This work shows that 4CL, an enzyme in monolignol biosynthesis, is found as a heterotetrameric complex of two isoforms in Populus trichocarpa. The activity of the heterotetramer can be described by a mathematical model that explains the effects of each isoform with mixtures of substrates and three types of inhibition, providing insights into the regulation of metabolic flux for this pathway.
Abstract
As a step toward predictive modeling of flux through the pathway of monolignol biosynthesis in stem differentiating xylem of Populus trichocarpa, we discovered that the two 4-coumaric acid:CoA ligase (4CL) isoforms, 4CL3 and 4CL5, interact in vivo and in vitro to form a heterotetrameric protein complex. This conclusion is based on laser microdissection, coimmunoprecipitation, chemical cross-linking, bimolecular fluorescence complementation, and mass spectrometry. The tetramer is composed of three subunits of 4CL3 and one of 4CL5. 4CL5 appears to have a regulatory role. This protein–protein interaction affects the direction and rate of metabolic flux for monolignol biosynthesis in P. trichocarpa. A mathematical model was developed for the behavior of 4CL3 and 4CL5 individually and in mixtures that form the enzyme complex. The model incorporates effects of mixtures of multiple hydroxycinnamic acid substrates, competitive inhibition, uncompetitive inhibition, and self-inhibition, along with characteristic of the substrates, the enzyme isoforms, and the tetrameric complex. Kinetic analysis of different ratios of the enzyme isoforms shows both inhibition and activation components, which are explained by the mathematical model and provide insight into the regulation of metabolic flux for monolignol biosynthesis by protein complex formation.
INTRODUCTION
Lignin, a phenolic structural polymer of plants, is essential for water transport, mechanical support, and protection against biotic and abiotic stresses (Sarkanen and Ludwig, 1971; Eriksson et al., 1990; Higuchi, 1997; Vanholme et al., 2010; Denness et al., 2011). Lignin is also a major barrier to wood processing either for production of pulp and paper or for the conversion of lignocellulosic biomass to biofuel (Chiang, 2002; Chen and Dixon, 2007; Hinchee et al., 2009). Improvement of biomass quality could result from directed modification of lignin and depends on our knowledge of its biosynthesis.
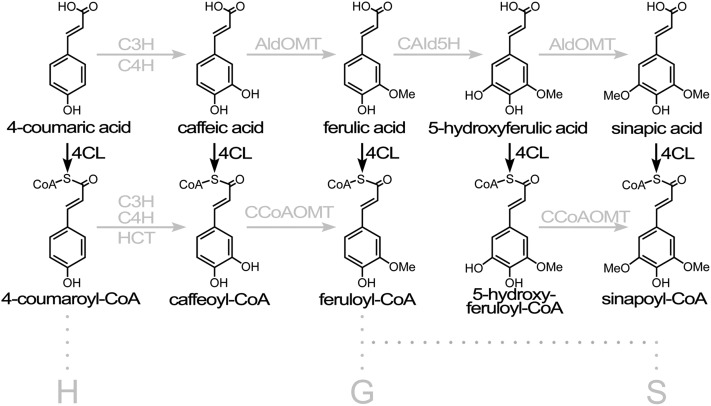
Three phenylpropanoid precursors, 4-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol, also known as the H, G, and S monolignols, respectively (Figure 1), are the predominant precursors for lignin. S and G subunits predominate in the lignin of dicots. In vascular tissue, the vessels, specialized elements for water conduction, have G-rich cell walls, while S subunits are more abundant in fiber cells specialized for mechanical support. Lignin polymers have extremely diverse combinations of subunit sequences and linkages (Ralph et al., 2004; Morreel et al., 2010; Vanholme et al., 2010). The structure and composition of the polymer depends on the composition of the monolignols delivered to the lignifying zone and on a combinatorial mode of polymerization (Higuchi, 1985; Sederoff et al., 1999; Ralph et al., 2004). In most flowering plants, 10 enzyme families are involved in the conversion of Phe to monolignols. These enzymes have been studied intensively in several plant species to infer their functions in vivo (Barrière et al., 2007; Vanholme et al., 2010; Shi et al., 2010; Lee et al., 2011). Many aspects of monolignol biosynthesis and the regulation of metabolic flux through the pathway are not yet sufficiently defined or quantified. The pathway continues to be revised as new enzyme activities and new types of lignin are discovered (Osakabe et al., 1999; Chen et al., 2011, 2012).
Figure 1.
The Roles of 4CL in Monolignol Biosynthesis.
4CL is able to ligate five cinnamic acids, but the main roles are in the conversion of 4-coumaric acid to 4-coumaroyl-CoA and in the conversion of caffeic acid to caffeoyl-CoA (Chen et al., 2013). 4-Coumaric acid may be hydroxylated by the enzyme complex (C4H and C3H) to form caffeic acid. Caffeic acid may be activated to its CoA derivative caffeoyl-CoA by 4CL. 4-Coumaric acid may be activated by 4CL to form 4-coumaroyl-CoA, which can be reduced to 4-coumaryl alcohol, the primary precursor for H units in lignin. 4-Coumaroyl-CoA can also form a shikimic acid ester through the activity of hydroxycinnamoyl-CoA shikimate hydroxycinnamoyl transferase. The 4-coumaroyl shikimic acid ester is converted by the C4H and C3H complex to the caffeoyl shikimic acid, ester which in turn is converted by hydroxycinnamoyl transferase to caffeoyl-CoA. Caffeoyl-CoA is methylated to form feruloyl-CoA by caffeoyl-CoA 3-O-methyltransferase, which is the main route for monolignol biosynthesis.
4-Coumaric acid:CoA ligase (4CL) (EC 6.2.1.12) catalyzes the formation of CoA thioesters of several hydroxycinnamic acids in the monolignol biosynthesis pathway (Figure 1). In this ligation reaction, for example, ATP, CoA, and 4-coumaric acid form the 4-coumaroyl CoA thioester plus AMP and diphosphate (Gross and Zenk, 1974). In the phenylpropanoid pathway, 4CL is the enzyme at the branch point for flavonoids and lignin biosynthesis (Hahlbrock and Scheel, 1989) and is also involved in the biosynthesis of isoflavonoids, coumarins, suberin, and cell wall–bound phenolics (Vanholme et al., 2012). Genomic sequencing has revealed a 4CL gene family with multiple 4CL and 4CL-like sequences (Allina et al., 1998). Four 4CL gene family members were found in Arabidopsis thaliana (Hamberger and Hahlbrock 2004) and Physcomitrella patens (Silber et al., 2008), five members in rice (Oryza sativa; Gui et al., 2011), and 17 in Populus trichocarpa (Shi et al., 2010). Distinct 4CLs display tissue and/or substrate specificity and are likely to function in different pathways (Hu et al., 1998; Ehlting et al., 1999). The substrate specificities of 4CLs may be determined by 12 amino acids lining the substrate binding pocket (Schneider et al., 2003).
Lignin-associated 4CL genes have been identified in poplars (Hu et al., 1998; Shi et al., 2010), Eucalyptus grandis (Naoki et al., 2011), tobacco (Nicotiana tabacum; Kajita et al., 1996), Arabidopsis (Raes et al., 2003), rice (Gui et al., 2011), and sorghum (Sorghum bicolor; Saballos et al., 2012). We identified one lignin-associated 4CL (4CL1) in quaking aspen (Populus tremuloides) (Hu et al., 1998). In P. trichocarpa, two xylem-specific 4CLs (4CL3 and 4CL5) encode active enzymes in monolignol biosynthesis (Chen et al., 2013). The enzyme kinetic parameters and inhibition specificity of Ptr-4CL3 is very similar to its aspen ortholog Pt-4CL1 (Hu et al., 1998). Both Pt-4CL1 and Ptr-4CL3 prefer 4-coumaric acid as substrate and are strongly inhibited by caffeic acid. Unlike Ptr-4CL3, Ptr-4CL5 prefers caffeic acid as substrate and exhibits competitive and uncompetitive inhibition, as well as substrate self-inhibition (Chen et al., 2013).
Protein–protein interactions affect the regulation of biosynthetic pathways and control metabolic flux (Srere, 1987) and are likely to be important in the monolignol biosynthetic pathway. A P. trichocarpa C4H1/C4H2/C3H3 (cinnamic acid 4-hydroxylase1/cinnamic acid 4-hydroxylase2/coumaric acid 3-hydroxylase3) protein complex provides direct 3-hydroxylation of 4-coumaric acid, an activity not detected with the individual enzymes (Chen et al., 2011). Quantitative information about such interactions is essential to optimize the inputs for systems analysis of this biological process.
We initiated a systems biology approach to the monolignol pathway in P. trichocarpa (Shi et al., 2010; Chen et al., 2011, 2013; Wang et al., 2014). We propose to quantify the biosynthetic components of the pathway at the genomic, transcriptomic, proteomic, and metabolomic levels to develop mathematical models. Mathematical models of the integrated pathway lead to the development of hypotheses and discovery of pathway components and provide a predictive understanding of the pathway for directed modification.
Mathematical modeling of enzyme kinetic data has been used previously to analyze, simulate, and predict metabolic flux through enzymatic pathways for well-defined biological processes (Schallau and Junker, 2010). While many of the enzymes of monolignol biosynthesis have been studied in detail, much of the work has been done in different species, some annual, perennial, woody, or herbaceous, with quantitative analysis done on materials varying from whole plants to specific wood-forming tissues. Some quantitative modeling of lignin biosynthesis has been performed to investigate the energetics of the pathway (Amthor, 2003), and models of the genetic regulation of monolignol biosynthesis have been presented for poplar (Lee and Voit, 2010) and Medicago (Lee et al., 2011). A comprehensive predictive metabolic flux model of the monolignol pathway in SDX of P. trichocarpa is presented in a companion article (Wang et al., 2014). Previous work has not incorporated protein complex formation into metabolic models of monolignol biosynthesis, although metabolic channeling based on proximal spatial orientation has been invoked (Lee et al., 2012).
Here, we provide evidence for the discovery and characterization of a protein–protein heterotetramer of P. trichocarpa 4CL3 and 4CL5, which may have a regulatory role. Five lines of evidence are presented to describe this complex: (1) laser microdissection (LMD), (2) mass spectrometry (MS), (3) coimmunoprecipitation (co-IP), (4) chemical cross-linking, and (5) bimolecular fluorescence complementation (BiFC; Hu et al., 2002). A quantitative mathematical model was constructed that describes the enzyme activity and the modifications of activity that result from the complex, taking into account the ratios of enzymes, the effects of multiple substrates, inhibitors, and their modes of inhibition.
RESULTS
Ligation Activity with Mixtures of 4CL3 and 4CL5 Indicate a Molecular Interaction
To better understand the CoA ligation rates in an environment containing multiple
4CL enzymes, we mixed P.
trichocarpa 4CL3 and 4CL5 in different ratios and measured the CoA
ligation rates using 4-coumaric acid as the substrate. The first case (solid line in
Figure 2A) measured the CoA ligation rates
when the 4CL3 molar concentration was held constant at 40 nM, while the 4CL5 molar
concentration was gradually increased from 0 to 40 nM, in increments of 10 nM. The
reciprocal case (solid line in Figure 2B)
measured the CoA ligation rates when 4CL5 was held constant at 40 nM and 4CL3 was
increased from 0 to 40 nM. The dashed lines in Figures 2A and 2B represent the
total CoA ligation rate if 4CL3 and 4CL5 acted independently, that is as the sum of
the individual activity, calculated by Equation 1: (νtot =
ν4CL3 + ν4CL5). Here, ν4CL3
and ν4CL5 follow basic Michaelis-Menten kinetics (Chen et al., 2013). Equation 1 predicts a total
rate (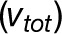 )
that increases linearly with increasing total enzyme concentration (4CL3 + 4CL5).
This linearity is expected because the individual rates for 4CL3 and 4CL5 were linear
and proportional to the total enzyme concentration when the substrate concentration
was in excess and constant. The experimentally derived ligation rate for a mixture of
4CL3 and 4CL5 proteins (solid lines in Figures
2A and 2B) does not conform to
expectation for the summed rate of individual enzymes (dashed lines in Figures 2A and 2B). This suggests a specific interaction between 4CL3 and 4CL5. If 4CL3
and 4CL5 are able to interact in vitro, it is important to demonstrate that both
enzymes are expressed in the same cells in vivo.
)
that increases linearly with increasing total enzyme concentration (4CL3 + 4CL5).
This linearity is expected because the individual rates for 4CL3 and 4CL5 were linear
and proportional to the total enzyme concentration when the substrate concentration
was in excess and constant. The experimentally derived ligation rate for a mixture of
4CL3 and 4CL5 proteins (solid lines in Figures
2A and 2B) does not conform to
expectation for the summed rate of individual enzymes (dashed lines in Figures 2A and 2B). This suggests a specific interaction between 4CL3 and 4CL5. If 4CL3
and 4CL5 are able to interact in vitro, it is important to demonstrate that both
enzymes are expressed in the same cells in vivo.
Figure 2.
