A dual biosynthesis that produces seed albumin and a protease-inhibiting peptide in sunflower was found to have evolved over 18 million years ago and has spawned a family of peptides diverse in sequence and three-dimensional structure. Using the gene sequences and an understanding of the peptide biosynthesis, the biochemical steps that allowed these buried peptides to evolve de novo were proposed.
Abstract
The de novo evolution of proteins is now considered a frequented route for biological innovation, but the genetic and biochemical processes that lead to each newly created protein are often poorly documented. The common sunflower (Helianthus annuus) contains the unusual gene PawS1 (Preproalbumin with SFTI-1) that encodes a precursor for seed storage albumin; however, in a region usually discarded during albumin maturation, its sequence is matured into SFTI-1, a protease-inhibiting cyclic peptide with a motif homologous to unrelated inhibitors from legumes, cereals, and frogs. To understand how PawS1 acquired this additional peptide with novel biochemical functionality, we cloned PawS1 genes and showed that this dual destiny is over 18 million years old. This new family of mostly backbone-cyclic peptides is structurally diverse, but the protease-inhibitory motif was restricted to peptides from sunflower and close relatives from its subtribe. We describe a widely distributed, potential evolutionary intermediate PawS-Like1 (PawL1), which is matured into storage albumin, but makes no stable peptide despite possessing residues essential for processing and cyclization from within PawS1. Using sequences we cloned, we retrodict the likely stepwise creation of PawS1’s additional destiny within a simple albumin precursor. We propose that relaxed selection enabled SFTI-1 to evolve its inhibitor function by converging upon a successful sequence and structure.
INTRODUCTION
The de novo evolution of genes and proteins is a principal driver of biological innovation (Tautz and Domazet-Lošo, 2011; Carvunis et al., 2012; Neme and Tautz, 2013). A growing number of reports document how a new protein may evolve by the creation of a new protein-coding gene directly out of noncoding DNA. This requires that if not already transcribed, the noncoding DNA must become transcriptionally active as well as acquire an open reading frame (ORF) that encodes a protein that confers some benefit. Published examples include genes from yeast (Cai et al., 2008), Plasmodium (Yang and Huang, 2011), Arabidopsis thaliana (Donoghue et al., 2011), Drosophila melanogaster (Levine et al., 2006), and humans (Wu et al., 2011). De novo evolution of proteins can also result from frameshifts in existing genes. An early example is an enzyme in Flavobacterium that resulted from a single nucleotide insertion that changed it to an alternative frame and ORF from the originally repetitive and Arg-rich sequence (Ohno, 1984). A related route specific to the evolution of proteins de novo is overprinting. This involves the use of a different frame, but the original frame may also be used so that effectively there become two genes that overlap. Overprinting has been observed in several species but is especially well documented in viruses (Pavesi et al., 2013). For most examples of de novo evolution, the focus is on the gene with little attention paid to the event or events that permit the protein to become a stable component of an organism’s proteome. Also, few examples of de novo evolved proteins include an understanding of the structure or biochemical function of the new protein, although these newly evolved, lineage-specific genes are said to be important for adaptation (Tautz and Domazet-Lošo, 2011).
The genetic and biosynthetic origin of a small, cyclic peptide from seeds called Sunflower Trypsin Inhibitor 1 (SFTI-1) was recently reported (Mylne et al., 2011), and it revealed an unusual biosynthesis and a potential case of de novo protein evolution. The 14-residue sequence for the cyclic trypsin inhibitor SFTI-1 was buried in a precursor protein 151 residues in length and most similar to precursors for seed storage albumins (Mylne et al., 2011). Seed storage albumins accumulate to high levels in seeds and upon germination are catabolized to become an initial nutritive source of nitrogen and sulfur for the developing seedling (Shewry and Pandya, 1999). The coding sequence for SFTI-1 was buried within the albumin precursor sequence between its N-terminal endoplasmic reticulum (ER) signal and the first of its two mature albumin subunits. Mature albumin encoded by this gene was purified and characterized by mass spectrometry, demonstrating that the precursor protein from this intronless gene was matured into both the small cyclic peptide SFTI-1 and a mature, heterodimeric albumin. The precursor was called PawS1 for Preproalbumin with SFTI-1 (Mylne et al., 2011). To be matured from PawS1, both SFTI-1 and PawS1 albumin were shown to be dependent upon asparaginyl endopeptidase (also known as vacuolar processing enzyme), a protease already well known for its importance in processing seed storage proteins (Shimada et al., 2003; Gruis et al., 2004).
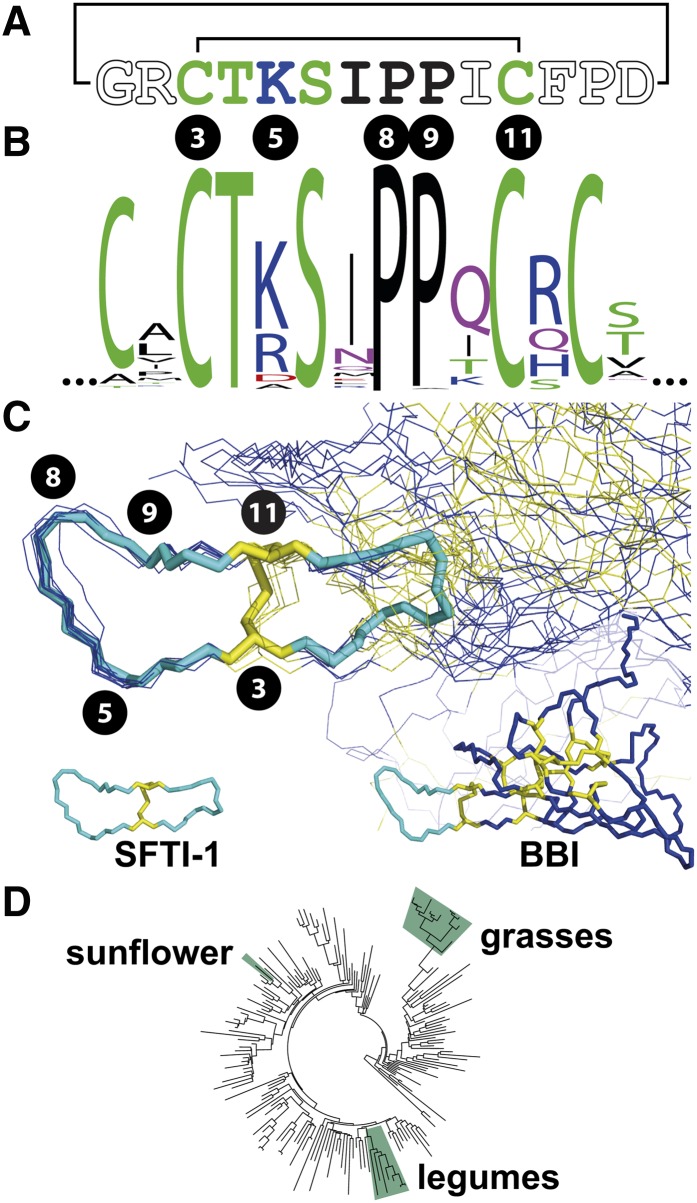
The biosynthetic origins of SFTI-1 being linked to albumin was a surprise as its sequence, tertiary structure, and function had meant SFTI-1 was considered the smallest member of the Bowman Birk Inhibitor (BBI) family of proteins (Luckett et al., 1999). BBIs are typically over 100 residues in length, are derived from a dedicated precursor protein (Birk, 1985), and are only known to be present in two distantly related plant families, the legumes (Fabaceae) and the grasses (Poaceae). Despite such different biosynthetic origins and phylogenetic distribution, SFTI-1 and the trypsin inhibitory loop of BBIs share similarity at three levels. First, at the sequence level, within CTKSIPPxC a string of seven consecutive residues as well as the ninth are shared by SFTI-1 and a BBI consensus (Figures 1A and 1B; Supplemental Figure 1 and Supplemental Data Set 1). The likelihood of any eight residues appearing in order by random chance is 1 in 25 billion or 1 in 70 billion if the eukaryotic amino acid frequency of this particular eight is considered (Tekaia and Yeramian, 2006). Second, both sequences adopt nearly identical 3D structures (Figure 1C). Third, they both function as potent Ser protease inhibitors, especially against trypsin (Birk, 1985; Luckett et al., 1999). This striking structural similarity and analogous function despite different biosynthetic origins and phylogenetic isolation (Figure 1D) raises the question how this extra peptide with its potent biochemical function arose inside an albumin precursor.
Figure 1.
Structural Similarity between SFTI-1 and the Inhibitory Arm of Bowman-Birk Inhibitors
(A) Sequence of the backbone cyclic and disulfide bonded SFTI-1 with the line joining Cys residues denoting a disulfide bond and the line joining Gly and Asp denoting the cyclic backbone. Circled numbers from SFTI-1 (Cys-3, Lys-5, Pro-8, Pro-9, and Cys-11) are aligned with the BBI WebLogo (B) and used to mark their locations within the structural overlay (C).
(B) WebLogo summary of 150 aligned short segments of BBIs.
(C) SFTI-1 (stick format) overlaid upon the inhibitory loops of 10 BBIs (line format).
(D) Angiosperm phylogeny based on rbcL sequences and adapted from Mylne et al. (2012). The families containing BBIs are shown (Fabaceae or legumes, and Poaceae or grasses). The phylogenetically separate position of the BBI-loop mimic sunflower SFTI-1 is also shown.
