This study presents a computational model for auxin transport based on actual root cell geometries and carrier subcellular localizations and tested using the DII-VENUS auxin sensor. The model shows that nonpolar AUX1/LAX influx carriers control which tissues have high auxin levels, whereas the polar PIN carriers control the direction of auxin transport within these tissues.
Abstract
Auxin is a key regulator of plant growth and development. Within the root tip, auxin distribution plays a crucial role specifying developmental zones and coordinating tropic responses. Determining how the organ-scale auxin pattern is regulated at the cellular scale is essential to understanding how these processes are controlled. In this study, we developed an auxin transport model based on actual root cell geometries and carrier subcellular localizations. We tested model predictions using the DII-VENUS auxin sensor in conjunction with state-of-the-art segmentation tools. Our study revealed that auxin efflux carriers alone cannot create the pattern of auxin distribution at the root tip and that AUX1/LAX influx carriers are also required. We observed that AUX1 in lateral root cap (LRC) and elongating epidermal cells greatly enhance auxin’s shootward flux, with this flux being predominantly through the LRC, entering the epidermal cells only as they enter the elongation zone. We conclude that the nonpolar AUX1/LAX influx carriers control which tissues have high auxin levels, whereas the polar PIN carriers control the direction of auxin transport within these tissues.
INTRODUCTION
The plant hormone auxin is an important regulator of plant growth and development (Benjamins and Scheres, 2008) and plays a key role in organ initiation and in tropic responses (Marchant et al., 1999; Benková et al., 2003). Auxin is transported through plant tissues via specialized carriers on the cell membranes; the cellular and subcellular location of these membrane proteins can create auxin maxima and directed fluxes on the organ scale (Swarup et al., 2005; Grieneisen et al., 2007).
Within the plant root, auxin moves in a rootward direction within the stele, redistributes at the root tip, and moves in a shootward direction through the root's outer layers, a flux pattern often referred to as a reversed fountain (Grieneisen et al., 2007). These fluxes are thought to be essential both for patterning the root tip, specifying the position of the quiescent center (QC) and developmental zones (Sabatini et al., 1999; Blilou et al., 2005; Jiang and Feldman, 2005; Grieneisen et al., 2007), and for controlling the gravitropic response, communicating an auxin asymmetry formed at the tip to the elongating tissues (Marchant et al., 1999; Rashotte et al., 2000; Ottenschläger et al., 2003; Swarup et al., 2005). How these organ-scale auxin fluxes are created by regulation at the cellular scale is an active research area. Studies have primarily focused on the role of the PIN membrane proteins that facilitate auxin efflux from the cytoplasm to the apoplast (Blilou et al., 2005). These proteins have tissue-specific subcellular locations; for example, PIN1 is observed on the rootward faces of the stele (Gälweiler et al., 1998) and PIN2 on the shootward faces of epidermal cells (Blilou et al., 2005) (see Figure 1A for the location of the different root tissues). Several modeling studies have shown that this PIN distribution can create the reversed fountain auxin flux pattern (Grieneisen et al., 2007, 2012; Stoma et al., 2008; Santuari et al., 2011; Mironova et al., 2012).
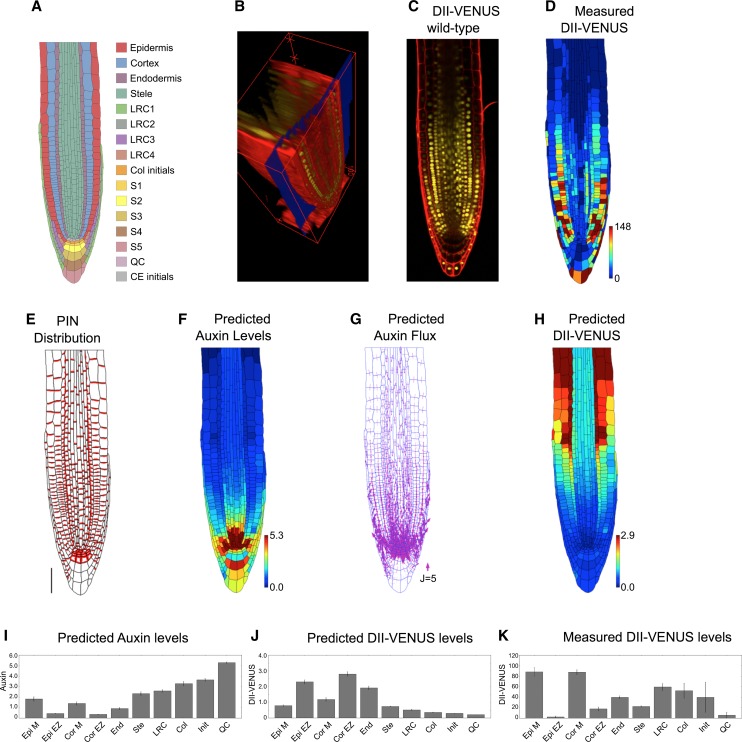
Figure 1.
Auxin Transport by the PIN Efflux Carriers Alone Does Not Produce the Auxin Distribution in the Root Tip.
(A) Schematic showing cell types labeled on the wild-type root geometry. CE, cortical/endodermal.
(B) 3D measuring plane (blue) through a confocal stack of images produced using SurfaceProject software.
(C) Output 2D interpolated slice, showing DII-VENUS (yellow) and cell geometries (red) within an Arabidopsis root tip (stained using propidium iodide).
(D) Measured DII-VENUS levels extracted from the confocal image shown in (C).
(E) Prescribed PIN distribution. Bar = 50 μm.
(F) Predicted auxin distribution.
(G) Predicted auxin fluxes (with arrow width scaling with flux).
(H) Predicted DII-VENUS distribution.
(I) Mean predicted auxin concentration in the different cell types.
(J) Mean predicted DII-VENUS concentration in the different cell types.
(K) Mean measured DII VENUS levels in the different cell types.
In (D), (F), and (H), the heat map upper limit is prescribed automatically to be the 95th percentile of the predicted/measured values in each case. In (I) to (K), error bars show 1 se. Epi, epidermis; Cor, cortex; End, endodermis; Ste, stele; Col, columella; Init, columella initials; M, meristem; EZ, elongation zone. Supplemental Table 1 lists the number of cells in each category.
Genetic studies suggest that the auxin distribution in the root is governed not only by the PINs but also by the AUX1/LAX membrane proteins, which facilitate anionic auxin influx from the apoplast to the cytoplasm (Kramer, 2004; Swarup and Péret, 2012). The presence of AUX1 is essential for the root's gravitropic response (Marchant et al., 1999). This finding was corroborated by a computational model of the three outer root layers of the elongation zone, which showed that AUX1 expression within the epidermis, but not the cortex and endodermis, causes the epidermal auxin concentration to be high, resulting in a shootward flux (Swarup et al., 2005). Models have also demonstrated the importance of AUX1 in controlling the auxin distribution during root hair initiation (Jones et al., 2009) and root nodule formation (Perrine-Walker et al., 2010). Despite these findings, the spatial distribution of AUX1/LAX proteins has not been incorporated into recent root tip models (Grieneisen et al., 2007, 2012; Stoma et al., 2008; Mironova et al., 2012). Other classes of membrane proteins have also been shown affect auxin transport; for example, members of the ABCB transporter family contribute to auxin efflux (Spalding, 2013), and nitrate and potassium transporters have been shown to facilitate auxin movement (Vicente-Agullo et al., 2004; Krouk et al., 2010).
In this study, we seek to develop realistic models of auxin transport within the root tip of the model plant Arabidopsis thaliana. Previous experimental work has suggested that the lateral root cap (LRC) is important in creating the shootward auxin fluxes (Swarup et al., 2005). However, this tissue has also not been included in most previous auxin transport models (Swarup et al., 2005; Grieneisen et al., 2007; Grieneisen et al., 2012; Mironova et al., 2012): These represented root cells (or subcellular compartments) by rectangular geometries and hence could not capture accurately cell-to-cell connectivities. To address these issues, we base our model on actual cell geometries segmented from confocal microscopy image data.