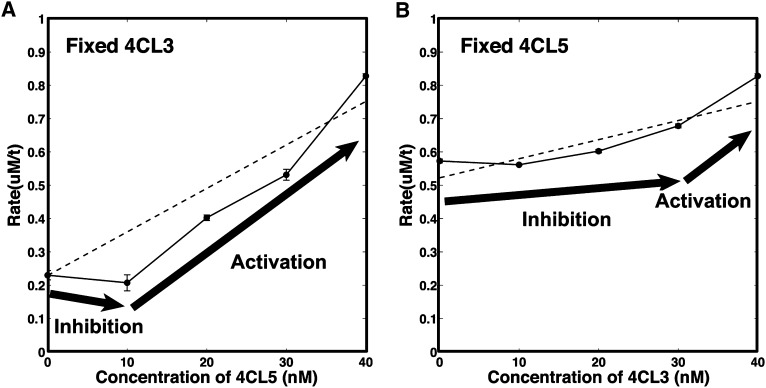
Impact of Enzyme Complex Formation on Product Formation.
To determine the effect of the complex on enzyme activity, P. trichocarpa 4CL3 and 4CL5 were mixed at different concentrations. 4-Coumaric acid (28.28 μM) was used as substrate. The dashed lines represent the simple sum of expected individual rates for 4CL3 and 4CL5 without enzyme complex effects. The solid line (with data points as dots) represents experimental data with 4-coumaric acid as a main substrate. The arrows predict inhibition and activation impacts by the enzyme complex. Error bars represent se of three replicates. In some cases, the precision is high, and error bars are smaller than the size of the data points.
(A) 4CL3 was fixed at 40 nM, and 4CL5 concentration was varied from 0 to 40 nM.
(B) 4CL3 concentration was varied from 0 to 40 nM, and the concentration of 4CL5 was fixed at 40 nM.
LMD Indicates the Coexpression of 4CL3 and 4CL5 Transcripts in SDX Fiber Cells
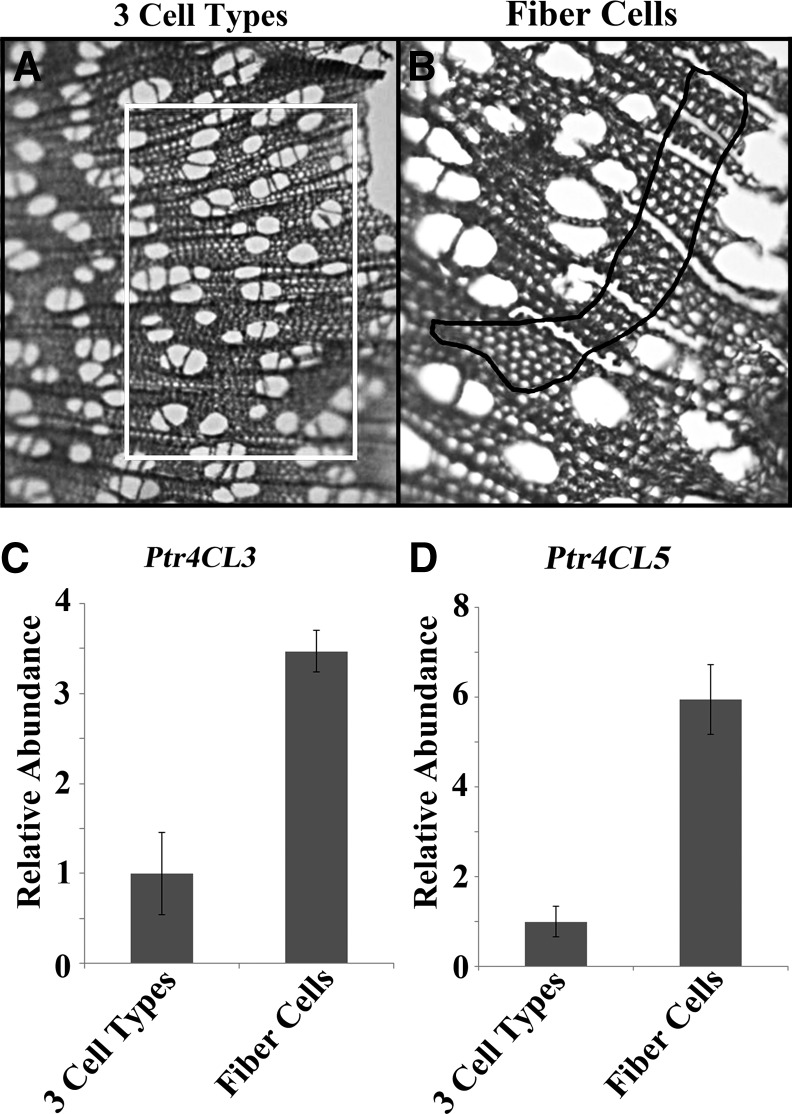
The stems of 6-month-old trees were used to collect different cell types from SDX by LMD. A cryostat was used to obtain stem cross sections. Three different types of samples were collected: (1) samples that included fibers, rays, and vessels; (2) samples of vessel cells only; and (3) fiber cells only. Ray cells proved to be too difficult to collect as an isolated cell type. In the three-cell-type sample, vessel, fiber, and ray cells are collected together (Figure 3A). In vessel or fiber cells only samples, we used the laser to burn out the ray cells and then selected only fiber or vessel cells (Figure 3B). The yields of total RNA from vessel cells were too low for further analysis. The transcript abundance of 4CL3 and 4CL5 was analyzed using quantitative RT-PCR and gene-specific primers (Shi et al., 2010). 4CL3 and 4CL5 are both expressed in fiber cells (Figures 3C and 3D). 4CL3 and 4CL5 are expressed 3 to 6 times higher in fiber cell samples compared with the transcript abundance in the samples containing fibers, rays, and vessels (Figures 3C and 3D). These data show that 4CL3 and 4CL5 are coexpressed in fiber cells, known to be highly active in lignin biosynthesis.
Figure 3.
LMD Indicates Coexpression of P. trichocarpa 4CL3 and 4CL5 Transcripts in Fiber Cells.
The quantitation of 4CL3 and 4CL5 transcripts from captured tissue sections containing three different cell types (fiber, vessel, and ray) and from sections containing only fiber cells indicate that both 4CL isoforms are coexpressed in fiber cells.
(A) The white rectangle shows the tissue sections containing all three cell types isolated by LMD.
(B) The black line surrounds the area of the fiber cells isolated by LMD where ray cells were burned away.
(C) The transcript abundance of 4CL3 in the two samples.
(D) The transcript abundance of 4CL5 in the two samples.
The 4CL3/4CL5 Complex Was Verified and Characterized by BiFC, Chemical Cross-Linking, Co-IP, and MS
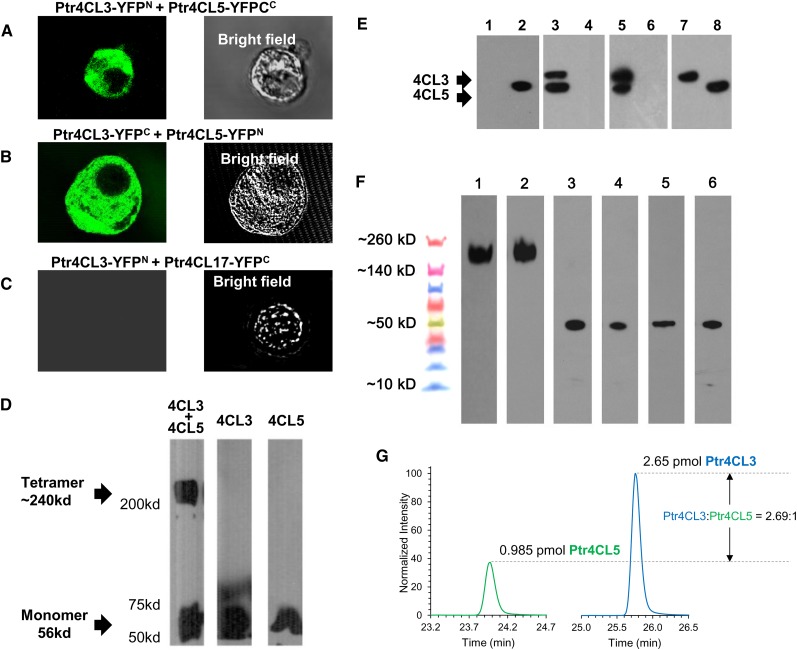
To test for protein–protein interactions between 4CL3 and 4CL5 using BiFC, different pairs of plasmids, each containing a target protein fused to one of the two complementing segments of yellow fluorescent protein (YFP), YFPN (amino acids 1 to 155) and YFPC (amino acids 156 to 239), were cotransformed into P. trichocarpa SDX protoplasts (Lin et al., 2013). When 4CL3-YFPN was coexpressed with 4CL5-YFPC, or the reciprocal, strong fluorescence signals were observed in the cytoplasm indicating 4CL3 and 4CL5 heterodimerization (Figures 4A and 4B). Strong fluorescence signals were observed when 4CL3-YFPN was coexpressed with 4CL3-YFPC, demonstrating that 4CL3 is able to form homodimers (Supplemental Figure 1). A fluorescent signal was not observed for coexpression of 4CL5-YFPN and 4CL5-YFPC, indicating that 4CL5 does not form homodimers (Supplemental Figure 1). We did not observe any fluorescence signals when the protoplasts were cotransfected with 4CL3-YFPN and 4CL17-YFPC (Figure 4C). These interactions suggest specific mechanisms affecting the composition of the protein complex and the pathway for its formation, if 4CL5 does not form homodimers.
Figure 4.
Physical Evidence for a P. trichocarpa 4CL3/4CL5 Protein Complex.
(A) and (B) BiFC. 4CL fusion proteins of N-terminal or C-terminal fragments of YFP. 4CL3-YFPN+4CL5-YFPC (A) and 4CL3-YFPC+4CL5-YFPN (B) expressed YFP signals.
(C) 4CL3-YFPN+4CL17-YFPC was used as a control and expressed no fluorescent signal.
(D) Chemical cross-linking of a 4CL3/4CL5 protein complex. Recombinant 4CL3 and 4CL5 were cross-linked by DSP. Cross-linking of 4CL3 or 4CL5 alone shows predominantly 4CL monomers. Mixing 4CL3 and 4CL5 together shows both tetramers and monomers.
(E) Co-IP of the 4CL3/4CL5 complex. Lane 1: Recombinant 4CL3 is not detected by 4CL5-specific antibody. Lane 2: Recombinant 4CL5 is detected by 4CL5-specific antibody. Lane 3: Native SDX protein pulled down by anti-4CL5 antibody shows a lower band, which is 4CL5, and a higher band, which is 4CL3. Lane 4: Native SDX protein pulled down by preimmune antibody. Lane 5: Xylem crude extract added to recombinant 4CL3-His and pulled down by anti-His antibody. The bottom band is 4CL5, and the top band is 4CL3. Lane 6: Control, which is xylem crude extract prepared following the same procedure as lane 3. Lane 7 is 4CL3 recombinant protein detected by 4CL general antibody, and lane 8 is 4CL5 recombinant protein also detected by 4CL antibody.
(F) A high molecular mass 4CL3/4CL5 protein complex was detected by 4CL3 (lane 1) and 4CL5 (lane 2) specific antibodies in an in vivo DSP cross-linked SDX sample. Non-cross-linked SDX and recombinant proteins were used as controls for the detection of monomeric 4CL3 (lanes 3 and 5) and 4CL5 (lanes 4 and 6).
(G) Quantification of the cross-linked 4CL protein complex by PC-IDMS. Extracted ion chromatograms for the native surrogate peptides of 4CL5 (left) and 4CL3 (right), which have been normalized to the intensity of their corresponding SIL peptides to allow for visualization of the relative surrogate peptide (i.e., protein) concentrations.
To test if 4CL3 and 4CL5 form a protein complex, we mixed the two recombinant proteins with the chemical cross-linker dithiobis(succinimidyl propionate) (DSP), which has a spacer arm equivalent to eight carbon linkages (Lomant and Fairbanks, 1976). Chemical cross-linking stabilizes protein–protein interactions (Lomant and Fairbanks, 1976). When 4CL3 recombinant protein at low concentrations (200 nM) was cross-linked with DSP and the product was size separated on SDS-PAGE, an immunoblot displayed predominantly monomers and some possible dimers (Figure 4D). Only monomers were observed when 4CL5 recombinant protein (200 nM) was cross-linked. The cross-linked mixture of 4CL3 and 4CL5 recombinant proteins at equal concentrations (200 nM) resulted in monomers and a protein band greater than 200 kD, a size consistent with a heterotetramer (Figure 4D).
To further verify the existence of a protein complex of 4CL3/4CL5 in SDX, we used a 4CL5-specific antibody (Figure 4E) to carry out co-IP. This antibody can only detect 4CL5, not 4CL3 in immunoblotting (lanes 1 and 2, Figure 4E). An antibody for one protein of the complex could coprecipitate both proteins from an SDX protein extract. The result of a coprecipitation test using anti-4CL5 antibody was analyzed on SDS-PAGE and an immunoblot (lane 3, Figure 4E). Both members of the proposed complex (4CL3/4CL5) were detected on the immunoblot with an anti-4CL antibody (Li et al., 2003), which detects both forms of 4CL (lanes 7 and 8, Figure 4E). A reciprocal experiment using 4CL3-specific antibody gave similar results. The specificity of the antibodies was verified in a previous publication (Chen et al., 2013). When preimmune serum was used to perform the co-IP, 4CL protein was not detected on the immunoblot (lane 4, Figure 4E). The co-IP evidence supports a 4CL3/4CL5 protein complex in native SDX.
