The gene regulatory network that regulates the timing, location, and level of anthocyanin pigmentation in petunia is characterized, establishing features of the network that are functionally conserved in eudicots. A model of the activation and repression mechanisms of the anthocyanin regulatory network is presented, which includes a role for mobile regulatory proteins for pigmentation patterning.
Abstract
Plants require sophisticated regulatory mechanisms to ensure the degree of anthocyanin pigmentation is appropriate to myriad developmental and environmental signals. Central to this process are the activity of MYB-bHLH-WD repeat (MBW) complexes that regulate the transcription of anthocyanin genes. In this study, the gene regulatory network that regulates anthocyanin synthesis in petunia (Petunia hybrida) has been characterized. Genetic and molecular evidence show that the R2R3-MYB, MYB27, is an anthocyanin repressor that functions as part of the MBW complex and represses transcription through its C-terminal EAR motif. MYB27 targets both the anthocyanin pathway genes and basic-helix-loop-helix (bHLH) ANTHOCYANIN1 (AN1), itself an essential component of the MBW activation complex for pigmentation. Other features of the regulatory network identified include inhibition of AN1 activity by the competitive R3-MYB repressor MYBx and the activation of AN1, MYB27, and MYBx by the MBW activation complex, providing for both reinforcement and feedback regulation. We also demonstrate the intercellular movement of the WDR protein (AN11) and R3-repressor (MYBx), which may facilitate anthocyanin pigment pattern formation. The fundamental features of this regulatory network in the Asterid model of petunia are similar to those in the Rosid model of Arabidopsis thaliana and are thus likely to be widespread in the Eudicots.
INTRODUCTION
Anthocyanins are flavonoids that range in color from red to blue and provide pigmentation to flowers and fruit where they act as visual cues to pollinators and seed distributors (Davies et al., 2012). Elaborate anthocyanin pigmentation patterns often occur in flowers (e.g., stripes and spots), which arise as a result of spatially regulated gene expression (Albert et al., 2011; Shang et al., 2011). In vegetative tissues, anthocyanins can perform photoprotective roles by providing a light-absorbing screen for photosynthetic cells (Hughes et al., 2005; Albert et al., 2009) and may also serve to scavenge reactive oxygen species during conditions of light stress (Gould, 2004). The accumulation of anthocyanins for photoprotection in vegetative organs is balanced against light-harvesting requirements for photosynthesis (Albert et al., 2009). To fulfill these varied roles for anthocyanin pigments in both leaves and flowers, stringent regulatory mechanisms must exist that respond to environmental and developmental cues.
The synthesis of anthocyanins and other flavonoids is well characterized, and there is an increasing understanding of the mechanisms that regulate their production (Feller et al., 2011; Hichri et al., 2011). Consequently, the regulatory networks governing anthocyanin accumulation have become important models for understanding how metabolic pathways are controlled in plants and how patterned gene expression can occur. The expression of the anthocyanin biosynthetic genes is regulated by transcription factors (TFs) that activate anthocyanin synthesis, and these have been identified in many model species, including maize (Zea mays), petunia (Petunia hybrida), snapdragon (Antirrhinum majus), and Arabidopsis thaliana (Cone et al., 1986; Paz-Ares et al., 1986, 1987; Ludwig et al., 1989; Goodrich et al., 1992; de Vetten et al., 1997; Quattrocchio et al., 1998, 1999; Walker et al., 1999; Spelt et al., 2000, 2002; Carey et al., 2004; Schwinn et al., 2006; Albert et al., 2011). The anthocyanin (and proanthocyanidin [PA]) biosynthetic genes are activated by a transcriptional activation complex, consisting of basic-helix-loop-helix (bHLH), R2R3-MYB, and WD-repeat (WDR) proteins (MBW complex) (Baudry et al., 2004; Koes et al., 2005; Ramsay and Glover, 2005), although the precise stoichiometry of proteins is not currently known. The patterning and spatial localization of anthocyanins are primarily determined by the activity of the R2R3-MYB factors in the complex, with individual gene-family members regulating separate patterns (Davies et al., 2012), which act with common bHLH and WDR factors (Schwinn et al., 2006; Gonzalez et al., 2008; Albert et al., 2011; Lowry et al., 2012).
Recently, additional levels of regulation have been identified that modulate the production of anthocyanins. These include the posttranslational modifications of the TFs that regulate anthocyanin biosynthesis (Xie et al., 2012; Ye et al., 2012; Maier et al., 2013; Patra et al., 2013), chromatin remodeling (Hernandez et al., 2007), and the identification of repressor proteins that limit expression of the anthocyanin biosynthetic genes (Aharoni et al., 2001; Dubos et al., 2008; Matsui et al., 2008; Yuan et al., 2013). Two distinct classes of MYB TFs have been identified that can limit anthocyanin production: R3-MYBs and R2R3-MYB repressors, which have one or two repeats of the MYB domain region, respectively. The R2R3-MYB repressors contain domains in their C terminus required for their repressive activity (Tamagnone et al., 1998; Jin et al., 2000), such as the ERF-associated amphiphilic repression (EAR) motif, which actively represses transcription (Ohta et al., 2001) and may involve the recruitment of chromatin remodeling factors (e.g., TOPLESS) (Kagale and Rozwadowski, 2011). R2R3-MYB proteins of this type that repress anthocyanin biosynthesis include Fa/Fc-MYB1 from strawberry (Fragaria spp) (Aharoni et al., 2001; Lin-Wang et al., 2011; Salvatierra et al., 2013). At-MYBL2 from Arabidopsis is an R3-MYB (containing a single MYB repeat) but appears functionally similar to subgroup 4 R2R3-MYB repressors (Kranz et al., 1998) as it contains a repression motif (TLLLFR) present in its C terminus that is required for its repressive activity (Dubos et al., 2008; Matsui et al., 2008). The petunia gene MYB27 encodes an R2R3-MYB protein that contains an EAR motif and has similar light-responsive expression as At-MYBL2, which led to a proposed role for it in repressing anthocyanin synthesis (Albert et al., 2011); however, functional data are still lacking. In a wider context, there is still relatively little known about the roles of these repressors in the regulation of anthocyanin synthesis, in particular with regard to how they network with the activator TFs.
The role of R3-MYBs in regulating flavonoid synthesis is less clear than for the R2R3-MYBs, although there are a range of observations supporting such a function for specific R3-MYB proteins. The R3-MYB factors contain a single MYB repeat and are typically small proteins that lack any repressive amino acid motifs (At-MYBL2 is an exception) but do contain the amino acid signature required to bind bHLH partners. They are thought to assert their repressive function through competition (passive repression) for bHLH partners with R2R3-MYB factors that activate anthocyanin synthesis (Kroon, 2004; Koes et al., 2005; Zhang et al., 2009). The most studied are CAPRICE (CPC) and TRIPTYCHON (TRY) of Arabidopsis, which principally inhibit the MBW complexes involved in trichome and root hair formation but also have an effect on flavonoid production (Tominaga et al., 2007; Wang et al., 2008; Wester et al., 2009). The cpc mutant in Arabidopsis is reported to have elevated levels of anthocyanins, suggesting CPC may normally contribute to anthocyanin regulation (Zhu et al., 2009). A petunia homolog, MYBx, has been identified that can repress the activities controlled by the bHLH factor ANTHOCYANIN1 (AN1), including anthocyanin synthesis and vacuolar pH (Kroon, 2004; Koes et al., 2005; Quattrocchio et al., 2006). The expression profile of MYBx in vegetative tissues suggests it provides feedback regulation to limit the accumulation of anthocyanins (Albert et al., 2011). In Arabidopsis, the expression of the R3-MYB factors involved in epidermal cell fate patterning are regulated by an MBW complex (Digiuni et al., 2008; Wang et al., 2008; Zhao et al., 2008), but it is not known if a similar mechanism exists for MYBx in petunia that could provide feedback regulation for anthocyanin synthesis.
Hierarchical and feedback mechanisms between the MYB and bHLH components of the MBW activation complex may be important features of flavonoid regulation. These may provide for gearing of the activation response for precise gene regulation during development or stress responses. In Arabidopsis, the bHLH factor TRANSPARENT TESTA8 (TT8) regulates its own expression as part of a positive feedback loop, through an MBW complex, that ultimately contributes to the regulation of anthocyanin and PA synthesis (Baudry et al., 2006; Xu et al., 2013). In petunia, ectopic expression of AN2 (R2R3-MYB activator) in leaves resulted in ectopic expression of the bHLH factor AN1 (Spelt et al., 2000). Similarly, AN1 transcript levels are severely reduced in the anthers of petunias that lack a functional AN4 allele (R2R3-MYB activator) (Spelt et al., 2000), suggesting that AN1 expression may be regulated by the R2R3-MYB factors in the MBW complex for anthocyanin gene regulation. However, the details of the hierarchical regulation of AN1 expression have not been determined, nor is it known if the putative MYB repressors participate in such feedback mechanisms.
Petunia offers an excellent model system for understanding how hierarchical, feedback, and repressor activities are integrated into controlling the activity of the MBW complex for stress-responsive and developmentally programmed production of anthocyanins in leaves and flowers, respectively. While significant understanding of individual regulatory components has come from studies in Arabidopsis, its flowers lack even simple pigmentation, making it an unsuitable model for understanding the relationship between the vegetative and floral regulatory systems and the developmental influence on pigmentation for pollinator attraction. Petunia and Arabidopsis represent model species from the two major groupings of dicotyledonous plants, the Asterids and Rosids, respectively. Studies of the role of the MBW complex in controlling trichome production suggest that the MYB/bHLH regulatory systems may have diverged since the evolutionary separation of these major plant groups, although the details of this are still not clear (Martin and Glover, 2007; Brockington et al., 2013).
Petunia is one of the original model species for understanding the regulation and synthesis of anthocyanins; consequently, there is a strong knowledge base for the anthocyanin-related MBW complex components. Petunia has both vegetative and complex floral pigmentation patterns, which are determined by individual R2R3-MYB activators (Quattrocchio et al., 1998, 1999; Kroon, 2004; Albert et al., 2011), and these function with the bHLH factor AN1 (Spelt et al., 2000) and the WDR protein AN11 (de Vetten et al., 1997). In addition, a second bHLH factor, JAF13, has been identified that may contribute to anthocyanin regulation (Quattrocchio et al., 1998). The preliminary characterization of the petunia repressors MYBx (R3-MYB) and MYB27 (R2R3-MYB) suggest that these may perform important roles within a regulatory network to determine the timing, level, and perhaps patterning of anthocyanin accumulation (Kroon, 2004; Albert et al., 2011).
Using this important model Asterid species, we characterize the putative R2R3-MYB repressor MYB27 and examine its role with the R3-MYB, R2R3-MYB, and bHLH proteins in regulating anthocyanin synthesis. The results show hierarchical and feedback regulation involving activators and both passive and active repressors. We present a model for the gene regulation network determining anthocyanin pigmentation in Eudicots that integrates the activities of activators, repressors, and mobile components in response to environmental and developmental cues.
RESULTS
MYB27 and R2R3-MYB Activators Bind bHLH Factors
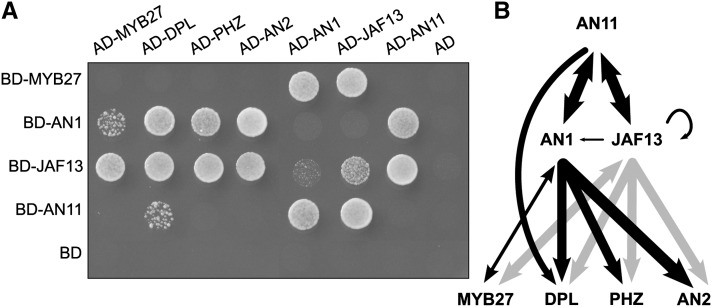
Previously, we showed that MYB27 expression in shade or light-stressed leaves was consistent with a role repressing anthocyanin pigment synthesis. Supporting this hypothesis, MYB27 is expressed in flowers during the active stages of anthocyanin accumulation (Spelt et al., 2000) (Supplemental Figure 1) and is a direct target of the bHLH anthocyanin regulator AN1 (AN1-GR fusion) (Spelt et al., 2000). The predicted MYB27 protein contains a putative repression motif DLNSPP that conforms to the DLNxxP type of EAR motif (Kagale et al., 2010) and contains the amino acid motif [D/E]Lx2[R/K]x3Lx6Lx3R, which is required for interaction with bHLH partners (Zimmermann et al., 2004). Yeast two-hybrid (Y2H) experiments were performed to determine if MYB27 could bind TFs involved in anthocyanin regulation (Figure 1A). The Y2H assays contained increasing concentrations (0 to 100 mM) of 3-amino-1,2,4-triazole (3-AT), which inhibits the action of the HIS3 reporter gene, and allowed the strength of protein interactions to be quantified. The molecular interactions between known R2R3-MYB activators (AN2, DEEP PURPLE [DPL], PURPLE HAZE [PHZ]) or MYB27 were performed with the bHLH proteins AN1 and JAF13, and the WDR protein AN11 (Figure 1A). A summary of the interactions and their relative strengths is shown in Figure 1B. Preliminary experiments revealed that when fused to the GAL4-BD DPL, PHZ and AN2 self-activated the reporter genes (MYB27 did not). Consequently, only the GAL4-AD fusions of these MYBs were used in the analysis. All the MYBs tested interacted with AN1 and JAF13, but with varying strengths; MYB27 shows a higher affinity for JAF13 than for AN1 (75 to 100 mM 3-AT versus 25 mM 3-AT); AN2 shows a higher affinity for AN1 than for JAF13 (Supplemental Table 1). AN11 has a strong affinity for both JAF13 and AN1, but among the MYBs tested, only DPL was shown to interact with AN11. Weak interactions between the bHLH factors AN1 and JAF13, and JAF13 with itself were observed.