When two protein motifs share strong sequence and 3D structural similarity, it is common for them to simply share a common ancestor (Ponting and Russell, 2002), but when two similar protein motifs are buried in completely different proteins such as the trypsin inhibitory loops of SFTI-1 and BBIs, their origins are less obvious (Lupas et al., 2001). Although a few long, seemingly convergent protein sequences have been documented (Lawn et al., 1997; Rey et al., 1998; Robson et al., 2000), motifs that are reported to have converged upon similar protein sequence and 3D structure are short and only moderately conserved in sequence. The best known examples of such similar motifs in different protein folds contain three to four conserved residues, often interspersed with one or two nonconserved residues (Lupas et al., 2001). For example, the Asp-box sequence (SxDGxxW) and structural motif has been found in at least three different proteins, including sialidases, chitobiase, and ribonucleases (Copley et al., 2001).
Within PawS1 preproalbumin, the sequence for SFTI-1 resides between the ER signal sequence and the small albumin subunit. In other preproalbumins, this spacer region is discarded during proteolytic maturation of albumin (Shewry and Pandya, 1999). Aligning the amino acid sequences of PawS1 and other preproalbumins revealed a gap forced in the alignment by the SFTI-1 sequence, suggesting genetic insertions created its additional peptide (Supplemental Figure 2). Using mass spectrometry, SFTI-1 was found exclusively in the Helianthus genus (Mylne et al., 2011), suggesting its origin was recent. This was consistent with in-gel trypsin inhibition assays using seed peptide extracts from members of the daisy family Asteraceae (Konarev et al., 2002), which could only find low molecular weight trypsin inhibitors like SFTI-1 in Helianthus species and their close relative Tithonia.
These observations led us to speculate how 3-fold similarity in sequence, structure, and function could be found in proteins with such disparate origins. A hypothetical origin for SFTI-1 could be a lateral genetic transfer (Bock, 2010) of ∼50 bp of DNA encoding a very small BBI motif into a preproalbumin. However, BBI genes are nuclear and sunflowers (Helianthus annuus) lack BBIs (Supplemental Methods), so for BBI DNA to be the origin of SFTI-1, the lateral genetic transfer would have to be from an unrelated, BBI-containing species. Although common in microbial systems, lateral genetic transfer of nuclear genes between unrelated plant species has only been demonstrated once for a gene between a parasitic plant and its host (Yoshida et al., 2010). Lateral gene transfer is more common in plants between organelles with cases documented for movement of organellar genes and even genomes (Won and Renner, 2003; Davis and Wurdack, 2004; Keeling and Palmer, 2008; Rice et al., 2013). A competing hypothesis we might propose for the striking similarity between SFTI-1 and the BBI inhibitory loop is that motif parallelism between SFTI-1 and BBIs evolved gradually from more ancient origins within the daisy family.
As an opportunity to document the de novo evolution of a protein, we sought to understand how PawS1 acquired its additional destiny, an interstitial peptide with novel biochemical functionality. We exploited the rapid speciation that has occurred in the Asteraceae to study PawS1 in sunflower relatives and to our surprise found peptides have been buried in preproalbumin in this way for over 18 million years. This led us to propose that the protease-inhibiting peptides like SFTI-1 evolved within the lineage leading to sunflower by convergent exploration of sequence space under relaxed selection to arrive at the protease inhibitory motif also seen in BBIs.
RESULTS
Identification of a Novel Seed Peptide Family
SFTI-1 was previously found only in Helianthus based on liquid chromatography–mass spectrometry (LC-MS) analysis of peptide extracts of 129 species spanning over 30 million years of evolution (Mylne et al., 2011). In this work, by screening an additional 138 Asteraceae species using the same LC-MS parameters, we found further evidence for SFTI-1 in Helianthus, but also discovered it in the close relative Aldama phenax (previously Viguiera phenax) (Supplemental Data Set 2). Using a PCR approach with primers designed to bind within the PawS1 ORF, we discovered a sequence encoding a peptide in Tithonia detected previously in-gel assays for trypsin inhibition (Konarev et al., 2002), but which we missed using LC-MS because a single amino acid change of Phe to Tyr changed its mass. Because this subtle variant of SFTI-1 is the third known PawS-derived peptide (PDP) after the sunflower cyclic peptides SFTI-1 and SFTI-Like 1 (SFT-L1), we named it PDP-3 following purification and sequencing (Supplemental Figure 3).
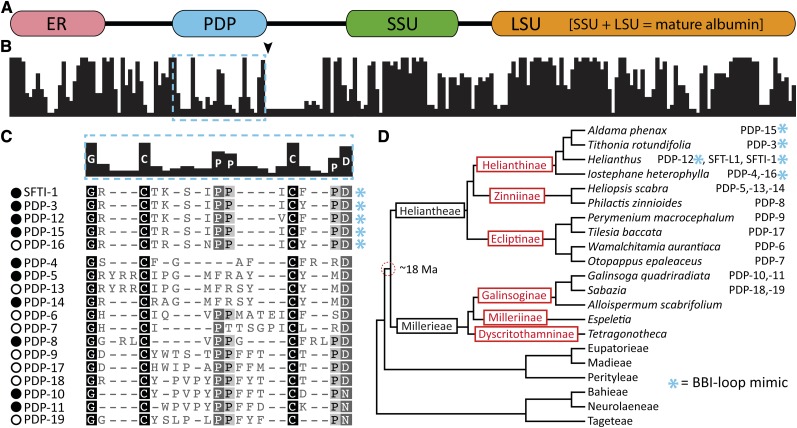
PDP-3 indicated that to follow the ancestry of PDPs, a gene-based approach was required that could be confirmed by peptide mass spectrometry, rather than being reliant upon it for discovery. We obtained the PawS-flanking genomic DNA sequence from a sunflower BAC (Supplemental Figure 4) and identified a primer pair that bound in flanking, noncoding DNA and could amplify full-length PawS1 ORFs from several Asteraceae species. We used these primers to amplify 25 PawS1 genes from genera in the subtribes Helianthinae, Zinniinae, Ecliptinae, and Galinsoginae (Figure 2; Supplemental Figure 5 and Supplemental Table 1). We typically amplified only one or, occasionally, two PawS1 genes per species. Their distribution in several tribes and subtribes suggested that the origin of the dual-fated PawS1-type genes was much more ancient than the Helianthus genus and encoded a novel peptide family (Figure 2D).
Figure 2.
PawS1 Proteins from the Daisy Family (Asteraceae) and Their Buried Peptides
(A) PawS1 domains include the ER signal, PDP region, and the small (SSU) and large subunits (LSU) of mature albumin based on sunflower PawS1 (Mylne et al., 2011).
(B) Percentage (0 to 100%) identity graph for 28 PawS1 protein sequences showing weaker conservation around the PDP region (dashed box). The arrowhead beneath the graph indicates the position where four PawS1 sequences have 100 amino acid internal expansions. See Supplemental Figure 5 for an alignment of full-length sequences.
(C) Alignment of selected in vivo confirmed (closed circles) and gene-predicted (open circles) PDPs showing little conservation except the N-terminal Gly, C-terminal Asp, a Cys-pair, and central Pro-Pro. The BBI-loop mimics are marked with asterisks.
(D) Phylogenetic tree (Supplemental Data Sets 3 and 4) including species used in this study and showing PDPs are common to the Heliantheae and Millerieae tribes estimated to have diverged (dashed circle) 18 million years ago (Ma) (Supplemental Figure 6). Tribe names are black, whereas subtribe names are in red/gray.
[See online article for color version of this figure.]
To establish the age of the PDP family, we developed a chronogram using a phylogram built from chloroplastic gene sequences for 25 genera calibrated using a previous Asteraceae chronogram (Kim et al., 2005). This showed that the subtribes containing PawS1 genes diverged ∼18 million years ago during the early Miocene era (Figure 2D; Supplemental Figure 6 and Supplemental Data Sets 3 to 6), defining the minimum age for this new peptide family. Based on the number of species evolving from the root node containing the tribes Millereae and Heliantheae, PDPs could be present in over 4700 species (Panero, 2007).
To confirm that these PawS1 genes encoded stable peptides (Figure 2C), we confirmed predicted peptide masses by LC-MS in species for which plant material was available and compared their tandem mass spectrometry (MS/MS) fragmentation patterns with those of a corresponding synthetic PDP. Some PDPs could be purified from plant material and sequenced directly by MS (Supplemental Table 1 and Supplemental Figures 3 and 7 to 14). For example, Iostephane heterophylla PawS1 encoded PDP-4, and we detected the predicted [M+1H]1+ and [M+2H]2+ ions at m/z 1345.6 and 673.3 in the LC-MS profile, then purified and sequenced PDP-4 (Supplemental Figure 8). Heliopsis helianthoides scabra PawS1 encodes PDP-5 for which we identified matching [M+3H]3+ and [M+4H]4+ ions, but in the absence of sufficient plant material for purification and sequencing, PDP-5 was produced by solid phase peptide synthesis, and we demonstrated that synthetic and native PDP-5 had matching retention times, charge states, and fragment ion spectra upon MS/MS (Supplemental Figure 9). Using these two approaches, we confirmed the existence of eight PDPs (PDP-3 to -5, -8, -10 to -12, and -14) at the peptide level (Supplemental Table 1 and Supplemental Figures 3 and 7 to 14).