Recombinant 4CL3 protein with a 6x His-tag was added into an SDX crude protein extract to test whether the complex (4CL3/4CL5) can be formed after the preparation of the SDX protein extract. If the recombinant 4CL3 and the endogenous 4CL5 produced a new complex in the extract, then anti-His monoclonal antibody will pull down the recombinant 4CL5. Co-IP, SDS-PAGE, and immunoblotting showed both 4CL3 and 4CL5 monomer bands, indicating that these two proteins formed a complex de novo in the extract (lane 5, Figure 4E). These results also showed that the native 4CL5 protein, as well as the recombinant protein, forms a complex with 4CL3 recombinant protein. A reciprocal experiment, using recombinant 4CL5 with a His-tag, also pulled down a 4CL3/CL5 complex. His-tags do not affect the kinetic parameters or specificity of these 4CL isoforms (Chen et al., 2013).
The 4CL3/4CL5 Complex Is Found in Vivo in SDX
To determine whether the 4CL3/4CL5 complex exists in vivo in SDX, we performed a cross-linking experiment using debarked stem segments of P. trichocarpa. Debarked stem segments were submerged in DSP, and the SDX tissue was subsequently scraped from the stem and ground into a powder using liquid nitrogen. The powder was then homogenized in a protein-denaturing buffer to produce a crude protein extract. The extract was then analyzed on SDS-PAGE and 4CL protein was detected by immunoblotting (Figure 4F). The 4CL protein was detected between the 140 and 260 kD molecular markers, consistent with the molecular size of the complexes detected for the cross-linking of the recombinant proteins. No bands were detected at sizes that would represent dimers or trimers. These results indicate that the complex detected with recombinant proteins is a reasonable representation of the native proteins and indicate that the majority of the 4CL protein exists as a tetrameric complex in vivo.
To determine the stoichiometry of the monomers in the complex, we separated the 4CL complex from the monomers by covalently cross-linking the protein complex using a thiol-specific reagent, 1,4-bis-maleimidobutane, and then removing the free 4CL monomers (∼60 kD) with a 100-kD centrifugal filter. The retained, high molecular mass fraction was then subjected to protein cleavage–isotope dilution mass spectrometry (PC-IDMS) quantification (Shuford et al., 2012) to determine the absolute concentration of each 4CL subunit (Supplemental Figure 2). The absolute quantities of 4CL3 and 4CL5 in the complex (Figure 4G) provide a ratio of normalized intensities of 2.69:1.
This sample was created with purified recombinant proteins, making it unlikely that large contaminants were copurified and cross-linked (>100 kD) that could bias quantification. The assay was determined to be free from interference because coelution and conserved selected reaction monitoring–mass spectrometry fragmentation patterns observed between the surrogate peptides and their stable isotope–labeled (SIL) counterparts confirmed the identity of the quantified peptides (Supplemental Figure 2). Bias in stoichiometry determination for protein complexes when utilizing synthetic SIL peptide standards (Schmidt et al., 2010) is generally low. Given the molecular mass of the complex was estimated to be ∼240 kD by SDS-PAGE (Figures 4D and 4F) and the mass of the monomers is ∼60 kD, the molecular mass of the complex (2.69:1) agrees well with a 4CL3: 4CL5 ratio of 3:1.
Mixed Enzyme Effect Analysis with Basic Mass Action Modeling
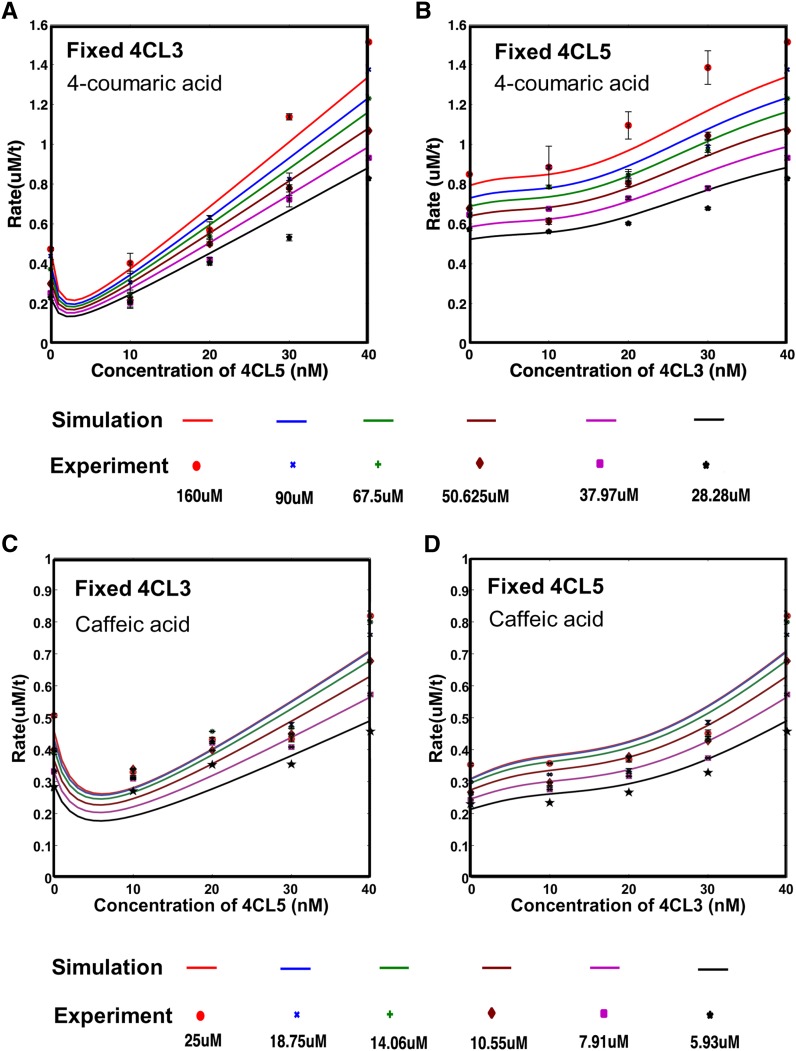
Attaining a more comprehensive understanding of CoA ligation and the function of the complex can come from developing a quantitative model, describing how the 4CL3/4CL5 complex affects the overall reaction rate. The expected baseline and experimental rates are shown for 4-coumaric acid as substrate at 28 μM (Figures 2A and 2B). The same experiments were conducted at concentrations of 160, 90, 68, 51, 38, and 28 μM with 4-coumaric acid, and a similar set was done with caffeic acid (Figure 6). All results confirm a nonlinear deviation of the experimental rates from the baseline rates. For a 4CL3 concentration of 40 nM and a 4CL5 concentration of 10 nM (ratio 4:1), the rate drops significantly below baseline followed by a rise for both the 40 nM:20 nM and 40 nM:30 nM (4CL3:4CL5 total concentration) conditions. Despite the subtle rise, the measured rates for the 40 nM:10 nM, 40 nM:20 nM, and 40 nM:30 nM experiments were primarily below the baseline rates. The experimental rates for the 40 nM:40 nM case (4CL3:4CL5) were significantly above the baseline rate and displayed a distinct change from lower 4CL3:4CL5 ratios. Similar results were observed in the reciprocal case (4CL5 fixed:4CL3 varied), although a weaker reduction was observed. Similar experiments were conducted using caffeic acid as substrate. The reaction with caffeic acid is more complex due to additional uncompetitive and self-inhibition.
Figure 6.
Comparison of Simulation and Experiment Using Either 4-Coumaric Acid or Caffeic Acid as Individual Substrates.
(A) and (B) Solid lines represent simulations based on the equation and the optimized parameters in Figure 5B. Dots represent experimental data with 4-coumaric acid as the main substrate. Colors represent different substrate concentrations. Fixed amounts of 4CL3 and increasing amounts of 4CL5 (A), and fixed amounts of 4CL5 and increasing amounts of 4CL3 (B). The error bars represent 1 se. The coefficient of determination (R2) is 0.94, and the RMSD is 0.079 for the goodness of fit of the model.
(C) and (D) Simulation results along with the experimental rates using caffeic acid as substrate. Solid lines represent simulation results based on the equation and optimized parameters in Supplemental Figure 3B. The red line (25 μM) and blue line (18.75 μM) in (C) closely overlap. Dots represent experimental data with caffeic acid as substrate. Colors represent different substrate concentrations in both cases: (C) shows a fixed amount of 4CL3 with increasing amounts of 4CL5, while (D) shows a fixed amount of 4CL5 with increasing amounts of 4CL3. The R2 value is 0.82, and the RMSD is 0.058 for the goodness of fit.
Figures 2 and 6 illustrate the impact of enzyme complex formation on product formation. The difference between the expected baseline rate and the experimental rates in Figures 2A and 2B indicates that low concentrations of 4CL5 mixed with higher concentrations of 4CL3 results in inhibition of product formation. This inhibitory effect may be attributed to the formation of the 4CL3/4CL5 complex, where 4CL3 is recruited by low concentrations of 4CL5, reducing the amount of 4CL3 that is available to generate product. Therefore, the recruitment of 4CL3 by low concentrations of 4CL5 results in a net decrease in the overall rate. High concentrations of both 4CL3 and 4CL5 display an activation effect where the rate of product formation is higher than the sum of the independent rates of 4CL3 and 4CL5. This paradoxical inhibition and activation can be explained by differential regulation, which is a function of different ratios of 4CL3 and 4CL5 where the amount of 4CL5 controls the extent of inhibition or activation. Next, we propose a mechanistic model leading to a mathematical description of the total CoA-ligation rate using 4-coumaric acid as substrate, which includes the inhibition and activation effects attributed to the formation of the 4CL3/4CL5 complex.
A Mechanistic Model of the Interaction of 4CL3 and 4CL5 Using 4-Coumaric Acid as Substrate
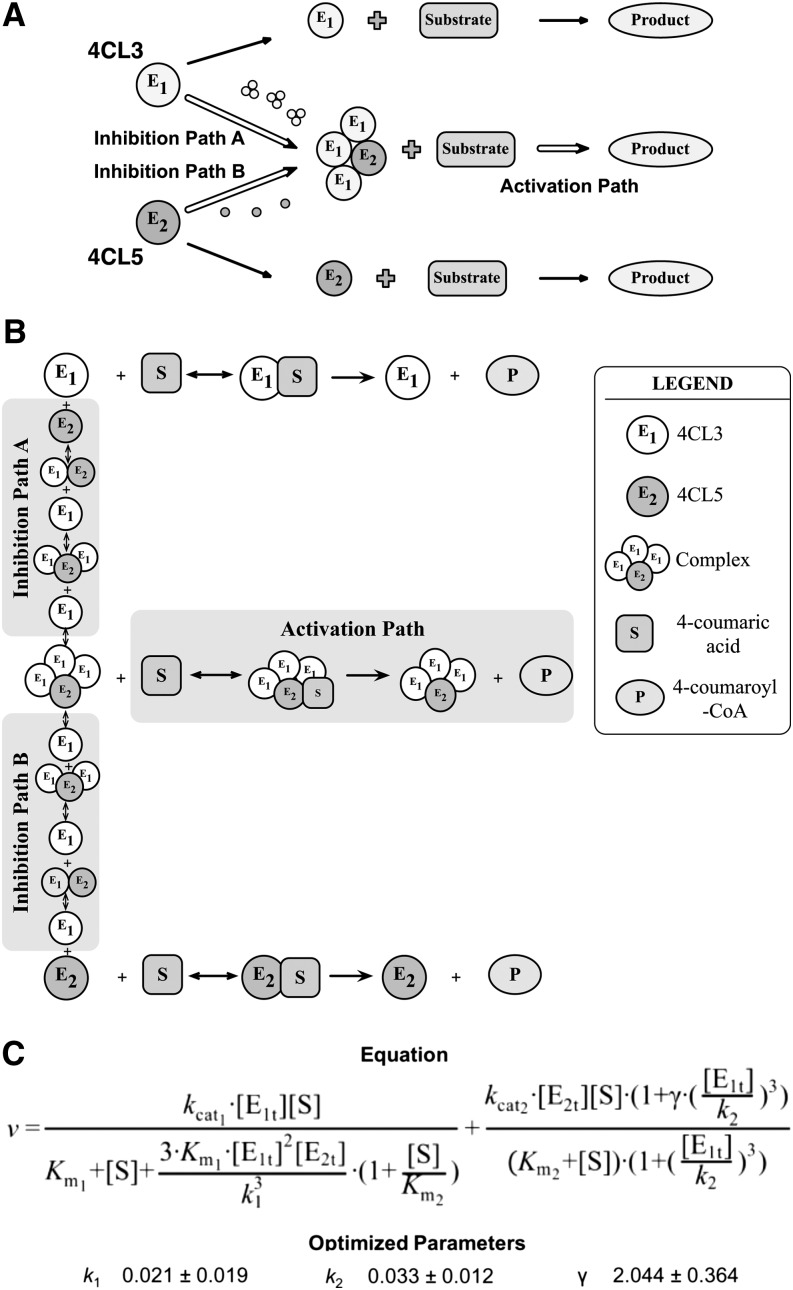
An interaction block diagram (Figure 5A) describes the effects of the enzyme complex formation on the total CoA-ligation rate associated with 4-coumaric acid. The total CoA ligation rate is a function of the activity of free 4CL3 (E1), free 4CL5 (E2), and their protein complex. The complex formed by 4CL3 and 4CL5 is a tetramer with a 3:1 ratio. The inhibition effects resulting from the formation of the tetramer and the activation path of the tetramer affect the rate of product formation. First, 4CL3 binds with available 4CL3, reducing the rate associated with 4CL3 (inhibition path A in Figure 5A) and then 4CL5 binds with 4CL3, leading to a reduction in rate associated with 4CL5 (inhibition path B in Figure 5A). Higher amounts of 4CL3 and 4CL5 result in the formation of high concentrations of the 4CL3/4CL5 complex, which can then bind to available substrate. The complex produces an alternative path toward product formation, resulting in a net increase in the product rate (activation path in Figure 5A).