To test our model predictions, we use the recently developed auxin sensor DII-VENUS (Santuari et al., 2011; Vernoux et al., 2011; Brunoud et al., 2012). Until recently, modelers have had to rely on the auxin response reporter DR5 (Ulmasov et al., 1997) for a qualitative verification of their predictions. However, as an output of the auxin signaling pathway, the DR5 response depends on the cell's level of intermediate proteins (namely, members of the Aux/IAA and ARF families). These intermediate protein levels are likely to vary significantly between different tissues, making quantitative comparisons subject to error (Wells et al., 2013). In contrast, DII-VENUS is closely related to auxin concentrations: Within each cell, auxin degrades Aux/IAA by first binding with the TIR1/AFB receptors, forming complexes that can bind with domain II of the Aux/IAA, promoting their degradation via the ubiquitination pathway (Supplemental Figure 1; Dharmasiri et al., 2005); with domain II of the Aux/IAA fused to the VENUS fluorescent protein, the DII-VENUS sensor does not depend on the levels of Aux/IAA and ARF proteins (Brunoud et al., 2012; Wells et al., 2013), enabling it to be used as a quantitative tool (Band et al., 2012). Again, in contrast with previous modeling studies, we explicitly incorporate part of the auxin signaling pathway in order predict the DII-VENUS distribution (using the parameterized model of Band et al., 2012). By comparing these predictions with DII-VENUS measurements from the same root geometry, we can test our predictions on a cell-by-cell basis, providing a quantitative assessment of the model results. This systems approach reveals that both influx and efflux carriers are essential to create the auxin distribution at the root tip, with the influx carriers determining the sites of auxin accumulation.
RESULTS
Creating a Virtual Root Model by Integrating Cell Geometries and Auxin Efflux Carrier Distributions
Recent models that use regular cell geometries have suggested that the pattern of PIN efflux carriers can create the auxin distribution at the root tip (Grieneisen et al., 2007, 2012; Stoma et al., 2008; Mironova et al., 2012) with an auxin maxima in the QC region (Petersson et al., 2009). We therefore first considered the role of the PINs and predicted what auxin distribution they create when modeling transport within a root tip comprised of actual root cell geometries. We used a static root tip geometry, which is reasonable because auxin-transport time scales are considerably shorter than the time scale of growth (see discussion in Band and King, 2012).
We used stacks of confocal microscopy images of DII-VENUS–expressing roots stained with propidium iodide to capture root cell geometries and reveal cellular organization (Figure 1B; see Methods). As expected, different cells were in focus in different planes within the image stack; therefore, to produce a 2D geometry suitable for our analysis, we developed a software tool, called SurfaceProject, to extract a plane from the stack (Figure 1B; Supplemental Figure 2; see Methods). Using this plane (Figure 1C), we segmented the multicellular root tip geometry and nuclear fluorescence channel using the CellSeT tool (Pound et al., 2012); these geometries (Supplemental Table 1) and fluorescence intensities (Figure 1D) were read into a vertex-based model, based on the OpenAlea modeling framework (Pradal et al., 2008).
We prescribed a PIN distribution on our virtual root tissue (Figure 1E) based on reports in the literature and our own observations using anti-PIN antibodies raised against the central loop of PIN1, 2, 3, 4, and 7 (Supplemental Figure 3, Supplemental Tables 2 and 3, and Supplemental Methods, specifying the carrier distributions). We specified PIN proteins to be on the shootward-facing membranes in the LRC, elongation-zone cortex, and elongation-zone and distal-meristem epidermis, on the rootward-facing membranes in the stele, endodermis and meristem cortex, and proximal-meristem epidermis, and on all faces of cells in the QC, columella initial, and S1 and S2 tiers of the columella (Friml et al., 2002a, 2002b; Peer et al., 2004; Blilou et al., 2005; Abas et al., 2006; Müller et al., 1998). Given that the relative rates of auxin transport by the different PIN proteins are unknown, we did not distinguish the different members of the PIN family and supposed that membrane faces that are observed to contain PIN proteins have the same PIN-mediated efflux permeability.
The model also incorporated a weak background efflux to account for any low levels of nonpolar PIN and for the presence of ABCB transporters. ABCBs appear to have nonpolar subcellular distributions with different members of the ABCB family collectively present within most root tissues (Spalding, 2013); we could therefore capture the ABCB-facilitated efflux by specifying a background efflux on the cell membranes.
We also included auxin production and degradation: Auxin has been shown to be synthesized in the root tip (Ljung et al., 2005), and high levels of auxin biosynthesis enzymes have been observed in the QC and columella initials (Stepanova et al., 2008). We therefore specified small production and degradation rates within every cell and higher auxin production in the QC and columella initials. The model also captured auxin diffusion through the apoplast, as measured by Kramer et al. (2007). Auxin diffusion within the cytoplasm is thought to be much quicker than that in the apoplast (Swarup et al., 2005; Kramer et al., 2007), and we treated auxin concentrations as uniform within each cell. Spatial auxin variations in the root tip can be considered negligible due to the small size of cells in this region (Band and King, 2012). This is in contrast with the larger cells further from the root tip, where subcellular variations in auxin have previously been considered (Laskowski et al., 2008; Payne and Grierson, 2009).
The resulting model can be described by a system of ordinary differential equations (ODEs) for the auxin concentrations within each cell and each segment of apoplast (Supplemental Methods, modeling auxin dynamics in the root tip); these equations depend on a number of model parameters, as detailed in Supplemental Tables 4 to 6. We represented the supply of auxin from the shoot by prescribing a non-zero auxin concentration within the stele cells at the boundary of the modeled tissue and allowed auxin to leave that tissue through the outer layers by setting the auxin concentrations in the epidermal, cortical, and endodermal cells at the boundary to zero. Starting from an initial condition in which all remaining concentrations equal zero, we simulated the ODEs until the concentrations and fluxes reached a steady state. In addition, we developed a graphical user interface (GUI), named SimuPlant: The Virtual Root to enable those interested to interact with the model (see Methods and Discussion below).
Auxin Distribution in the Root Tip Depends on Mechanisms in Addition to the Efflux Carriers
In agreement with previous studies that used rectangular cell geometries (Grieneisen et al., 2007, 2012), the model predicted a hormone maxima at the root tip, with auxin levels reducing in a shootward direction (Figures 1F and 1I). We predicted the reversed fountain flux pattern, with rootward fluxes through the stele and shootward fluxes through the roots outer layers (Figure 1G).
To test quantitatively our predicted auxin distributions, we also modeled the DII-VENUS auxin sensor. The dynamic relationship between auxin and DII-VENUS has recently been characterized using a parameterized mathematical model (Band et al., 2012), and we used this model to predict the DII-VENUS level within each cell (Figures 1H and 1J). These predictions were compared with observed DII-VENUS levels extracted using the CellSeT tool (Figures 1D and 1K). Interestingly, there were significant differences between the predicted and measured DII-VENUS distributions. We predicted DII-VENUS levels to be low close to the QC and to increase in a shootward direction (Figures 1H and 1J) (the reverse of the auxin pattern), whereas our measurements revealed that DII-VENUS is high in the meristematic epidermal and cortical cells, reducing as these cells enter the elongation zone (Figures 1D and 1K) (Brunoud et al., 2012).