Figure 1.
MYB27 Binds Members of the MBW Activation Complex.
(A) Protein–protein interactions between members of the MBW regulatory complex and MYB27 were determined by Y2H assays. The regulators tested were AN1 and JAF13 (bHLH), AN2, DPL, and PHZ (R2R3-MYB activators), AN11 (WDR), and MYB27 (R2R3-MYB repressor). The relative interaction strengths were determined by increasing the concentration of 3-AT. The assay shown represents growth on selective media lacking Leu, Trp, and His, supplemented with 25 mM 3-AT. AD, GAL4 activation domain; BD, GAL4 DNA binding domain.
(B) Summary of interactions between MBW regulators. The relative strength of the interactions (as measured in the yeast system) is indicated by the thickness of the arrows. Arrows indicate protein–protein interactions and do not indicate hierarchy/sequence of regulation.
MYB27 Is a Direct Repressor of Anthocyanin Synthesis
The interaction of MYB27 with known members of the anthocyanin regulatory complex suggests it functions as a repressor of anthocyanin synthesis. To determine if MYB27 acts in planta to repress anthocyanin synthesis in petunia, stable transgenic petunias were generated to knock down (RNA interference [RNAi]) or overexpress (OE) MYB27 from a cauliflower mosaic virus 35S (CaMV35S) promoter (35Spro). The transgenic lines were made in the Mitchell petunia (MP) background, which produces anthocyanins in vegetative tissues in response to light (Albert et al., 2009, 2011). Under ambient greenhouse conditions (in New Zealand), the stems and pedicel are typically bronze due to anthocyanin accumulation, and sepals are also pigmented with a stripe and blush of anthocyanin (Figures 2A and 3). The flowers of MP are white due to null alleles of the R2R3-MYB factors an2 and an4, but anthocyanin venation patterning is present in the flower tube. Light-regulated bud blush patterning is also present in MP, albeit a very subtle phenotype in this background (Figure 2A). Venation is regulated by the R2R3-MYB factor DPL, and bud blush is determined by PHZ (Albert et al., 2011). The light-regulated patterns in stems, leaves, and sepals are somewhat variable and are regulated by the light-responsive R2R3-MYB PHZ and bHLH AN1.
Figure 2.
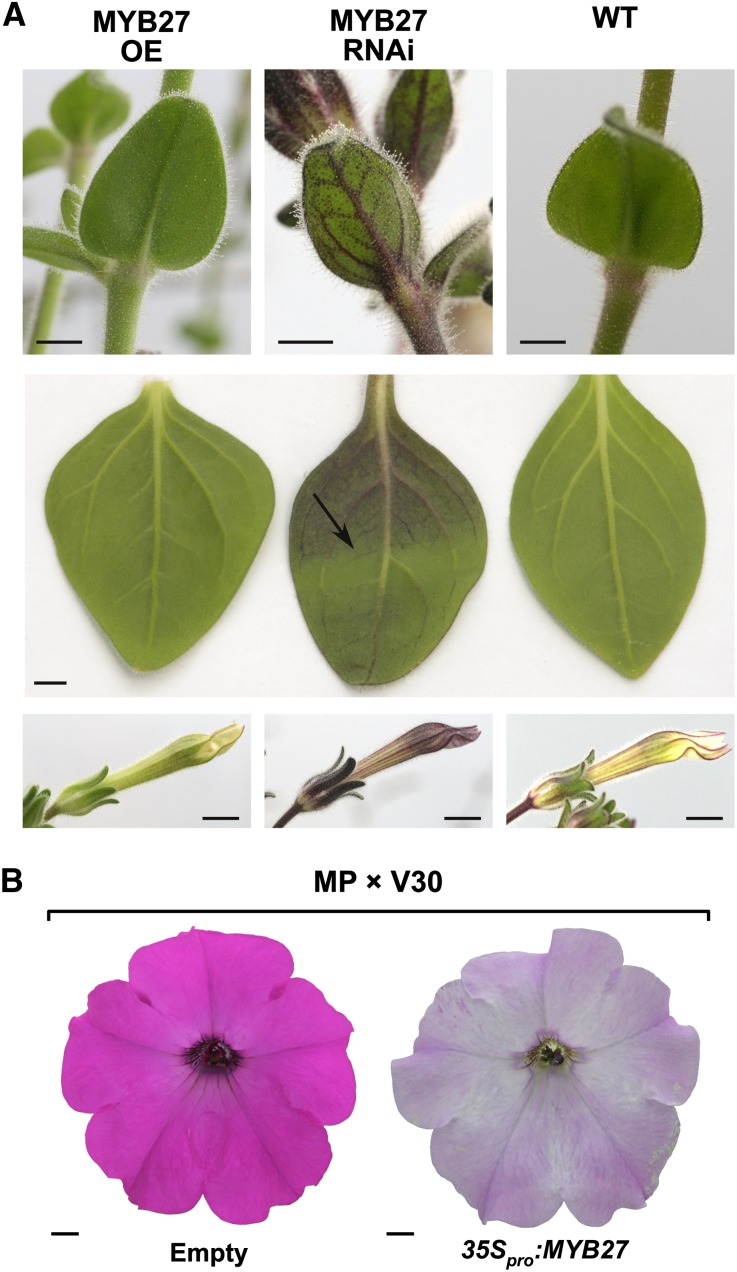
MYB27 Is a Repressor of Anthocyanin Pigmentation.
(A) Pigmentation phenotypes of leaves, stems, and flowers of stable transgenic petunias overexpressing MYB27 (MYB27 OE and 35Spro:MYB27), MYB27 silenced (MYB27 RNAi), or wild-type MPs. The top panel shows young leaves, middle panel shows mature fully expanded leaves, and the bottom panel shows developing flower buds. The arrow indicates where a wire support cage shaded a portion of the abaxial leaf surface from a MYB27 RNAi plant.
(B) Floral pigmentation phenotypes of representative progeny from a 35Spro:MYB27 (MP background; an2−) × V30 (AN2+) petunia cross.
Bars = 0.5 cm.
Figure 3.
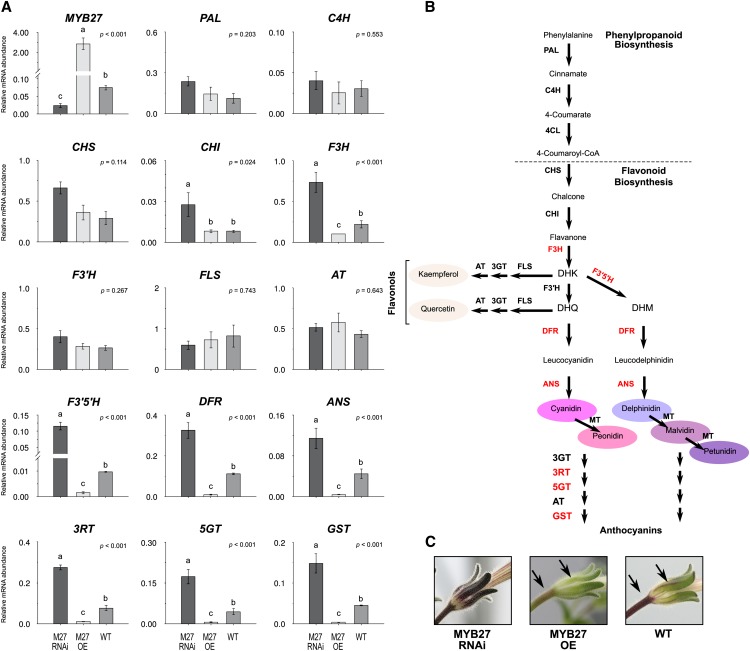
Phenylpropanoid and Flavonoid Gene Expression in Sepals of MYB27 Overexpression and Suppression Petunias.
(A) Transcript abundance for MYB27 or phenylpropanoid/flavonoid biosynthesis genes was determined by qRT-PCR. Means ± se are shown for MYB27 RNAi or MYB27 OE transgenic lines and wild-type MP controls: n = 3 biological replicates consisting of independent lines; n = 2 for the wild type. Statistical significance was determined by one-way ANOVA (P values reported); significant differences between means (LSD, P = 0.05) are indicated where letters (a, b, and c) above the bar differ.
(B) The position of each gene examined is shown in the diagram of the flavonoid pathway. Genes targeted by MYB27 are highlighted in red.
(C) Pigmentation phenotypes of the sepal tissues analyzed from MYB27 RNAi, MYB27 OE, or wild-type MP. Arrows indicate anthocyanin pigmentation on the pedicel and main vein of the sepal.
Representative MYB27 RNAi lines had increased anthocyanin accumulation, while OE lines had reduced anthocyanin pigmentation, as would be predicted if MYB27 were a repressor of anthocyanin synthesis. These phenotypes were reproducible across independent transgenic lines and clonal copies. Twelve independent RNAi lines were generated and assessed, four of which showed increased anthocyanin accumulation. Five MYB27 OE lines were generated, of which three had a reduced anthocyanin phenotype. The consistent MYB27 RNAi phenotype was an enhancement of the normal pigmentation patterns observed in MP (Figure 2A). The stems and pedicels were dark purple due to anthocyanin, and flowers had enhanced petal blush and vein patterning on the abaxial petal epidermis. The enhancement to sepal pigmentation was particularly striking, as they were darkly colored with anthocyanin (Figures 2A and 3). Young leaves also had enhanced levels of anthocyanins, commonly on the leaf margins and in a venation pattern on the abaxial leaf surface. Mature (fully expanded) leaves of MYB27 RNAi plants also displayed enhanced anthocyanin accumulation; this too was light responsive, as shading a portion of a leaf prevented pigmentation (Figure 2A, arrow). The 35Spro:MYB27 petunias displayed reduced pigmentation throughout the plant (Figure 2A). While anthocyanins were never completely absent, stems, leaves, pedicels, and sepals were considerably less pigmented than those of control MP plants. The flower buds also had less anthocyanin on the abaxial veins and the petals appeared white, losing the usual weak blush of anthocyanin. However, the open flowers on 35Spro:MYB27 plants retained venation patterning in the flower tube.
Three independent transgenic lines from the MP primary transformants containing MYB27 RNAi or MYB27 OE constructs and two wild-type MP plants were selected for further molecular and biochemical characterization. Independent transgenic lines with reproducible pigmentation phenotypes between clonal copies were selected for analysis. The sepals were chosen as a representative tissue to assay in detail, as although phenotypic changes were observed throughout the plants, the most striking changes were observed in the sepals, probably as they are the most pigmented tissues of wild-type plants. Furthermore, the sepals allow for a well-defined tissue with regard to location on the plant and consistency of developmental age. The anthocyanin content of sepals of MP and MYB27 RNAi and OE plants, as measured using HPLC (Supplemental Table 2), matched the visual phenotypes; RNAi lines had an increase of 50-fold compared with the wild type, while in the MYB27 OE lines anthocyanins were not detected. The HPLC profiles of anthocyanins showed that total levels, rather than specific anthocyanin species, were altered. The level of flavonols (the predominant flavonoids present in vegetative petunia tissues) was not affected by altered MYB27 expression. Seed from self-fertilized MYB27 OE and RNAi plants were produced to examine the effect on seed coat (proanthocyanidin) pigmentation. The seed coat pigmentation, as visually assayed, remained similar in all lines, with perhaps a small reduction in pigment intensity in the MYB27 OE seed (Supplemental Figure 2).
To confirm that the phenotypes were heritable and to evaluate further the effect of MYB27 OE, the hemizygous MYB27 RNAi and 35Spro:MYB27 lines were crossed to the V30 petunia line (AN2 and AN4), which has a full pigmentation phenotype (having dark magenta petals). The F1 populations displayed cosegregation of the transgene with vegetative enhanced anthocyanin accumulation in sepals and abaxial leaf venation in MYB27 RNAi×V30 and reduced stem and sepal anthocyanin accumulation in MYB27 OE×V30 (Supplemental Figure 3), confirming the anthocyanin phenotypes were caused by the MYB27 transgene constructs. The crosses to V30 also allowed the effect of MYB27 OE to be assessed in a plant background with a functional allele of AN2 (R2R3-MYB activator). Clear suppression of the normally strong petal pigmentation of V30 occurred in the F1 population of MYB27 OE×V30 plants (Figure 2B), and this cosegregated with the reduced vegetative pigmentation and the presence of the transgene (Supplemental Figure 3). The MYB27 RNAi×V30 plants did not show any visible enhancement of petal pigmentation when compared with the control MP×V30 plants, despite cosegregation of the enhanced vegetative pigmentation with the presence of the RNAi transgene.
