 to 87% for maize from visual scoring (Oldenburg et al., 2006; Shaver et al., 2006, 2008; Zheng et al., 2011). These data indicate that the absence of detectable DAPI-DNA fluorescence in some plastids is not due to an artifact arising from DNA degradation by nucleases during plastid isolation. Leaky plastid membranes were also suggested by Golczyk et al. to be caused by polyvinyl pyrrolidone and by high salt, but neither data nor references were provided to support this speculation. In sum, Golczyk et al. alternately invoke both impermeable and extremely permeable membranes to conclude that the chloroplasts without DAPI-DNA staining resulted from artifacts in our experimental procedures. However, our voluminous data (and some of their own micrographs), a more accurate reading of the cited literature, and data presented below refute their conclusion.
Golczyk et al. also claim that a “lack of appropriate controls checking the biochemistry (was) used” for our DAPI-DNA imaging and quantification of ptDNA. The DNA specificity of DAPI has been well documented (Coleman, 1979; Lawrence and Possingham, 1986; Miyamura et al., 1986). Nonetheless, we employed numerous controls for our fluorescence microscopic imaging of DAPI-stained ptDNA. These include measuring the levels of DAPI fluorescence in unstained plastids and fixed plastids pretreated with DNase and then stained with DAPI. We also determined optimal exposure times, since DAPI-DNA signal intensity is generally higher for proplastids than mature chloroplasts (Supplemental Figure 3). We switched from using a broad-band DAPI filter to a narrow-band filter to reduce plastid autofluorescence from chlorophyll and accounted for any background fluorescence in our calculations of plastid DAPI-DNA values. We also demonstrated that chlorophyll autofluorescence does not mask or quench the plastid DAPI-DNA signal (Zheng et al., 2011). Thus, we conclude that DAPI-DNA quantification is reliable for determining ptDNA mass.
DAPI-DNA IN MAIZE TISSUE SECTIONS, PROTOPLASTS, AND ISOLATED PLASTIDS
The fluorescent microscopy images of plastid nucleoids presented by Golczyk et al. (2014) were obtained using tissues that were treated with fixative prior to protoplasting, squashing, and DAPI staining of the tissues, whereas analogous images we reported previously were obtained by sectioning leaves and immediately plunging the tissue into fixative (Shaver et al., 2006; Rowan et al., 2009). Thus, it is possible that the lack of detectable DAPI-stained, punctate nucleoids we reported for mature tissues of Arabidopsis and maize could have been due to amounts of ptDNA below the detection limit, as was our interpretation, or nuclease activity in the mature (but not younger) tissue that degraded the ptDNA before the fixative inactivated the nuclease, as suggested by Golczyk et al.
We vacuum-infiltrated maize tissues with fixative, prepared tissue sections, and performed DAPI staining as described in Methods. A tissue section from the mature first leaf blade is shown in Figure 2 that contains both a region where DAPI-DNA nucleoids are clearly visible in the plastids and an adjacent region containing plastids with no detectable DAPI-DNA fluorescence (additional images are shown in Supplemental Figure 1). Examples of protoplasts (Figure 3) and isolated chloroplasts (Supplemental Figures 3G and G°) with and without detectable DAPI-DNA fluorescence are also shown. We classified the chloroplasts into three types with respect to DAPI-DNA signal and then scored the number of cells (Supplemental Figure 2; see Methods), protoplasts, and isolated chloroplasts in each category (Table 1). These results demonstrate that plastids without detectable DAPI-DNA are present in the mature leaf blade and indicate that our inability to detect punctate forms of ptDNA in some chloroplasts cytologically is not attributable to a DNase artifact, as suggested by Golczyk et al., but to a decrease in ptDNA content and/or molecular integrity as leaves develop, as we concluded previously (Oldenburg and Bendich, 2004; Rowan et al., 2004, 2009; Oldenburg et al., 2006; Shaver et al., 2006, 2008; Zheng et al., 2011). This progression of proplastid-to-chloroplast development is shown in Supplemental Figure 3, illustrating a visible reduction in DAPI-DNA intensity and decreased genome equivalents per plastid for the mature chloroplasts (Table 2). We report zero genome equivalents for one plastid (pt13 in Table 2; Supplemental Figure 3G), although our interpretation of a plastid with “no detectable DAPI fluorescence…does not imply…absolutely no DNA within the plastid” (Oldenburg and Bendich, 2004). For the images shown in Golczyk et al. (2014), it is important to ascertain the degree to which an image of a cell with punctate chloroplast nucleoids (e.g., Figure 2I in Golczyk et al.) represents all cells at stage III/IV of maize leaf development and whether that degree changes as the leaf develops. To obtain this information, statistical data of the type we provide in Table 1 must be presented, but such data are lacking in the Golczyk et al. Commentary.