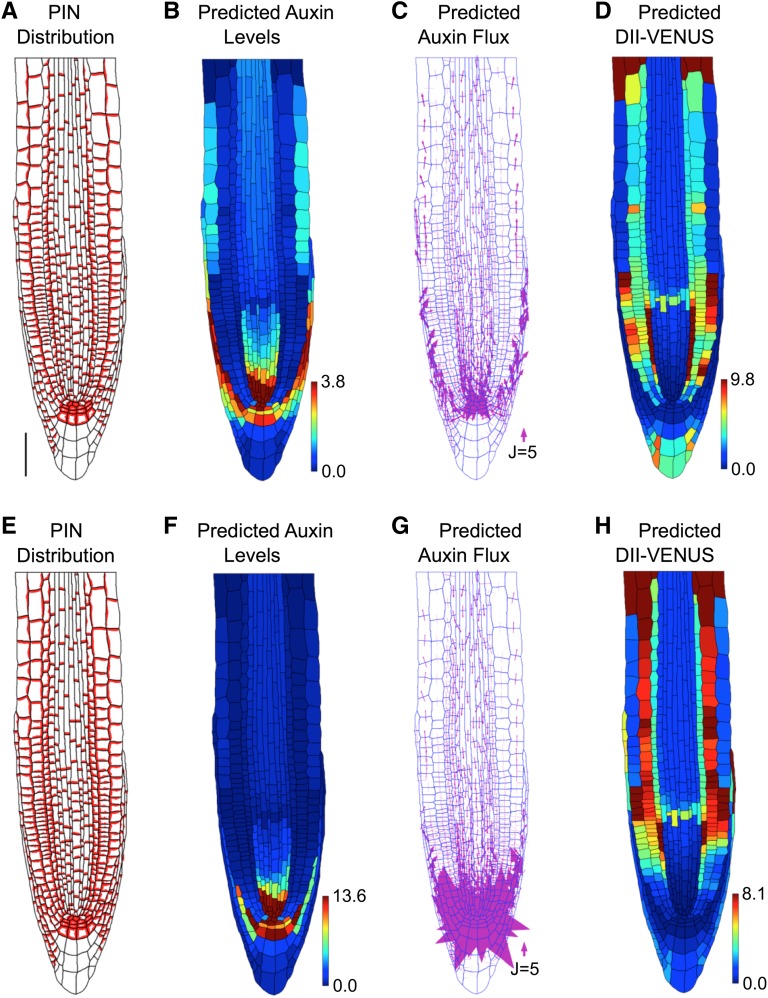
To explore the robustness of the model results, we considered several modifications to our model. We predicted how the auxin distribution is affected by (1) different cases of wild-type cell geometries (Supplemental Figure 4), (2) the presence of PINs on the inner periclinal membranes (i.e., the membranes facing the root center) of the epidermal, cortical, and LRC cells (Supplemental Figure 5), (3) removing auxin production and degradation (Supplemental Figure 6), and (4) removing the nonpolar auxin efflux (Supplemental Figure 7). We found that these changes modified the gradient of auxin away from the QC but did not qualitatively affect the predicted auxin and DII-VENUS distributions. We concluded that carrier-mediated efflux alone cannot reproduce the auxin distribution at the root tip.
The Interplay between PIN/ABCB-Mediated Efflux and AUX1/LAX-Mediated Influx Can Capture the Auxin Distribution at the Root Tip
Since the efflux carriers alone could not recreate the auxin distribution, we extended our model to incorporate active influx via the nonpolar AUX1/LAX membrane proteins. As described in the literature and shown in Supplemental Figure 8, we took AUX1 to be present in the LRC, elongation zone epidermis, and S1, S2, and S3 tiers of the columella (Swarup et al., 2001, 2005), LAX2 to be present in the QC, columella initials, and rootward half of the meristematic stele (Péret et al., 2012), and LAX3 to be present only in the S2 tier of the columella (Swarup et al., 2008; Péret et al., 2012).
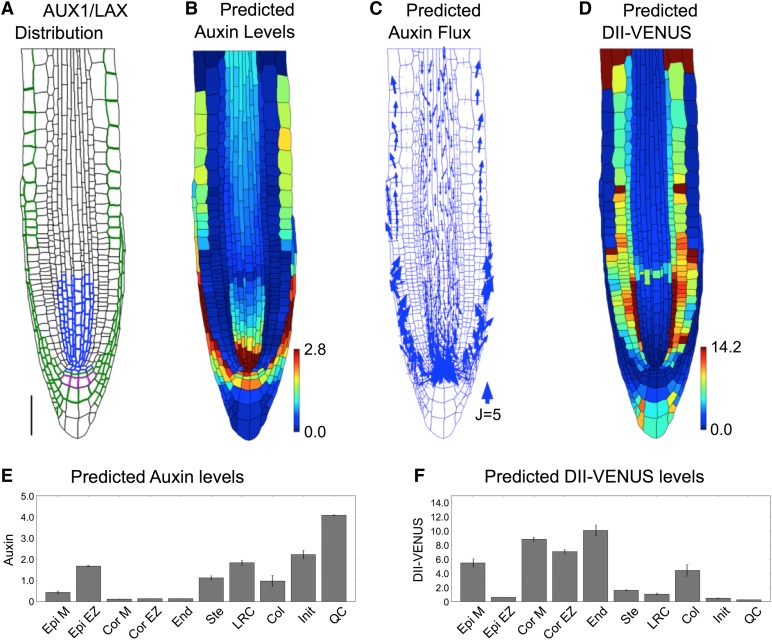
Prescribing the observed AUX1/LAX distributions on our virtual root tissue (Figure 2A; Supplemental Table 7 and Supplemental Methods, specifying the carrier distributions) and simulating the model dynamics, we predicted the steady state distributions and fluxes (Figures 2B to 2F), obtaining high auxin levels within the QC, LRC, and elongating epidermal cells (Figures 2B and 2E). Auxin flows toward the QC through the stele and away from the QC though the LRC, flowing into the epidermis at the end of the LRC, as cells enter the elongation zone (Figure 2C).
Figure 2.
The Auxin Distribution in the Root Tip Is Due to the Interplay between PIN and AUX1/LAX-Mediated Active Transport.
(A) Prescribed distributions of AUX1 (green and purple), LAX2 (blue), and LAX3 (purple). Bar = 50 μm.
(B) Predicted auxin distribution.
(C) Predicted auxin fluxes (with arrow width scaling with flux).
(D) Predicted DII-VENUS distribution.
(E) Mean predicted auxin concentration in the different cell types.
(F) Mean predicted DII-VENUS concentration in the different cell types.
The predicted DII-VENUS distribution is in reasonably good agreement with our measurements (compare Figures 2D and 2F to Figures 1D and 1K): DII-VENUS is predicted to be low in the QC region and LRC, whereas in the epidermis, DII-VENUS is predicted to be high in the meristem but to reduce significantly as cells emerge from under the LRC to enter the elongation zone (Figure 2D). Tissue-specific auxin measurements using cell-sorting techniques have also revealed the epidermal auxin concentration to be low in the meristem (Petersson et al., 2009), consistent with our model predictions (Figures 2B and 2E) and DII-VENUS measurements (Figures 1D and 1K) (see Supplemental Figure 9 for replicates). We conclude that influx carriers play crucial roles in controlling the auxin distribution at the root tip.