Quantitative RT-PCR (qRT-PCR) was used to determine MYB27 transcript levels in sepals of MYB27 RNAi, MYB27 OE, and control plants and to identify which phenylpropanoid and flavonoid biosynthetic genes were repressed (Figure 3). As expected, MYB27 transcript levels were reduced in the RNAi lines and greatly increased in the OE lines, relative to control lines. Transcript levels for the general phenylpropanoid pathway genes PAL and C4H, the flavonoid pathway genes CHS-A and F3′H (HT1), the flavonol-specific gene FLS, and the AT gene were not significantly affected by altered MYB27 expression levels. The flavonoid genes CHI-A and F3H (AN3) had higher transcript levels in MYB27 RNAi lines but no clear difference between MYB27 OE and control plants. Transcript levels for the anthocyanin biosynthetic genes F3′5′H (HF1-1), DFR-A (AN6), ANS, 3RT (RT), 5GT, and GST (AN9) were lower in MYB27 OE lines and higher in MYB27 RNAi lines than control plants.
MYB27 Participates in a Regulatory Network
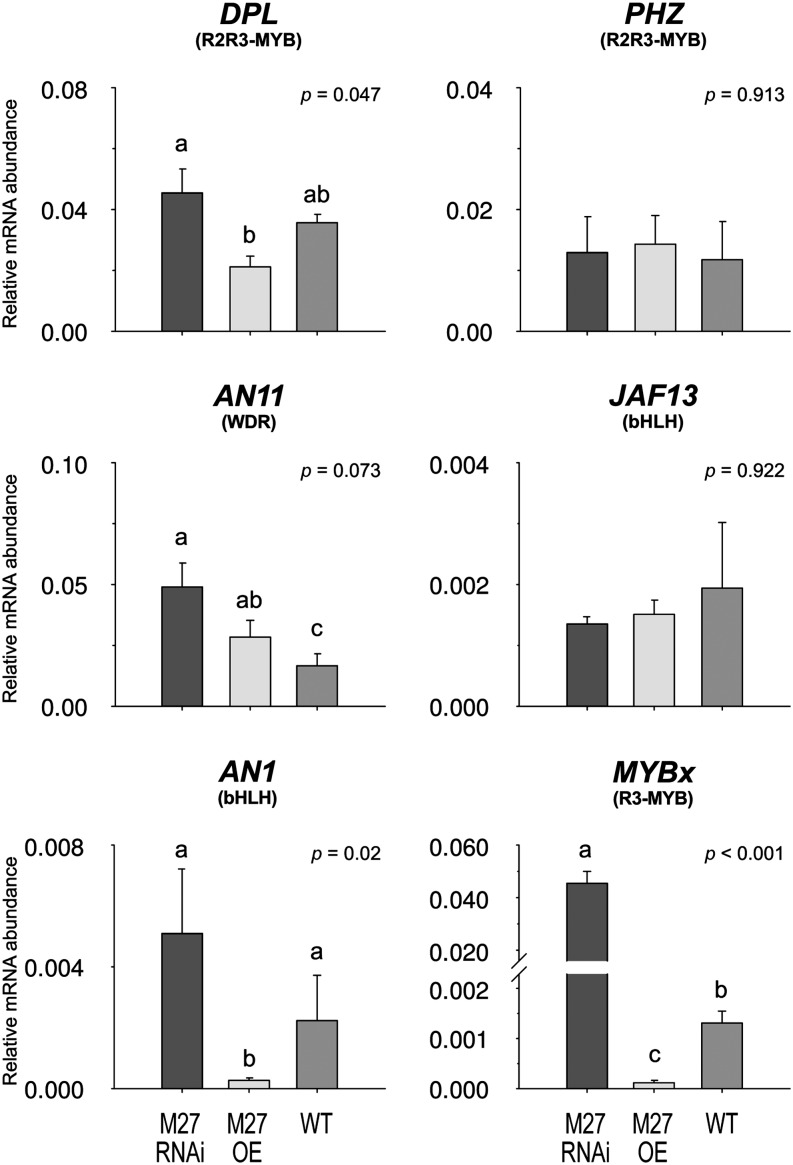
Evidence from Arabidopsis and petunia suggest that the key bHLH genes (TT8/AN1) may themselves be regulated by the MBW complex, which may contain the same bHLH and result in self-regulation (Spelt et al., 2000; Baudry et al., 2006; Xu et al., 2013). Transcript levels for the bHLH, MYB, and WDR components of the petunia MBW complex were assayed in the MYB27 RNAi, OE, and control lines (Figure 4). The transcript levels for DPL, PHZ (R2R3-MYB activators), JAF13 (bHLH), and AN11 (WDR) did not appear to be directly affected by MYB27 suppression or OE. However, AN1 (bHLH) transcript levels were enhanced in the MYB27 RNAi lines and suppressed in the OE lines. Transcript levels for the R3-MYB repressor MYBx were affected substantially by MYB27 expression levels: MYB27 RNAi plants had substantially higher MYBx levels (∼35-fold, 3500%), and MYB27 OE lines had greatly reduced levels (0.1-fold, 10%) compared with control plants.
Figure 4.
MYB27 Regulates AN1 and MYBx Expression in Transgenic Petunias.
Transcript abundance for anthocyanin regulators in sepal tissue determined by qRT-PCR. Means ± se are shown for MYB27 RNAi or MYB27 OE transgenic lines and wild-type MP controls: n = 3 biological replicates consisting of independent lines; n = 2 for the wild type. Statistical significance was determined by one-way ANOVA (P values reported); significant differences between means (LSD, P = 0.05) are indicated where letters (a, b, and c) above the bar differ.
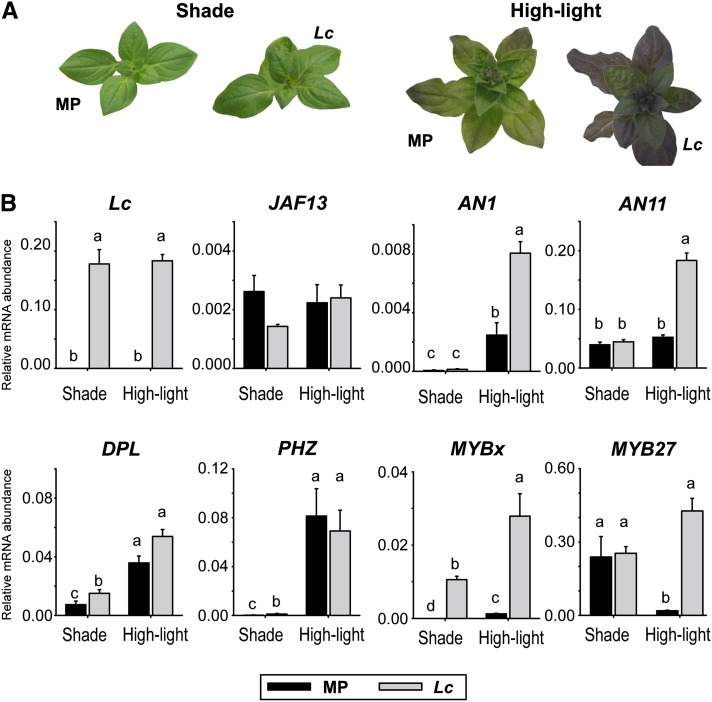
Because the MYB27 transgenic results suggested hierarchical and feedback regulation among the anthocyanin regulators, the analysis was extended to previously generated petunia lines that ectopically express anthocyanin activator TFs, specifically the maize bHLH Leaf color (35Spro:Lc) and the petunia R2R3-MYB activators DPL and PHZ in the MP background (Albert et al., 2009, 2011). OE of Lc enhances the light-induced pigmentation phenotype of MP, while DPL and PHZ OE lines accumulate high levels of anthocyanins in their leaves independent of high light, with DPL OE lines consistently giving a dark leaf phenotype and PHZ a bronze phenotype (Albert et al., 2011). The light-inducible Lc OE phenotype (Figure 5A) enabled the changes in expression of the endogenous anthocyanin regulators in response to excess bHLH protein to be assayed in the absence or presence of anthocyanin accumulation (Figure 5B). Expression of Lc driven by the CaMV35S promoter did not change with light conditions. Transcripts for the R2R3-MYB activators DPL and PHZ were induced by high light, particularly PHZ, which is considered the primary regulator of light-induced pigmentation (Albert et al., 2011). AN1 transcripts were also induced by light stress in MP plants, and this was considerably enhanced in Lc petunia under high light. AN11 transcript levels were similar between shade and high-light treatment in MP and shade-grown Lc petunia but were higher in high-light-treated Lc petunias. MYBx transcripts were not detected in shade-grown MP leaves but were present at high levels in Lc petunias under shade conditions. Under high-light conditions, a low level of MYBx transcripts was detected in MP leaves, but in Lc petunia, transcript abundance for MYBx was 23-fold higher than in MP. MYB27 transcripts were expressed at similar levels in shade-grown MP and Lc petunias. However, in high-light-grown MP leaves, MYB27 transcripts dropped significantly, while Lc petunias had even higher transcript levels than under shade conditions. JAF13 expression was unaffected by Lc OE.
Figure 5.
Expression of Anthocyanin Regulators in 35Spro:Lc Petunias.
(A) Anthocyanin pigmentation phenotypes of wild-type MP or plants expressing the maize bHLH anthocyanin regulator Lc from a CaMV35S promoter, grown under shade (50 to 350 µmol m−2 s−1) or high-light (750 µmol m−2 s−1) conditions.
(B) Transcript abundance for the Lc transgene or endogenous anthocyanin regulators in MP or Lc petunias. Means ± se are shown (n = 4 biological replicates); statistical significance was determined by one-way ANOVA (P < 0.001); significant differences between means (LSD, P = 0.05) are indicated where letters (a, b, c, and d) above the bar differ.
The DPL and PHZ OE lines provide plants with different levels of vegetative anthocyanin pigmentation (DPL > PHZ > MP) (Supplemental Figure 4). Transcript levels for TFs involved in anthocyanin regulation were determined by qRT-PCR in independent transgenic lines (Supplemental Figure 4). AN1 and MYBx transcripts were significantly higher in the DPL and PHZ OE lines than MP, with DPL OE having the highest transcript levels (matching the higher expression of the R2R3-MYB activator and pigmentation level). MYB27 transcript levels were slightly increased in both DPL and PHZ OE lines compared with MP, a trend also observed for AN11. JAF13 expression was relatively unaffected.
The temporal expression patterns of the anthocyanin-related TFs were examined across petal development. In flowers of line V26 (Supplemental Figure 1A), transcript levels for the AN1, AN2, DFR-A (a biosynthetic gene target of AN1/AN2), and MYB27 shared a similar pattern of expression; they were low at stage 1 and rose to a peak at stage 4, prior to flower maturation (Supplemental Figure 1B). MYBx expression was limited to a small window at stages 3 and 4, with only low levels present at stage 5, when AN1 and MYB27 are still relatively strongly expressed. In developing MP flowers, which lack the R2R3-MYB activator AN2, the pattern of expression for MYBx and MYB27 is altered, and transcript levels are severely reduced (Supplemental Figure 5) compared with V26 petunia (Supplemental Figure 1B).
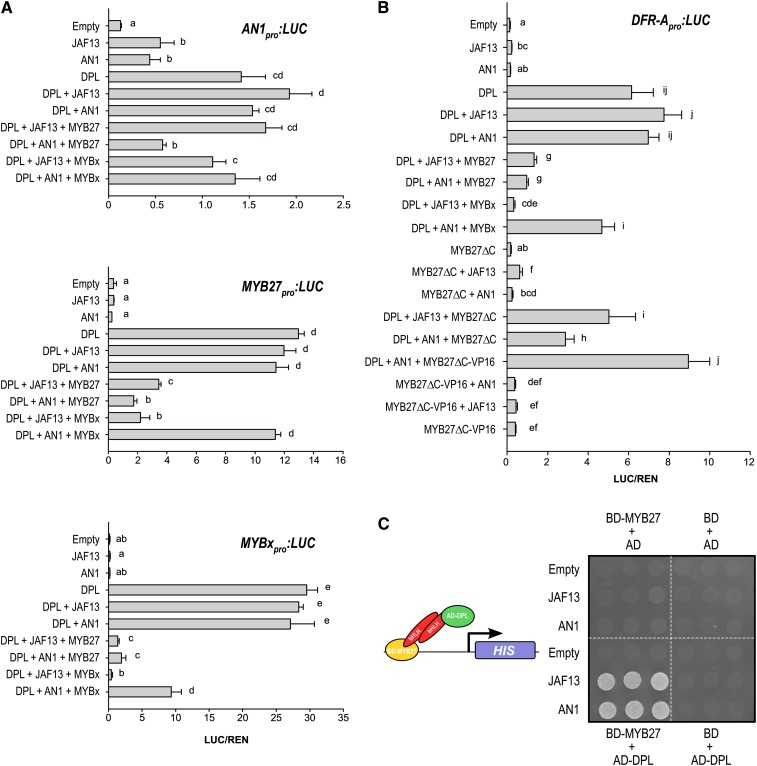
The hierarchical regulation among the MBW factors was further examined by assaying promoter activation, with a dual luciferase system in Agrobacterium tumefaciens–infiltrated Nicotiana benthamiana leaves (Figure 6A). The promoter and 5′ untranslated region (UTR) of AN1 (1.4 kb), MYBx (0.95 kb), and MYB27 (1.55 kb) were isolated and used in the promoter constructs. DPL was used as a representative R2R3-MYB activator for these experiments, although similar responses have been observed with PHZ and AN2 in preliminary experiments. The AN1 promoter was activated by the R2R3-MYB DPL alone (using endogenous bHLH and WDR proteins present in N. benthamiana leaves) or in combination with either bHLH factor JAF13 or AN1. This activation was repressed by MYB27, even when bHLH factors were not limited (+ AN1). Similarly, the promoters of MYBx and MYB27 were activated by DPL alone or in combination with either AN1 or JAF13; this activation was repressed by coexpressing MYB27. MYBx was able to suppress activation of the MYBx and MYB27 promoters by DPL, but the suppressive effect was stronger against DPL/JAF13 than DPL/AN1 complexes. MYBx had a modest suppressive effect on the AN1 promoter activity.