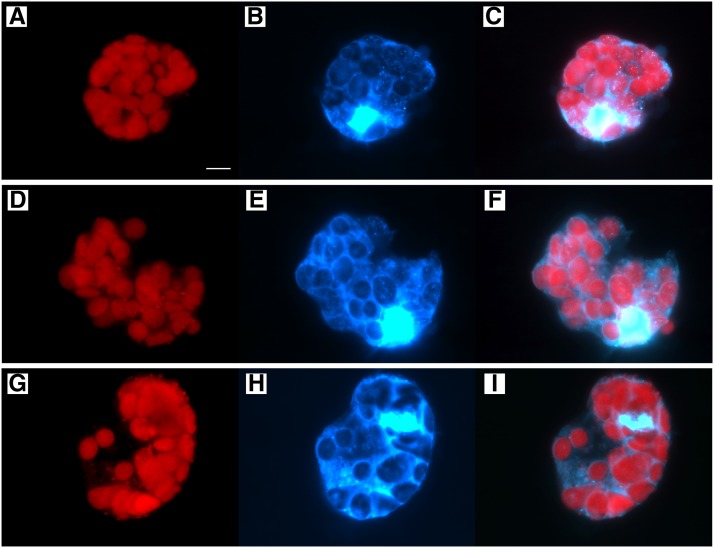
Figure 3.
Fluorescence Microscopy Images of Protoplasts from the Mature First Leaf Blade of Maize 10-d-Old Seedlings.
The left panels show chlorophyll autofluorescence, middle panels show DAPI fluorescence, and right panels are merged images. The protoplasts in (A) to (C) contain plastids in Category 1 (discrete DAPI-DNA nucleoids), (D) to (F) plastids in Category 2 (diffuse pattern and discrete DAPI-DNA nucleoids), and (G) to (I) plastids in Category 3 (undetectable DAPI-DNA). Bar in (A) is 10 µm for all panels.
Table 2. Size and Genome Copy Number for Individual Maize Plastids.
| Size (µm2) | Genome Copy No. | ||
|---|---|---|---|
| Base of stalk | pt1 | 6.7 | 209 |
| pt2 | 3.6 | 113 | |
| pt3 | 7.1 | 307 | |
| pt4 | 8.7 | 251 | |
| Middle of stalk | pt5 | 17.0 | 331 |
| pt6 | 13.1 | 302 | |
| pt7 | 22.7 | 334 | |
| pt8 | 20.9 | 387 | |
| Top of stalk | pt9 | 22.6 | 727 |
| pt10 | 30.1 | 652 | |
| pt11 | 16.0 | 160 | |
| pt12 | 26.7 | 650 | |
| Leaf 1 blade | pt13 | 24.3 | 0 |
| pt14 | 14.0 | 10 | |
| pt15 | 83.5 | 241 | |
| pt16 | 48.1 | 218 |
The plastid size and copy number are given for some of the plastids shown in Supplemental Figure 3. The number of plastids measured, average copy numbers ± se, and ranges for the complete data set were: 55, 113 ± 9 (11 to 307) for base of stalk; 54, 252 ± 23 (42 to 766) for middle of stalk; 43, 241 ± 33 (0 to 1047) for top of stalk; and 52, 106 ± 13 (0 to 351) for the first leaf blade (Zheng et al., 2011).
Golczyk et al. state that “undetectability of stainable DNA…is per se not a valid criterion to postulate the absence of DNA or to assess nature and impact of changes of in-gel DNA structures remaining after lysis of embedded chloroplasts.” We employed assays that quantify ptDNA mass (DAPI-DNA per plastid, blot hybridization, and qPCR) and monitor ptDNA molecular size/structure (PFGE and in-gel DNA movies) during plastid development, in addition to the less informative (and qualitative) presence/absence assay for DAPI-stained nucleoids. Our data reveal a developmental decline in both ptDNA amount and quality, leading to the conclusion that the lack of visible DAPI-DNA nucleoids in mature plastids is due to ptDNA degradation and fragmentation to less-than-genome-sized molecules that occurs in vivo.