Figure 5.
Mechanistic Description of the Inhibition and Activation Effects on the Rates of Product Formation Using 4-Coumaric Acid as Substrate.
(A) Proposed interactions of 4CL3 and 4CL5 and the effects on product formation. The formation of the 4CL3/4CL5 complex with a 3:1 ratio leads to decreasing amounts of free enzymes and causes a rate reduction (Inhibition Path A and Inhibition Path B). The 4CL3/4CL5 complex is then involved in product formation and results in an increase in rate (Activation Path).
(B) The model described in (A) is extended for one
substrate, 4-coumaric acid, using experimentally derived kinetic parameters,
and deriving rate estimates at each step. Each enzymatic reaction for 4CL3,
4CL5, and the 4CL3/4CL5 complex is based on Michaelis-Menten kinetics.
 and
and  are kinetic parameters of
the 4CL3 enzymatic reaction.
are kinetic parameters of
the 4CL3 enzymatic reaction.  and
and
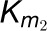 are kinetic parameters of the 4CL5 enzymatic reaction.
are kinetic parameters of the 4CL5 enzymatic reaction.
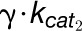 and
and 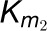 are assumed as kinetic
parameters of 4CL3/4CL5 complex, where
are assumed as kinetic
parameters of 4CL3/4CL5 complex, where 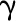 represents activation
effects of the complex on the rate. The interaction rate between 4CL3 and 4CL5
is assumed to be
represents activation
effects of the complex on the rate. The interaction rate between 4CL3 and 4CL5
is assumed to be 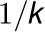 . The interactions occur for the formation of the
4CL3/4CL5 tetramer in succession, which leads to inhibition of the rate.
. The interactions occur for the formation of the
4CL3/4CL5 tetramer in succession, which leads to inhibition of the rate.
(C) A mathematical model is shown for multiple enzymes and
4-coumaric acid as a single substrate. The equation represents the rate of
total product formation associated with 4CL3, 4CL5, and the 4CL3/4CL5 complex,
where [E1t] and [E2t] are the total amounts of 4CL3 and
4CL5 respectively, [S] is the 4-coumaric acid concentration.
k1, k2, and
γ are unknown parameters defined for the
enzyme–enzyme interaction. k1 is
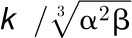 and k2 is
and k2 is
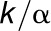 , where
, where 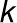 is the
association/disassociation rate between enzyme and enzyme complex in
(A). α and β
are the proportions of 4CL3 and 4CL5 involved with each interaction between
enzymes (see Supplemental Methods for the derivation).
γ represents the product rate of the enzyme complex.
The optimized values of the unknown parameters are fitted by hybrid
optimization using MATLAB. k1,
k2, and γ values
represent the mean ± sd of 100 optimized values.
is the
association/disassociation rate between enzyme and enzyme complex in
(A). α and β
are the proportions of 4CL3 and 4CL5 involved with each interaction between
enzymes (see Supplemental Methods for the derivation).
γ represents the product rate of the enzyme complex.
The optimized values of the unknown parameters are fitted by hybrid
optimization using MATLAB. k1,
k2, and γ values
represent the mean ± sd of 100 optimized values.
A plausible mechanistic description of the inhibition and activation block diagram
(Figure 5A) is shown in Figure 5B for 4-coumaric acid. Inhibition occurs
due to interactions between free 4CL3 and 4CL5 that lead to the formation of the
4CL3/4CL5 tetramer. All interactions involving free 4CL3, free 4CL5, and other enzyme
complex intermediates have an equilibrium constant of k, where
k =
kd/ka. Here,
kd is the dissociation rate of the enzymes and
ka is the association rate of the enzymes. Alternative
causes of inhibition involving the interaction of free enzymes with enzyme substrate
complexes are not supported by other plausible model structures. A mechanistic model
for activation in enzymatic reactions (Saboury,
2009; Fontes et al., 2000) was
used here to describe the activation effect of the 4CL3/4CL5 complex (Figure 5B). The kinetic parameters associated
with 4CL3 (E1) and 4CL5 (E2) are the same as the kinetics of Chen et al. (2013). Given that the results described above and
in Chen et al. (2013) indicate dominant 4CL5
kinetics, we set the kinetic parameters associated with the 4CL3/4CL5 complex to
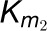 and
and
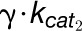 ,
respectively.
,
respectively.
A mathematical model representing the rate of total product formation associated with
4CL3, 4CL5, and the 4CL3/4CL5 complex (Figure
5C) was derived from Figure 5B. The
derivation was based on the Michaelis-Menten assumption of quasi-equilibrium where
the association and disassociation of enzymes, enzyme complexes, and their
intermediates are in binding equilibrium. The additional assumption is that the
tetrameric complex plays a more significant role in CoA ligation than dimers and
trimers, as these intermediates were undetected in mixtures containing both 4CL3 and
4CL5. Hence, the dimer and trimer formation are assumed to be transient in the
reversible reaction. This allows us to make the quasi-equilibrium assumption (see
Supplemental Methods for full derivation). These derivations lead to a
combined rate equation for 4-coumaric acid as substrate (Figure 5C). The assumption of quasi-equilibrium allows us to
write the concentrations of free 4CL3, free 4CL5, and all enzyme complex
intermediates in terms of proportional amounts of known total 4CL3 and 4CL5
concentrations. The quasi-equilibrium reaction rates between 4CL3 and 4CL5 for all
enzyme complex intermediates are assumed to be the unknown parameter
k. The formation of the 4CL3/4CL5 complex does not depend on the
presence of the substrates (Figure 4D).
Although the results of MS support a ratio of
3:1 for a 4CL3/4CL5 tetramer, a more precise estimate of the proportion of 4CL3
(α) and the proportion of 4CL5 (β) is yet to be determined. Thus, these
unknowns are combined with the parameter k to form
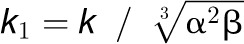 and
and  ,
leading to three unknown values in the equation, k1,
k2, and γ (Figure 5C).
,
leading to three unknown values in the equation, k1,
k2, and γ (Figure 5C).
Experimental rates of product formation under different total 4CL3 and 4CL5 concentrations (Figures 6A and 6B) were used to optimize k1, k2, and γ. An objective function based on least mean square error (Widrow and Hoff, 1960) was used to assess the goodness of fit between the experimental data and the mathematical model in Figure 5C. This objective function, in combination with a Hybrid optimization approach (Xia and Wu, 2005), was used to estimate the values of k1, k2, and γ (Figure 5C). The hybrid optimization algorithm is based on a search routine that uses both global optimization and local optimization to efficiently optimize large-scale problems within a complex search space. Genetic algorithms (Goldberg, 1989) were used as the global optimization, and Fmincon (Mathworks; Optimization Toolbox, version 3, User’s Guide, 2007) was used for the local optimization. One hundred runs using the hybrid optimization scheme were performed with random initial conditions over the range [0 to 0.1] for k1, [0 to 0.1] for k2, and [1 to 3] for γ. Wider ranges for these parameter values did not lead to any significant change in the optimized value.
The resulting mean values and standard deviations of the optimized parameters are shown in Figure 5C. These mean values were incorporated in the equation in Figure 5C and used to produce simulated total product formation rates for case 1 (4CL3 fixed:4CL5 varied) and case 2 (4CL3 varied:4CL5 fixed). Figures 6A and 6B display these simulated values along with the experimentally measured rates for cases 1 and 2. The model fits the data well as shown by the scatter (experimentally measured rates) and line (simulated rates) plots, with a mean squared error of 0.0062 (considering all experiments at all concentrations). R2 values and root mean standard deviations (RMSDs) used to describe the goodness of fit of the model further confirmed the accuracy of the model prediction (Figures 6A and 6B). The model describes the prominent dip in Figures 2A and 2B that occurs due to the inhibition of the product formation rate at high levels of 4CL3 and low levels of 4CL5. The model also reflects the activation at high levels of both 4CL3 and 4CL5. Overall, the model provides an adequate representation of the metabolic rate involving 4CL3, 4CL5, and the 4CL3/4CL5 complex for 4-coumaric acid as substrate.
A Mechanistic Model of the Interaction of the 4CL3/4CL5 Complex Using Caffeic Acid as Substrate
The mechanistic model derived for 4-coumaric acid cannot be applied directly to caffeic acid because caffeic acid exhibits substrate self-inhibition in the 4CL5 reaction (Chen et al., 2013). 4-Coumaric acid does not show self-inhibition with either 4CL3 or 4CL5. A mechanistic description of the self-inhibition of caffeic acid with 4CL5 is shown in Supplemental Figure 3A. A mathematical model quantifying the rate of total product formation associated with caffeic acid as a substrate was again derived based on the Michaelis-Menten assumptions of quasi-equilibrium used previously for 4-coumaric acid. The inclusion of self-inhibition in the combined mechanistic model for caffeic acid modifies the equation in Figure 5C slightly, resulting in the equation in Supplemental Figure 3B.
Hybrid optimization was performed for 4-coumaric acid to assess the goodness of fit of the equation in Supplemental Figure 3B with measured product formation rates for varying amounts of total 4CL3, 4CL5, and caffeic acid concentrations. One hundred runs of optimization were performed with random initial conditions over the range [0 to 0.1] for k1, [0 to 0.1] for k2, and [1 to 6] for γ to assess variation in estimated parameters. While the range of values for k1 and k2 did not change from 4-coumaric acid to caffeic acid, it was expected that the range for γ might change due to differences between enzyme substrate interactions. Larger ranges did not reveal any significant difference in the optimized results. The resulting mean values of the optimized parameters are shown in Supplemental Figure 3B.
Both case 1, 4CL3 fixed:4CL5 varied, and case 2, 4CL5 fixed:4CL3 varied, were simulated for caffeic acid using the mean values of k1, k2, and γ (Figures 6C and 6D). Comparison of the observed and simulated data for the models shows a good fit (Figures 6C and 6D). Both simulated cases fit the data well as indicated by a minimum mean square error of 0.0034. The proposed models in Figure 5B and Supplemental Figure 3A capture an important characteristic that differentiates the experimentally measured product formation rates for 4-coumaric acid and caffeic acid, respectively. Product formation rates associated with 4-coumaric acid increase steadily as the concentration of 4-coumaric acid increases (Figures 6A and 6B). However, experimental rates with caffeic acid (Figures 6C and 6D) show that at some enzyme concentration ratios, increased concentrations of caffeic acid result in a slowdown of the product formation rate, due to the self-inhibition exhibited by caffeic acid and 4CL5. The model of the total product formation in the equation in Supplemental Figure 3B captures this reduction in the rate. This consistency of the model and the experimental results emphasizes that self-inhibition should be included when modeling and predicting CoA ligation rates for caffeic acid as substrate.
A Mechanistic Model of the 4CL3/4CL5 Interaction Using Multiple Substrates with 4-Coumaric Acid as the Main Substrate
To model the total rate associated with CoA ligation so that it more closely represents what occurs in vivo, we now include how this rate may change with multiple hydroxycinnamic acids. The independent kinetics of 4CL3 and 4CL5 in the presence of 4-coumaric acid, caffeic acid, and ferulic acid substrates showed various levels of competitive (4CL3 and 4CL5) and uncompetitive (only 4CL5) inhibitions (Chen et al., 2013). We used this knowledge to create a block diagram of the expected interactions when 4CL3 and 4CL5 are mixed with 4-coumaric acid, caffeic acid, and ferulic acid (Figure 7). In this case, we are interested in 4-coumaric acid as substrate and the rate of formation of the product, the 4-coumaroyl-CoA thioester.
Figure 7.
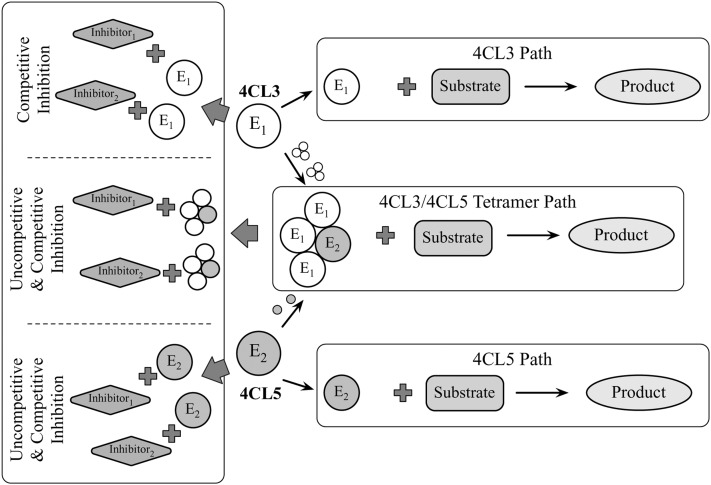
Block Diagram Showing the Effects of the Interaction of P. trichocarpa 4CL3 and 4CL5 with Multiple Inhibitors on Product Formation.
We consider multiple inhibition effects of other substrates. The 4CL3 enzymatic reaction has only competitive inhibition. The 4CL5 enzymatic reaction has competitive and uncompetitive inhibition. The enzymatic reaction of the 4CL3/4CL5 complex also considers competitive and uncompetitive inhibition.