Figure 6.
MYB27 Is a Corepressor of the MBW Activation Complex.
(A) and (B) Activation and repression assays were performed upon the promoters of the anthocyanin regulatory genes AN1pro, MYB27pro, and MYBxpro (A) or the anthocyanin biosynthetic gene DFR-Apro (B) using a dual luciferase assay. Agrobacteria containing the reporter and effector constructs were coinfiltrated into N. benthamiana leaves. The effector constructs express each TF from a CaMV35S promoter: DPL (R2R3-MYB activator), JAF13 and AN1 (bHLH), MYB27 (R2R3-MYB repressor), and MYBx (R3-MYB repressor). Additionally, two modified versions of MYB27 were assayed: MYB27ΔC (EAR motif removed) and MYB27ΔC-VP16 (EAR motif removed + viral activation domain). Firefly luciferase (LUC) values are reported relative to the Renilla luciferase (REN) control; means ± se (n = 4). Statistical significance was determined by one-way ANOVA (P < 0.001); significant differences between means (LSD, P = 0.05) are indicated where letters (a, b, c, etc.) above the bar differ.
(C) Y3H assay of BD-MYB27, AD-DPL, JAF13, or AN1. The assay shown is grown on selective media lacking Leu, Trp, uracil, and His. The diagram indicates the protein–protein interactions required to activate the HIS reporter gene.
[See online article for color version of this figure.]
To test whether MYB27 requires the EAR motif to repress target genes, a C-terminal deletion construct that removed the EAR motif (MYB27ΔC) was made, and its effect on activation of the DFR-A promoter (1.86 kb) by the MBW complex was compared with that of the intact protein and to MYBx (Figure 6B). DPL and AN1 activated the DFR-A promoter, and this was repressed when intact MYB27 was included. MYB27ΔC did not cause the same repression of the promoter activation as seen when using intact MYB27. MYBx had a strong suppressive effect upon DFR-A promoter activation when coinfiltrated with DPL/JAF13, but only modestly suppressed activation by DPL/AN1.
To test whether MYB27 function requires direct DNA binding, a construct was made (MYB27ΔC-VP16) that fused the strong activation domain VP16 to MYB27ΔC (Figure 6B). If MYB27 binds the DFR-A promoter directly, fusing the activation domain to this MYB27 version that lacks the EAR domain could result in promoter activation. Additionally, if MYB27 binds a TF complex, such as the DPL/AN1 complex, then inclusion of MYB27ΔC-VP16 together with DPL and AN1 should result in increased promoter activation. MYB27ΔC-VP16 was unable to activate the DFR-A promoter, with or without a bHLH partner. However, cotransformation of MYB27ΔC-VP16 with DPL and AN1 resulted in greater activation than DPL+AN1+MYB27ΔC and was slightly higher than DPL+AN1 alone. The hypothesis that MYB27 acts as a corepressor that binds MBW complexes was further examined by yeast three-hybrid (Y3H) assays. Y3H assays demonstrate that an interaction between MYB27 and DPL can be bridged by either JAF13 or AN1 (Figure 6C), supporting the proposed role for MYB27 as a corepressor.
Inter- and Intracellular Movement of MYB27 and MYBx
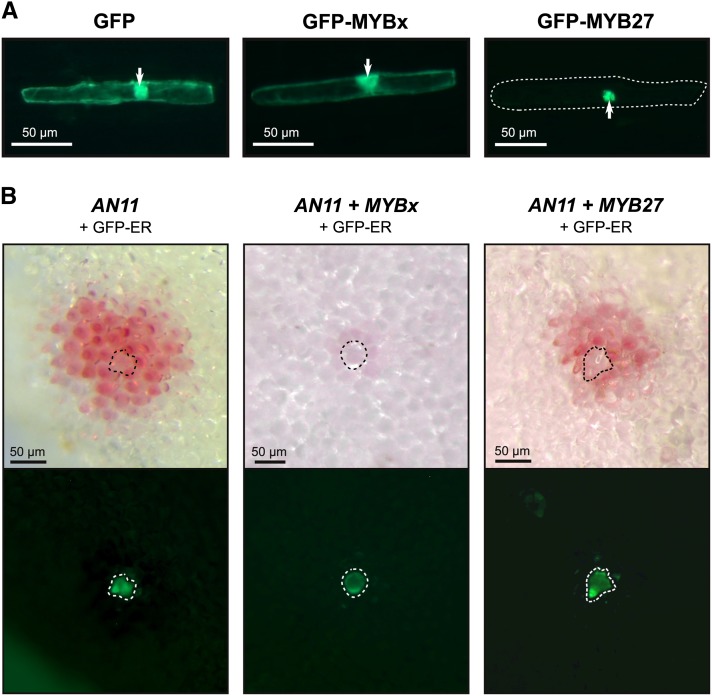
The cellular localization and the ability for MYB27 and MYBx to move between cells was investigated using GFP-MYB27 and GFP-MYBx fusions. These fusion proteins were expressed from a CaMV35S promoter and biolistically transformed into onion (Allium cepa) epidermal cells (Figure 7A) or into petals of a Cymbidium orchid cultivar (Supplemental Figure 6) previously used to study anthocyanin TF function (Albert et al. 2010). GFP, GFP-MYBx, and GFP-MYB27 all produced foci consisting of a single GFP fluorescent cell. GFP (26.9 kD) and GFP-MYBx (36.3 kD) are both below the nuclear diffusion barrier (40 kD) (Grebenok et al., 1997; Hanson and Köhler, 2001; Merkle, 2004M) and exhibit cell-wide localization, while GFP-MYB27 (48.1 kD), which exceeds this limit, does not accumulate in the cytoplasm and is localized to the nucleus. Similar localizations were observed in both onion and Cymbidium epidermal cells. The GFP signal produced from GFP-MYB27 transformation was only transiently present, being detected 1 d following bombardment but not present after 2 d in all cell types tested, contrasting with free GFP or GFP-MYBx.
Figure 7.
Localization and Intercellular Movement of MYB Repressors and AN11.
(A) Biolistic transformation of onion bulb scale epidermis with 35Spro:GFP, 35Spro:GFP-MYBx, or 35Spro:GFP-MYB27 viewed with blue light on a dissecting microscope; arrows indicate the nucleus and the cell margins are indicated (dotted boundary) for GFP-MYB27.
(B) Biolistic transformation of petunia W134 (an11) petals with 35Spro:AN11, 35Spro:AN11 + 35Spro:MYBx, or 35Spro:AN11 + 35Spro:MYB27. A 35Spro:GFP-ER construct was included in all transformation as an internal control to label the transformed cells; GFP was viewed with blue light. The transformed cell is indicated (dotted boundary).
A transient transformation assay was developed to further test the ability of MYBx and MYB27 to move between cells and repress anthocyanin synthesis, without the use of GFP fusions that may prevent passive movement. Petunia cv W134 (an11) was used, as it lacks a functional AN11 allele (WDR), preventing the formation of the MBW complex and resulting in acyanic flowers (de Vetten et al., 1997). Biolistic transformation with the control 35Spro:GFP-ER or free GFP construct did not produce anthocyanin pigmented cells, but GFP positive single-celled foci were apparent (Supplemental Figure 7). Complementation by biolistic transformation with 35Spro:AN11 resulted in multicellular foci that produced anthocyanins, with the internal positive control GFP-ER being retained in the central transformed cell of each focus (Figure 7B). This contrasts with the single-cell pigmented foci that form when AN2 (R2R3-MYB activator; 29.0 kD) is transformed into an2− (cv MP) petals or AN1 (bHLH; 74.1 kD) is transformed into an1− (cv W225) petals (Supplemental Figure 8).The formation of multicellular colored foci demonstrates that AN11 (37.8 kD) is capable of intercellular movement in the petal epidermis. This capacity for AN11 to move between cells allowed the intercellular movement and repressive action of MYBx or MYB27 to also be assayed, through cotransformation of AN11 and the repressors. Transformation with AN11 + MYB27 + GFP-ER closely resembled AN11 complementation alone, although the central transformed cell may have slightly reduced pigmentation (Figure 7B; Supplemental Figure 7). By contrast, transformation with AN11+ MYBx + GFP-ER resulted in weakly pigmented foci and significantly reduced the proportion of colored foci that formed (16.5%), compared with AN11 alone (66.2%) (Supplemental Table 3).
DISCUSSION
In petunia, and all other plant species studied to date, the timing, level, and spatial distribution of anthocyanin pigmentation is determined by an MBW (R2R3MYB-bHLH-WDR) activation complex, with the R2R3-MYBs being key to specifying the action of the complex. In this study, we examined in more detail the regulatory network that surrounds this core MBW complex, in particular with regard to hierarchical and feedback regulation and the role of R3-MYB and R2R3-MYB proteins that can either competitively inhibit the formation of the MBW complex or transform it so that it actively represses transcription. This has enabled an integrated model of anthocyanin regulation in petunia to be developed and comparison of the data for this important Asterid model to the system described for the Rosid model of Arabidopsis.
MYB27 Is an Anthocyanin Repressor
Based on the results for MYB27 (R2R3-MYB) and MYBx (R3-MYB), MYB repressor proteins have a central role in the regulation of anthocyanin production in both floral and vegetative tissues of petunia. The transgenic OE and RNAi results combined with the promoter activation assays show that MYB27 has a repressive action at multiple levels within the anthocyanin regulatory network. It not only acts to limit the expression of the anthocyanin biosynthesis genes directly (Figures 3 and 6B) but also represses the expression of the gene for the key bHLH factor of the MBW activation complex (AN1) (Figures 4 and 6A). Thus, MYB27 has a potential double-lockdown mechanism for reducing anthocyanin production; first, by preventing MBW complex formation and then by incorporating or binding to MBW activation complexes, converting them into repressive complexes.
Fa-MYB1 from strawberry was the first anthocyanin-associated R2R3-MYB repressor identified, when it was shown that it reduced anthocyanin accumulation when ectopically expressed in tobacco (Nicotiana tabacum; Aharoni et al., 2001). Subsequently, other repressor MYB factors have been identified as candidate anthocyanin regulators (Gao et al., 2011; Lin-Wang et al., 2011), although a mechanism of action has only been reported for At-MYBL2 (R3-MYB) (Dubos et al., 2008; Matsui et al., 2008). Notably, At-MYBL2 differs from MYB27 (and other studied R2R3-MYB repressors) in that it lacks an intact R2-MYB repeat and contains a “TLLLFR” motif that is essential for the repressive function, rather than the more common EAR motif present in Fa-MYB1 and MYB27 (Matsui et al., 2008). While At-MYBL2 has been demonstrated to repress anthocyanin production, it also has a role in brassinosteroid signaling (Ye et al., 2012). It is not yet clear whether R2R3-MYBs with similar mechanisms of action to At-MYBL2 are common across Asterid and Rosid species, for example, with regard to action on multiple target pathways. The results for MYB27 suggest it has some functional commonalities with At-MYBL2, but also shares characteristics with At-MYB4 of Arabidopsis, the well-characterized R2R3-MYB that represses early steps of phenylpropanoid metabolism in response to UV stress (Jin et al., 2000).