PHOTOSYNTHESIS WITHOUT ptDNA?
Golczyk et al. (2014) make the argument that chloroplasts would not be able to conduct photosynthesis after leaf maturation without ptDNA and support their argument with “the maximum mRNA half-life reported for (barley) psbA are in the range of 40 h.” In fact, the reported half-life was >40 h, the mRNA level did not change over a 30-h period, and mRNA stability increased at least 5-fold during chloroplast development (Kim et al., 1993). Furthermore, Baumgartner et al. (1993) reported an increase in psbA mRNA stability of >100-fold during development of young barley (Hordeum vulgare) seedlings. Unfortunately, no long-term studies of plastid mRNA and protein stability have been reported that could support or refute the possibility that photosynthesis can continue for months without the coding function of ptDNA. Nonetheless, it is generally accepted that the expression of photosynthetic genes in ptDNA is modulated primarily at the posttranscriptional, translational, and posttranslational levels by nucleus-encoded factors (Eberhard et al., 2002; Barkan, 2011; Mulo et al., 2012).
CONCLUDING REMARKS
Long molecules of ptDNA are found in immature, nongreen plastids in stalk tissue of light-grown maize seedlings (Oldenburg and Bendich, 2004), and the amount of ptDNA decreases 2- to 3-fold to ∼100 genome equivalents per plastid in the green leaf blade (Zheng et al., 2011). However, molecular integrity declines to the point at which most ptDNA is present as less-than-genome-sized fragments or ptDNA is no longer recognized in DNA movies or PFGE (Oldenburg and Bendich, 2004; Oldenburg et al., 2006). Some ptDNA molecules may be fragmented to a size less than that required to encode the function served at an earlier stage of plastid development, even if those ptDNA fragments can be detected by standard qPCR and DAPI-DNA fluorescence. Thus, ptDNA “copy number” can appear similar at different stages of leaf development, even as the contribution of ptDNA to plastid coding function declines.
METHODS
Preparation of Maize Tissue, Cells, and Plastids for Fluorescence Microscopy
Seedlings of maize (Zea mays, inbred line B73) were sown in Sunshine mix #4 and grown under 16-h/8-h light/dark cycles in a controlled growth room for 9 to 12 days. Whole seedlings were harvested, washed in 0.5% sarkosyl, and rinsed in distilled water.
For the fixed-then-sectioned tissue shown in Figure 2 and Supplemental Figure 1, whole seedlings (11 d old) were immediately immersed and fixed in 0.8% glutaraldehyde (v/v) in 0.33 M sorbitol, 20 mM HEPES, pH 7.6, 2 mM EDTA, and 0.1% BSA (w/v). Fixation was performed by vacuum infiltration of the whole seedling for ∼3 h, by which time no more air bubbles were observed to surface from the seedling tissue. Fixed tissues not immediately used for imaging were stored at 4°C. Sectioned-then-fixed tissue (Table 1) from the first leaf blade of 10-d seedlings was prepared as described (Shaver et al., 2006).
For the protoplasts shown in Figure 3, the first leaf blade (unfixed) was digested in PrB#5 (1% cellulysin [w/v], 0.1% macerase [w/v], 0.5 M sorbitol, 10 mM MES, pH 5.7, 1 mM CaCl2, 1 mM MgCl2, 0.1% BSA [w/v], and 1.4 mM β-mercaptoethanol) as follows. The leaf tissue was cut into small pieces using a razor blade, placed in PrB#5, vacuum infiltrated for 20 min, and incubated for 2 h at room temperature with gentle shaking (40 rpm) and then at 80 rpm for 30 min to release protoplasts (all steps performed in the dark). The digestion solution containing the protoplasts was filtered through a 100-μm mesh, centrifuged at 150g to pellet the protoplasts, washed twice with PrB#5 (without enzymes), then resuspended, and glutaraldehyde was added to give a final concentration of 0.8% (v/v). The fixed protoplasts were directly prepared for imaging or stored at 4°C.