Structural and Functional Characterization of PDPs
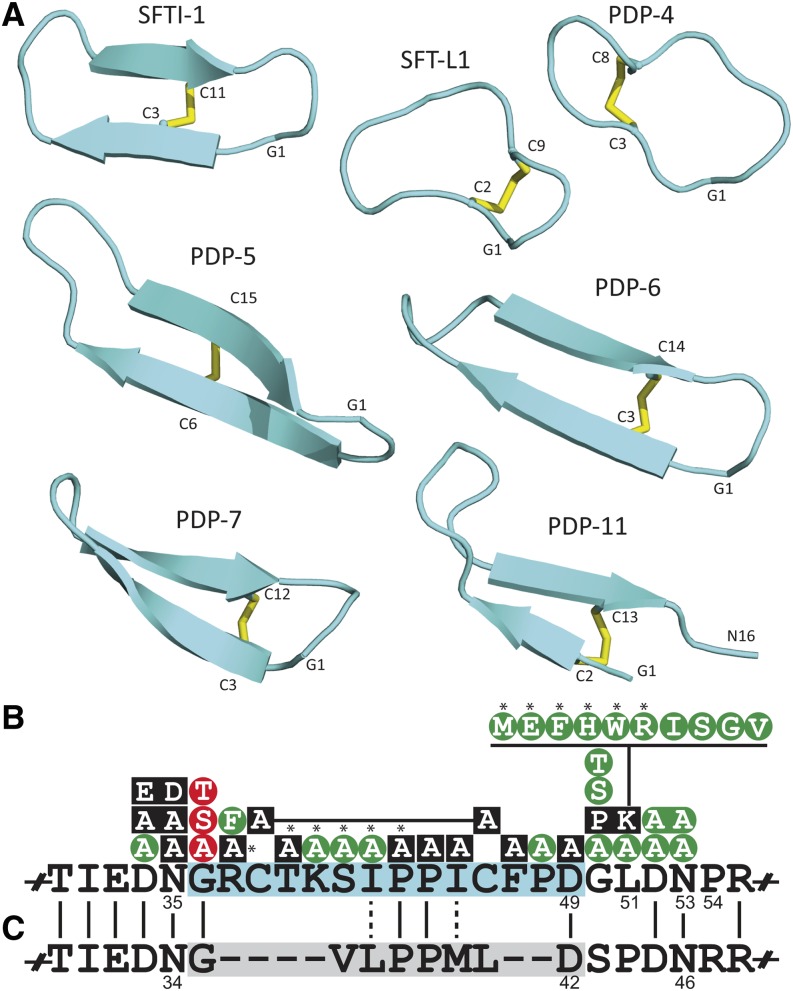
To establish whether these peptides structurally mimic the BBI inhibitory loop or have trypsin inhibitory activity, we chemically synthesized seven PDPs and studied them by NMR spectroscopy (Figure 3A; Supplemental Table 2). PDP-3 and PDP-12 were so similar to SFTI-1, based on chemical shift comparisons, that no structure was pursued (Supplemental Figure 15). In contrast, the other five PDPs varied in structure (Figure 3A; Supplemental Figures 16 to 18) as well as in physicochemical properties (Supplemental Table 3). PDP-4 stands apart, as it has no β-strands like most other PDPs. PDP-5, -6, -7, and -11 form β-hairpin motifs like SFTI-1, with two to four residues in each strand. PDP structures are primarily stabilized by hydrogen bonds and the disulfide bond, which “staples” the two strands together. The chemical shifts for Hα, Cα, and Cβ, which generally deviate significantly from random coil values, indicate that these new PDPs are highly ordered structures (Supplemental Figure 16). Common to most PDPs is a central Pro-Pro as well as an absolutely conserved Gly and Asp or Asn that, in all but the Galinsoga peptides, link to form the cyclic peptide backbone.
Figure 3.
Structures of PDPs and in Vivo Peptide-Processing Constraints for SFTI-1.
(A) Ribbon representation of NMR-derived structures for novel PDPs alongside sunflower SFTI-1 (Korsinczky et al., 2001) and SFT-L1 (Mylne et al., 2011).
(B) In vivo mutagenesis data from previous work (Mylne et al., 2011) and this study (asterisked) reveals which changes are tolerated biologically (green), perturb peptide processing (black), or specifically perturb the cyclization reaction (red).
(C) PawL1 from A. montana shares many of the critical residues for peptide maturation but lacks Cys residues and does not make a stable peptide.
The BBI-Mimicking PDPs Are Restricted to a Single Subtribe
Of the tested PDPs, only SFTI-1 and the subtle variants PDP-3 and PDP-12 inhibited trypsin (Supplemental Figures 15 and 19). Knowing this, we used mass and retention time to screen 267 Asteraceae seed peptide LC-MS profiles (237 from subfamily Asteroideae and 27 from other Asteraceae subfamilies) for evidence of these BBI loop-mimicking peptides (Supplemental Data Set 2). The species containing BBI loop-mimicking peptides were found only in genera closely related to Helianthus and, thus, specifically within subtribe Helianthinae (Tithonia, Simsia, Iostephane, Pappobolus, and Aldama) and not in subtribes Zinniinae, Ecliptinae, or the tribe Millereae that contain PDPs, but none that inhibit trypsin.
Based on PawS1 sequences and searching LC-MS profiles, we could only find BBI loop-mimicking trypsin inhibitory PDPs in subtribe Helianthinae. These findings are also functionally supported by an extensive series of in-gel trypsin inhibitory assays by Konarev et al. (2002), who found no evidence of low molecular weight trypsin inhibitors in two subfamilies of the Asteraceae and seven tribes of subfamily Asteroideae (Supplemental Table 4). In subfamily Asteroideae tribe Heliantheae, negatives were common (18 genera and 27 species), with the only two genera to test positive for low molecular weight trypsin inhibitors being Tithonia and Helianthus from subtribe Helianthinae (Konarev et al., 2002).
The PawS1 sequences, searches of LC-MS data, and work of Konarev et al. (2002) provide three layers of evidence that trypsin inhibitory PDPs are restricted to species from just one subtribe. As this is a ≥18-million-year-old peptide family that spans at least two Asteraceae tribes, this suggests the BBI-like sequence and functional fold evolved specifically within the lineage leading to sunflower.
SFTI-1 Blocks Insect Trypsins
All previous studies that show SFTI-1 to be a potent trypsin inhibitor have used bovine trypsin, bringing into question the relevance of its inhibitory function. The specificity of an inhibitor for a particular trypsin was first questioned by Feeney et al. (1969) using a comparison of Bowman-Birk Inhibitors with human and bovine trypsin, and this was followed by further studies of BBI inhibitory specificity discussed by Birk (1985). To ensure the inhibitory activity of SFTI-1 was of broader relevance than to just bovine trypsin, we tested SFTI-1 against insect whole mid gut extracts from cotton bollworm (Helicoverpa armigera) and used chromogenic substrates to monitor the ability of SFTI-1 to inhibit H. armigera trypsin activity and chymotrypsin activity (Supplemental Figure 20) in a similar manner to experiments with bovine β-trypsin (Supplemental Figure 19). We included as a negative control the sunflower cyclic peptide SFT-L1 that lacks Lys/Arg residues and was shown not to inhibit trypsin (Mylne et al., 2011). Additionally, we tested the cyclic peptide PDP-4 from I. heterophylla as it differed in structure from SFTI-1, but contains two Arg residues, and therefore had the potential to inhibit trypsin. We also included some highly potent inhibitors as positive controls; namely, the small chemical inhibitor leupeptin and a recombinant trypsin inhibitor originally isolated from Nicotiana alata flowers (rT4) (Atkinson et al., 1993) for the trypsin inhibition assay, and for the chymotrypsin assay we used the chemical inhibitor chymostatin and the recombinant chymotrypsin inhibitor (StPin1A) discovered in potato (Solanum tuberosum) leaves (Dunse et al., 2010). Of the PDPs, SFTI-1 inhibited H. armigera trypsin activity but required a higher concentration than the most potent known inhibitors leupeptin and rT4 (Supplemental Figure 20A). None of the PDPs inhibited H. armigera chymotrypsin activity (Supplemental Figure 20B). A comparison of SFTI-1 inhibition of H. armigera trypsins versus bovine trypsin was similar (Supplemental Figure 20C), confirming the inhibitory activity of SFTI-1 is not specific to bovine trypsin.
Within PawS1 Genes, Both the PDP and Albumin Regions Are Evolving Rapidly
To better understand the evolution of PDPs, we analyzed nucleotide sequence alignments of PawS1 genes and their encoded proteins. The PDP region appears hypervariable relative to the major subunits of the adjacent napin-type albumin (Supplemental Figure 5). These albumins typically vary greatly between species with the main conserved features being an ER targeting signal sequence, richness in Gln, and eight conserved Cys residues (Shewry, 1999). Sunflower contains at least six highly variable albumin-coding genes Ha-G5 (Allen et al., 1987), Ha-BA1, SUNFLOWER ALBUMIN8 (SFA8) (Kortt et al., 1991), and pHAO (Thoyts et al., 1996), along with PawS1 and PawS2 (Mylne et al., 2011). Genetic redundancy provided by large multigene families permits rapid evolution, an example being the great number of highly variable peptides found within venoms of predatory animals (Puillandre et al., 2010; Rokyta et al., 2011), but for the unique PawS1 gene structure, we saw very little or no genetic redundancy. Using our PCR approach, from any one species we typically cloned a single PawS1 gene, and we did not find additional PawS1 mRNA from a de novo–assembled transcriptome of sunflower (Supplemental Table 5 and Supplemental Data Set 7).
Without the complication of genetic redundancy, we analyzed the rate of synonymous to nonsynonymous change to test whether natural selection has acted upon PawS1 and detected residues evolving rapidly within the ER signal and albumin region (Supplemental Tables 6 and 7). However, this standard method collapses regions of alignments where gaps exist (including the PDP region), leading to underestimates of sequence divergence. We therefore studied the genetic insertion-deletion (indel) rates themselves. Estimating how fast insertions and deletions occurred and expanded in each PawS1 region indicated that the peptide region of PawS1 is evolving rapidly. The average insertion rate for the PDP region is at least twice as high as other regions of the protein (Supplemental Table 8).