Repression by MYB27 requires the EAR motif and the formation of the MBW complex, as evidenced by the Y2H and promoter activation studies with the intact or EAR deletion (MYB27ΔC) proteins (Figure 6B). For the EAR motif to mediate transcriptional repression (active repression), MYB27 must bind to the promoter of target genes, either directly itself or as part of a DNA binding complex (Ohta et al., 2001; Kagale et al., 2010). The enhanced activation of the DFR-A promoter by cotransforming MYB27ΔC-VP16 with DPL and AN1 (Figure 6B), together with the Y3H data (Figure 6C), suggest that MYB27 acts as a corepressor that is incorporated into or binds MBW complexes (e.g., DPL/PHZ/AN2 + AN1 + AN11), rather than directly binding DNA. The recruitment of MYB27 to MBW complexes may be facilitated through dimerized bHLH factors present within the MBW complex as either JAF13 or AN1 can bridge the interaction between DPL and MYB27 (Figure 6C), and these bHLH factors weakly interact in Y2H experiments (Figure 1). Dimerization of bHLH factors within anthocyanin MBW complexes in maize has recently been demonstrated to be important for the targeting of MBW complexes to target cis-elements (Kong et al., 2012).
This action of R2R3-MYBs as corepressors within the anthocyanin-related MBW DNA binding complex is likely common across species, as action of both At-MYBL2 and Fa-MYB1 requires binding to bHLH factors and the C-terminal repressive motifs (Aharoni et al., 2001; Matsui et al., 2008). However, it should be noted that there are other R2R3-MYBs with a dominant repressive action on anthocyanin biosynthesis that do not require the EAR motif. These are commonly truncated alleles of the activator R2R3-MYBs that have lost their activation function but retain the ability to bind the bHLH partner and target DNA motifs. These include C1-I (R2R3-MYB) in maize (Paz-Ares et al., 1990) and MYB114Col-0 (Gonzalez et al., 2008) and an artificial MYB90 (PAP2) variant (Velten et al., 2010) in Arabidopsis. The ability of MYB27ΔC to moderately inhibit MBW activation suggests MYB27 may also titrate free bHLH factors (Figure 6B), in addition to active EAR motif–mediated repression upon MBW complexes.
MYB27 targets the same genes as those targeted by the anthocyanin MBW activation complex (Figures 3 and 6). This suggests that MYB27 is incorporated or binds to MBW complexes, changing the complex activity from activation to repression. The action on common regulatory targets is not just through the reduction of the bHLH (AN1) levels, as there is also the direct activity on the DFR-A promoter (Figure 6B). The MYB27 data are in agreement with that for Fa-MYB1; based on timing of gene expression and Y2H studies, Fa-MYB1 is thought to be specifically associated with anthocyanin production in strawberry fruit and not PA biosynthesis (Schaart et al., 2013), yet when overexpressed in Lotus corniculatus, it repressed the PA biosynthetic pathway genes normally activated by the endogenous MBW complex (Paolocci et al., 2011). These observations in both petunia and strawberry suggest the R2R3-MYB repressors bind MBW complexes and that the targeting to promoters is likely determined by the R2R3-MYB activator.
Based on the localization studies with the GFP fusion proteins and the inability of 35Spro:MYB27 to repress anthocyanin production in adjacent cells in biolistic transformation assays, MYB27 is a nuclear localized protein that acts cell autonomously (Figure 7). Again, MYB27 is similar in this aspect to the R2R3-MYB activators, as various studies have shown they act cell autonomously (Supplemental Figure 8; Quattrocchio et al., 1998, 1999; Schwinn et al., 2006; Albert et al., 2010, 2011). Whether this is common for R2R3-MYB repressors across species is not known, as data are lacking for proteins such as Fa-MYB1 or At-MYBL2. However, the larger size of these R2R3-MYBs compared with the small R3-MYB proteins suggests they would be retained within a cell unless actively transported out. The loss of GFP signal for the GFP-MYB27 fusion a day after transformation was in contrast with results for other proteins, such as GFP, GFP-ER, or GFP-MYBx, which are detectable for several days, and suggests that MYB27 is subject to active posttranslational degradation. Recent studies have shown that posttranslational regulation is an important component in regulating the action of the anthocyanin-related MBW activator complex, typically mediated via the ubiquitin/proteasome (CONSTITUTIVE PHOTOMORPHOGENESIS1) pathway (Maier et al., 2013; Patra et al., 2013). Furthermore, phosphorylation of At-MYBL2 may increase its stability and longevity, preventing its degradation through the proteasome (Ye et al., 2012).
MYBx Is a Repressor
The data show that the R3-MYB MYBx is also part of the MBW regulatory network for anthocyanin biosynthesis in petunia but that it has a distinct mechanism of action to that of MYB27. MYBx is expressed in light-stressed leaves (Figure 5) and has been proposed to provide feedback repression upon anthocyanin biosynthesis (Albert et al., 2011). MYBx was originally identified as a protein that bound the bHLH AN1 in a Y2H screen, and preliminary studies showed that AN1 activity was inhibited in 35Spro:MYBx petunia plants, as indicated by reduced anthocyanin pigmentation and altered vacuolar pH (Kroon, 2004). The ability of MYBx OE to reduce anthocyanin pigmentation was also shown here in biolistic transient assays in petunia petals (Figure 7B). Moreover, MYBx inhibited the ability of the MBW complex to activate the DFR-A promoter (Figure 6B). MYBx has high sequence similarity to the R3-MYB proteins from Arabidopsis (e.g., CPC and TRY) that regulate epidermal cell fate patterning (Koes et al., 2005). These are small proteins that contain the amino acid signature to bind bHLH proteins but lack active repression domains and do not bind DNA directly. They have been characterized in Arabidopsis as competitive inhibitors that act by binding bHLH proteins (GL3 and EGL3) required for the formation of MBW complexes (with GL1 and TTG1) for regulation of trichome development (Pesch and Hülskamp, 2004; Wester et al., 2009). This passive repressive mechanism, where essential TFs are depleted, is also known as “squelching” (Gill and Ptashne, 1988). Squelching activities within the MBW regulatory network for anthocyanin biosynthesis likely extend beyond the R3-MYBs, as the SBP-box protein SPL9 and various JA-ZIM domain proteins can also limit anthocyanin production in Arabidopsis by inhibiting the formation of active MBW complexes via binding to the R2R3-MYB and/or bHLH factors (Gou et al., 2011; Qi et al., 2011).
A biolistic transient transformation assay developed using AN11 complementation of the an11 mutant line enabled non-cell-autonomous activity of MYBx in inhibiting anthocyanin biosynthesis to be demonstrated (Figure 7B; Supplemental Table 3). Complementation with 35Spro:AN11 results in multicellular colored foci around the single transformed cell (as indicated by cotransformed 35Spro:GFP-ER). The addition of 35Spro:MYBx inhibited the formation of these multicellular foci, while inclusion of 35Spro:MYB27 did not. This movement of MYBx between cells is more likely by diffusion through plasmodesmata than by active transport. When a GFP-fusion tag was added to MYBx, which greatly increased the overall protein size from 9.4 to 39 kD, no intracellular movement was seen, but rather general localization across the single transformed cell (Figure 7A). This contrasts with results for GFP-CPC in Arabidopsis, where movement between cells of the fusion protein was observed (Kurata et al., 2005; Digiuni et al., 2008). However, in that case, a specific amino acid motif was required for movement to occur, and a gated transport mechanism through plasmodesmata was suggested (Kurata et al., 2005; Wang et al., 2008; Rim et al., 2011). Notably, however, CPC variants lacking the motif were capable of intercellular movement in roots, suggesting passive diffusion as a secondary transport mechanism (Rim et al., 2011). An alternative interpretation of the biolistic data is that MYBx prevented AN11 from moving; however, this is unlikely given that the MYBs are not effective binders of WDR proteins (Figure 1; Payne et al., 2000; Baudry et al., 2004, 2006; Chiu et al., 2010; An et al., 2012).
The intracellular movement of AN11, demonstrated for the first time in these experiments, matches the previously reported ability of the Arabidopsis WDR TTG1 to move between cells during regulation of trichome (and root hair) formation. The mobility of TTG1 and the R3-MYBs is important in the mechanism for determining trichome spacing. TTG1 moves across leaf epidermal cells but becomes trapped by the partner bHLH factors expressed in trichoblasts; this depletes the cells surrounding the trichome of the WDR required for the MBW activation complex (Bouyer et al., 2008; Balkunde et al., 2011). The MBW activation complex that is formed in the trichoblasts promotes the production of the R3-MYB repressors, which then move to neighboring cells and further repress potential trichome initiation (Digiuni et al., 2008; Zhao et al., 2008). The movement of AN11 and MYBx between cells in the petal epidermis raises the possibility that they may have an analogous role to the trichome factors but in intercellular regulation of anthocyanin biosynthesis. Anthocyanin pigments often accumulate in complex patterns within flowers, such as stripes and spots, with spots often being surrounded by a halo of colorless/pale cells. The mechanisms that determine the identity of pigmented versus nonpigmented epidermal cells in these patterns are poorly understood but seem to reflect patterned expression of the R2R3-MYB genes (Albert et al., 2011; Shang et al., 2011; Davies et al., 2012; Lowry et al., 2012). Taking into account the mobility of the WDR and R3-MYB repressors may help further explain pattern formation. First, spot halos could be formed by either the depletion of WDR proteins from neighboring cells and/or the export the R3-MYB repressors into them (Davies et al., 2012). Second, the short-range acting activators and long-range acting inhibitors (repressors) fit many of the requirements of the theoretical models proposed to explain the formation of ordered 2D patterns (Turing, 1952; Gierer and Meinhardt, 1972; Davies et al., 2012). In these models, the activators activate the repressors, the repressors inhibit the activators, the activator activates itself, and the repressors move between cells (Gierer and Meinhardt, 1972; Meinhardt and Gierer, 1974; Pesch and Hülskamp, 2004). It has previously been observed that the MBW regulatory network for trichome formation fits these models (Pesch and Hülskamp, 2004), and our data suggest that the petunia anthocyanin-related MBW network does also: AN1 (a cell-autonomous activator) activates MYBx; AN1 activates its own expression; MYBx inhibits the activity of AN1; and MYBx is a repressor capable of intercellular lateral inhibition. While theoretical in nature and based on assumptions such as diffusion-based movement that may not apply in planta, the models can provide useful directions for experimental exploration, such as investigating the genetic basis of the various patterning mutant lines available in species such as petunia and Antirrhinum.
TFs Operate in a Hierarchy
The results from transgenic overexpression and promoter activation experiments confirm that the anthocyanin regulatory network in petunia is organized in a hierarchical manner. Transcript abundance for AN1, MYB27, and MYBx was increased when DPL and PHZ were overexpressed in leaves (Supplemental Figure 4), and MYB27 and MYBx expression was reduced in petals of an2− petunias (Supplemental Figure 5). These three genes also had higher transcript levels in transgenic petunia expressing the maize bHLH activator Lc, particularly if the expression of PHZ was also induced using high-light conditions (Figure 5). AN1, MYBx, and MYB27 itself were also identified as target genes of MYB27 from analysis of OE and RNAi suppression lines (Figure 4). Together, the data suggest AN1, MYB27, and MYBx are subject to both activation and repression by the MBW complex. For all three genes, these indirect observations were supported by promoter activation assays using both the activator and repressor proteins (Figure 6). The findings fit with a variety of previous observations that also indicated hierarchy within the regulatory complex. Specifically, ectopic expression of AN2 in petunia leaves resulted in ectopic expression of AN1 (Quattrocchio et al., 1998), AN1 expression is reduced in anthers of an4 lines (AN4 is an R2R3-MYB activator) (Spelt et al., 2000), MYBx expression is reduced in flowers of an1 and an11 lines (Kroon, 2004), and MYB27 was found to be a direct target of an AN1-glucocorticoid receptor fusion protein (Spelt et al., 2000).
Hierarchical regulation is also established for the MBW complex for regulating flavonoid biosynthesis/trichome production in Arabidopsis. First, the bHLH factor TT8 (AN1 homolog) gene is activated by MBW complexes that include TT8 itself or the bHLH factors GL3 and EGL3 (JAF13 homologs) (Baudry et al., 2006; Xu et al., 2013). Second, members of the R3-MYB repressor family are regulated by the MBW complexes (GL1/GL3/TTG1) that initiate trichome development (Morohashi et al., 2007; Wang et al., 2008; Morohashi and Grotewold, 2009). Third, MYBL2 represses expression of TT8, in addition to directly repressing the anthocyanin biosynthetic genes (Matsui et al., 2008).
Our studies have not resolved fully the role in flavonoid regulation of the petunia bHLH factor JAF13, which belongs to the phylogenetic clade that includes anthocyanin regulators in other species such as R (maize), Delila (snapdragon), and GL3/EGL3 (Arabidopsis) (Quattrocchio et al., 1998; Davies et al., 2012). JAF13 can activate the promoters of various anthocyanin biosynthesis genes (Quattrocchio et al., 1998; Spelt et al., 2000; Figure 6B) and AN1, MYBx, and MYB27 genes (Figure 6A) when an R2R3-MYB partner is supplied. Binding of MYBx was found to both JAF13 and AN1 in Y2H assays (Kroon, 2004), and the repressive effect of MYBx against DFR-A, AN1, MYB27, and MYBx promoters was far stronger when JAF13 was included than when AN1 was included (Figures 6A and 6B). Moreover, a recently identified unstable JAF13 mutant produces flowers with sectors in which anthocyanin levels are reduced by ∼50% (Tornielli et al., 2008). Together, this evidence suggests that JAF13 does participate in regulating anthocyanin production. However, JAF13 does not compensate for the loss of AN1, as an1 mutants completely lack anthocyanins despite expressing JAF13 (Spelt et al., 2000). It is possible that this reflects specialization for the roles of the two bHLH clade partners in petunia, perhaps in a regulatory hierarchy. Consistent with this, petunias expressing the maize Lc gene (JAF13-clade bHLH) have enhanced expression of AN1 (Figure 5).