Fluorescence Microscopy
Our general procedures for microscopy imaging have been described in detail previously (Oldenburg and Bendich, 2004; Shaver et al., 2006; Rowan and Bendich, 2011; Zheng et al., 2011). Some specifics are as follows. For imaging, the samples were stained in 1 μg/mL DAPI with 1% β-mercaptoethanol (v/v) added to reduce fading. Multiple images were recorded at exposure times from 0.1 to 2 s with a narrow-band DAPI filter (360 excitation; 450/50 emission) and at several focal planes (z axis and z-stacks). Plastid autofluorescence images were recorded using a G1B filter (546 excitation; 590 emission). Although not shown, white light images were also recorded. A ×20 or ×40 objective was used for tissue sections and ×40 or ×60 oil emersion objective for protoplasts and isolated plastids.
Digital images were acquired using a QImaging Retiga 1300 camera and OpenLab software. The images shown in the figures were modified to optimize visualization of DAPI-DNA signal in the plastids. Openlab and Adobe Photoshop were used to refine the images by (1) merging multiple images from a z-stack into a single image, (2) subtracting the background fluorescence, and (3) adjusting the brightness and contrast. For the merged z-stack, deconvolution was tested, but did not improve the images produced using Photoshop. In fact, deconvolution created a color change that improved neither image quality nor detection of DAPI-DNA fluorescence (Supplemental Figure 2). The original gray-scale images were colorized to show plastids in red for chlorophyll autofluorescence, DAPI staining of DNA in blue-white for plastids and nuclei, and for merging of the two images to show overlap of ptDNA within plastids. An example of some original images and the subsequent steps used to produce the final image is presented in Supplemental Figure 2.
Classification of DAPI-DNA Fluorescence in Maize Plastids
Recorded images were examined visually to classify maize plastids with respect to their DAPI-DNA fluorescence. We report three types of DAPI-DNA signals in Table 1: Category 1, plastids with many discrete DAPI-DNA nucleoids; Category 2, a diffuse DAPI signal throughout the plastids and in some, but not all, cases a few discrete nucleoids were also visible; Category 3, plastids with no detectable DAPI-DNA fluorescence. The images of chlorophyll autofluorescence were used to identify plastids and to verify the colocalization of the DAPI-DNA signal. Multiple DAPI-stained images were examined that were recorded at different exposure times of the same field of view for tissue sections, protoplasts, and isolated plastids. For tissue sections and protoplasts, DAPI-stained nuclei were clearly evident, indicating that DAPI entered the fixed cells. For tissue sections, unambiguous identification of individual cells (even after merging of z-stacks) was generally not achieved because cell boundaries are not always visible and there were several layers of cells. However, a few bundle sheath cells were clearly identifiable on both sides of the autofluorescent vascular tissue (example in Supplemental Figure 1). Thus, for the purpose of quantification, a “cell” was assigned to a region containing one or more clusters of plastids and a DAPI-staining nucleus (a few regions without nuclei were also counted). An example is shown in Supplemental Figure 2H, where each “cell region” has been circled. Both the original single focal planes and composite images were examined in order to estimate the number of cells within a tissue section, as well as the presence or absence of DAPI-DNA signal in plastids. For isolated plastids, the same images used for determination of DNA copy number per plastid (Zheng et al., 2011) were used here for assignment into Category 1, 2, or 3 (Table 1), and examples shown in Supplemental Figure 3 with copy numbers given in Table 2. The procedures used to quantify genome copy number per plastid from DAPI-DNA fluorescence were described by Zheng et al. (2011).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Fluorescent Microscopy Images of Tissue Sections from the Middle of the Mature First Leaf Blade of 11-d-Old Maize Seedlings.
Supplemental Figure 2. Process Used to Optimize Visualization of DAPI-DNA Signal in Plastids.
Supplemental Figure 3. Fluorescent Microscopy Images of Isolated Plastids from Maize Seedlings.
Supplementary Material
AUTHOR CONTRIBUTIONS
D.J.O. performed the experimental work and data analysis. D.J.O. and A.J.B. wrote the article. B.A.R. and R.A.K. contributed ideas to the article.
Footnotes
Online version contains Web-only data.