Figure 7 shows the three paths for product formation via E1 (4CL3), E2 (4CL5), and the 4CL3/4CL5 tetramer. Caffeic and ferulic acids trigger an additional level of inhibition for each path. The inhibition of E1 (4CL3) by caffeic acid and ferulic acid is expected to show competitive inhibition (Chen et al., 2013). Similarly, the inhibition of E2 (4CL5) by caffeic acid and ferulic acid is expected to show competitive and uncompetitive inhibition (Chen et al., 2013). Because 4CL5 is the controlling enzyme, we expect the complex to show the same inhibition as 4CL5.
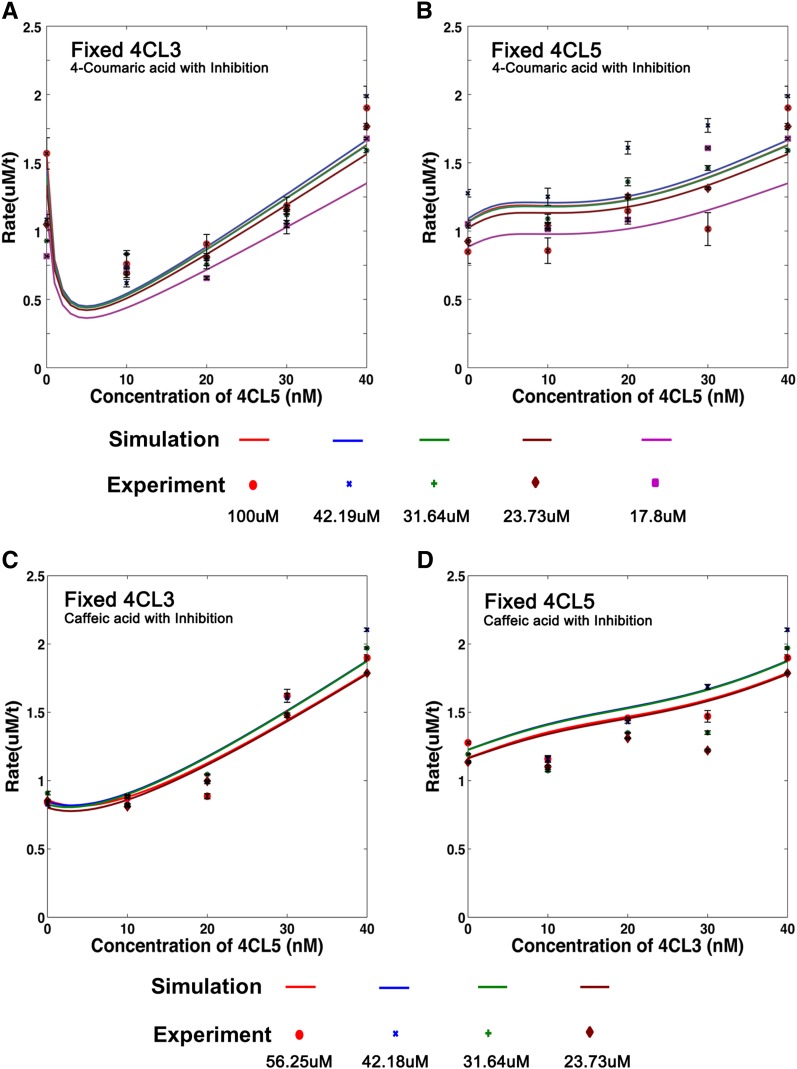
Figure 8A is a mechanistic model of interactions between 4-coumaric acid, caffeic acid, ferulic acid, 4CL3, 4CL5, the 4CL3/4CL5 complex, and the corresponding intermediates with 4-coumaric acid being the main substrate. All kinetic constants and associated inhibition constants except k and γ are obtained from in vitro kinetics of 4CL3 and 4CL5 (Chen et al., 2013). The quasi-equilibrium assumption was again used to derive a mathematical model of the total product formation rate associated with 4-coumaric acid in the presence of 4CL3, 4CL5, the 4CL3/4CL5 complex, caffeic acid, and ferulic acid (equation in Figure 8B; see Supplemental Methods for the full derivation). Measured product formation rates under conditions of varying total 4CL3, 4CL5, 4-coumaric acid, caffeic acid, and ferulic acid concentration were used in combination with hybrid optimization to estimate k1, k2, and γ. One hundred runs were performed with random initial conditions over the range [0 to 0.1] for k1, [0 to 0.1] for k2, and [1 to 3] for γ, and optimized values for k1, k2, and γ are shown in Figure 8B. Both experimental cases (4CL3 fixed:4CL5 varied and 4CL5 fixed:4CL3 varied) were simulated based on the equation in Figure 8B using the mean values of k1, k2, and γ (Figures 9A and 9B). Figures 9A and 9B show that the model fits the data well, with the mean squared error equal to 0.0077.
Figure 8.
Mechanistic Description of Product Formation with Kinetic Parameters and Multiple Inhibition Effects Using 4-Coumaric Acid as the Main Substrate.
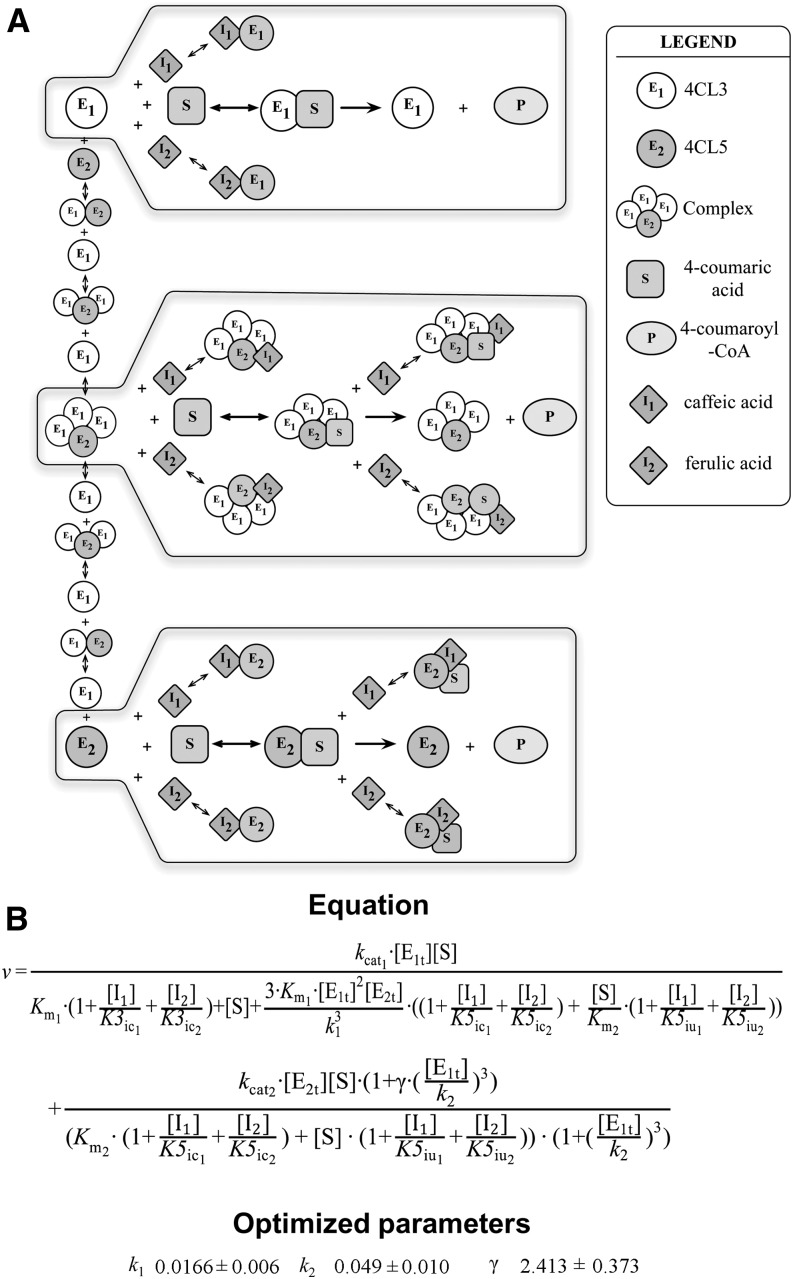
(A) In addition to the reactions represented in Figure 5B, we now consider reactions
between enzymes and inhibitors. The top gray box represents the 4CL3 enzymatic
reaction with two inhibitors. 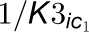 and
and
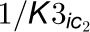 are the competitive inhibition rates of inhibitors on
4CL3. The bottom gray box represents the 4CL5 enzymatic reaction with the same
two inhibitors.
are the competitive inhibition rates of inhibitors on
4CL3. The bottom gray box represents the 4CL5 enzymatic reaction with the same
two inhibitors. 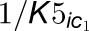 and
and 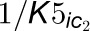 are competitive inhibition
rates of these inhibitors on 4CL5 and
are competitive inhibition
rates of these inhibitors on 4CL5 and 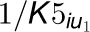 and
and
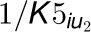 are uncompetitive inhibition rates of the inhibitors on
4CL5. The middle gray box represents the 4CL3/4CL5 complex enzymatic reaction
with two inhibitors. The inhibition rates in the 4CL5 enzymatic reaction are
used for the reaction of the complex. Each enzymatic reaction by 4CL3, 4CL5,
and the 4CL3/4CL5 complex is based on Michaelis-Menten kinetics.
are uncompetitive inhibition rates of the inhibitors on
4CL5. The middle gray box represents the 4CL3/4CL5 complex enzymatic reaction
with two inhibitors. The inhibition rates in the 4CL5 enzymatic reaction are
used for the reaction of the complex. Each enzymatic reaction by 4CL3, 4CL5,
and the 4CL3/4CL5 complex is based on Michaelis-Menten kinetics.
 and
and 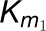 are kinetic parameters of
the 4CL3 enzymatic reaction.
are kinetic parameters of
the 4CL3 enzymatic reaction.  and
and
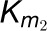 are kinetic parameters of the 4CL5 enzymatic reaction.
are kinetic parameters of the 4CL5 enzymatic reaction.
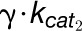 and
and 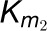 are assumed as kinetic
parameters of the 4CL3/4CL5 complex, where γ represents
activation effects of the complex on the rate. The interaction rate between
4CL3 and 4CL5 is assumed to be
are assumed as kinetic
parameters of the 4CL3/4CL5 complex, where γ represents
activation effects of the complex on the rate. The interaction rate between
4CL3 and 4CL5 is assumed to be 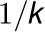 . The interactions occur for
the formation of the 4CL3/4CL5 tetramer in succession, which leads to
inhibition.
. The interactions occur for
the formation of the 4CL3/4CL5 tetramer in succession, which leads to
inhibition.
(B) A mathematical model is shown for multiple enzymes and
multiple substrates (4-coumaric acid as the main substrate and caffeic acid and
ferulic acid as inhibitors). The equation represents the rate of total product
formation associated with 4CL3, 4CL5, and the 4CL3/4CL5 complex including the
effect of multiple inhibitors, where 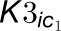 and
and
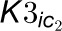 are competitive inhibition rate constants of inhibitors
on 4CL3.
are competitive inhibition rate constants of inhibitors
on 4CL3. 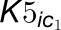 and
and 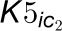 are the competitive
inhibition rate constants of inhibitors on 4CL5 and the 4CL3/4CL5 complex, and
are the competitive
inhibition rate constants of inhibitors on 4CL5 and the 4CL3/4CL5 complex, and
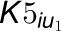 and
and 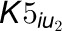 are the uncompetitive
inhibition rate constants of inhibitors on 4CL5 and the 4CL3/4CL5 complex. The
definitions of other variables and parameters are the same as those in Figure 5C. The optimized values of the
unknown parameters are fitted by hybrid optimization using MATLAB.
k1, k2, and
γ values represent the mean ± sd of
100 optimized values.
are the uncompetitive
inhibition rate constants of inhibitors on 4CL5 and the 4CL3/4CL5 complex. The
definitions of other variables and parameters are the same as those in Figure 5C. The optimized values of the
unknown parameters are fitted by hybrid optimization using MATLAB.
k1, k2, and
γ values represent the mean ± sd of
100 optimized values.
Figure 9.
Simulations and Experimental Rates Using 4-Coumaric Acid as the Main Substrate, with Caffeic Acid and Ferulic Acid as Inhibitors.
(A) and (B) Solid lines represent simulation results based on the equation and optimized parameters in Figure 8B. Dots represent experimental data. Colors represent different substrate concentrations.
(A) Activity with a fixed amount of 4CL3 and increasing amounts of 4CL5.
(B) A fixed amount of 4CL5 with increasing amounts of 4CL3. The R2 value is 0.60, and the RMSD is 0.088 for the goodness of fit.
(C) and (D) Simulation results are shown compared with the experimental rates for caffeic acid as the main substrate and 4-coumaric acid and ferulic acid as inhibitors. Solid lines represent simulation results based on the equation and optimized parameters in Supplemental Figure 4B. Dots represent experimental data. Colors represent different substrate concentrations.
(C) Results with a fixed amount of 4CL3 and increasing amounts of Ptr-4CL5.
(D) A fixed amount of 4CL5 with increasing amounts of 4CL3. The R2 value is 0.79, and the RMSD is 0.15 for the goodness of fit.