AUX1 Is Essential to Create the Auxin Distribution at the Root Tip
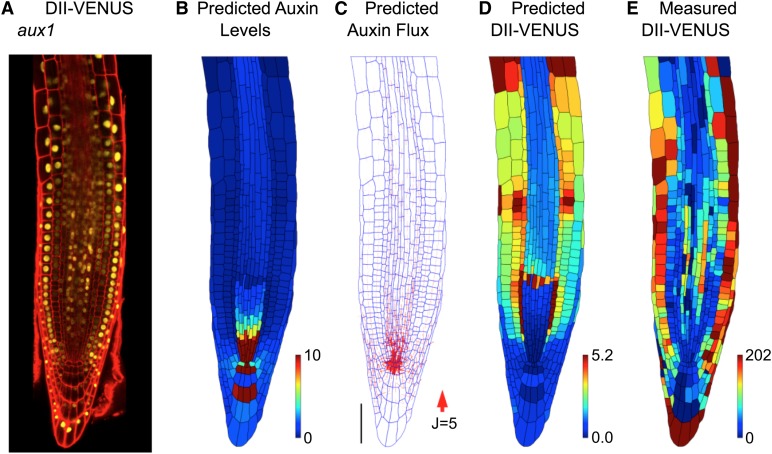
Although extending our model to incorporate the AUX1/LAX influx carriers enabled us to predict the observed wild-type DII-VENUS distribution, this does not rule out other explanations. We tested our model predictions further by considering an aux1 null mutant by crossing the aux1-22 allele (Swarup et al., 2004) with the DII-VENUS reporter. Using root cell geometries extracted from an image of this line (Figure 3A), we predicted that removing AUX1 would result in a buildup of auxin within the QC and surrounding cells, with little difference between the auxin levels in the different tissue layers and only small shootward fluxes through the root's outer layers (Figures 3B and 3C). The predicted and measured DII-VENUS levels were in reasonably good agreement, with low DII-VENUS (corresponding to the pool of high auxin) surrounding the QC (Figures 3D and 3E) (see Supplemental Figure 10 for replicates). We conclude that AUX1 is necessary to produce the observed auxin distributions.
Figure 3.
AUX1 Significantly Affects the Auxin Distribution at the Root Tip.
(A) Confocal image of an aux1 null mutant root tip, reporting DII-VENUS (yellow) with propidium iodide background staining (red).
(B) to (E) Predictions/measurements for the aux1 null mutant.
(B) Predicted auxin distribution.
(C) Predicted auxin fluxes (with arrow width scaling with flux). Bar = 50 μm.
(D) Predicted DII-VENUS distribution.
(E) Measured DII-VENUS levels.
To investigate further the role of the PIN efflux carriers, we simulated the auxin dynamics with no PINs present (Supplemental Figure 11). The predicted auxin distributions are similar both with and without polar PINs (Figure 2B; Supplemental Figure 11A); however, omitting the polar PINs led to predicted auxin fluxes that are considerably smaller (Supplemental Figure 11B). These results suggested that the polar PINs create the directed auxin fluxes but have little effect on the auxin distribution. We conclude that the AUX1/LAX influx carriers control which tissues have high auxin levels, whereas the polar PIN carriers control the direction of auxin transport within these tissues.
AUX1 in LRC and Elongation Zone Epidermal Cells Is Required for the Auxin Dynamics
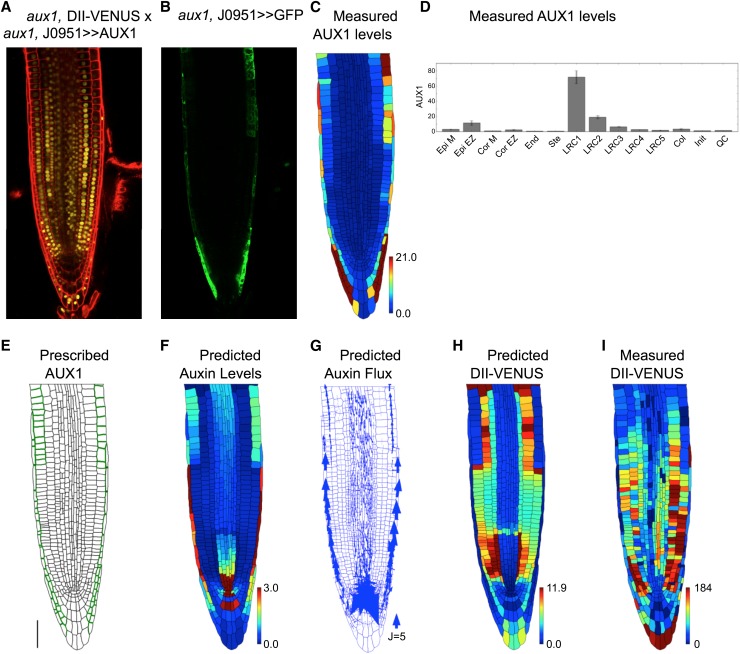
AUX1 within LRC and elongating epidermal cells has been shown to be essential for the gravitropic response (Swarup et al., 2005). To investigate the role of these tissues further, we crossed the aux1, DII-VENUS reporter line with aux1, J0951>>AUX1 (Swarup et al., 2005), which expresses AUX1 solely in the outer LRC and epidermal cells (Figures 4A to 4D). Considering this case in our model (Figure 4E), we predicted high auxin levels within the LRC and elongating epidermal cells (Figure 4F), consistent with the observed DII-VENUS pattern (Figures 4H and 4I). Furthermore, AUX1 in just these tissues reinstates the shootward auxin fluxes through the root's outer layers (Figure 4G) (explaining their role in the gravitropic response). Results of replicate experiments are shown in Supplemental Figures 12 and 13.
Figure 4.
AUX1 in LRC and Elongation Zone Epidermal Cells Is Required for the Auxin Dynamics.
(A) Confocal image of aux1, DII-VENUS × aux1, J0951>>AUX1, reporting DII-VENUS (yellow) with propidium iodide background staining (red).
(B) Green channel from confocal image in (A) showing J0951>>GFP (and hence AUX1), confirming AUX1 expression solely in the LRC and epidermis.
(C) AUX1 fluorescence level for each cell extracted from confocal image in (B); heat map shows the average fluorescence calculated as the total fluorescence in the cell divided by the length of the cell's wall.
(D) Mean AUX1 fluorescence in the different cell types.
(E) Prescribed AUX1 distribution reflecting aux1, J0951>>AUX1 pattern ([C] and [D]). Bar = 50 µm.
(F) Predicted auxin distribution.
(G) Predicted auxin fluxes (with arrow width scaling with flux).
(H) Predicted DII-VENUS distribution.
(I) Measured DII-VENUS levels extracted from the confocal image in (A).
Within the epidermis, AUX1 is present mainly in the elongation zone cells and appears as cells emerge from under the LRC (Swarup et al., 2001, 2005) (Supplemental Figure 8A). This pattern is key to predicting epidermal auxin levels that are low in meristem and increase significantly as cells enter the elongation zone: We predicted that this auxin gradient is not present if AUX1 is expressed in all epidermal cells and moves position if more rootward epidermal cells express AUX1 (Supplemental Figure 14). We conclude that AUX1 within the LRC and elongating epidermal cells is crucial for predicting the auxin distribution and creating significant shootward auxin fluxes through the root's outer layers, consistent with the stabilization of DII-VENUS observed in these tissues in aux1 (Figure 3A).
AUX1 Prevents Significant Flux from the Shootward Stream through the Outer Layers to the Rootward Stream in the Stele
The simulations suggested that AUX1 maintains auxin within the LRC and elongating epidermal cells, resulting in no auxin flux from the outer layers into the stele (Figure 1G) (henceforth referred to as reflux). However, it has previously been suggested that this periclinal flux plays a significant role in creating the auxin distribution and is created by PINs on the inner periclinal membranes of the epidermal cells (Grieneisen et al., 2007, 2012). To test whether the placement of these PINs affected our conclusions, we simulated the model dynamics with additional PINs on the inner periclinal membranes of either the epidermal cells only (Figures 5A to 5D) or on the epidermal, cortical, and LRC cells (Figures 5E to 5H). In each case, no significant reflux was predicted (Figures 5C and 5G). The importance of reflux has been advocated in previous studies; however, these models either omitted the spatial distribution of AUX1 (Grieneisen et al., 2007, 2012) or did not include the stele tissue (Swarup et al., 2005). We conclude that AUX1 prevents significant flux from the epidermal shootward auxin stream to the rootward stream within the stele.
Figure 5.
Effect of Periclinal PINs on Predicted Wild-Type Distributions and Fluxes with Auxin Transport Mediated by Both Influx and Efflux Carriers.