Although storage proteins evolve rapidly (Kreis and Shewry, 1989), we found a 2-fold elevated insertion rate for the PDP region relative to the albumin region that suggests the peptide region is evolving faster than its adjacent storage protein (Supplemental Table 8). Another consideration when interpreting PDP rates of evolution is the critical processing residues in the peptide region and its flanks. By making additional PawS1 mutants for in vivo expression, we completed a full Ala scan of the entire SFTI-1 region (Figure 3B; Supplemental Figure 21). Specifically, when sunflower PawS1 is expressed in Arabidopsis seeds, mutation to Gly1, the disulfide bond, the central Pro-Pro, terminal Asp, as well as several other residues prevents production of stable cyclic peptide. The molecular evolutionary data and the deleterious effect of many in vivo mutations together indicate that both the albumin and PDP regions are evolving rapidly, but in different ways. Considering processing constraints, 2-fold higher evolution of the PDP region is exceptional. The PDP region has also been subject to internal tandem duplications, an example being PawS1 from Galinsoga, which encodes multiple peptides (Supplemental Figures 11 and 12).
A PawS-Like Gene That Makes No Peptide Represents a Potential Ancestor of PawS1
The BBI loop-mimicking PDPs from Helianthinae and the noninhibitory PDPs all share conserved features. These features begin with the residues preceding the PDP region and, in particular, the absolutely conserved Asn residue, which is the target of asparaginyl endopeptidase (Mylne et al., 2011). All PDP regions begin with a Gly residue, contain a disulfide pair, and end with either an Asp, or in the PawS1 genes from the Galinsoginae subtribe, an Asn. Most PDP regions also have a central Pro-Pro sequence. Trailing the PDP region is the conserved sequence Gly-Leu-Asp-Asn (GLDN), which itself precedes the small albumin subunit that often starts with a Pro. This conservation between peptides of different function but also the demonstration that the PDP region is evolving rapidly begs the question: Where did the PDP sequences come from if not a BBI? The PDP region causes a break in protein sequence alignments of seed storage albumin precursors that do not encode a peptide or possess the GLDN tail. This implies that the additional residues causing this break in alignment are encoded by DNA that appeared as one or more DNA insertions. If this is the case, we might expect to find such evolutionary intermediates between a typical albumin precursor and PawS1.
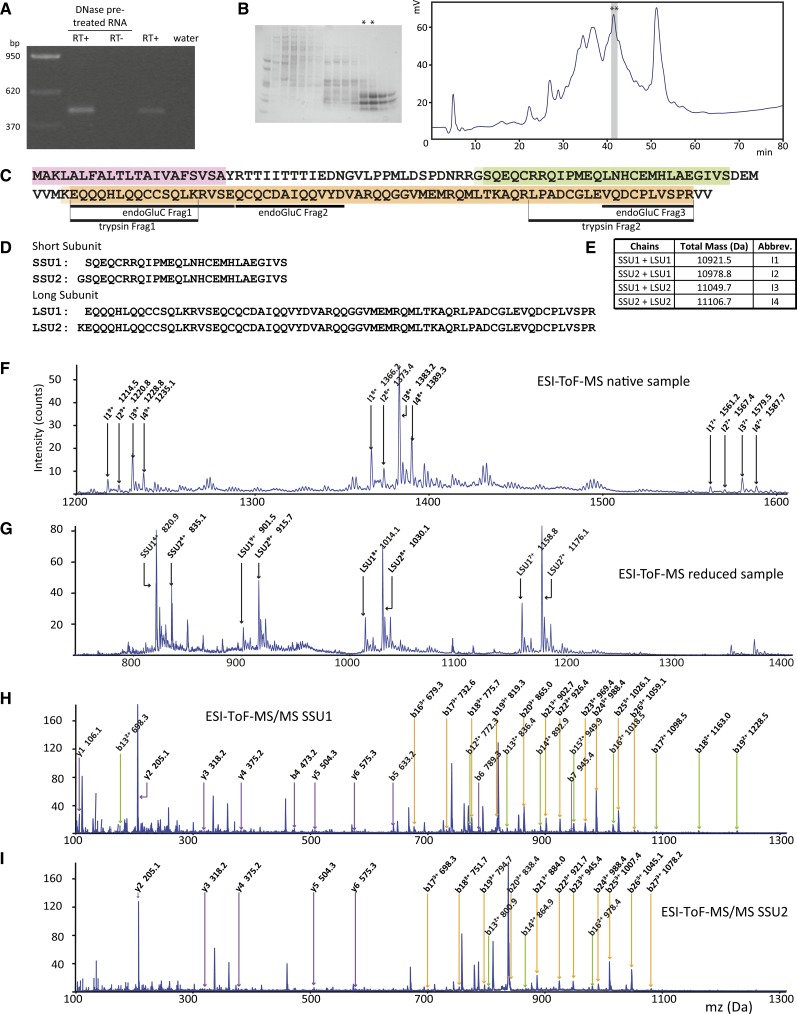
Using our PCR approach with DNA from Arnica montana, a species of the Madieae tribe, we amplified a product that when sequenced was found to encode a protein similar to PawS1, but its PDP region was only eight residues in length and had no Cys residues (Figure 3C). The protein encoded by this PawS-Like gene still had many features shared by PawS1 genes, including (1) the sequence preceding the PDP region including the absolutely conserved Asn; (2) the Gly residue at the proto-N terminus of PDPs; (3) the central Pro-Pro common to most PDPs; (4) the proto-C-terminal Asp conserved in most PDPs; and (5) the SSU-releasing Asp-Asn of the PawS1 GLDN tail. Because this PCR product was amplified from genomic DNA, we confirmed this gene, A. montana PawS-Like1 (PawL1), was expressed at the mRNA level (Figure 4A; Supplemental Data Set 8), and by targeted proteomics and MS/MS we detected mature, heterodimeric PawL1 albumin in its protein extracts (Figures 4B to 4I; Supplemental Tables 10 and 11). However, there was no evidence for masses that indicated a cyclic or linear GVLPPMLD peptide (Supplemental Figure 22). This suggests that despite sharing many features with PawS1 genes, PawL1 is matured into an albumin, but not a second stable peptide. Five other full-length PawL1 genes were found by the PCR approach in species from the Heliantheae and Millereae; therefore, PawL1 is widespread. We also found a partial PawL1 gene in the assembled de novo transcriptome of sunflower (Supplemental Data Set 7) and cloned the full-length transcript by 5′ and 3′ rapid amplification of cDNA ends (RACE). When aligned, PawS1 and PawL1 proteins grouped separately with differences at several regions clearly separating the two different types of gene, most noticeably the PDP region, but also in the ER signal and albumin subunits (Supplemental Figure 23). These data suggest PawL1 represents an independent biochemical sister lineage to typical albumin and peptide-making PawS genes. Based on the current data, the presence of PawL1 and PawS1 genes in Heliantheae and Millereae does not allow us to conclusively say which best represents the ancestral form.
Figure 4.
The Am-PawL1 Gene Is Transcribed, Translated, and Processed into Four Subtly Different Heterodimeric Albumins.
(A) RT-PCR confirmation of Am-PawL1 expression.
(B) Gel image of fast protein liquid chromatography separation of an albumin-rich extract from A. montana seeds. Two fast protein liquid chromatography fractions (asterisks) were further separated by HPLC, and two fractions at 41 min (asterisks) were identified by endo-GluC or trypsin digestion and MS/MS to contain Am-PawL1 mature albumin. For all MS/MS, see Supplemental Tables 10 and 11 for a complete list of ions.
(C) Endo-GluC and tryptic peptide fragments (underlined) mapped onto Am-PawL1. The predicted Am-PawL1 ER signal is highlighted in pink, the mature albumin SSU in green, and the mature albumin LSU in orange.
(D) Am-PawL1 SSU and LSU sequences.
(E) Expected masses for the four SSU/LSU combinations and the abbreviations for each.
(F) ESI-TOF-MS spectrum of the fractions containing native Am-PawL1 albumin. Ions corresponding to the expected masses for each conformation are indicated.
(G) ESI-TOF-MS spectrum of the reduced and alkylated fraction containing the Am-PawL1 albumin.
(H) ESI-TOF-MS/MS spectrum of the peak corresponding to SSU1 (820.9 D). 1+ (purple), 2+ (green), and 3+ (orange) b and y ions are indicated.
(I) ESI-TOF-MS/MS spectrum of the peak corresponding to SSU2 (835.1 D).