We tested the plausibility of the multisubstrate mechanistic model and resulting mathematical model in Figure 8B to assess whether inhibition is needed to better describe changes in the total rate of product formation seen in Figures 9A and 9B. We performed this test by fitting the equation in Figure 5C (no integrated inhibition) to experimental rates in Figures 9A and 9B. Without including inhibition, multiple runs of hybrid optimization provided estimates of k1, k2, and γ that yield a mean square error of 2.4, which is four orders of magnitude higher than the mean square error of 0.0077 produced using the model that includes substrate inhibition (Figure 8B). The mechanistic description of the rate of product formation using 4-coumaric acid does not adequately describe situations that are closer to in vivo conditions without considering interactions between caffeic acid and ferulic acid, 4CL3, and 4CL5.
A Mechanistic Model of the 4CL3/4CL5 Interaction with Multiple Substrates and Caffeic Acid as the Main Substrate
The substrate self-inhibition exhibited by caffeic acid on the 4CL5 reaction should be incorporated into the mechanistic model to better predict the rate of product formation resulting from caffeic acid in the presence of multiple substrates. As before, we modeled the self-inhibition on 4CL5 as uncompetitive (Supplemental Figures 4A and 4B). Experimental rates and optimized values of k1, k2, and γ were obtained as described in previous sections, with random initial conditions of k1, k2, and γ extending over [0 to 0.1], [0 to 0.1], and [1 to 6], respectively. The optimized values for k1, k2, and γ are shown in Supplemental Figure 4B. Figures 8C and 8D illustrate both the experimental rates (case 1, 4CL3 fixed:4CL5 varied; and case 2, 4CL3 varied:4CL5 fixed) along with the simulated rates calculated using equation Supplemental Figure 4B with mean values of k1, k2, and γ. We calculate a mean square error of 0.0233, showing that the model adequately fits the experimental product formation rates.
To further evaluate this caffeic acid model, we asked whether the mechanistic description of the multisubstrate inhibitions of 4-coumaric acid and ferulic acid and substrate self-inhibition of caffeic acid were needed to adequately describe the experimental rates. We explored the plausibility of equation in Supplemental Figure 4B by assessing the following characteristics independently: (1) inhibitions associated with the presence of 4-coumaric acid and ferulic acid substrates and (2) substrate self-inhibition associated with caffeic acid and 4CL5. We assess point 1 by identifying if the multiple enzyme/single substrate mechanistic model for caffeic acid that lacked multisubstrate inhibitions (Supplemental Figure 3B) presents an equally plausible model to describe the data shown in Figures 9C and 9D. The fit of equation in Supplemental Figure 3B to these data resulted in a mean square error of 0.1076, which was substantially greater than the mean square error (0.0233) associated with the multiple enzyme/multiple substrate model for caffeic acid (Supplemental Figure 4B).
We assess point 2 by investigating the impact of self-inhibition on the experimental rates. Comparing the experimental rates in Figures 9A and 9B (4-coumaric acid: no known self-inhibition) and Figures 9C and 9D (caffeic acid: known self-inhibition), we see more tightly grouped measurements for experimental rates in Figures 9C and 9D, as the substrate concentration is increased for each 4CL3:4CL5 combination. Similar characteristics are seen in Figures 6C and 6D, which we attribute to substrate self-inhibition. We assess whether a mechanistic description of substrate self-inhibition better describes these data by fitting a multiple enzyme/multiple substrate model for caffeic acid without self-inhibition to the data in Figures 9C and 9D. The fit resulted in a mean square error of 0.074, which is higher than 0.023, the mean square error of the model that incorporates substrate self-inhibition into the multiple enzyme/multiple substrate model (see equation in Supplemental Figure 4B). Based on these results, we conclude that both a mechanistic description of 4-coumaric acid and ferulic acid inhibition, along with a mechanistic description of caffeic acid self-inhibition, better describes CoA ligation associated with caffeic acid in the presence of 4-coumaric acid and ferulic acid.
DISCUSSION
The experimental results presented here advance the mechanistic understanding of CoA ligation in monolignol biosynthesis. This pathway has been commonly described as a collection of independent enzymes (Boerjan et al., 2003; Vanholme et al., 2012). However, we obtained strong physical and biochemical evidence for protein–protein interactions of 4CL3 and 4CL5 in the formation of a heterotetramer, based on BiFC, co-IP, chemical cross-linking, and MS. We have also shown that the interactions of 4CL3 and 4CL5 have a functional role, affecting the kinetic behavior of this step in the pathway. This result is similar to the protein–protein interactions identified in our earlier article for C4H and C3H where the activity of both 3- and 4-hydroxylation is modified by a C4H1/C4H2/C3H3 protein complex (Chen et al., 2011). The extent of functional protein–protein interactions for the entire monolignol biosynthetic pathway in P. trichocarpa has yet to be fully elucidated.
Some exceptions to the concept on enzymes acting independently have been proposed related to the possibility of metabolic channeling. Stafford (1974) suggested that an enzyme complex performed flavonoid biosynthesis. Hrazdina and Wagner (1985) proposed that Phe ammonia-lyase, the first enzyme in the pathway for the biosynthesis of both flavonoids and monolignols, was attached to the endoplasmic reticulum by binding to cytochrome P450 reductase. Others suggested that Phe ammonia-lyase and C4H might interact to create metabolic channeling for the early steps in monolignol biosynthesis (Czichi and Kindl, 1977; Rasmussen and Dixon, 1999; Winkel-Shirley, 1999). Protein complex formation and metabolic channeling was also proposed for two other monolignol biosynthetic enzymes, caffeic acid O-methyl transferase and caffeoyl CoA 3-O-methyltransferase, to direct synthesis to either S or G subunits in lignin (Lee et al., 2012). However, biochemical evidence of substrate flux does not support metabolic channeling (Chen et al., 2011; Wang et al., 2014). A predictive kinetic metabolic flux model presented by Wang et al. (2014) (companion article) indicates that the reaction and inhibition kinetic parameters of the individual monolignol pathway enzymes are sufficient to explain major features of metabolic flux through the pathway.
Mathematical Description of CoA Ligation
We proposed mathematical models derived from experimentally verified mechanistic interactions between enzymes and substrates that participate in CoA ligation of 4-coumaric acid and caffeic acid in P. trichocarpa. These models (Figure 8B; Supplemental Figure 4B) were developed to mimic conditions expected in vivo (multiple enzymes and multiple substrates). These rate equations provide the same benefits as Michaelis-Menten kinetics, which have been used to describe the product rate formation for simpler enzymatic reactions (Segel, 1975). Conceptually, Figure 8B and Supplemental Figure 4B can be illustrated using a nonlinear function block model whose output (product formation rate) is dependent on the inputs (initial substrate concentrations and total enzyme concentrations), the functional form of the rate equation, and internal kinetic constants (Figure 10A). Such a mathematical construct enables us to predict how variation in these substrates and enzymes influence the rate of product formation.
Figure 10.
Model Description for CoA Ligation by 4CL.
(A) The white box represents the model concept with all rate equations and kinetic parameters. The inputs of the model are initial substrate concentrations and total enzyme amounts in the left gray box. The output of the model is the rate of product formation. For a given hydroxycinnamic acid as substrate, other hydroxycinnamic acids may function as inhibitors.
(B) and (C) The predicted fraction of the total rate attributed to each of the enzyme entities (multiple enzymes and single substrate model: 4-coumaric acid as substrate). The black solid lines and dots represent rate simulations using the equation and optimized values in Figure 5C and experimental data (4-coumaric acid concentration of 50.6 μM). The total rate represented by the black line is the sum of rates catalyzed by the enzymes 4CL3 (red line), 4CL5 (green line), and the 4CL3/4CL5 complex (blue line) in both cases. A fixed amount of 4CL3 with increasing amounts of 4CL5 (B) and a fixed amount of 4CL3 with increasing amounts of 4CL5 (C).
(D) and (E) The predicted fraction of the total rate attributed to each of the enzyme entities (multiple enzymes and multiple substrate model: caffeic acid as a main substrate and 4-coumaric acid and ferulic acid as inhibitors). The black solid line and dots represent rate simulation results using the equation and optimized values in Supplemental Figure 4B and experimental data (caffeic acid concentration of 23.7 μM). The total rate represented by the black line is the sum of rates catalyzed by the enzymes 4CL3 (red line), 4CL5 (green line), and the 4CL3/4CL complex (blue line) in both cases. A fixed amount of 4CL3 with increasing amounts of 4CL5 (D) and a fixed amount of 4CL5 with increasing amounts of 4CL3 (E).
Knowledge of these and other individual reaction rates in the monolignol biosynthetic pathway provides a basis for predicting how the interplay between multiple metabolites impact pathway-wide substrate consumption and product formation. The development of specific reaction kinetics, such as this model of CoA ligation, is the initial step to constructing a multireaction pathway model consisting of a system of equations that can predict changes in metabolite products and their substrate intermediates in response to changes in enzyme regulation, inhibition characteristics, and other enzymatic properties critical to metabolic engineering.
Effects of the 4CL3/4CL5 Enzyme Complex on Reaction Rates of Individual Enzymes (Single Substrate): 4-Coumaric Acid
The mathematical models presented here allow us to assess how reaction rate changes with variation in the amounts of each of the individual enzymes or the complex. This free enzyme and enzyme complex pathway analysis for 4CL allows us to assess plausible mechanistic interactions that impact the overall reaction rate through individual enzyme states. This ability to analyze the contribution of the individual reaction rates to the total rate is currently difficult due to the inability to isolate and purify the functional 4CL3-4CL5 complex. Figures 10B and 10C display the results of using Figure 5C to compute the fraction of the total rate that can be attributed to each of the enzymes and the enzyme complex for any combination of individual enzyme concentrations.
As 4CL5 is added to 4CL3, the rate associated with 4CL3 decreases (Figure 10B). This reduction in rate associated with free 4CL3 can be attributed to the recruitment of 4CL3 by 4CL5 into the enzyme complex, thus reducing the amount of free 4CL3 that is available to bind to the substrate. This free 4CL3 rate reduction may lead to the initial slowdown or inhibition that was observed with the total rate when small amounts of 4CL5 are introduced (Figure 2A). As more 4CL5 is introduced, the rate associated with 4CL3 continues to decrease rapidly, while the simulated rate associated with the 4CL3-4CL5 complex increases rapidly. Under this model, free 4CL5 and the 4CL3/4CL5 complex are the primary drivers of the total rate and are responsible for the increased activation seen at equal levels of 4CL3 and 4CL5. The reduction in rate associated with 4CL3 in the model fits well with the experimental results.
In Figure 10C, as we introduce small amounts of 4CL3 to fixed amounts of 4CL5, we see that the simulated rate associated with the 4CL3-4CL5 complex starts to increase slowly. This slow increase indicates that high concentrations of 4CL5 and low concentrations of 4CL3 result in relatively low concentrations of the 4CL3-4CL5 complex. The simulated rate associated with free 4CL3 does not increase, likely meaning that available 4CL3 has been recruited into the 4CL3-4CL5 complex. As the initial concentration of 4CL3 increases, the simulated rate associated with 4CL3 remains relatively low. This low free 4CL3 rate would suggest that the 4CL3 enzyme is not available to bind to the substrate but instead, continues to bind to the free 4CL5, resulting in a greater concentration of the 4CL3-4CL5 complex and an increased rate associated with the complex. We see a decrease in the simulated rate associated with 4CL5 in Figure 10C but not as significant as the decrease seen with 4CL3 in Figure 10B. Several factors can be inferred from the simulation in Figures 10B and 10C. First, CoA ligation can be manipulated in the plant by a nonlinear control that is not proportional to the individual expression of 4CL3 and 4CL5. Second, an initial control of CoA ligation could come from the manipulation of 4CL5 concentration when both enzymes are present. Any metabolic engineering of CoA ligation should focus on the manipulation of 4CL5 because it appears to be the primary controller of the total reaction rate.
Effects of 4CL3/4CL5 Enzyme Complex on Reaction Rates of Individual Enzymes: Multiple Substrates and Caffeic Acid
We further investigated how inhibition impacts the individual rates associated with 4CL3, 4CL5, and the 4CL3-4CL5 complex by analyzing the model in Supplemental Figure 4B. Figures 10D and 10E display the simulated and experimental total reaction rates for caffeic acid in the presence of 4-coumaric acid and ferulic acid, along with the simulated individual rates associated with each enzyme entity (4CL3, 4CL5, and enzyme complex).
In Figure 10D, there is a reduction in the 4CL3 rate when small amounts of 4CL5 are introduced. This reduction is less than that seen in Figures 10B and 10C. We infer that the inhibition (multiple substrate inhibition of 4-coumaric acid and ferulic acid and/or substrate self-inhibition of caffeic acid) impacts the recruitment of 4CL3 by 4CL5 to form the enzyme complex, increasing the amounts of free 4CL3 and, hence, free 4CL5 that are available to bind to the substrate. We also see that the simulated rate of 4CL5 is greater than the simulated rate of the 4CL3-4CL5 complex, which is different from the results seen in Figure 10B. This effect supports the hypothesis that the inhibitions identified in the 4CL pathway either (1) reduce the concentration of available 4CL3-4CL5 complex or (2) reduce the reaction rate of the complex or both. The simulated individual rates (Figure 10E), confirm that the introduction of increasing initial concentrations of 4CL3 to a fixed concentration of 4CL5 results in increased rates associated with the 4CL3-4CL5 complex. The simulated rates of free 4CL3 and 4CL5 are still relatively large compared with those in Figure 10C, supporting the hypothesis that more free 4CL3 and 4CL5 enzymes are available. Thus, altering the inhibition characteristics of the enzymes may mitigate the rate of product formation from the 4CL3/4CL5 enzyme complex. This change in priority of flux through the pathway under inhibition provides another potential avenue for engineering nonlinear control of CoA ligation.