(A) to (D) With inner periclinal PINs on epidermal cell membranes. Bar = 50 μm.
(E) to (H) With inner periclinal PINs on epidermal, cortical, and LRC cell membranes.
(A) and (E) Prescribed PIN distribution.
(B) and (F) Predicted auxin distribution.
(C) and (G) Predicted auxin flux (with arrow width scaling with flux).
(D) and (H) Predicted DII-VENUS distribution.
AUX1 in the Elongating Cortical Cells Significantly Modifies the Auxin Distribution
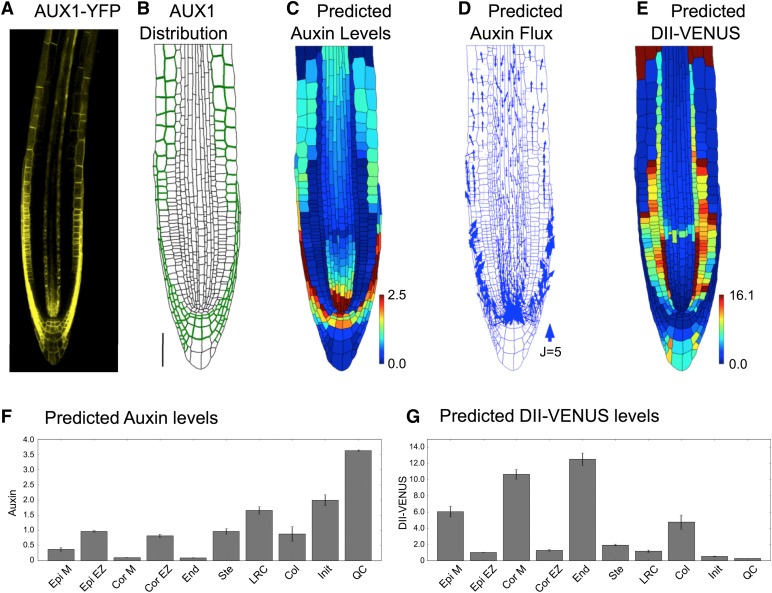
Although our above model predictions were in reasonably good agreement with the observed DII-VENUS distribution, there were some inconsistencies (in particular in the predictions for the elongating cortical cells; Figures 1D, 1K, 2D, and 2F). This led us to question our assumptions about the AUX1 distribution: Since AUX1 appears to play an essential role in determining the auxin distribution, additional cells presenting AUX1 could significantly change that distribution. Using a newly created genomic AUX1-Ypet line that includes the 14-kb promoter sequence, we found AUX1 also to be expressed within elongating cortical cells (Figure 6A; Supplemental Figure 15), suggesting a novel location for AUX1.
Figure 6.
AUX1 Expression within Elongating Cortical Cells Significantly Modifies the Auxin Distribution.
(A) Localization of AUX1 using AUX1-Ypet line reveals AUX1 in the cortex of wild-type Arabidopsis root tips.
(B) Prescribed AUX1 positions based on the AUX1-Ypet image (shown in [A]). Bar = 50 μm.
(C) to (G) Predictions for a wild-type root tip, using the AUX1 distribution shown in (B).
(C) Predicted auxin distribution.
(D) Predicted auxin fluxes (with arrow width scaling with flux).
(E) Predicted DII-VENUS distribution.
(F) Mean predicted auxin concentration in the different cell types.
(G) Mean predicted DII-VENUS concentration in the different cell types.
Including AUX1 within the elongating cortical cells (Figure 6B), we resimulated the dynamics in the wild-type plant root (Figures 6C to 6G). The addition of AUX1 to these cortical cells increases their auxin concentration (compare Figures 6C and 6F to Figures 2B and 2E) and led to a predicted DII-VENUS pattern that is in much better agreement with that observed experimentally (Figures 1D, 1K, 6E, and 6G; see Supplemental Figure 16 for replicates). We conclude that AUX1 expression within elongating cortical cells significantly modifies the auxin distribution.
DISCUSSION
The auxin distribution in the root tip controls tropic responses and specifies the developmental zones. We report that the AUX1/LAX auxin influx carriers play crucial roles in creating this auxin distribution. By developing and testing a model of auxin dynamics, our study suggested that the nonpolar influx carriers control which tissues have high auxin levels, whereas the polar PIN carriers control the direction of auxin transport within these tissues. Previous models of auxin transport within the root tip have focused primarily on PIN distribution and have investigated how the PINs create the auxin maxima close to the QC and the reversed-fountain auxin flux pattern (Grieneisen et al., 2007, 2012; Stoma et al., 2008; Santuari et al., 2011; Mironova et al., 2012). While these studies have significantly improved our understanding of auxin transport within the root tip, new techniques and tools (namely, detection via DII-VENUS, segmentation using CellSeT, and modeling based on the OpenAlea framework) motivated our further investigation. Our simulations suggested that the PIN carriers are not alone in strongly influencing the auxin distribution, as reported by both DII-VENUS (Brunoud et al., 2012) and direct measurements using cell-sorting methods (Petersson et al., 2009).
Our results demonstrated the importance of the LRC, showing that AUX1 causes the shootward auxin flux to be channeled through the LRC, entering the epidermis only at the start of the elongation zone. Interestingly, this flux results in a significant increase in epidermal auxin levels at approximately the place where cells cease division and begin rapid elongation, providing evidence against the hypothesis that a gradual reduction in auxin levels away from the QC specifies the root's developmental zones (Blilou et al., 2005; Grieneisen et al., 2007). Although not addressed here, investigating how the auxin distributions revealed in this study affect cell growth will significantly impact understanding of root development.
The current model predicts many of the features of the observed DII-VENUS distribution; however, there are still some discrepancies. In particular, the predicted endodermal DII-VENUS levels are higher than those observed, suggesting that additional components are required to increase the predicted endodermal auxin concentrations. The qualitative features of the predicted auxin distribution appear to be relatively robust to the specific choice of parameter values, although further experimental efforts to characterize key model parameters (and their temporal and spatial variations) would enable additional model refinement. Detailed measurements of the permeabilities associated with the different influx and efflux carriers would improve the accuracy of the model and enable us to investigate the roles of the different family members further (for example, enabling us to assess whether the different members of the PIN family facilitate different rates of auxin transport). Similarly, the relative roles of the carrier-mediated and passive transport components depend on pH measurements: For example, a larger apoplastic pH would lead to a higher proportion of auxin being anionic and entering the cells via the influx carriers. Thus, increases/decreases in apoplastic pH leads to the influx carriers having larger/smaller effects on the predicted distributions (Supplemental Figures 17 and 18).
Auxin distributions are also affected by spatial variations in auxin production and degradation. We prescribed high auxin production in the QC and in the columella initials (reflecting observed biosynthesis enzyme distributions; Stepanova et al., 2008); increasing/decreasing the production rate in these tissues increases/decreases their auxin levels (Supplemental Figures 19 and 20). Furthermore, simulations suggested that apoplastic diffusion and nonpolar efflux (as mediated by nonpolar PINs and/or ABCBs, for example) promote periclinal auxin transport, enabling auxin to move between epidermal, cortical, and endodermal tissues; removing these fluxes reduces the agreement between our predicted and observed DII-VENUS levels (Supplemental Figures 21 and 22). Measurements of apoplast thicknesses and characterization of the mechanisms underlying nonpolar transport would allow further fine-tuning of our predictions and understanding.
Our study also highlights the benefits of developing and modeling fluorescent reporters (Wells et al., 2013). Until recently, the auxin response has been visualized using the DR5 reporter (Ulmasov et al., 1997). Although observed DR5 patterns qualitatively agree with those for DII-VENUS, the latter provides a more detailed readout of the auxin distribution, enabling us to test model predictions quantitatively on a cell-by-cell basis. Since using DII-VENUS in conjunction with state-of-the-art segmentation tools enabled us to gain a unique perspective on this key plant hormone, we envisage the procedure being applied to auxin studies in other plant organs and species, with similar integrative approaches benefiting the use of fluorescently labeled hormone variants developed for gibberellin (Shani et al., 2013) and brassinosteroids (Irani et al., 2012).