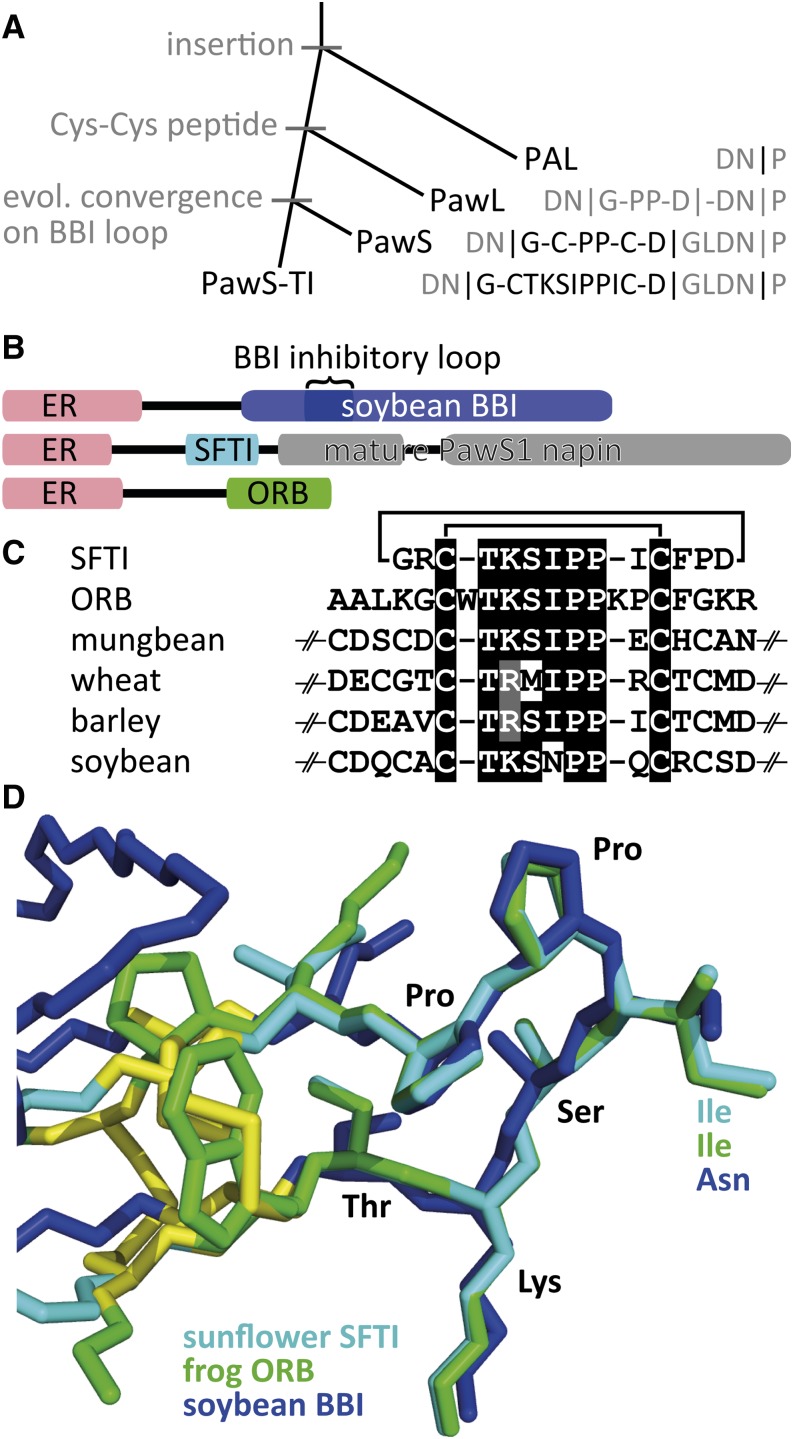
With the genes we cloned, the phylogenetic history of Asteraceae, and what we know of the distribution of PDPs, we hypothesize the following sequence of events for the appearance of the BBI inhibitory motif within PawS1 (Figure 5A). The ancestral protein is the widespread napin-type preproalbumin (PAL), which typically has a proalbumin region ending with an Asn residue that precedes the Pro cap of the albumin small subunit; Asn-Pro bonds being the typical target of cleavage for asparaginyl endopeptidase (Hara-Hishimura et al., 1993; Shimada et al., 2003; Gruis et al., 2004). We hypothesize an insertion event between this Asn and the small albumin subunit created the PawL1 gene that had many of the sequences later important for stable peptide production, asparaginyl endopeptidase targeting, and peptide cyclization. The evolution of a pair of Cys residues flanking both sides of the already present central Pro-Pro created the first PawS genes and stable (probably cyclic) peptide. Specifically in the lineage leading to sunflower, these stable cyclic peptides evolved one loop that we recognize as BBI’s trypsin inhibitory arm. More work is required to fully elucidate the sequence of genetic events that led to a separate peptide being produced within a seed storage albumin precursor. More broadly, the PawL and PawS genes and their products present an opportunity to study the de novo genesis of a peptide.
Figure 5.
Model for PawS1 Evolution and the Evolutionary Recurrence of the BBI Inhibitory Motif.
(A) A model proposed for the progressive evolution of the BBI mimicking peptide SFTI-1 from within a standard preproalbumin (PAL) with the key sequence changes shown on the right.
(B) Precursor proteins highlighting the location of the inhibitory loop within the mature BBI (XP_003533609), SFTI-1 and its adjacently matured napin, and the mature ORB peptide from amphibian frog skin (DQ672940). ER denotes the ER signal sequence.
(C) Sequences of SFTI-1, ORB, and four partial BBIs.
(D) Structural overlay of SFTI-1 (1SFI), frog ORB lacking two N-terminal and one C-terminal residues (2O9Q), and a BBI from soybean (1BBI).
DISCUSSION
Advances in the field of natural evolution of protein structure have come from bioinformatic analyses of structures in the Protein Data Bank that have documented the convergent evolution of domain architectures (Gough, 2005; Forslund and Sonnhammer, 2012), evolutionary divergence of fold (Grishin, 2001), and even offered a substitute for DNA based phylogenies (Yang and Bourne, 2009). Advancing from bioinformatic analyses of convergent evolution of protein sequence and structure (Russell, 1998; Copley et al., 2001; Lupas et al., 2001), we sought the evolutionary origin of one particular protein motif experimentally. While pursuing the evolutionary origin of SFTI-1, we discovered a novel peptide family, the PDPs, which like SFTI-1 are encoded within a precursor protein that also encodes a napin-type seed storage albumin. All the PDPs are <20 amino acids in length and possess a disulfide bond. Most of the PDPs, like SFTI-1, are backbone cyclized, but some, like those of Galinsoga, are acyclic. The phylogenetic distribution of the various PDPs in Millereae and Heliantheae plant families infers the peptide family to be at least 18 million years old. Despite the similarity shared by SFTI-1 and BBIs, SFTI-1 is just one member of a large peptide family, most of which do not mimic the BBI inhibitory loop or inhibit trypsin.
Before its biosynthetic origin was known, SFTI-1 was classed in the same protein family as BBIs based on structural similarity and shared function. The different biosynthetic origin and phylogenetically distinct position of sunflower relative to legumes and cereals are strong evidence that SFTI-1 evolved independently from BBIs. Why this sequence as well as the structure of this inhibitory loop is being favored by evolution is unknown, especially as a structural analysis of a range of co-complexes between trypsin and a peptidic inhibitor (Supplemental Figure 24) shows that there appears to be no sequence or structural constraints on trypsin inhibitors outside the three residues (P3-P1) known to be key to successful protease recognition (Tyndall et al., 2005). Nevertheless, the Bowman-Birk inhibitory fold has potentially evolved three times; within the skin of amphibious frogs there are peptides, such as ORB (Li et al., 2007), that share similar sequence and fold (Figures 5B and 5C), but only become potent trypsin inhibitors when truncated. The ORB precursor protein is unrelated to anything in plants and consists only of an ER signal sequence, a small pro-region, and the ORB peptide at its C-terminal end (Figure 5B). A structural overlay of ORB, SFTI-1, and BBIs (Figure 5C) is homologous over a six-residue stretch (TKSIPP) but is disrupted in its alignment by two residues in ORB either side of this region that extend the Cys-clamped loop. The longest stretches of similar sequence and structure said to have arisen by convergent evolution are no longer than eight residues and contain three to four conserved residues (Copley et al., 2001; Lupas et al., 2001; Ponting and Russell, 2002). Additional experiments that chart the evolution of the highly conserved CTKSIPPIC sequence could therefore confirm one of the most remarkable cases of convergent evolution on protein structure, sequence, and function.
In addition to pursuing the evolutionary origin of the Bowman-Birk inhibitory motif within SFTI-1, we sought to understand the origin of PawS1’s interstitial peptide. We discovered a novel and widespread gene family that is closely related to PawS1, the PawS-Like (PawL) genes that, based on evidence acquired for A. montana PawL1, are transcribed and translated into a precursor protein that is matured into albumin, but not an additional peptide. Like PawS1, when the predicted proteins encoded by PawL1 genes are aligned to other preproalbumins, they force a gap in the alignment in the same place as SFTI-1, suggesting it too has been subjected to substantial genetic insertion(s). This region in PawL1 genes also encodes many features essential for peptide maturation in PawS1 genes. Combined, these findings make the widespread PawL1 an appealing representative of an ancestral form of PawS1, but a more detailed picture of the distribution of PawS1 and PawL1 genes is needed to prove the genetic sequence of events that enabled a peptide to arise within an albumin precursor.
Although ubiquitous and abundant, little is known about the evolution of seed storage proteins. As seed storage proteins provide nutrition to embryos, their evolution is not constrained as it is for many other proteins where natural selection acts to maintain active sites, protein interaction motifs, or particular structural properties (Kreis and Shewry, 1989). PawS1 is not the only storage protein precursor to have extra peptides buried within it. In pumpkin (Cucurbita maxima), a precursor protein PV100 is matured into the seed storage protein vicilin as well as an array of buried peptides. One of these buried peptides, called C2, was shown to inhibit trypsin (Yamada et al., 1999). These examples draw attention to the ubiquitous storage proteins as potential “hotbeds” of evolution and a source of bioactive peptides. Most mRNA sequencing projects avoid profiling seeds as the transcripts in this tissue lack diversity as they are predominately storage proteins. For example, within the table of 1328 sequenced plant samples held by The 1000 Plants Initiative (www.onekp.com), only three of the samples list their tissue type as seeds, the remainder being mostly leaves or seedlings.
Evidence of relaxed selection for albumins includes the weak conservation of protein sequence among albumin precursors (Supplemental Figure 2), apart from the eight Cys residues and a general richness in Glu. Even within sunflower, there are highly variable albumin genes such as Ha-G5 (Allen et al., 1987), HaBA1 (GenBank AJ275962), pHAO (Thoyts et al., 1996), and SFA8 (Kortt et al., 1991). Our own attempts to find rapidly evolving and constrained sites in PawS1 were hampered by the high observed frequency of insertions and deletions. We directly calculated rates of insertion and deletion to measure rates of evolution throughout PawS1, which led us to hypothesize that relaxed selection on albumin sequence and redundancy for its albumin function provided by other albumin genes, allowed PawS1 to explore sequence space. The manner in which PawS1 has evolved a trypsin-inhibitory peptide for its PDP region within the Helianthinae subtribe implies natural selection has also played a role in the origin of the trypsin-inhibitory function. Seed protease inhibitors are thought to provide protection from gramnivores (Shewry, 1999), with some Ser protease inhibitors demonstrated to protect plants from insects (Dunse et al., 2010; Hartl et al., 2010). The age of the PDP family and the appearance of the protease inhibitory motif independently in plant and animal kingdoms suggests the trypsin-inhibitory biochemical function arose by convergent exploration of sequence space under the relaxed selection permitted by albumin.