Elucidation of Plausible Mechanisms That Control CoA Ligation in the Lignin Biosynthesis Pathway
The purpose of these experiments was to understand the characteristics of each specific enzyme step in monolignol biosynthesis. In the course of these investigations, we found that two 4CL isomers, 4CL3 and 4CL5, involved in the CoA ligation of hydroxycinnamic acids during wood formation and lignin biosynthesis did not act independently. This result prompted studies of their interaction and the construction of a mathematical model to predict the behavior of the enzymes during monolignol biosynthesis by describing the factors affecting flux through the pathway. The characteristics of the individual enzymes, identified through in vitro analysis, provided insight into plausible mechanisms controlling the rate of hydroxycinnamic acid CoA ligation. Specifically, we tested whether a mechanistic model that incorporated the competitive (4CL3 and 4CL5) and uncompetitive inhibitions (4CL5) in the presence of multiple substrates and the substrate self-inhibition between caffeic acid and 4CL5 was necessary to best fit the experimental rates. To verify how these mechanistic interactions control the overall rate of product formation associated with CoA ligation, transgenic perturbation will be performed where the activity of each enzyme will be reduced through different levels, and the consequent effects will be evaluated according to predictions of the mathematical model. We do not yet know the specific amino acids involved in the interaction of 4CL3 and 4CL5. Some insights may come from analysis of purified tetramers and information on 4CL crystal structure. A crystal structure of monomeric 4CL1 from Populus tomentosa (Pto-4CL1), a plausible ortholog of Ptr-4CL3, has been obtained (Hu et al., 2010). Hu et al., (2010) identified three residues essential for 4CL catalytic activity (Lys-438, Gln-443, and Lys-523) and five for substrate binding (Tyr-236, Gly-306, Gly-331, Pro-337, and Val-338). The size of the binding pocket had the greatest influence on substrate specificity, as suggested by Schneider et al. (2003). In Arabidopsis, the crystal structure of At-4CL2 (also an ortholog of Ptr-4CL3) has been determined (Morita et al., 2011).
Finally, these results indicate an unanticipated level of complexity for the 4CL catalyzed CoA ligation of hydroxycinnamic acids in monolignol biosynthesis. Genome sequencing (Tuskan et al., 2006) led to the discovery of 4CL5 in P. trichocarpa (Shi et al., 2010; Chen et al., 2013). The work described here indicates how P. trichocarpa 4CL5 forms a tetrameric complex with 4CL3 and regulates its activity. Even though the additional complexity of CoA ligation in monolignol biosynthesis in P. trichocarpa is considerable, nevertheless, this complex reaction can now be described mathematically and the predicted metabolic flux could be incorporated into more comprehensive mathematical models of the pathway.
4CL is a key enzyme in phenylpropanoid metabolism, providing activated CoA thioesters precursors for many products including flavonoids and anthocyanins in addition to monolignols (Vanholme et al., 2012). The regulation of metabolism for these products depends on developmental specificity and the response to many environmental stimuli. How the mechanisms of 4CL function presented here relate to the wider functions of 4CL remains to be investigated. Why do plants such as Arabidopsis synthesize monolignols using a single 4CL enzyme and forego the 4CL complexity of P. trichocarpa? Perhaps this difference is associated with differences between woody and herbaceous dicots.
METHODS
Enzymology, Co-IP, and BiFC
These studies have been performed with recombinant proteins containing His-tags (Hochuli et al., 1988), which greatly aid purification and characterization. The His-tags do not affect the substrate specificity and relative reaction rates (Chen et al., 2013).
Chemical sources, preparation of recombinant proteins and SDX extracts, and synthesis of hydroxycinnamic acids, SIL hydroxycinnamic acids, and CoA thioesters followed that of Chen et al. (2013). MS was used to confirm the purity and identity of all synthesized products. Similarly, the methods for enzyme reactions, HPLC analysis, co-IP, immunoblotting, and protein cross-linking have also been previously described (Chen et al., 2011, 2013). Preparation and analysis of SDX protoplasts for transfection and bimolecular fluorescence microscopy follow Chen et al. (2011) and Lin et al. (2013).
Protein-specific peptide sequences, AGEVPVAFVVKSEKS and SGEIPVAFVIKSENS, were selected from the predicted amino acid sequences of 4CL3 and 4CL5, synthesized, conjugated to carrier protein keyhole limpet hemocyanin, and used as antigenic epitopes to raise polyclonal antibodies in rabbits (Antagene). The specificity of the antibodies for 4CL3 and 4CL5 has been verified in a previous publication (Chen et al., 2013).
LMD
Sections from internodes between 15 and 20 of 6-month-old Populus trichocarpa grown in a greenhouse were debarked, cut into 0.5-mm segments, and frozen in liquid nitrogen. The stem segments were attached to a chuck using optimal cutting temperature compound at −20°C for 20 min to solidify the optimal cutting temperature and to prevent the stem fragments from detachment. The 10-µm-thick cross sections were cut using a cryostat at −20°C. Six to eight cross sections were attached to a glass slide. The slide was dipped in 95% ethanol for 2 min, transferred into 100% xylene, and air-dried for 15 min. Different cell types were collected using a Laser Specifications Leica LMD7000, and the collected tissue was dipped directly into RNeasy lysis buffer (Qiagen) for RNA extraction. Tissue with three cell types (vessel, fiber, and ray), fiber cells only, and vessel cells only was collected. For fiber cell collection, ray cells were burned away first using the laser, and then vessel cells were avoided. Total RNA was isolated using a RNeasy Plant RNA isolation kit (Qiagen) as described (Li et al., 2012), and the quality of the RNA was examined using an Agilent 2100 bioanalyzer and Agilent RNA 6000 Pico Assay chips. Quantitative RT-PCR for 4CL3 and 4CL5 transcript abundance detection was performed as described (Shi et al., 2010) in three cell types (control) and fiber cell samples.
MS
Absolute quantification of the cross-linked 4CL3 and 4CL5 was performed by PC-IDMS as described (Shuford et al., 2012). Briefly, the high molecular mass fraction (>100 kD) of the cross-linked enzymes was reduced in the presence of 50 mM DTT (30 min, 56°C), alkylated with 200 mM iodoacetamide (1 h, 37°C), buffer exchanged three times in a 10-kD Amicon 0.5-mL centrifugal filter (Millipore) with enzyme digestion buffer (2 M urea and 10 mM CaCl2 in 50 mM Tris-HCl, pH 8.0), and finally digested within the 10-kD filter unit using 400 µg/mL bovine trypsin (12 h, 37°C). Each surrogate peptide derived from digestion of 4CL3 (FDIGTLLGLIEK) and 4CL5 (FEIGSLLGLIEK) was quantified by spiking in 611 fmol of the analogous SIL synthetic peptide (Mayo Clinic Proteomics Research Center) at the start of the digestion to serve as an internal standard. The trypsin digestion was quenched by adding 1% formic acid containing 0.001% zwittergent 3-16 (Calbiochem) to the filter unit, and the tryptic and SIL peptides were eluted through the filter via centrifugation (15 min, 14,000g). Detection of the natural surrogate and SIL peptides by nano-flow liquid chromatography–selected reaction monitoring and data analysis was performed as reported (Shuford et al., 2012).
Optimization of Parameters for the 4CL3/4CL5 Interaction
The rate equations we developed have unknown parameters related to the enzyme
complex, such as k1, k2, and
γ (see Supplemental Methods for modeling). It is possible to infer the
parameters from the experimental data. Our parameter optimization, based on
experimental data, considers the values of unknown parameters in minimizing the
objective function following Equation 2: 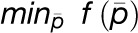 where
where
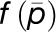 is
the objective function (see objective function) and
is
the objective function (see objective function) and 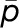 represents the parameters to be
estimated such as k1, k2, and
γ. The optimization process used a hybrid optimization
algorithm (Xia and Wu, 2005) for parameter
estimation, which includes global and local optimization for effective searches in
complex nonlinear systems. The genetic algorithm, used as the global optimization
method, is a search method that mimics natural evolution. Its random nature of the
various starting points is the reason that the genetic algorithm is not contained by
local optima. The genetic algorithm uses the probabilistic selection rule with
parallel searching, which is attractive to evaluating large search space and solving
nonlinear problems with a large number of parameters (global optimization problems).
Fmincon (find the minimum of constrained nonlinear multivariable function), used as
the local optimization, is a function for minimizing a scalar function of several
variables within a constrained region (Mathworks; Optimization Toolbox, version 3,
User’s Guide, 2007). Fmincon is suitable for solving problems with nonlinear
constraints and several variables such as 4CL
enzyme kinetic modeling. Fmincon is available in the Optimization Toolbox of MATLAB.
The optimization process was simulated by the optimization toolbox in MATLAB 7.11.
One hundred runs according to substrates were performed. The mean value of 100
optimized solutions was used in the model equation. The scatter of optimized values
was computed using an estimate of the sd:
represents the parameters to be
estimated such as k1, k2, and
γ. The optimization process used a hybrid optimization
algorithm (Xia and Wu, 2005) for parameter
estimation, which includes global and local optimization for effective searches in
complex nonlinear systems. The genetic algorithm, used as the global optimization
method, is a search method that mimics natural evolution. Its random nature of the
various starting points is the reason that the genetic algorithm is not contained by
local optima. The genetic algorithm uses the probabilistic selection rule with
parallel searching, which is attractive to evaluating large search space and solving
nonlinear problems with a large number of parameters (global optimization problems).
Fmincon (find the minimum of constrained nonlinear multivariable function), used as
the local optimization, is a function for minimizing a scalar function of several
variables within a constrained region (Mathworks; Optimization Toolbox, version 3,
User’s Guide, 2007). Fmincon is suitable for solving problems with nonlinear
constraints and several variables such as 4CL
enzyme kinetic modeling. Fmincon is available in the Optimization Toolbox of MATLAB.
The optimization process was simulated by the optimization toolbox in MATLAB 7.11.
One hundred runs according to substrates were performed. The mean value of 100
optimized solutions was used in the model equation. The scatter of optimized values
was computed using an estimate of the sd: 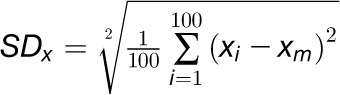 , where
, where
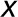 represents one of the estimated unknown parameters, k1,
k2, or γ.
represents one of the estimated unknown parameters, k1,
k2, or γ.
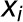 represents the estimated parameter on the ith run, and
represents the estimated parameter on the ith run, and
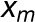 represents the arithmetic mean over all 100 runs.
represents the arithmetic mean over all 100 runs.
Objective Function
The objective function in the optimization process indicates how we define the suitability of the model. The objective function was targeted at representing the experiment data as closely as possible, for which we wanted to minimize the distance between the expression data and the simulation results. One common objective function to assess the goodness-fit modeling is the mean square error, which is defined as
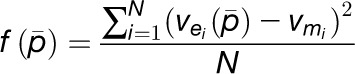 |
(3) |
(Wackerly
et al. 2008). In Equation 3,
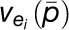 is the ith estimated reaction rate by the developed model,
is the ith estimated reaction rate by the developed model,
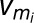 is the ith measured reaction rate, and N is the
number of experimental data samples. Mean square error values that approach zero
represent improved model fit of the experimental data.
is the ith measured reaction rate, and N is the
number of experimental data samples. Mean square error values that approach zero
represent improved model fit of the experimental data.
Mathematical Model Development
A more complete description of the assumptions and derivations of the 44 equations used in the development of the mathematical model are described in Supplemental Methods.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Ptr-4CL3, EU603298; and Ptr-4CL5, EU603299.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Bimolecular Fluorescence Complementation of Homomeric 4CL3 and 4CL5.
Supplemental Figure 2. Quantification of the 4CL3 and 4CL5 Proteins by PC-IDMS.
Supplemental Figure 3. Mechanistic Description of the Inhibition and Activation Effects on the Rate of Product Formation Using Caffeic Acid as Substrate.
Supplemental Figure 4. Mechanistic Description and Mathematical Model Including the Multiple Inhibition Effects on Product Formation with Caffeic Acid as the Main Substrate.
Supplemental Methods. Mathematical Model Definitions, Details, Kinetic Parameters, and Model Derivation.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation (USA), Plant Genome Research Program Grant DBI-0922391 (to V.L.C.). We also wish to acknowledge the support from the NC State University Jordan Family Distinguished Professor Endowment and the NC State University Forest Biotechnology Industrial Research Consortium.
AUTHOR CONTRIBUTIONS
H.-C.C., J.S., J.P.W., J.D., C.W., D.C.M., R.R.S., and V.L.C created the experimental design. H.-C.C., J.P.W., (biochemistry and cell biology), C.M.S. (protein MS), J.L. (biochemistry), Y.C.L., A.N. (LMD), and Q.L. (biochemistry) carried out experiments. D.C.M., R.R.S., and V.L.C. supervised the experiments. J.S. carried out modeling. J.D. and C.W. supervised the modeling. H.-C.C., J.S., J.P.W., R.S., J.D., C.W., F.I., and V.L.C. performed data analysis and statistical evaluation. H.-C.C., J.S., J.P.W., R.S., J.D., C.M.S., D.C.M., C.W., R.R.S., and V.L.C. prepared the article.
Glossary
- 4CL
4-coumaric acid:CoA ligase
- LMD
laser microdissection
- co-IP
coimmunoprecipitation
- BiFC
bimolecular fluorescence complementation
- MS
mass spectrometry
- DSP
dithiobis(succinimidyl propionate)
- PC-IDMS
protein cleavage–isotope dilution mass spectrometry
- SIL
stable isotope–labeled
- RMSD
root mean standard deviation
Footnotes
Online version contains Web-only data.