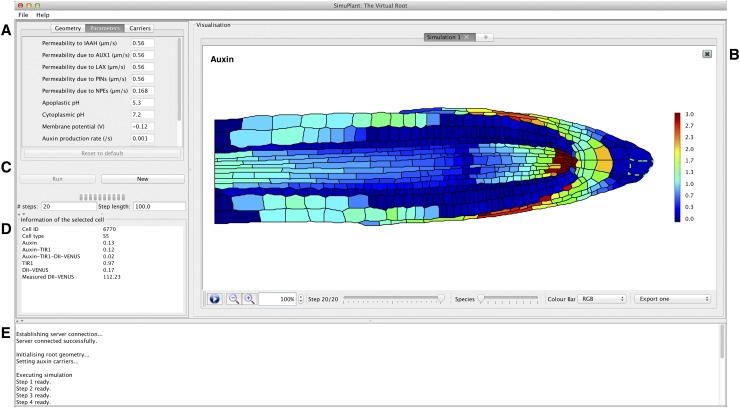
Finally, to enable researchers to interact with our models, we developed a GUI, SimuPlant: The Virtual Root (Figure 7). Through the GUI, one can explore how the dynamic auxin and DII-VENUS distributions are affected by the model parameter values and by the presence and distributions of the auxin carriers.
Figure 7.
The SimuPlant GUI Consists of Five Main Components.
(A) Model specification, which allows different cell geometries to be selected, model parameters to be altered, and carrier classes to be included or excluded from cell files.
(B) Simulation visualization, showing the output for the modeled species in the cell geometry for all time steps.
(C) Simulation Control, which allows the simulation on the server to be started or stopped and to control the number and duration of modeled time steps.
(D) Simulation cell output, which shows the model output for any cell selected in the simulation visualization (indicated by green dashed lines around the cell wall).
(E) Task window, which lists progress of tasks sent to and received from the server.
METHODS
Plant Material and Growth Conditions
All Arabidopsis thaliana lines were in the Columbia-0 background. Six-day-old DII-VENUS seedlings were grown from surface-sterilized seeds as described previously (Holman et al., 2010). To generate reporter plants with tissue-specific AUX1 expression (Figures 4A and 4B), the aux1, DII-VENUS line (Figure 3A) was crossed with the aux1, J0951>>AUX1 line (Swarup et al., 2005), in which AUX1 expression is targeted to the LRC and expanding epidermal tissues in the aux1 mutant background using a GAL4-based transactivation expression approach. The Ypet fusion to AUX1 (Figure 6A) was generated by employing a recombineering approach (Zhou et al., 2011). In brief, a PCR fragment containing the Ypet coding sequence was recombined in frame into the AUX1 genomic sequence (encoded on a TAC plasmid in Escherichia coli) to create an AUX1-VENUS protein fusion whose expression was driven by the native 14-kb promoter sequence.
Immunolocalization and Imaging
Four-day-old seedlings were fixed, and immunolocalization experiments were performed as described previously (Péret et al., 2012) and visualized using confocal laser scanning microscopy. Primary antibodies were normally used at 1:100 to 1:200; Alexa Fluor–coupled secondary antibodies (Invitrogen) were used at 1:200 dilution. Live imaging was performed using an SP5 spectral detection confocal laser scanning microscope (Leica). Roots were stained with propidium iodide (Sigma-Aldrich) to visualize apoplast organization. Microscope settings were as detailed by Pound et al. (2012).
Segmentation
Image stacks were taken about the midline of roots to include all nuclei required for quantification (typically 30 steps at 1-µm intervals). 2D planes were constructed from the 3D stacks using the SurfaceProject plug-in for the Fiji image processing package (Schindelin et al., 2012). Details of SurfaceProject can be found in the Supplemental Methods, and the software and user documentation are provided at www.simuplant.org. Cell geometries and DII-VENUS and GFP fluorescence intensities were extracted from the confocal images using the CellSeT segmentation tool, as described by Pound et al. (2012).
Modeling
The geometrical and fluorescence measurements from CellSeT were read into an OpenAlea data structure (Pradal et al., 2008). Using the resulting multicellular tissue structure, auxin transport was modeled using a vertex-based approach (Supplemental Figure 23). The auxin and DII-VENUS dynamics described in the main text were captured by a system of ODEs, which depend on a number of parameter values (Supplemental Tables 4 to 6), which were sourced from the literature (Scott and Allen, 1999; Sze et al., 1999; Fasano et al., 2001; Swarup et al., 2005; Heisler and Jönsson, 2006; Kramer et al., 2007). Simulations were performed in the Python programming language (www.python.org) using the ODE solver LSODES from the ODEPACK suite (Hindmarsh, 1983) to approximate numerically solutions of the system of differential equations. This solver exploits the sparsity of the Jacobian of the system of equations; the sparsity pattern of the Jacobian was supplied to the numerical solver to improve the speed of the simulations. Simulations were run until the distributions and fluxes reached their steady states. These steady state solutions were then presented using the Python library matplotlib (Hunter, 2007). Plotting the steady state concentration distributions required us to choose a range for the heat map, which we prescribed automatically to minimize subjectivity. Plotting histograms of the predicted and measured concentrations of the different cells (for example, Supplemental Figure 24), we observed that the lower limit of the concentration distribution is (unsurprisingly) zero for both auxin and DII-VENUS and that the distributions are negatively skewed. Although we could have chosen the upper limit of the heat map range to be maximum predicted/measured concentration in each case, outliers in the concentration distribution would significantly affect the appearance of the presented distribution. We therefore prescribed the heat map upper limit to be the 95th percentile of the concentration values. In the main text and supplemental figures, we present our predicted distributions of auxin and DII-VENUS. The model also predicted the concentration distribution of the free and bound TIR1/AFB receptors; in Supplemental Figure 25, we show sample predictions of these that correspond to the simulations in Figures 3 and 6 (see Supplemental Methods for further details).
SimuPlant GUI
A GUI called SimuPlant was developed to enable those interested to interact directly with the simulations presented in this study. SimuPlant is written in Java 7, which is easily installable on most modern operating systems (OSX/Windows/Linux). The client connects to a simulation server, where all the computationally intensive operations are performed. The default behavior is for the client to connect to the server provided by the University of Nottingham, available via www.simuplant.org. However, for intensive use, it is also possible to install the server locally. In this case, the software is available from Sourceforge; links to this software can be found on the GUI website www.simuplant.org. The client permits the user to choose between the nine different root tip geometries presented herein, each acquired from confocal images using SurfaceProject and CellSeT; three of these are from wild-type plants (Figure 1C; Supplemental Figures 4A and 4G), three from aux1 null mutant plants (Figure 3A; Supplemental Figures 10A and 10F), and three from the aux1, DII-VENUS × aux1, J0951>>AUX1 line (Swarup et al., 2005) in which AUX1 is expressed solely in the outer LRC and epidermal cells (Figure 4A; Supplemental Figures 12A and 13A). The parameters in the auxin transport model may be varied by the user, and the user can select different options for the placement of the auxin influx and efflux carriers. Default settings correspond to the parameter values listed in Supplemental Tables 4 and 6 and the carrier distributions detailed in Supplemental Tables 3 and 7. Simulation outputs can be displayed in the GUI. The GUI shows the temporal evolution of the solution toward a steady state, allowing one to visualize the flux of auxin through the root tissues. Simulation outputs can be saved in a portable network graphics format, and one can select individual cells to view the predicted concentrations within that cell. Further information about the GUI is provided in the user documentation available at www.simuplant.org.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Network of Interactions That Leads to Auxin-Mediated Degradation of DII-VENUS.