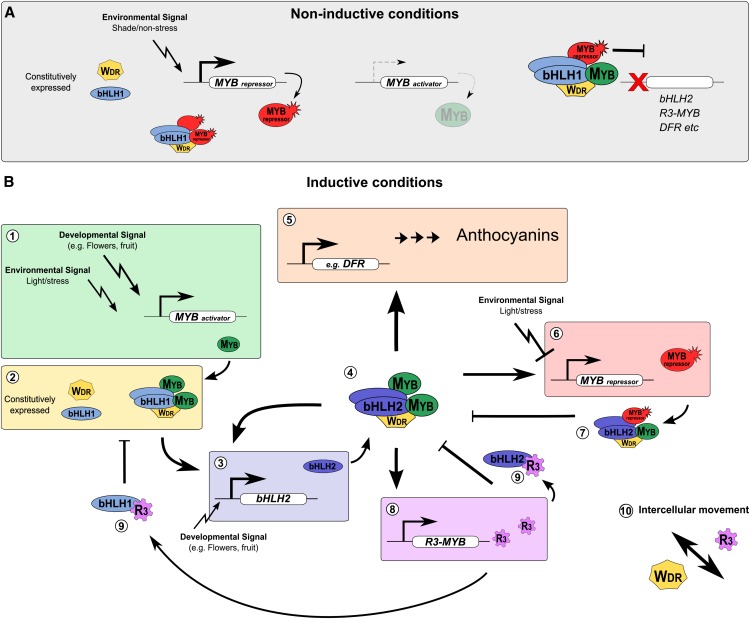
An Integrated Model for Anthocyanin Regulation in Eudicots
Despite significant progress identifying and characterizing individual regulatory components in many species, a model is lacking that integrates this cross-species data for the actions of the activator and repressor components of the MBW regulatory system for anthocyanin pigmentation. Based on our findings here for petunia, as well as other species (particularly Arabidopsis), we present a model highlighting common features of the regulatory network that determines anthocyanin synthesis in Eudicots (Figure 8). During noninductive conditions (Figure 8A), such as leaves of plants growing under shade conditions, low levels of R2R3-MYB activator are present, while the MYB repressor is expressed at high levels and the bHLH1 (Ph-JAF13/At-EGL3 clade) and WDR proteins are constitutively expressed. The MYB repressor may inhibit MBW complex formation by binding free bHLH proteins, as well as conferring a repressive action to any MBW complexes that do form through the action of the repression motif (EAR or TLLLFR), preventing activation of target genes, such as bHLH2 (Ph-AN1/At-TT8 clade), R3-MYB, and the anthocyanin biosynthetic genes (e.g., DFR) (Figure 8A).
Figure 8.
Model of the Anthocyanin Gene Regulation Network in Eudicots.
(A) During noninductive conditions, active MYB repressors (typically R2R3-MYB) are expressed at high levels, and WDR proteins and bHLH1 (Ph-JAF13/At-EGL3 clade bHLH) are constitutively expressed. Repressor MBW complexes may form, titrating bHLH and WDR proteins. MYB repressors are in excess of R2R3-MYB activators, allowing recruitment of the repressor to target genes by MBW complexes containing an R2R3-MYB activator. Transcription is actively repressed through repression motifs (e.g., EAR domain).
(B) (1) Anthocyanin synthesis is initiated in stressed leaves or developing flowers/fruit by activating the expression of the R2R3-MYB activator. (2) The R2R3-MYB activator, WDR, and bHLH1 (Ph-JAF13/At-EGL3 clade bHLH) proteins form an MBW activation complex that (3) activates the expression of bHLH2 (Ph-AN1/At-TT8 clade bHLH). (4) A core MBW activation complex containing bHLH2 proteins forms; this activates the expression of bHLH2 (reinforcement) and activates the expression of the anthocyanin biosynthesis genes (5), ultimately resulting in anthocyanin accumulation. The MBW complex also activates the expression of the MYB repressor (typically R2R3-MYB) (6), although this activation appears to be overruled by environmental signals in vegetative tissues. The inclusion of the repressor into the MBW complex (7) results in active transcriptional repression of target promoters of the core MBW complex (anthocyanin biosynthetic genes, such as DFR, bHLH2, R3-MYB, and MYB repressor). Feedback inhibition is provided by R3-MYB repressors; these are activated by the MBW complex (8) and inhibit the formation of new MBW complexes (9) by titrating bHLH factors. (10) The R3-MYB repressors and WDR proteins are capable of intercellular movement, which may contribute to pigmentation patterning.
During inductive conditions (Figure 8B), such as light stress in vegetative tissues or in maturing flowers and fruits, R2R3-MYB activator genes are expressed. MBW complexes containing bHLH1 form that activate expression of bHLH2. The bHLH2 is then able to form MBW complexes with the R2R3-MYB activator and WDR proteins, which activate transcription of the target genes encoding anthocyanin biosynthetic genes (e.g., DFR), the bHLH2, MYB repressor, and the competitive R3-MYB. The activation of genes encoding the MYB repressor (typically R2R3-MYB) and the competitive R3-MYB repressor by the MBW activation complex allows for feedback repression. In leaves, environmental signals appear to prevent the activation of the repressor by the MBW activation complex.
The activation of bHLH2 genes appears to serve as a reinforcement mechanism to intensify the response, ensuring there is sufficient bHLH to promote anthocyanin synthesis. While this feature is present in both petunia and Arabidopsis (Baudry et al., 2006; Xu et al., 2013), this does not seem to be necessary in Arabidopsis for pigmentation to occur, as GL3/EGL3 (bHLH1) can act redundantly with TT8 (bHLH2) for anthocyanin synthesis (Zhang et al., 2003). By contrast, petunia (Spelt et al., 2000, Albert et al., 2011), Ipomoea sp (Park et al., 2004), pea (Pisum sativa; Hellens et al., 2010), and Dahlia variabilis (Ohno et al., 2011) bHLH2 mutants result in a complete or severe loss of anthocyanin pigmentation, despite the continued expression of bHLH1 having been shown in the case of petunia, Ipomoea, and Dahlia. Thus, it appears that the requirement of bHLH2 activity for anthocyanin synthesis, and the hierarchical regulation between bHLH components, varies throughout the Eudicots and likely reflects functional specialization of the bHLH factor roles between genera.
The active MYB repressors act as corepressors of the MBW complex and repress the genes targeted by the MBW activation complex, through repressive domains such as the EAR or TLLLFR motif. This mode of action was first proposed for At-MYBL2 because a MYBL2-VP16 fusion protein failed to activate a DFR promoter, suggesting it could not directly bind DNA (Matsui et al., 2008). We also observed similar results using a MYB27-VP16 construct (Figure 6B). Importantly, however, we also provide direct experimental evidence for this mechanism by demonstrating that an interaction between MYB activators and MYB repressors can be bridged by bHLH factors (Figure 6C). Such an interaction is required if the MYB repressor acts as a corepressor of the MBW activation complex.
We have also demonstrated that active MYB repressors (Ph-MYB27) can be regulated by the MBW activation complex, which has not previously been demonstrated for At-MYBL2 or R2R3-MYB repressors involved in anthocyanin synthesis. This finding provides a mechanism that explains the expression pattern of MYB repressors in developing petunia flowers and during fruit ripening of strawberry (Aharoni et al., 2001; Lin-Wang et al., 2010) and grape (Vitis vinifera; Huang et al., 2014). Similarly, the regulation of R3-MYB repressors by the MBW complex regulating anthocyanin synthesis, demonstrated for MYBx in petunia, likely holds true across plants. Recently, the Rose Intensity1 (ROI1) locus in Mimulus, which affects floral pigmentation intensity, was shown to encode a competitive R3-MYB repressor (Yuan et al., 2013). The expression profile of ROI1 in developing flowers is similar to MYBx in petunia and likely reflects regulation by the MBW activation complex. While R3-MYBs have been shown to be activated by MBW complexes involved in trichome and root hair formation in Arabidopsis, this has not been demonstrated for MBW complexes involved in anthocyanin or PA synthesis (containing PAP1 or TT2) nor have these genes been shown to be targeted by MYBL2.
The commonalities identified between the systems in the Asterid petunia and the Rosid Arabidopsis suggest that key regulatory components are strongly conserved across at least the Eudicots, specifically an MBW activator complex, R2R3-MYB active repressors, mobile R3-MYB competitive inhibitors, and hierarchical regulation among these factors. Moreover, specific details of the regulatory mechanism appear to be conserved, such as the double-lockdown repression through action of repressors on both the biosynthetic genes and the activator TF genes. The ability of MYB27 to repress its own expression and that of the competitive inhibitor (MYBx) has not yet been described for similar proteins in other species, although At-MYB4 represses its own expression in Arabidopsis (Zhao et al., 2007). It has yet to be shown whether the TFs involved in anthocyanin regulation are posttranslationally regulated in petunia as they are in Arabidopsis. However, the observations on GFP-MYB27 stability, along with the finding that phosphorylation of At-MYBL2 in Arabidopsis prevents its degradation (Ye et al., 2012), suggest protein levels of the active repressors are subject to strong posttranslational regulation. Posttranslational regulation has also been reported for the flavonoid related bHLH and R2R3-MYB activators of Arabidopsis (Maier et al., 2013; Patra et al., 2013) and for an anthocyanin related bHLH in apple (Malus domestica) (Xie et al., 2012), indicating cross-species conservation of this aspect of the regulatory network.
A notable difference between petunia and Arabidopsis is the strong pigmentation of petunia flowers, which allows control of stress-regulated vegetative pigmentation to be compared with that of developmentally regulated floral pigmentation. Substantial overlap between the vegetative and floral regulatory networks was found, with the major difference being specialization of the members of the R2R3-MYB activator multigene family in terms of spatial expression patterns. Although the origin of the MBW activator complex for anthocyanins has not been resolved, the occurrence of at least some common TF components in gymnosperms and angiosperms (Wilkins et al., 2009; Feller et al., 2011) indicates it preceded the development of colorful flowers and so likely arose initially for vegetative pigmentation regulation. Several of the pigmentation characters defined by the R2R3-MYB activators in flowers are indeed also shared with the vegetative tissues. Light-induced floral bud blush is regulated by PHZ, a principal role of which is vegetative pigmentation in response to light. Venation in petunia flowers is determined by DPL. DPL is also expressed in leaves, and the strong leaf venation phenotype of the MYB27 RNAi plants is likely due to DPL activity. Thus, many floral pigmentation regulators may have been co-opted from more ancient roles in vegetative pigmentation. Similarly, petal venation is thought to be an older trait than full-red pigmentation in the Antirrhinum genus (Shang et al., 2011).
Biological Significance of This Regulatory Mechanism
A sophisticated regulatory mechanism is required to enable the plant to produce anthocyanins in the correct spatial location and at the appropriate levels to meet the differing demands of plant development (e.g., floral signals) and environmental response. The gene regulatory network we characterized in petunia facilitates stringent regulation of not only the activation of the pathway but also its repression. Moreover, the hierarchical and feedback regulation by activators and repressors should enable cell-specific production of anthocyanin for developmental purposes (e.g., pigmentation patterns) and the fine-gearing needed to prevent inappropriate anthocyanin levels becoming limiting to photosynthesis, while retaining the plasticity to respond rapidly to changing environmental conditions. Given the many commonalities identified between the Asterid model of petunia and the Rosid model of Arabidopsis, it is likely that the fundamentals of the regulatory network identified are widespread in the Eudicots.
METHODS
Yeast Two- and Three-Hybrid Assays
Full-length cDNAs of the candidate genes were cloned in the ProQuest Y2H vectors pDEST22 and pDEST32 (Invitrogen). AD constructs and BD constructs were transformed in the yeast strains PJ69-4a and pJ69-4α, respectively (James et al., 1996). Strains containing each individual construct were mated with one another on YPDA solid media, and diploid PJ69-4 strains were selected on SD media lacking Leu and Trp and were replicated on SD media lacking Leu, Trp, and His and supplemented by increasing amounts of 3-AT (0 to 100 mM). Growth was scored after 2 d at 30°C. For Y3H experiments, AN1 and JAF13 were cloned into the pGW6/U vector, a Gateway-enabled version of vector pTFT1 (Egea-Cortines et al., 1999) in which the ADE2 selection marker was changed to URA3, and transformed into the diploid strains already containing BD-MYB27 and AD-DPL or the corresponding empty vector combinations. Growth was scored after 2 d at 30°C on SD media lacking Leu, Trp, uracil and His and supplemented by increasing amounts of 3-AT (0 to 100 mM).