References
- Allina S.M., Pri-Hadash A., Theilmann D.A., Ellis B.E., Douglas C.J. (1998). 4-Coumarate:coenzyme A ligase in hybrid poplar. Properties of native enzymes, cDNA cloning, and analysis of recombinant enzymes. Plant Physiol. 116: 743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amthor J.S. (2003). Efficiency of lignin biosynthesis: A quantitative analysis. Ann. Bot. (Lond.) 91: 673–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrière Y., Riboulet C., Méchin V., Pichon M., Cardinal A., Lubberstedt T., Lapierre C. (2007). Genetics and genomics of lignification in grass cell walls based on maize as a model system. Genes Genomes Genomics 1: 133–156. [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54: 519–546. [DOI] [PubMed] [Google Scholar]
- Chen F., Dixon R.A. (2007). Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25: 759–761. [DOI] [PubMed] [Google Scholar]
- Chen F., Tobimatsu Y., Havkin-Frenkel D., Dixon R.A., Ralph J. (2012). A polymer of caffeyl alcohol in plant seeds. Proc. Natl. Acad. Sci. USA 109: 1772–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.C., Li Q., Shuford C.M., Liu J., Muddiman D.C., Sederoff R.R., Chiang V.L. (2011). Membrane protein complexes catalyze both 4- and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc. Natl. Acad. Sci. USA 108: 21253–21258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.C., et al. (2013). Monolignol pathway 4-coumaric acid:coenzyme A ligases in Populus trichocarpa: Novel specificity, metabolic regulation, and simulation of coenzyme A ligation fluxes. Plant Physiol. 161: 1501–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang V.L. (2002). From rags to riches. Nat. Biotechnol. 20: 557–558. [DOI] [PubMed] [Google Scholar]
- Czichi U., Kindl H. (1977). Phenylalanine ammonia lyase and cinnamic acid hydroxylases as assembled consecutive enzymes on microsomal membranes of cucumber cotyledons: Cooperation and subcellular distribution. Planta 134: 133–143. [DOI] [PubMed] [Google Scholar]
- Denness L., McKenna J.F., Segonzac C., Wormit A., Madhou P., Bennett M., Mansfield J., Zipfel C., Hamann T. (2011). Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis. Plant Physiol. 156: 1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J., Büttner D., Wang Q., Douglas C.J., Somssich I.E., Kombrink E. (1999). Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 19: 9–20. [DOI] [PubMed] [Google Scholar]
- Eriksson, K.E.L., Blanchette, R.A., and Ander, P. (1990). Microbial and Enzymatic Degradation of Wood and Wood Components. (Berlin, Heidelberg, New York: Springer-Verlag). [Google Scholar]
- Fontes R., Ribeiro J.M., Sillero A. (2000). Inhibition and activation of enzymes. The effect of a modifier on the reaction rate and on kinetic parameters. Acta Biochim. Pol. 47: 233–257. [PubMed] [Google Scholar]
- Goldberg, D. (1989). Genetic Algorithms in Search, Optimization and Machine Learning. (Reading, MA: Addison-Wesley Professional). [Google Scholar]
- Gross G.G., Zenk M.H. (1974). Isolation and properties of hydroxycinnamate: CoA ligase from lignifying tissue of Forsythia. Eur. J. Biochem. 42: 453–459. [DOI] [PubMed] [Google Scholar]
- Gui J., Shen J., Li L. (2011). Functional characterization of evolutionarily divergent 4-coumarate:coenzyme a ligases in rice. Plant Physiol. 157: 574–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahlbrock K., Scheel D. (1989). Physiology and molecular biology of phenylpropanoid metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40: 347–369. [Google Scholar]
- Hamberger B., Hahlbrock K. (2004). The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc. Natl. Acad. Sci. USA 101: 2209–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, T. (1985). Biosynthesis of lignin. In Biosynthesis and Biodegradation of Wood Components, T. Higuchi, ed (New York: Academic Press), pp. 141–160. [Google Scholar]
- Higuchi, T. (1997). Biochemistry and Molecular Biology of Wood. (New York: Springer). [Google Scholar]
- Hinchee M., Rottmann W., Mullinax L., Zhang C., Chang S., Cunningham M., Pearson L., Nehra N. (2009). Short-rotation woody crops for bioenergy and biofuels applications. In Vitro Cell. Dev. Biol. Plant 45: 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochuli E., Bannwarth W., Döbeli H., Gentz R., Stüber D. (1988). Genetic approach to facilitate purification of recombinant proteins with a novel metal chelate adsorbent. Nat. Biotechnol. 6: 1321–1325. [Google Scholar]
- Hrazdina G., Wagner G.J. (1985). Compartmentation of plant phenolic compounds; sites of synthesis and accumulation. In The Biochemistry of Plant Phenols, C.F. Van Sumere and P.J. Lea, eds (Oxford, UK: Oxford University Press), pp. 119–133. [Google Scholar]
- Hu C.D., Chinenov Y., Kerppola T.K. (2002). Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9: 789–798. [DOI] [PubMed] [Google Scholar]
- Hu W.J., Kawaoka A., Tsai C.J., Lung J., Osakabe K., Ebinuma H., Chiang V.L. (1998). Compartmentalized expression of two structurally and functionally distinct 4-coumarate:CoA ligase genes in aspen (Populus tremuloides). Proc. Natl. Acad. Sci. USA 95: 5407–5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Gai Y., Yin L., Wang X., Feng C., Feng L., Li D., Jiang X.N., Wang D.C. (2010). Crystal structures of a Populus tomentosa 4-coumarate:CoA ligase shed light on its enzymatic mechanisms. Plant Cell 22: 3093–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajita S., Katayama Y., Omori S. (1996). Alterations in the biosynthesis of lignin in transgenic plants with chimeric genes for 4-coumarate:coenzyme A ligase. Plant Cell Physiol. 37: 957–965. [DOI] [PubMed] [Google Scholar]
- Lee Y., Voit E.O. (2010). Mathematical modeling of monolignol biosynthesis in Populus xylem. Math. Biosci. 228: 78–89. [DOI] [PubMed] [Google Scholar]
- Lee Y., Chen F., Gallego-Giraldo L., Dixon R.A., Voit E.O. (2011). Integrative analysis of transgenic alfalfa (Medicago sativa L.) suggests new metabolic control mechanisms for monolignol biosynthesis. PLOS Comput. Biol. 7: e1002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Escamilla-Treviño L., Dixon R.A., Voit E.O. (2012). Functional analysis of metabolic channeling and regulation in lignin biosynthesis: A computational approach. PLOS Comput. Biol. 8: e1002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhou Y., Cheng X., Sun J., Marita J.M., Ralph J., Chiang V.L. (2003). Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc. Natl. Acad. Sci. USA 100: 4939–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Lin Y.C., Sun Y.H., Song J., Chen H., Zhang X.H., Sederoff R.R., Chiang V.L. (2012). Splice variant of the SND1 transcription factor is a dominant negative of SND1 members and their regulation in Populus trichocarpa. Proc. Natl. Acad. Sci. USA 109: 14699–14704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-C., Li W., Sun Y.-H., Kumari S., Wei H., Li Q., Tunlaya-Anukit S., Sederoff R.R., Chiang V.L. (2013). SND1 transcription factor-directed quantitative functional hierarchical genetic regulatory network in wood formation in Populus trichocarpa. Plant Cell 25: 4324–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomant A.J., Fairbanks G. (1976). Chemical probes of extended biological structures: Synthesis and properties of the cleavable protein cross-linking reagent [35S]dithiobis(succinimidyl propionate). J. Mol. Biol. 104: 243–261. [DOI] [PubMed] [Google Scholar]
- Morita H., Mori T., Wanibuchi K., Kato R., Sugio S., Abe I. (2011). Crystallization and preliminary X-ray analysis of 4-coumarate:CoA ligase from Arabidopsis thaliana. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67: 409–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K., Dima O., Kim H., Lu F., Niculaes C., Vanholme R., Dauwe R., Goeminne G., Inzé D., Messens E., Ralph J., Boerjan W. (2010). Mass spectrometry-based sequencing of lignin oligomers. Plant Physiol. 153: 1464–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoki, N., Nanto, K., Hayashi, K., Onogi, S., and Kawaoka, A. (2011). Wood quality-related gene expressions of Eucalyptus globulus grown in a greenhouse. BMC Proc. 5: P115. [Google Scholar]
- Osakabe K., Tsao C.C., Li L., Popko J.L., Umezawa T., Carraway D.T., Smeltzer R.H., Joshi C.P., Chiang V.L. (1999). Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc. Natl. Acad. Sci. USA 96: 8955–8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J., Rohde A., Christensen J.H., Van de Peer Y., Boerjan W. (2003). Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133: 1051–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J., Lundquis K., Brunow G., Lu F., Kim H., Schatz P.F., Marita J.M., Hatfield R.D., Ralph S.A., Christensen J.H., Boerjan W. (2004). Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem. Rev. 3: 29–60. [Google Scholar]
- Rasmussen S., Dixon R.A. (1999). Transgene-mediated and elicitor-induced perturbation of metabolic channeling at the entry point into the phenylpropanoid pathway. Plant Cell 11: 1537–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saballos A., Sattler S.E., Sanchez E., Foster T.P., Xin Z., Kang C., Pedersen J.F., Vermerris W. (2012). Brown midrib2 (Bmr2) encodes the major 4-coumarate:coenzyme A ligase involved in lignin biosynthesis in sorghum (Sorghum bicolor (L.) Moench). Plant J. 70: 818–830. [DOI] [PubMed] [Google Scholar]
- Saboury A.A. (2009). Enzyme inhibition and activation: A general theory. J. Iran. Chem. Soc. 6: 219–229. [Google Scholar]
- Sarkanen, K.V., and Ludwig, C.H. (1971). Lignins: Occurrence, Formation, Structure and Reactions, Vol. 1. (New York: John Wiley & Sons). [Google Scholar]
- Schallau K., Junker B.H. (2010). Simulating plant metabolic pathways with enzyme-kinetic models. Plant Physiol. 152: 1763–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C., Lenz C., Grote M., Lührmann R., Urlaub H. (2010). Determination of protein stoichiometry within protein complexes using absolute quantification and multiple reaction monitoring. Anal. Chem. 82: 2784–2796. [DOI] [PubMed] [Google Scholar]
- Schneider K., Hövel K., Witzel K., Hamberger B., Schomburg D., Kombrink E., Stuible H.P. (2003). The substrate specificity-determining amino acid code of 4-coumarate:CoA ligase. Proc. Natl. Acad. Sci. USA 100: 8601–8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sederoff R.R., MacKay J.J., Ralph J., Hatfield R.D. (1999). Unexpected variation in lignin. Curr. Opin. Plant Biol. 2: 145–152. [DOI] [PubMed] [Google Scholar]
- Segel, I.H. (1975). Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. (New York: John Wiley & Sons). [Google Scholar]
- Shi R., Sun Y.H., Li Q., Heber S., Sederoff R., Chiang V.L. (2010). Towards a systems approach for lignin biosynthesis in Populus trichocarpa: Transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol. 51: 144–163. [DOI] [PubMed] [Google Scholar]
- Shuford C.M., Li Q., Sun Y.H., Chen H.C., Wang J., Shi R., Sederoff R.R., Chiang V.L., Muddiman D.C. (2012). Comprehensive quantification of monolignol-pathway enzymes in Populus trichocarpa by protein cleavage isotope dilution mass spectrometry. J. Proteome Res. 11: 3390–3404. [DOI] [PubMed] [Google Scholar]
- Silber M.V., Meimberg H., Ebel J. (2008). Identification of a 4-coumarate:CoA ligase gene family in the moss, Physcomitrella patens. Phytochemistry 69: 2449–2456. [DOI] [PubMed] [Google Scholar]
- Srere P.A. (1987). Complexes of sequential metabolic enzymes. Annu. Rev. Biochem. 56: 89–124. [DOI] [PubMed] [Google Scholar]
- Stafford H.A. (1974). Possible multi-enzyme complexes regulating the formation of C6-C3 phenolic compounds and lignins in higher plants. Rec. Adv. Phytochem. 8: 53–79. [Google Scholar]
- Tuskan G.A., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604. [DOI] [PubMed] [Google Scholar]
- Vanholme R., Demedts B., Morreel K., Ralph J., Boerjan W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153: 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R., Storme V., Vanholme B., Sundin L., Christensen J.H., Goeminne G., Halpin C., Rohde A., Morreel K., Boerjan W. (2012). A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24: 3506–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackerly, D., Mendenhall, W., and Scheaffer, R.L. (2008). Mathematical Statistics with Applications, 7th ed. (Pacific Grove, CA: Duxbury Press). [Google Scholar]
- Wang J.P., et al. (2014). Complete proteomic-based enzyme reaction and inhibition kinetics reveal how monolignol biosynthetic enzyme families affect metabolic flux and lignin. Plant Cell 26: 894–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widrow, B., and Hoff, M.E. (1960). Adaptive switching circuits. IRE WESCON Convention Record 4: 96–104. [Google Scholar]
- Winkel-Shirley B. (1999). Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol. Plant. 107: 142–149. [Google Scholar]
- Xia W.J., Wu Z.M. (2005). An effective hybrid optimization approach for multiobjective flexible job-shop scheduling problems. Comput. Ind. Eng. 48: 409–425. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.