Supplemental Figure 2. Constructing a 2D Plane from a 3D Confocal Image Stack Using SurfaceProject.
Supplemental Figure 3. In Situ Immunodetection of PIN Proteins in the Arabidopsis Root Tip.
Supplemental Figure 4. Replicates of Results for the Wild-Type Root Tip Considering Efflux Carrier–Mediated Transport Only (i.e., with No Influx Carriers Present).
Supplemental Figure 5. Effect of Periclinal PINS on the Predicted Distributions and Fluxes with Efflux Carrier–Mediated Transport only (i.e., with No Influx Carriers Present).
Supplemental Figure 6. Effect of Omitting Auxin Synthesis and Degradation on the Predicted Distributions and Fluxes with Efflux Carrier–Mediated Transport Only (i.e., with No Influx Carriers Present).
Supplemental Figure 7. Effect of Omitting Nonpolar Efflux (as Mediated by Nonpolar PINs and/or ABCBs) on the Predicted Distributions and Fluxes with Efflux Carrier–Mediated Transport Only (i.e., with No Influx Carriers Present).
Supplemental Figure 8. Expression Patterns of AUX1/LAX Auxin Transporters in the Arabidopsis Root Tip.
Supplemental Figure 9. Replicates of Predictions for the Wild-Type Plant Root with Auxin Transport Mediated by Both Efflux and Influx Carriers.
Supplemental Figure 10. Replicates of Results for the aux1 Null Mutant Plant Root.
Supplemental Figure 11. Effect of Omitting the Polar PINs on the Predicted Wild-Type Distributions and Fluxes (with Auxin Transport Mediated by Both Influx and Efflux Carriers).
Supplemental Figure 12. Replicate 1 Results for the aux1 J0951>>AUX1 Mutant Plant Root in Which AUX1 Is Expressed Solely in the Outer LRC and Epidermal Cells.
Supplemental Figure 13. Replicate 2 Results for the aux1 J0951>>AUX1 Mutant Plant Root in Which AUX1 Is Expressed Solely in the Outer LRC and Epidermal Cells.
Supplemental Figure 14. Effect of AUX1 Being Expressed in Meristematic Epidermal Cells on the Predicted Wild-Type Distributions and Fluxes (with Auxin Mediated by Both Influx and Efflux Carriers).
Supplemental Figure 15. Replicates for the AUX1-Ypet Line.
Supplemental Figure 16. Replicates of Simulations for the Wild-Type Root Tip with AUX1 Expressed in the Elongating Cortical Cells.
Supplemental Figure 17. Effect of Apoplastic pH on the Predicted Distributions and Fluxes in the Wild-Type Root Tip.
Supplemental Figure 18. Effect of Apoplastic pH on the Predicted Distributions and Fluxes in the aux1 Null Mutant Root Tip.
Supplemental Figure 19. Effect of Auxin Production in the QC and Columella Initials on the Predicted Distributions and Fluxes in the Wild-Type Root Tip.
Supplemental Figure 20. Effect of Auxin Production in the QC and Columella Initials on the Predicted Distributions and Fluxes in the aux1 Null Mutant Root Tip.
Supplemental Figure 21. Effect of Omitting Cell Wall Diffusion and/or Nonpolar Efflux on the Predicted Distributions and Fluxes in Wild-Type Root Tip.
Supplemental Figure 22. Effect of Omitting Apoplastic Diffusion and/or Nonpolar Efflux on the Predicted Distributions and Fluxes in an aux1 Null Mutant Root Tip.
Supplemental Figure 23. Schematics Illustrating the Vertex-Based Interpretation of the Multicellular Plant Tissues.
Supplemental Figure 24. Histograms Showing the Distributions of Predicted/Measured Auxin and DII-VENUS Levels.
Supplemental Figure 25. Predicted Distributions of the Free and Bound TIR1/AFB Receptors.
Supplemental Table 1. Number of Each Cell Type in Each Cell Geometry.
Supplemental Table 2. Observed Distributions of Different Members of the PIN Family, Based on the Images Shown in Supplemental Figure 3.
Supplemental Table 3. Rules Used to Prescribe the PIN Distribution on Our Virtual Root Geometries.
Supplemental Table 4. Parameter Values Used in the Auxin-Transport Model.
Supplemental Table 5. Description of Parameters in the DII-VENUS Network Model (See Equation 3.1 in Supplemental Methods).
Supplemental Table 6. Parameters Used in the Reduced DII-VENUS Network Model (See Equation 3.11 in Supplemental Methods).
Supplemental Table 7. Distributions of the AUX1/LAX Influx Carriers, Based on the Images Shown in Supplemental Figure 8.
Supplementary Material
Acknowledgments
This project was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) and Engineering and Physical Sciences Research Council funding to the Centre for Plant Integrative Biology. In addition, we acknowledge the support of the Leverhulme Trust (L.R.B.), the Al-Tajir World of Islam Trust (H.I.H.), National Science Foundation Grant DBI0820755 (J.M.A.), the Royal Society and Wolfson Foundation (J.R.K.), and BBSRC Professorial Research Fellowship funding (M.J.B.). We thank Antoine Larrieu, Caroline Howells, and Edward Venison for assistance in generation of the AUX1:VENUS line.
AUTHOR CONTRIBUTIONS
L.R.B. and D.M.W. designed the research, performed the research, contributed new tools, analyzed data, and wrote the article. J.A.F. performed research, contributed new tools, and analyzed data. A.P.F. and M.P.P. performed research and contributed new tools. T.G., M.H.W., L.Y., W.L., H.I.H., J.O., S.P.P., M.A.P.-A., J.Y., J.M.A., C.G., and T.V. contributed new tools. E.K. and T.C.H. designed the research. T.P.P. designed the research and contributed new tools. R.S. designed the research, performed research, and contributed new tools. J.R.K. and M.J.B. designed the research, analyzed data, and wrote the article. All authors discussed the results and commented on the article.