References
- Barkan A. (2011). Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 155: 1520–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner B.J., Rapp J.C., Mullet J.E. (1993). Plastid genes encoding the transcription/translation apparatus are differentially transcribed early in barley (Hordeum vulgare) chloroplast development: Evidence for selective stabilization of psbA mRNA. Plant Physiol. 101: 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman A.W. (1979). Use of the fluorochrome 4’6-diamidino-2-phenylindole in genetic and developmental studies of chloroplast DNA. J. Cell Biol. 82: 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard S., Drapier D., Wollman F.A. (2002). Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 31: 149–160. [DOI] [PubMed] [Google Scholar]
- Evans I.M., Rus A.M., Belanger E.M., Kimoto M., Brusslan J.A. (2010). Dismantling of Arabidopsis thaliana mesophyll cell chloroplasts during natural leaf senescence. Plant Biol. (Stuttg.) 12: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golczyk H., Greiner S., Wanner G., Weihe A., Bock R., Börner T., Hermann R.G. (2014). Chloroplast DNA in mature and senescing leaves: A reappraisal. Plant Cell 26: 847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Christopher D.A., Mullet J.E. (1993). Direct evidence for selective modulation of psbA, rpoA, rbcL, and 16S RNA stability during barley chloroplast development. Plant Mol. Biol. 22: 447–463. [DOI] [PubMed] [Google Scholar]
- Lawrence M.E., Possingham J.V. (1986). Direct measurement of femtogram amounts of DNA in cells and chloroplasts by quantitative microspectrofluorometry. J. Histochem. Cytochem. 34: 761–768. [DOI] [PubMed] [Google Scholar]
- Li W., Ruf S., Bock R. (2006). Constancy of organellar genome copy numbers during leaf development and senescence in higher plants. Mol. Genet. Genomics 275: 185–192. [DOI] [PubMed] [Google Scholar]
- Miyamura S., Nagata T., Kuroiwa T. (1986). Quantitative fluorescence microscopy on dynamic changes of plastid nucleoids during wheat development. Protoplasma 133: 66–72. [Google Scholar]
- Mulo P., Sakurai I., Aro E.M. (2012). Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: From transcription to PSII repair. Biochim. Biophys. Acta 1817: 247–257. [DOI] [PubMed] [Google Scholar]
- Oldenburg D.J., Bendich A.J. (2004). Changes in the structure of DNA molecules and the amount of DNA per plastid during chloroplast development in maize. J. Mol. Biol. 344: 1311–1330. [DOI] [PubMed] [Google Scholar]
- Oldenburg D.J., Rowan B.A., Zhao L., Walcher C.L., Schleh M., Bendich A.J. (2006). Loss or retention of chloroplast DNA in maize seedlings is affected by both light and genotype. Planta 225: 41–55. [DOI] [PubMed] [Google Scholar]
- Rowan B.A., Bendich A.J. (2009). The loss of DNA from chloroplasts as leaves mature: Fact or artefact? J. Exp. Bot. 60: 3005–3010. [DOI] [PubMed] [Google Scholar]
- Rowan B.A., Bendich A.J. (2011). Isolation, quantification, and analysis of chloroplast DNA. Methods Mol. Biol. 774: 151–170. [DOI] [PubMed] [Google Scholar]
- Rowan B.A., Oldenburg D.J., Bendich A.J. (2004). The demise of chloroplast DNA in Arabidopsis. Curr. Genet. 46: 176–181. [DOI] [PubMed] [Google Scholar]
- Rowan B.A., Oldenburg D.J., Bendich A.J. (2009). A multiple-method approach reveals a declining amount of chloroplast DNA during development in Arabidopsis. BMC Plant Biol. 9: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selldén G., Leech R.M. (1981). Localization of DNA in mature and young wheat chloroplasts using the fluorescent probe 4′-6-diamidino-2-phenylindole. Plant Physiol. 68: 731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaver J.M., Oldenburg D.J., Bendich A.J. (2006). Changes in chloroplast DNA during development in tobacco, Medicago truncatula, pea, and maize. Planta 224: 72–82. [DOI] [PubMed] [Google Scholar]
- Shaver J.M., Oldenburg D.J., Bendich A.J. (2008). The structure of chloroplast DNA molecules and the effects of light on the amount of chloroplast DNA during development in Medicago truncatula. Plant Physiol. 146: 1064–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., Oldenburg D.J., Bendich A.J. (2011). Independent effects of leaf growth and light on the development of the plastid and its DNA content in Zea species. J. Exp. Bot. 62: 2715–2730. [DOI] [PubMed] [Google Scholar]
- Zoschke R., Liere K., Börner T. (2007). From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J. 50: 710–722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.