Generation of Stable Transgenic Lines
The 35S:MYB27 construct was generated by cloning a MYB27 cDNA (amplified from MP) into pART7 and subcloned into pART27. The MYB27 RNAi construct was generated by cloning the C-terminal and 3′UTR sequence of MYB27 into the RNAi vector pDAH2 and subcloned into pART27. Stable transgenic petunia (Petunia hybrida) plants harboring 35Spro:MYB27 or MYB27 RNAi constructs were generated in MP petunia by Agrobacterium tumefaciens–mediated (strain GV3101, MP-90) leaf-disc transformation, essentially as described by Conner et al. (2009). MYB27ΔC was generated using PCR (primers NA192/ NA286; listed in Supplemental Table 4) to remove the EAR motif; MYB27ΔC-VP16 was synthesized by GenScript (www.genscript.com). Effector constructs expressing anthocyanin TFs from a CaMV35S promoter were made by cloning cDNAs into pART7/pART27 and these binary constructs were transformed into Agrobacterium GV3101 (MP90).
Plant Material and Growth Conditions
MP ([P. hybrida × P. axillaris] × P. axillaris) (an2, AN1, and AN11), P. hybrida cv V26 (AN2, an4, AN1, and AN11), cv V30 (AN2, AN4, AN1, and AN11), cv W134 (AN2, an4, AN1, and an11), cv W225 (AN2, an4, an1, and AN11), 35Spro:MYB27, and MYB27 RNAi plants were grown in greenhouses that were heated at 15°C and vented at 25°C with ambient lighting. The sepals from six stage 1 flower buds, per biological replicate, were collected from MP, MYB27 OE, and MYB27 RNAi plants for RNA extraction, and sepals from approximately six to eight stage 4 flower buds were collected for anthocyanin and flavonoid analyses. Anthocyanin and flavonoids were extracted from 100 mg dry weight sepal tissue and quantified by HPLC (Albert et al., 2011).
qRT-PCR
RNA was extracted from sepals of MYB27 OE, RNAi, or MP plants or from the corolla tissue from stage 1 to 5 flower buds of V26 petunia by acid guanidinium/phenol extraction (Chomczynski and Sacchi, 1987) and treated with DNaseI (Roche) prior to RT. qRT-PCR was performed as described by Albert et al. (2011), using Transcriptor reverse transcriptase, oligo dT12-18, and LightCycler 480 SYBR Green I Master reagents (Roche). The primer sequences are listed in Supplemental Table 4. Primers for C4H1 and PAL1 (Colquhoun et al., 2011) and EF1α and ACT2 (Snowden et al., 2005) have been described previously. Transcript abundance was calculated relative to the geometric mean of EF1α and ACT2 (Pfaffl, 2001).
Dual Luciferase Promoter Activation Assays
The promoters for AN1 and MYBx were isolated using the Universal GenomeWalker kit (Clontech) from MP genomic DNA libraries using the gene-specific primers NA76, NA77, NA129, and NA130, respectively. The MYB27 promoter sequence was from Murr (1995). The promoters + 5′UTR were amplified from MP genomic DNA with the primer sets DFR-A (NA121/NA122), MYB27 (NA166/NA167), AN1 (NA115/NA116), and MYBx (NA112/NA133) and cloned into the XhoI-NcoI sites within the dual luciferase plasmid p0800LUC (Hellens et al., 2005). Promoter assays were performed in Agrobacterium-infiltrated Nicotiana benthamiana leaves, as described by Espley et al. (2009).
Statistical Validation
Statistical significance was determined by one-way ANOVA upon log-transformed data using Genstat (version 15). Log transformations were performed due to unequal variance. LSD values were calculated using P = 0.05 and used to compare treatment means.
Biolistic Transformation Assays
N-terminal GFP fusions, expressed from a CaMV35S promoter, were made with MYBx and MYB27 by cloning cDNAs in frame with GFP in an expression vector. One microgram was transformed into onion (Allium cepa) bulb scale epidermis or Cymbidium petals and viewed by fluorescence microscopy, as described by Albert et al. (2010). Petunia cv W134 (an11) petals were biolistically transformed with 5 µg of 35Spro:AN11 plasmid together with 2 µg 35Spro:GFP-ER, with or without 5 µg of 35Spro:MYBx or 35Spro:MYB27. The ability for MYBx to inhibit AN11 complementation was determined 48 h after bombardment. GFP was viewed with blue light, and anthocyanin producing cells were viewed with white light on a Leica dissecting microscope (Leica M205FA), and images were taken with a Leica digital camera (Leica DF550). The color balance and brightness of images were adjusted after capture to reflect what was observed through the microscope eyepiece.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MYBx, KF985022; MYB27, KF985023; DPL, HQ116169; PURPLE HAZE, HQ116170; AN1, AF260918; AN2, AF146702; AN11, U94748; JAF13, AF020545; and DFR-A, X79723.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Transcript Abundance for Anthocyanin Regulators during Flower Development in V26 Petunia.
Supplemental Figure 2. Seed Coat Pigmentation Phenotypes in MYB27 OE and RNAi Lines.
Supplemental Figure 3. Pigmentation Phenotypes Cosegregate with MYB27 Transgenes in MYB27 OE×V30 and MYB27 RNAi×V30 Crosses.
Supplemental Figure 4. Transcript Abundance for Anthocyanin Regulators in 35Spro:DPL and 35Spro:PHZ Petunias.
Supplemental Figure 5. MYB27 and MYBx Expression Is Reduced in Mitchell Petunia (an2).
Supplemental Figure 6. Localization of GFP-MYBx and GFP-MYB27 in epidermal cells of Cymbidium × hybrida petals.
Supplemental Figure 7. GFP and MYB27 Are Retained Cell Autonomously in Petunia Petals.
Supplemental Figure 8. AN1 and AN2 Act Cell Autonomously in Petunia Petals.
Supplemental Table 1. Yeast Two-Hybrid Interaction Strength.
Supplemental Table 2. Anthocyanin and Flavonoid Concentrations in MYB27 OE and RNAi Petunias.
Supplemental Table 3. Biolistic Repression Assays in Petunia W134 (an11).
Supplemental Table 4. Oligonucleotide Primers Used.
Supplementary Material
Acknowledgments
We thank Michael McManus for providing academic supervision to N.W.A. during his PhD, Steve Arathoon for technical assistance, Ian King and Julie Ryan for care of plants, Duncan Hedderley for statistics assistance, Ronald Koes for petunia cv W134, and Cathie Martin, Andrew Allan, and Roger Hellens for valuable discussions, encouragement, and critique. N.W.A. was supported by a Bright Futures Top Achiever Doctoral Scholarship awarded by the Tertiary Education Commission of New Zealand. This work was funded by the Ministry of Business, Innovation, and Employment contract “From Tools to Traits: Functional Genomics for Metabolite Accumulation” (C0ZX0805).
AUTHOR CONTRIBUTIONS
N.W.A., K.M.D., D.H.L., H.Z., P.E.J., and K.E.S. conceived of the experiments. N.W.A. conducted the experimentation and analysis of data, with the following exceptions: M.R.B. generated the MYB27 transgenic petunias; H.N. performed the AN11 biolistic assays; H.Z. performed the GFP-fusion experiments; D.H.L. performed the metabolite analysis; M.M. performed the promoter activation assays; and C.B. performed and analyzed Y2H and Y3H experiments. N.W.A., K.M.D., and K.E.S. wrote the article, with assistance from all coauthors.
Glossary
- TF
transcription factor
- bHLH
basic-helix-loop-helix
- EAR
ERF-associated amphiphilic repression
- Y2H
yeast two-hybrid
- 3-AT
3-amino-1,2,4-triazole
- RNAi
RNA interference
- OE
overexpress
- MP
Mitchell petunia
- qRT-PCR
quantitative RT-PCR
- UTR
untranslated region
- Y3H
yeast three-hybrid
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Aharoni A., De Vos C.H.R., Wein M., Sun Z.K., Greco R., Kroon A., Mol J.N.M., O’Connell A.P. (2001). The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28: 319–332. [DOI] [PubMed] [Google Scholar]
- Albert N.W., Arathoon S., Collette V.E., Schwinn K.E., Jameson P.E., Lewis D.H., Zhang H., Davies K.M. (2010). Activation of anthocyanin synthesis in Cymbidium orchids: variability between known regulators. Plant Cell Tissue Organ Cult. 100: 355–360. [Google Scholar]
- Albert N.W., Lewis D.H., Zhang H., Irving L.J., Jameson P.E., Davies K.M. (2009). Light-induced vegetative anthocyanin pigmentation in Petunia. J. Exp. Bot. 60: 2191–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert N.W., Lewis D.H., Zhang H., Schwinn K.E., Jameson P.E., Davies K.M. (2011). Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 65: 771–784. [DOI] [PubMed] [Google Scholar]
- An X.H., Tian Y., Chen K.Q., Wang X.F., Hao Y.J. (2012). The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol. 169: 710–717. [DOI] [PubMed] [Google Scholar]
- Balkunde R., Bouyer D., Hülskamp M. (2011). Nuclear trapping by GL3 controls intercellular transport and redistribution of TTG1 protein in Arabidopsis. Development 138: 5039–5048. [DOI] [PubMed] [Google Scholar]
- Baudry A., Caboche M., Lepiniec L. (2006). TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 46: 768–779. [DOI] [PubMed] [Google Scholar]
- Baudry A., Heim M.A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39: 366–380. [DOI] [PubMed] [Google Scholar]
- Bouyer D., Geier F., Kragler F., Schnittger A., Pesch M., Wester K., Balkunde R., Timmer J., Fleck C., Hülskamp M. (2008). Two-dimensional patterning by a trapping/depletion mechanism: The role of TTG1 and GL3 in Arabidopsis trichome formation. PLoS Biol. 6: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockington S.F., Alvarez-Fernandez R., Landis J.B., Alcorn K., Walker R.H., Thomas M.M., Hileman L.C., Glover B.J. (2013). Evolutionary analysis of the MIXTA gene family highlights potential targets for the study of cellular differentiation. Mol. Biol. Evol. 30: 526–540. [DOI] [PubMed] [Google Scholar]
- Carey C.C., Strahle J.T., Selinger D.A., Chandler V.L. (2004). Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana. Plant Cell 16: 450–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu L.W., Zhou X., Burke S., Wu X., Prior R.L., Li L. (2010). The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol. 154: 1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156–159. [DOI] [PubMed] [Google Scholar]
- Colquhoun T.A., Kim J.Y., Wedde A.E., Levin L.A., Schmitt K.C., Schuurink R.C., Clark D.G. (2011). PhMYB4 fine-tunes the floral volatile signature of Petunia x hybrida through PhC4H. J. Exp. Bot. 62: 1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone K.C., Burr F.A., Burr B. (1986). Molecular analysis of the maize anthocyanin regulatory locus C1. Proc. Natl. Acad. Sci. USA 83: 9631–9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner, A.J., Albert, N.W., and Deroles, S.C. (2009). Transformation and regeneration of petunia. In Petunia: Evolutionary, Developmental and Physiological Genetics, T. Gerats and J. Strommer, eds (New York: Springer), pp. 395–409. [Google Scholar]
- Davies K.M., Albert N.W., Schwinn K.E. (2012). From landing lights to mimicry: The molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 39: 619–638. [DOI] [PubMed] [Google Scholar]
- de Vetten N., Quattrocchio F., Mol J., Koes R. (1997). The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 11: 1422–1434. [DOI] [PubMed] [Google Scholar]
- Digiuni S., Schellmann S., Geier F., Greese B., Pesch M., Wester K., Dartan B., Mach V., Srinivas B.P., Timmer J., Fleck C., Hulskamp M. (2008). A competitive complex formation mechanism underlies trichome patterning on Arabidopsis leaves. Mol. Syst. Biol. 4: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C., Le Gourrierec J., Baudry A., Huep G., Lanet E., Debeaujon I., Routaboul J.-M., Alboresi A., Weisshaar B., Lepiniec L. (2008). MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 55: 940–953. [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M., Saedler H., Sommer H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18: 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espley R.V., Brendolise C., Chagné D., Kutty-Amma S., Green S., Volz R., Putterill J., Schouten H.J., Gardiner S.E., Hellens R.P., Allan A.C. (2009). Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21: 168–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A., Machemer K., Braun E.L., Grotewold E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66: 94–116. [DOI] [PubMed] [Google Scholar]
- Gao J.J., Shen X.F., Zhang Z., Peng R.H., Xiong A.S., Xu J., Zhu B., Zheng J.L., Yao Q.H. (2011). The myb transcription factor MdMYB6 suppresses anthocyanin biosynthesis in transgenic Arabidopsis. Plant Cell Tissue Organ Cult. 106: 235–242. [Google Scholar]
- Gierer A., Meinhardt H. (1972). A theory of biological pattern formation. Kybernetik 12: 30–39. [DOI] [PubMed] [Google Scholar]
- Gill G., Ptashne M. (1988). Negative effect of the transcriptional activator GAL4. Nature 334: 721–724. [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Zhao M., Leavitt J.M., Lloyd A.M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53: 814–827. [DOI] [PubMed] [Google Scholar]
- Goodrich J., Carpenter R., Coen E.S. (1992). A common gene regulates pigmentation pattern in diverse plant species. Cell 68: 955–964. [DOI] [PubMed] [Google Scholar]
- Gou J.-Y., Felippes F.F., Liu C.-J., Weigel D., Wang J.-W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23: 1512–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould K.S. (2004). Nature's Swiss army knife: The diverse protective roles of anthocyanins in leaves. J. Biomed. Biotechnol. 2004: 314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebenok R.J., Pierson E., Lambert G.M., Gong F.-C., Afonso C.L., Haldeman-Cahill R., Carrington J.C., Galbraith D.W. (1997). Green-fluorescent protein fusions for efficient characterization of nuclear targeting. Plant J. 11: 573–586. [DOI] [PubMed] [Google Scholar]