Glossary
- QC
quiescent center
- LRC
lateral root cap
- ODE
ordinary differential equation
- GUI
graphical user interface
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abas L., Benjamins R., Malenica N., Paciorek T., Wiśniewska J., Moulinier-Anzola J.C., Sieberer T., Friml J., Luschnig C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8: 249–256. Erratum. Nat. Cell Biol. 8: 424. [DOI] [PubMed] [Google Scholar]
- Band L.R., King J.R. (2012). Multiscale modelling of auxin transport in the plant-root elongation zone. J. Math. Biol. 65: 743–785. [DOI] [PubMed] [Google Scholar]
- Band L.R., et al. (2012). Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc. Natl. Acad. Sci. USA 109: 4668–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R., Scheres B. (2008). Auxin: The looping star in plant development. Annu. Rev. Plant Biol. 59: 443–465. [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. [DOI] [PubMed] [Google Scholar]
- Brunoud G., Wells D.M., Oliva M., Larrieu A., Mirabet V., Burrow A.H., Beeckman T., Kepinski S., Traas J., Bennett M.J., Vernoux T. (2012). A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. [DOI] [PubMed] [Google Scholar]
- Fasano J.M., Swanson S.J., Blancaflor E.B., Dowd P.E., Kao T.H., Gilroy S. (2001). Changes in root cap pH are required for the gravity response of the Arabidopsis root. Plant Cell 13: 907–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J., Benková E., Blilou I., Wisniewska J., Hamann T., Ljung K., Woody S., Sandberg G., Scheres B., Jürgens G., Palme K. (2002b). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673. [DOI] [PubMed] [Google Scholar]
- Friml J., Wiśniewska J., Benková E., Mendgen K., Palme K. (2002a). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809. [DOI] [PubMed] [Google Scholar]
- Gälweiler L., Guan C., Müller A., Wisman E., Mendgen K., Yephremov A., Palme K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230. [DOI] [PubMed] [Google Scholar]
- Grieneisen V.A., Scheres B., Hogeweg P., M Marée A.F. (2012). Morphogengineering roots: Comparing mechanisms of morphogen gradient formation. BMC Syst. Biol. 6: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieneisen V.A., Xu J., Marée A.F., Hogeweg P., Scheres B. (2007). Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013. [DOI] [PubMed] [Google Scholar]
- Heisler M., Jönsson H. (2006). Modeling auxin transport and plant development. J. Plant Growth Regul. 25: 302–312. [Google Scholar]
- Hindmarsh, A.C. (1983). ODEPACK: A systematized collection of ODE solvers. In Scientific Computing: IMACS Transactions on Scientific Computation, Vol. 1, R.S. Stepleman, ed (North-Holland, Amsterdam: IMACS), pp. 55–64. [Google Scholar]
- Holman T.J., Wilson M.H., Kenobi K., Dryden I.L., Hodgman T.C., Wood A.T., Holdsworth M.J. (2010). Statistical evaluation of transcriptomic data generated using the Affymetrix one-cycle, two-cycle and IVT-Express RNA labelling protocols with the Arabidopsis ATH1 microarray. Plant Methods 6: 9–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter J.D. (2007). Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 9: 90–95. [Google Scholar]
- Irani N.G., et al. (2012). Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat. Chem. Biol. 8: 583–589. [DOI] [PubMed] [Google Scholar]
- Jiang K., Feldman L.J. (2005). Regulation of root apical meristem development. Annu. Rev. Cell Dev. Biol. 21: 485–509. [DOI] [PubMed] [Google Scholar]
- Jones A.R., Kramer E.M., Knox K., Swarup R., Bennett M.J., Lazarus C.M., Leyser H.M., Grierson C.S. (2009). Auxin transport through non-hair cells sustains root-hair development. Nat. Cell Biol. 11: 78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer E.M. (2004). PIN and AUX/LAX proteins: Their role in auxin accumulation. Trends Plant Sci. 9: 578–582. [DOI] [PubMed] [Google Scholar]
- Kramer E.M., Frazer N.L., Baskin T.I. (2007). Measurement of diffusion within the cell wall in living roots of Arabidopsis thaliana. J. Exp. Bot. 58: 3005–3015. [DOI] [PubMed] [Google Scholar]
- Krouk G., et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18: 927–937. [DOI] [PubMed] [Google Scholar]
- Laskowski M., Grieneisen V.A., Hofhuis H., Hove C.A.T., Hogeweg P., Marée A.F.M., Scheres B. (2008). Root system architecture from coupling cell shape to auxin transport. PLoS Biol. 6: e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K., Hull A.K., Celenza J., Yamada M., Estelle M., Normanly J., Sandberg G. (2005). Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17: 1090–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A., Kargul J., May S.T., Muller P., Delbarre A., Perrot-Rechenmann C., Bennett M.J. (1999). AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 18: 2066–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironova V.V., Omelyanchuk N.A., Novoselova E.S., Doroshkov A.V., Kazantsev F.V., Kochetov A.V., Kolchanov N.A., Mjolsness E., Likhoshvai V.A. (2012). Combined in silico/in vivo analysis of mechanisms providing for root apical meristem self-organization and maintenance. Ann. Bot. (Lond.) 110: 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A., Guan C., Gälweiler L., Tänzler P., Huijser P., Marchant A., Parry G., Bennett M., Wisman E., Palme K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17: 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenschläger I., Wolff P., Wolverton C., Bhalerao R.P., Sandberg G., Ishikawa H., Evans M., Palme K. (2003). Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc. Natl. Acad. Sci. USA 100: 2987–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne R.J.H., Grierson C.S. (2009). A theoretical model for ROP localisation by auxin in Arabidopsis root hair cells. PLoS ONE 4: e8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer W.A., Bandyopadhyay A., Blakeslee J.J., Makam S.N., Chen R.J., Masson P.H., Murphy A.S. (2004). Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16: 1898–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B., et al. (2012). AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 24: 2874–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrine-Walker F., et al. (2010). Auxin carriers localization drives auxin accumulation in plant cells infected by Frankia in Casuarina glauca actinorhizal nodules. Plant Physiol. 154: 1372–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson S.V., Johansson A.I., Kowalczyk M., Makoveychuk A., Wang J.Y., Moritz T., Grebe M., Benfey P.N., Sandberg G., Ljung K. (2009). An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21: 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pound M.P., French A.P., Wells D.M., Bennett M.J., Pridmore T.P. (2012). CellSeT: Novel software to extract and analyze structured networks of plant cells from confocal images. Plant Cell 24: 1353–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradal C., Dufour-Kowalski S., Boudon F., Fournier C., Godin C. (2008). OpenAlea: A visual programming and component-based software platform for plant modeling. Funct. Plant Biol. 35: 751–760. [DOI] [PubMed] [Google Scholar]
- Rashotte A.M., Brady S.R., Reed R.C., Ante S.J., Muday G.K. (2000). Basipetal auxin transport is required for gravitropism in roots of Arabidopsis. Plant Physiol. 122: 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472. [DOI] [PubMed] [Google Scholar]
- Santuari L., Scacchi E., Rodriguez-Villalon A., Salinas P., Dohmann E.M., Brunoud G., Vernoux T., Smith R.S., Hardtke C.S. (2011). Positional information by differential endocytosis splits auxin response to drive Arabidopsis root meristem growth. Curr. Biol. 21: 1918–1923. [DOI] [PubMed] [Google Scholar]
- Scott A.C., Allen N.S. (1999). Changes in cytosolic pH within Arabidopsis root columella cells play a key role in the early signaling pathway for root gravitropism. Plant Physiol. 121: 1291–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E., Weinstain R., Zhang Y., Castillejo C., Kaiserli E., Chory J., Tsien R.Y., Estelle M. (2013). Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proc. Natl. Acad. Sci. USA 110: 4834–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding E.P. (2013). Diverting the downhill flow of auxin to steer growth during tropisms. Am. J. Bot. 100: 203–214. [DOI] [PubMed] [Google Scholar]
- Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.Y., Dolezal K., Schlereth A., Jürgens G., Alonso J.M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191. [DOI] [PubMed] [Google Scholar]
- Stoma S., Lucas M., Chopard J., Schaedel M., Traas J., Godin C. (2008). Flux-based transport enhancement as a plausible unifying mechanism for auxin transport in meristem development. PLOS Comput. Biol. 4: e1000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10: 946–954. [DOI] [PubMed] [Google Scholar]
- Swarup R., Péret B. (2012). AUX/LAX family of auxin influx carriers-an overview. Front Plant Sci 3: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Friml J., Marchant A., Ljung K., Sandberg G., Palme K., Bennett M. (2001). Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 15: 2648–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., et al. (2004). Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 16: 3069–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Kramer E.M., Perry P., Knox K., Leyser H.M., Haseloff J., Beemster G.T., Bhalerao R., Bennett M.J. (2005). Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat. Cell Biol. 7: 1057–1065. [DOI] [PubMed] [Google Scholar]
- Sze H., Li X., Palmgren M.G. (1999). Energization of plant cell membranes by H+-pumping ATPases. Regulation and biosynthesis. Plant Cell 11: 677–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T., et al. (2011). The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Agullo F., Rigas S., Desbrosses G., Dolan L., Hatzopoulos P., Grabov A. (2004). Potassium carrier TRH1 is required for auxin transport in Arabidopsis roots. Plant J. 40: 523–535. [DOI] [PubMed] [Google Scholar]
- Wells D.M., Laplaze L., Bennett M.J., Vernoux T. (2013). Biosensors for phytohormone quantification: challenges, solutions, and opportunities. Trends Plant Sci. 18: 244–249. [DOI] [PubMed] [Google Scholar]
- Zhou R., Benavente L.M., Stepanova A.N., Alonso J.M. (2011). A recombineering-based gene tagging system for Arabidopsis. Plant J. 66: 712–723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.