PDP biosynthesis as a system to better understand the de novo evolution of a protein is appealing because (1) the system is highly evolvable with the albumin under relaxed selection and the peptide evolving rapidly and able to adopt variable sequences and structures; (2) this is a natural system and so permits the observation of the genesis of a peptide in nature; (3) rapid speciation particularly over the last 2 to 18 million years for the Asteraceae (Barreda et al., 2010) provides the appropriate raw material for comparative evolutionary studies; (4) both the peptide and albumin are abundant, either readily purified (albumin) or synthesized (PDP) and protein structures for both can be solved; and finally (5) the widespread PawL1 genes, which have almost all the hallmarks of PawS1 genes, but whose PDP region appears biologically unstable, tantalizingly suggest this particular protein sequence may have arisen in a stepwise series of genetic insertion events. In addition to its potential use in understanding the de novo evolution of a protein, the PDP biosynthesis also presents an opportunity to study the natural evolution of a novel biochemical function, namely, trypsin inhibition. The ability of PDPs to inhibit trypsin is only possessed by a subset of PDPs, whereas the majority of PDPs do not. The successful use here of de novo transcriptomics to find PawS1 and PawL1 genes without reliance on homology-based PCR approaches offers a viable route to test this model for both the de novo evolution of this peptide family within the daisy (Asteraceae) family and the evolutionary emergence of specific protease inhibition.
METHODS
Cloning of PawS1 Genes from the Daisy Family (Asteraceae)
The sunflower (Helianthus annuus) BAC library (Bouzidi et al., 2006) held by The French Plant Genomic Resource Centre (INRA, CNRGV, Toulouse) was screened by PCR of 4D pools for PawS1 and PawS2 containing BACs. A 400-bp region of PawS1 was amplified using JM204 and JM192 (Supplemental Table 9). A 373-bp region of PawS2 was amplified using JM218 and JM192. Three positives, 117O20, 122C14, and 217D9, were found to be PCR positive for both PawS1 and PawS2, suggesting both genes were chromosomal neighbors. Fingerprinting of these BACs by restriction enzymes suggested all three were overlapping BACs containing an identical region of genomic DNA.
To obtain the genomic DNA sequence surrounding the PawS genes, BAC 122C14 was digested with a suite of restriction enzymes, separated on a long agarose gel, chemically treated for DNA gel blot analysis, and transferred to Hybond N+ nitrocellulose membrane. The fragmented BAC was then probed with a 32P-labeled 430-bp PCR product amplified from the BAC 122C14 using JM262 and JM73 (Supplemental Table 9) that covers the conserved region of both genes. The ∼2.7-kb positive HindIII band was cloned and sequenced (GenBank accession number JX910422). PawS1 was confirmed within the 2564-bp sequence, and this region contained 1.3 kb of upstream and 0.8 kb of genomic sequence downstream from the PawS1 ORF. The ∼1.1-kb positive DraI band was also cloned and sequenced (GenBank accession number JX910423). PawS2 was confirmed within the 1082-bp sequence, and this region contained 310 bp of upstream and 364 bp of genomic sequence downstream from the PawS2 ORF.
Several dozen primers were designed to the PawS1 and PawS2 flanking regions and used in combination in heterologous PCR with the template being genomic DNA extracted from other Asteraceae (daisy family) species. One primer combination was especially successful at amplifying PawS genes from other Asteraceae species and these were AE51 and AE54 (Supplemental Table 9). AE51 binds 188 bp upstream of sunflower PawS2, whereas AE54 binds 141 bp downstream from the PawS2 stop codon and inside its 3′ untranslated region. The AE51-AE54 primer pair amplified PawS1 genes from genomic DNA from Asteraceae species previously used in peptide analysis and found negative for SFTI-1 and SFT-L1 (Mylne et al., 2011).
Genomic DNA for an additional 193 species of the Asteraceae was obtained by extracting samples collected during fieldwork in Tennessee and Texas in October, 2010, destructive sampling of Knoxville and Austin herbarium specimens, and by taking aliquots of DNA purified by J.L. Panero (University of Texas at Austin) with caesium chloride gradients after collection from Southern United States and Mexico. When DNA extraction was required, genomic DNA was purified using a DNeasy Plant Mini Kit (Qiagen).
The amplified PawS1 genes and their GenBank accession numbers are listed in Supplemental Table 2. From the gene sequence and prior knowledge of asparaginyl endopeptidase–mediated PawS processing, the sequence of their potential cyclic peptides was predicted. Using these masses the LC-MS data from these species reanalyzed to identify ions that were consistent in mass with the predicted peptides.
Sequence Alignment
All sequences were preliminarily aligned using Geneious v6.5.4 (Kearse et al., 2012) and adjusted manually based on the known domain structure of PawS1 from sunflower (Supplemental Figure 5). This alignment was used for the substitution and indel rate analyses.
Phylogenetic Tree and Chronogram
Divergence times of lineages were inferred from chloroplast sequence data assuming a relaxed molecular clock. The data matrix of 22 Heliantheae alliance and three outgroup taxa in tribes Athroismeae and Inuleae included 12 genes (accD, atpB, matK, ndhC, ndhD, ndhF, ndhI, ndhJ, ndhK, rbcL, rpoB, and rpoC1 exon 1) as well as the noncoding trnL intron and trnL-trnF spacer region (trnL and trnF). Sequences were aligned manually. As expected in Asteraceae, and flowering plants in general, these data do not conform to a strict molecular clock model. Our Bayesian statistical approach used a Metropolis-coupled Markov Chain Monte Carlo (MCMC) algorithm in BEAST version 1.7.4 (Drummond and Rambaut, 2007) to generate posterior distributions; this was performed at the CIPRES Science Gateway (www.phylo.org; Miller et al., 2010). A general time-reversible model with gamma-distributed rates among sites was specified (Tavaré, 1986), allowing four single-nucleotide substitution categories. The molecular rate was allowed to vary among lineages, around an average value, by enforcing an uncorrelated lognormal clock. A Yule tree was placed prior to modeling a constant lineage birth rate for each branch. A normal prior distribution was placed on the mean and 95% distribution for the most recent common ancestor between tribes Athroismeae and the other tribes of the Heliantheae alliance (29.8 confidence interval = 24 to 35) based on a chronogram for the family Asteraceae (Kim et al., 2005). The MCMC chain was run for 100 million posterior iterations sampling every 10,000 posterior iterations. Tracer 1.5 (http://tre.bio.ed.ac.uk/software/tracer) was used to assess effective sample sizes and tree likelihood stationarity. After the initial 10% of samples was discarded (burn-in), a maximum clade credibility tree was constructed in TreeAnnotator version 1.4.8 (Drummond and Rambaut, 2007), depicting the maximum sum of posterior clade probabilities. The maximum clade credibility tree was visualized in FigTree version 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree), and a time scale and bars for the 95% highest posterior densities for all branches with >0.90 posterior probabilities were added.
Peptide Extraction and Confirmation
Peptides were extracted by grinding seeds in liquid nitrogen and extracting in 0.4 mL methanol and 0.4 mL dichloromethane, with separation of the phases by the addition of 0.1 mL 0.05% trifluoroacetic acid in water before lyophilizing the supernatant. Peptide pellets were dissolved in 1% formic acid and run on LC-MS as described (Mylne et al., 2011). Ions matching calculated predictions from gene sequences were confirmed directly by purification and MS/MS sequencing (when sufficient material was available) or by comparison of its LC-MS and MS/MS properties to synthetic peptides (where plant material was limited). For MS/MS sequencing, the mass of interest was purified by reverse-phase HPLC and reduced, alkylated, and digested to form a linear product that could be sequenced via MALDI-TOF-MS/MS and/or ESI-TOF-MS/MS. For the comparative approach, the retention time of the mass of interest and its fragmentation were compared with those of the synthetic peptide.
NMR Structural Studies of PDPs
Peptides for structural studies were made as previously described (Mylne et al., 2011) or obtained from GL Biochem. Samples were prepared for 1H NMR measurements in 80% water, 10% D2O, and 10% deuterated DMSO (v/v) at pH 5.0 to 6.1 at 2 to 4 mg/mL for 1D, TOCSY, NOESY, and DQF-COSY experiments (one-dimensional, total correlation spectroscopy, nuclear Overhauser effect spectroscopy, and double-quantum filtered correlation spectroscopy, respectively). Additional samples were prepared in 90% D2O and 10% deuterated DMSO (v/v) at pH 5.0 to 6.1 at 2 to 4 mg/mL for deuterium exchange measurements (series of 1D and TOCSY), as well as ECOSY and 13C HSQC experiments. The spectra were recorded using methods outlined by Daly et al. (2006) and assigned using established techniques (Wüthrich, 1986). Data were collected at temperatures that were deemed optimal on a case by case basis and ranged from 280 to 298K. Additionally, for PDP-5, 20 mM sodium phosphate with 10% D2O was preferred over water for differentiating between conformations, and it was common to compare data sets with varied 2D mixing times 100/200/300 ms. The peptides ranged in mass from 1344.6 D (PDP-4, 12 residues) to 2237.0 D (PDP-5, 18 residues). Structural restraints that were derived from the data included interproton distances from NOESY cross peak intensities, backbone dihedral angles derived from TALOS+ (Shen et al., 2009) analysis of the chemical shifts, side-chain dihedral angles based on coupling constant and intraresidual NOE patterns (Wagner, 1990), and hydrogen bonds based on exchange data. Structures were calculated using CYANA (Güntert, 2004) and a stereochemical analysis performed by MolProbity (Chen et al., 2010) to assess the quality of the peptide geometry compared with previously published structures. The 20 lowest energy structures had good structural and energetic statistics in each case, with no residues falling in the disallowed regions of Ramachandran analysis and each peptide having a backbone root mean square deviation below 0.6 Å (Supplemental Table 2). The 1H and 13C random coil chemical shifts were plotted for each PDP and compared with published shifts (Wishart et al., 1995) to compare one PDP to another in relation to sheet formation and disulfide bond positioning (Supplemental Figure 16). PROMOTIF (Hutchinson and Thornton, 1996) was used to describe disulfide bond conformation and identify secondary structure, and the ExPASy ProtParam tool (Artimo et al., 2012) was used to calculate the physiochemical properties of the PDPs (Supplemental Table 3).