- Hanson M.R., Köhler R.H. (2001). GFP imaging: Methodology and application to investigate cellular compartmentation in plants. J. Exp. Bot. 52: 529–539. [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., et al. (2010). Identification of Mendel’s white flower character. PLoS ONE 5: e13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez J.M., Feller A., Morohashi K., Frame K., Grotewold E. (2007). The basic helix loop helix domain of maize R links transcriptional regulation and histone modifications by recruitment of an EMSY-related factor. Proc. Natl. Acad. Sci. USA 104: 17222–17227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hichri I., Barrieu F., Bogs J., Kappel C., Delrot S., Lauvergeat V. (2011). Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62: 2465–2483. [DOI] [PubMed] [Google Scholar]
- Huang Y.F., Vialet S., Guiraud J.L., Torregrosa L., Bertrand Y., Cheynier V., This P., Terrier N. (2014). A negative MYB regulator of proanthocyanidin accumulation, identified through expression quantitative locus mapping in the grape berry. New Phytol. 201: 795–809. [DOI] [PubMed] [Google Scholar]
- Hughes N.M., Neufeld H.S., Burkey K.O. (2005). Functional role of anthocyanins in high-light winter leaves of the evergreen herb Galax urceolata. New Phytol. 168: 575–587. [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H.L., Cominelli E., Bailey P., Parr A., Mehrtens F., Jones J., Tonelli C., Weisshaar B., Martin C. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19: 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S., Links M.G., Rozwadowski K. (2010). Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 152: 1109–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S., Rozwadowski K. (2011). EAR motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6: 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R., Verweij W., Quattrocchio F. (2005). Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10: 236–242. [DOI] [PubMed] [Google Scholar]
- Kong Q., Pattanaik S., Feller A., Werkman J.R., Chai C., Wang Y., Grotewold E., Yuan L. (2012). Regulatory switch enforced by basic helix-loop-helix and ACT-domain mediated dimerizations of the maize transcription factor R. Proc. Natl. Acad. Sci. USA 109: E2091–E2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz H.D., et al. (1998). Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16: 263–276. [DOI] [PubMed] [Google Scholar]
- Kroon, A. (2004). Transcription Regulation of the Anthocyanin Pathway in Petunia hybrida. PhD dissertation (Amsterdam: Vrije Universiteit). [Google Scholar]
- Kurata T., et al. (2005). Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132: 5387–5398. [DOI] [PubMed] [Google Scholar]
- Lin-Wang K., Bolitho K., Grafton K., Kortstee A., Karunairetnam S., McGhie T.K., Espley R.V., Hellens R.P., Allan A.C. (2010). An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Wang K., Micheletti D., Palmer J., Volz R., Lozano L., Espley R., Hellens R.P., Chagnè D., Rowan D.D., Troggio M., Iglesias I., Allan A.C. (2011). High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ. 34: 1176–1190. [DOI] [PubMed] [Google Scholar]
- Lowry D.B., Sheng C.C., Lasky J.R., Willis J.H. (2012). Five anthocyanin polymorphisms are associated with an R2R3-MYB cluster in Mimulus guttatus (Phrymaceae). Am. J. Bot. 99: 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig S.R., Habera L.F., Dellaporta S.L., Wessler S.R. (1989). Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc. Natl. Acad. Sci. USA 86: 7092–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A., Schrader A., Kokkelink L., Falke C., Welter B., Iniesto E., Rubio V., Uhrig J.F., Hülskamp M., Hoecker U. (2013). Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 74: 638–651. [DOI] [PubMed] [Google Scholar]
- Martin C., Glover B.J. (2007). Functional aspects of cell patterning in aerial epidermis. Curr. Opin. Plant Biol. 10: 70–82. [DOI] [PubMed] [Google Scholar]
- Matsui K., Umemura Y., Ohme-Takagi M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55: 954–967. [DOI] [PubMed] [Google Scholar]
- Meinhardt H., Gierer A. (1974). Applications of a theory of biological pattern formation based on lateral inhibition. J. Cell Sci. 15: 321–346. [DOI] [PubMed] [Google Scholar]
- Merkle T. (2004). Nucleo-cytoplasmic partitioning of proteins in plants: implications for the regulation of environmental and developmental signalling. Curr. Genet. 44: 231–260. [DOI] [PubMed] [Google Scholar]
- Morohashi K., Grotewold E. (2009). A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet. 5: e1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K., Zhao M., Yang M., Read B., Lloyd A., Lamb R., Grotewold E. (2007). Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol. 145: 736–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr, L. (1995). Characterization of Members of the myb Gene Family of Transcription Factors from Petunia hybrida. PhD dissertation (Amsterdam: Vrije Universiteit). [Google Scholar]
- Ohno S., Hosokawa M., Hoshino A., Kitamura Y., Morita Y., Park K.I., Nakashima A., Deguchi A., Tatsuzawa F., Doi M., Iida S., Yazawa S. (2011). A bHLH transcription factor, DvIVS, is involved in regulation of anthocyanin synthesis in dahlia (Dahlia variabilis). J. Exp. Bot. 62: 5105–5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolocci F., Robbins M.P., Passeri V., Hauck B., Morris P., Rubini A., Arcioni S., Damiani F. (2011). The strawberry transcription factor FaMYB1 inhibits the biosynthesis of proanthocyanidins in Lotus corniculatus leaves. J. Exp. Bot. 62: 1189–1200. [DOI] [PubMed] [Google Scholar]
- Park K.I., Choi J.D., Hoshino A., Morita Y., Iida S. (2004). An intragenic tandem duplication in a transcriptional regulatory gene for anthocyanin biosynthesis confers pale-colored flowers and seeds with fine spots in Ipomoea tricolor. Plant J. 38: 840–849. [DOI] [PubMed] [Google Scholar]
- Patra B., Pattanaik S., Yuan L. (2013). Ubiquitin protein ligase 3 mediates the proteasomal degradation of GLABROUS 3 and ENHANCER OF GLABROUS 3, regulators of trichome development and flavonoid biosynthesis in Arabidopsis. Plant J. 74: 435–447. [DOI] [PubMed] [Google Scholar]
- Payne C.T., Zhang F., Lloyd A.M. (2000). GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares J., Ghosal D., Saedler H. (1990). Molecular analysis of the C1-I allele from Zea mays: A dominant mutant of the regulatory C1 locus. EMBO J. 9: 315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares J., Ghosal D., Wienand U., Peterson P.A., Saedler H. (1987). The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6: 3553–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares J., Wienand U., Peterson P.A., Saedler H. (1986). Molecular cloning of the c locus of Zea mays: a locus regulating the anthocyanin pathway. EMBO J. 5: 829–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch M., Hülskamp M. (2004). Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Curr. Opin. Genet. Dev. 14: 422–427. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T., Song S., Ren Q., Wu D., Huang H., Chen Y., Fan M., Peng W., Ren C., Xie D. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F., Verweij W., Kroon A., Spelt C., Mol J., Koes R. (2006). PH4 of Petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 18: 1274–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F., Wing J., van der Woude K., Souer E., de Vetten N., Mol J., Koes R. (1999). Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11: 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F., Wing J.F., van der Woude K., Mol J.N.M., Koes R. (1998). Analysis of bHLH and MYB domain proteins: Species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 13: 475–488. [DOI] [PubMed] [Google Scholar]
- Ramsay N.A., Glover B.J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10: 63–70. [DOI] [PubMed] [Google Scholar]
- Rim Y., Huang L., Chu H., Han X., Cho W.K., Jeon C.O., Kim H.J., Hong J.C., Lucas W.J., Kim J.Y. (2011). Analysis of Arabidopsis transcription factor families revealed extensive capacity for cell-to-cell movement as well as discrete trafficking patterns. Mol. Cells 32: 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatierra A., Pimentel P., Moya-León M.A., Herrera R. (2013). Increased accumulation of anthocyanins in Fragaria chiloensis fruits by transient suppression of FcMYB1 gene. Phytochemistry 90: 25–36. [DOI] [PubMed] [Google Scholar]
- Schaart J.G., Dubos C., Romero De La Fuente I., van Houwelingen A.M.M.L., de Vos R.C.H., Jonker H.H., Xu W., Routaboul J.-M., Lepiniec L., Bovy A.G. (2013). Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 197: 454–467. [DOI] [PubMed] [Google Scholar]
- Schwinn K., Venail J., Shang Y., Mackay S., Alm V., Butelli E., Oyama R., Bailey P., Davies K., Martin C. (2006). A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18: 831–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Venail J., Mackay S., Bailey P.C., Schwinn K.E., Jameson P.E., Martin C.R., Davies K.M. (2011). The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytol. 189: 602–615. [DOI] [PubMed] [Google Scholar]
- Snowden K.C., Simkin A.J., Janssen B.J., Templeton K.R., Loucas H.M., Simons J.L., Karunairetnam S., Gleave A.P., Clark D.G., Klee H.J. (2005). The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17: 746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelt C., Quattrocchio F., Mol J., Koes R. (2002). ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. Plant Cell 14: 2121–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelt C., Quattrocchio F., Mol J.N.M., Koes R. (2000). anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12: 1619–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagnone L., Merida A., Parr A., Mackay S., Culianez-Macia F.A., Roberts K., Martin C. (1998). The AmMYB308 and AmMYB330 transcription factors from antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10: 135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga R., Iwata M., Okada K., Wada T. (2007). Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell 19: 2264–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornielli, G., Koes, R., and Quattrocchio, F. (2008). The genetics of flower color. In Petunia: Evolutionary, Developmental and Physiological Genetics, T. Gerats and J. Strommer, eds (New York: Springer), pp. 269–299. [Google Scholar]
- Turing A.M. (1952). The chemical basis of morphogenesis. Philos. Trans. R. Soc. Lond. Series B 237: 37–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velten J., Cakir C., Cazzonelli C.I. (2010). A spontaneous dominant-negative mutation within a 35S:AtMYB90 transgene inhibits flower pigment production in tobacco. PLoS ONE 5: e9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A.R., Davison P.A., Bolognesi-Winfield A.C., James C.M., Srinivasan N., Blundell T.L., Esch J.J., Marks M.D., Gray J.C. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Hubbard L., Chang Y., Guo J., Schiefelbein J., Chen J.G. (2008). Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol. 8: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester K., Digiuni S., Geier F., Timmer J., Fleck C., Hülskamp M. (2009). Functional diversity of R3 single-repeat genes in trichome development. Development 136: 1487–1496. [DOI] [PubMed] [Google Scholar]
- Wilkins O., Nahal H., Foong J., Provart N.J., Campbell M.M. (2009). Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 149: 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X.-B., Li S., Zhang R.-F., Zhao J., Chen Y.-C., Zhao Q., Yao Y.-X., You C.-X., Zhang X.-S., Hao Y.-J. (2012). The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 35: 1884–1897. [DOI] [PubMed] [Google Scholar]
- Xu W., et al. (2013). Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis. New Phytol. 198: 59–70. [DOI] [PubMed] [Google Scholar]
- Ye H., Li L., Guo H., Yin Y. (2012). MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 20142–20147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y.-W., Sagawa J.M., Young R.C., Christensen B.J., Bradshaw H.D., Jr (2013). Genetic dissection of a major anthocyanin QTL contributing to pollinator-mediated reproductive isolation between sister species of Mimulus. Genetics 194: 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C.T., Lloyd A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869. [DOI] [PubMed] [Google Scholar]
- Zhang W., Ning G., Lv H., Liao L., Bao M. (2009). Single MYB-type transcription factor AtCAPRICE: a new efficient tool to engineer the production of anthocyanin in tobacco. Biochem. Biophys. Res. Commun. 388: 742–747. [DOI] [PubMed] [Google Scholar]
- Zhao J., Zhang W., Zhao Y., Gong X., Guo L., Zhu G., Wang X., Gong Z., Schumaker K.S., Guo Y. (2007). SAD2, an importin -like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell 19: 3805–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Morohashi K., Hatlestad G., Grotewold E., Lloyd A. (2008). The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135: 1991–1999. [DOI] [PubMed] [Google Scholar]
- Zhu H.-F., Fitzsimmons K., Khandelwal A., Kranz R.G. (2009). CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol. Plant 2: 790–802. [DOI] [PubMed] [Google Scholar]
- Zimmermann I.M., Heim M.A., Weisshaar B., Uhrig J.F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40: 22–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.