De Novo Transcriptomes
RNA was extracted from dehulled seeds of sunflower and whole seeds of Arnica montana using a hot phenol method scaled down from a method used by Botella et al. (1992) and which was fully described by Mylne et al. (2012). The total RNA solution was purified further using an RNA column (Machery-Nagel) and submitted to BGI Genomics for de novo transcriptome assembly. The total RNA solutions were enriched by BGI Genomics for poly(A) mRNA using oligo(dT) beads, and the mRNA was fragmented and used as a template for first-strand cDNA synthesis using random hexamers. The second-strand cDNA was synthesized, short fragments were purified with a QIAQuick PCR extraction kit (Qiagen), and the ends were repaired and then connected to sequencing adapters. After a final size selection, the library was sequenced using an Illumina HiSequation 2000. For sunflower, 40,742,686 clean paired-end reads containing a total of 3,666,841,740 clean nucleotides were obtained. These data could be assembled into 81,344 Unigenes with an average length of 649 bp. 49,077 Unigenes could be annotated using with the databases of NR, SwissProt, COG, KEGG, and GO. Searching for PawS genes in the de novo–assembled transcriptome, only PawS1 and PawS2 in sunflower were found (Supplemental Data Set 7). For A. montana, 27,516,042 clean paired-end reads containing a total of 2,476,443,780 clean nucleotides were obtained. These data could be assembled into 73,281 Unigenes (45,500 could be annotated) with an average length of 630 bp (Supplemental Data Set 8).
PawS1 Mutagenesis for in Planta Expression
Mutations were introduced into PawS1 by PCR, and after subcloning the mutant fragments into binary vectors, they were transformed into Arabidopsis thaliana as described (Mylne et al., 2011). Mutations were introduced by PCR to the PawS1 gene using PfuUltra polymerase (Agilent). The following primers were used to create mutant versions of PawS1: PawS1-L51E by JM508 and JM509; PawS1-L51M by JM510 and JM511; PawS1-L51R by JM512 and JM513; PawS1-L51W by JM514 and JM515; PawS1-L51H by JM516 and JM517; PawS1-L51F by JM518 and JM519; PawS1-R37A by JM520 and JM521; PawS1-T39A by JM522 and JM523; PawS1-K40A by JM524 and JM525; PawS1-S41A by JM526 and JM527; PawS1-I42A by JM528 and JM529; PawS1-P43A by JM530 and JM531 (Supplemental Table 9). Mutated versions of PawS1 were ligated with an OLEOSIN promoter into a binary vector conferring tetracycline (bacterial) and Basta herbicide (to plant) resistance that was moved from Escherichia coli into the LBA4404 strain of Agrobacterium tumefaciens by triparental mating and transformed by floral dip into Arabidopsis wild-type Columbia as described (Mylne et al., 2011). T2 transgenic Arabidopsis seed peptide extracts were prepared and analyzed by MALDI-TOF-MS as described (Mylne et al., 2011).
Bovine Trypsin Inhibitory Assays
Trypsin inhibitory assays were conducted using the chromogenic substrate (Nα-benzoyl-l-arginine 4-nitroanilide hydrochloride; Sigma-Aldrich) described by Erlanger et al. (1961) in a manner detailed by Mylne et al. (2011). The concentration range tested was 0.39 μM to 0.25 mM. The concentrations of PDP-5 and PDP-11 were calculated by UV absorbance at 280 nm. The concentrations of PDP-6 and PDP-7 were calculated by UV absorbance at 214 nm with their extinction coefficient calculated by the equation ε214 = 2846 × (nAA-1 + nN + nQ) + 7200 × nF + 6309 × nH + 22,735 × nW + 5755 × nY. For peptides for which an extinction coefficient could not be calculated (SFTI-1, SFT-L1, PDP-3, PDP-4, and PDP-12), the reverse-phase HPLC peak area was compared with a quantified, amino acid analyzed standard.
Insect Trypsin and Chymotrypsin Inhibitory Assays
Peptides (as quantified above) were incubated at various concentrations (range from 2.5 nM to 7.5 μM) with 5 μg of Helicoverpa armigera fourth instar total gut extract prepared as described by Stevens et al. (2013). Trypsin activity was measured using the commercially available substrate Benzoyl-FVR-pNA (Sigma-Aldrich), and chymotrypsin activity was measured using Succ-AAPFpNA (Sigma-Aldrich). All pipetting was automated using a Tecan liquid handling robot. Commercially available chemical protease inhibitors were used as comparative controls, including trypsin inhibitor leupeptin (Sigma-Aldrich) and the chymotrypsin inhibitor chymostatin (Roche Applied Sciences). Additional control recombinant protein inhibitors rT4 (trypsin inhibition) and StPin1A (chymotrypsin inhibition) were kindly provided by Hexima (Atkinson et al., 1993; Dunse et al., 2010). The extracts were preincubated for 30 min at 25°C in 50 mM CAPS buffer, pH 10.0, before the addition of substrate (1 mM) in a final volume of 100 μL. Substrate hydrolysis was measured at 405 nm using a Spectramax 250 plate reader (Molecular Devices). Inhibition curves were the result of measuring the percentage of remaining protease activity after incubation.
Indel Rate Analysis
A visual observation of the sequence alignment (Supplemental Figure 5) shows PawS1 to be rich in indels (insertions/deletions). While substitution rates are routinely measured and used to detect natural selection, little attention has been paid to the analogous use of indel rates. Here, ProtPal (Westesson et al., 2012) was used to calculate the indel rate observed within the PawS1 gene family. The species tree and inferred branch length with the maximum likelihood method implemented in Phylogenetic Analysis by Maximum Likelihood (PAML) were used (Supplemental Table 7). ProtPal is able to normalize indel rates using branch length information. Indel analysis results were supplied (Supplemental Table 8).
Substitution Analysis
To detect residues under positive selection, molecular evolutionary analyses were performed using different models implemented in PAML4 (Yang, 2007) to albumin genes (including the small peptide region) and different functional components of albumin (namely, ER region, SSU, LSU, the small peptide region, and albumin gene excluding the small peptide region). To detect sites under selection different site-specific models, which assume variable selective pressure across sites and over lineages, were used. To detect residues under positive selection, PAML4 (Yang, 2007) was used. PAML can calculate the proportion of sites with different dN/dS ratios defined under a variety of models. The following models were tested: Model M0, or a null model assuming single dN/dS ratio across all sites; Model M1a or the “neutral model,” assuming two categories of sites fixed with dN/dS ratio of either 0 or 1 and a third category of estimated dN/dS ratio; Model M2a, or the “positive selection model,” which allows an additional class of sites with positive selection signatures compared with model M1a; Model M3, or the “discrete model,” allowing dN/dS ratio for each site varying freely; Model M7, or the “beta model,” defining eight categories of site, with eight dN/dS ratios ranging from 0 to 1, drawn from a discrete approximation of the beta distribution; and Model M8, the “beta plus omega” model, which contains eight categories from M7 plus an additional one allowing sites with dN/dS that is free to vary from 0 to >1. To determine if particular models are better than alternative ones, likelihood ratio tests (−2[log likelihood ratio1 – log likelihood ratio2]) with critical values of χ2 distribution under a certain degree of freedom were performed (Yang, 1998). Probabilities of sites under positive selection were obtained using Bayesian approaches (Nielsen and Yang, 1998; Yang et al., 2005) implemented in PAML.
A. montana PawL1, Transcript Confirmation, and Characterization of PawL1 Albumin
To clone the genomic DNA sequence for PawL1, DNA was extracted from 100 mg of A. montana seeds (Jelitto) using the DNeasy Plant Mini Kit (Qiagen). A region of genomic DNA containing PawL1 was weakly amplified by PCR using primers AE51 and AE54. To confirm PawL1 is transcribed, total RNA was extracted from 100 mg of A. montana seeds as described (Mylne et al., 2012). Total RNA samples were pretreated with DNaseI as per the manufacturer’s instructions (New England Biolabs) before reverse transcription using SuperScript III (Invitrogen) and an oligo(dT)20 primer. The cDNA was used in PCR with the PawL1-specific primers CD3 that faces downstream and binds the start ATG and CD4 that faces upstream and binds to the stop codon (Supplemental Table 9). The amplified PawL1 ORF clones were sequenced to confirm the PawL1-coding sequence was correct. To confirm PawL1 is translated and produces mature albumin, 8 g of A. montana seeds was ground to a fine powder under liquid nitrogen. The seed meal was mixed with hexane and poured through filter paper (Whatman) to remove lipids and oils. The meal was washed twice with 50 mL hexane and left to air-dry on the filter paper. The dry meal was scraped from the filter paper, 70 mL water was added, and the mixture was sonicated for 1 min before centrifugation at 2600g for 10 min at 4°C. The clear supernatant was removed and centrifuged again. The supernatant was dialyzed against 5 liters of water overnight. The sample was lyophilized, and about half (70 mg) of the resulting crude albumin extract was dissolved in 5 mL 50 mM sodium phosphate buffer, pH 7.0, 150 mM sodium chloride, 1 mM DTT, and 5 mM EDTA and loaded onto a Sephadex 75 column for size exclusion chromatography. The albumin-rich fractions were separated by HPLC, searched for PawL1 albumin by targeted proteomics, and sequenced by MS/MS as described for PawS1 and PawS2 (Mylne et al., 2011).
Cloning Sunflower PawL1
Based on the sequence of a partial PawL1 gene in the de novo–assembled transcriptome of H. annuus, tentative gene-specific primers AJ1 and AJ3 were designed (Supplemental Table 9). One microgram of the same total RNA used for the sunflower de novo transcriptome was used to prepare 5′ and 3′ RACE ready cDNA using the SMARTer RACE cDNA amplification kit (Clontech). Primer AJ3 was used for 5′ RACE and AJ1 in 3′ RACE. The amplified fragments were cloned into the pGEM-T Easy vector (Promega) and sequenced. To clone full-length sunflower PawL1, the 5′ RACE-ready cDNA was used as the template for PCR with the primers AJ5, which binds the 3′ end of the Clontech UPM primer, and AJ6 (Supplemental Table 9), which binds the end of the sunflower PawL1 3′ untranslated region. The full-length product was cloned into pGEM-T Easy as above and sequenced.
Accession Numbers
The 25 PawS1 genes are GenBank IDs JX262717 to JX262741, sunflower PawS1 and PawS2 with flanking genomic DNA sequence JX910422 to JX910423, A. montana PawL1 JX518486, sunflower PawL1 KF574811, five other PawL1 genes KF574812 to KF574816, and Ha-BA1 AJ275962. The SFTI-1 Protein Data Bank ID is 1SFI. NMR data are available at Biological Magnetic Resonance Bank ID/PDB, 18643/2LWS (PDP-4), 18644/2LWT (PDP-5), 18646/2LWV (PDP-6), 18645/2LWU (PDP-7), and 18641/2LWQ (PDP-11). The raw transcriptomics data for sunflower and A. montana are being held at the National Center for Biotechnology Information under BioSample accession numbers SAMN02569067 and SAMN02569068, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Structural Similarity between SFTI-1 and the Inhibitory Arm of Bowman-Birk Inhibitors.
Supplemental Figure 2. A Multiple Sequence Alignment of PawS1 with Preproalbumins from Sunflower and Compared with Other Plant Species.
Supplemental Figure 3. In Planta Confirmation of PDP-3 in Seed Peptide Extracts of Tithonia rotundifolia.
Supplemental Figure 4. Fingerprinted Helianthus annuus Bacterial Artificial Chromosome 122C14 Using a Probe That Would Bind Both PawS1 and PawS2.
Supplemental Figure 5. Alignment of Predicted Protein Sequences for 27 PawS1 Genes Aligned against PawS1 from Helianthus annuus.
Supplemental Figure 6. Chronogram for 25 Genera of Asteraceae Subfamily Asteroideae.
Supplemental Figure 7. In Planta Confirmation of PDP-12 within Peptide Extracts from Helianthus schweinitzii Seeds.
Supplemental Figure 8. In Planta Confirmation of PDP-4 in Seed Peptide Extracts of Iostephane heterophylla seeds.
Supplemental Figure 9. In Planta Confirmation of PDP-5 within Peptide Extracts Made from Heliopsis helianthoides scabra Seeds.
Supplemental Figure 10. In Planta Confirmation of PDP-14 within Peptide Extracts Made from Heliopsis helianthoides scabra Seeds.
Supplemental Figure 11. In Planta Confirmation of PDP-10 within Peptide Extracts Made from Galinsoga quadriradiata Seeds.
Supplemental Figure 12. In Planta Confirmation of PDP-11 within Peptide Extracts Made from Galinsoga quadriradiata Seeds.
Supplemental Figure 13. In Planta Confirmation of PDP-8 within Peptide Extracts Made from Philactis zinnioides Seeds.
Supplemental Figure 14. In Planta Confirmation of PDP-8 within Peptide Extracts Made from Philactis nelsonii seeds.
Supplemental Figure 15. Structure and Activity Comparison for SFTI-1 and Variant Peptides.
Supplemental Figure 16. PDP Secondary Chemical Shifts.
Supplemental Figure 17. Stereo View of the Family of 20 Structures with Highest MOLPROBITY Score for Each PDP.
Supplemental Figure 18. Molecular Surface Representation and Comparison of PDPs.
Supplemental Figure 19. Trypsin Inhibitory Assays.
Supplemental Figure 20. Inhibition of Insect Proteases.
Supplemental Figure 21. In Vivo Processing of PawS1 Mutants.
Supplemental Figure 22. The Peptide GVLPPMLD Encoded by Am-PawL1 Is Not Detectable in Peptide Extracts of Arnica montana Seeds.
Supplemental Figure 23. Alignment of Predicted Protein Sequences for PawL1 and Selected PawS1 Genes.
Supplemental Figure 24. Structural Similarity between Trypsin Inhibitors from a Variety of Inhibitor Families.
Supplemental Table 1. Range of Predicted and Confirmed Peptides from PawS1 Genes.
Supplemental Table 2. NMR Structure Statistics.
Supplemental Table 3. PDP Physicochemical Properties.
Supplemental Table 4. Summary of Findings by Konarev et al. (2002) in-Gel Trypsin Inhibition Assays.
Supplemental Table 5. Output Statistics of Sequencing and Assembly Quality for the de Novo Transcriptomes of Helianthus annuus and Arnica montana.
Supplemental Table 6. Models and Output Statistics of Positive Selection Analysis.
Supplemental Table 7. Newick Trees Used in PAML Analyses of PawS1 Regions.
Supplemental Table 8. Exact Indel Rates and Extension Probabilities of the Different Albumin Protein Regions.
Supplemental Table 9. Primers Used in This Study.
Supplemental Table 10. ESI-TOF-MS/MS Product Ions for Endo-GluC and Trypsin Fragments That Correspond to Am-PawL1.
Supplemental Table 11. ESI-TOF-MS/MS Product Ions for the Am-PawL1 Small Subunit.
Supplemental Methods. Searching Heliantheae GenBank Entries for BBIs.
The following materials have been deposited in the DRYAD repository under accession number http://dx.doi.org/10.5061/dryad.j326j.
Supplemental Data Set 1. BBI Sequences Used to Generate BBI WebLogos.
Supplemental Data Set 2. Screening for SFTI-1 and PDPs.
Supplemental Data Set 3. Voucher Information and GenBank Accession Numbers for Sequences Used to Generate the Phylogeny in Figure 2D.
Supplemental Data Set 4. NEXUS Format Text File of the Sequences and Alignment Used to Generate the Phylogeny in Figure 2D.
Supplemental Data Set 5. Voucher Information and GenBank Accession Numbers for Sequences Used to Generate the Phylogeny in Supplemental Figure 6.
Supplemental Data Set 6. NEXUS Format Text File of the Sequences and Alignment Used to Generate the Phylogeny in Supplemental Figure 6.
Supplemental Data Set 7. FASTA Format of de Novo Seed Transcriptome of Helianthus annuus Translated into Protein Sequences from CDS Predicted on BLAST Results.
Supplemental Data Set 8. FASTA Format de Novo Seed Transcriptome of Arnica montana Translated into Protein Sequences from CDS Predicted on BLAST Results.
Supplementary Material
Acknowledgments
We thank A. Argyros for technical assistance and A. Jones for help with mass spectrometry. A.G.E. held an Australian Postgraduate Award Scholarship. K.J.R. was supported by a National Health and Medical Research Council (NHMRC) Career Development Award. D.O.-B. was supported by Australian Research Council (ARC) Grant DP0986175. D.J.C. is an NHMRC Professorial Fellow (569603). J.S.M. was supported by an ARC Queen Elizabeth II Fellowship (DP0879133) and is an ARC Future Fellow (FT120100013). This work was supported by ARC Grant DP12103369 to J.S.M. and K.J.R. as well as DP130101191 to J.S.M., J.L.P., and E.E.S.
AUTHOR CONTRIBUTIONS
J.S.M. designed research. A.G.E, C.D., H.L., Z.P., A.C., J.L.P, A.S.J., M.L.C., K.M.D., and J.S.M. performed research. A.G.E, C.D., H.L., Z.P., K.J.R., D.O.-B., J.L.P, M.L.C., and J.S.M. analyzed data. M.A.A., D.J.C., J.L.P., and E.E.S contributed new reagents/analytic tools. A.G.E., D.O.-B., and J.S.M. wrote the article with input from all authors.
Glossary
- ORF
open reading frame
- ER
endoplasmic reticulum
- LC-MS
liquid chromatography–mass spectrometry
- PDP
PawS-derived peptide
- MS/MS
tandem mass spectrometry
- RACE
rapid amplification of cDNA ends
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Allen R.D., Cohen E.A., Vonder Haar R.A., Adams C.A., Ma D.P., Nessler C.L., Thomas T.L. (1987). Sequence and expression of a gene encoding an albumin storage protein in sunflower. Mol. Gen. Genet. 210: 211–218. [DOI] [PubMed] [Google Scholar]
- Artimo P., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40: W597–W603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson A.H., Heath R.L., Simpson R.J., Clarke A.E., Anderson M.A. (1993). Proteinase inhibitors in Nicotiana alata stigmas are derived from a precursor protein which is processed into five homologous inhibitors. Plant Cell 5: 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreda V., Palazzesi L., Tellería M.C., Katinas L., Crisci J.V. (2010). Fossil pollen indicates an explosive radiation of basal Asteracean lineages and allied families during Oligocene and Miocene times in the Southern Hemisphere. Rev. Palaeobot. Palynol. 160: 102–110. [Google Scholar]
- Birk Y. (1985). The Bowman-Birk inhibitor. Trypsin- and chymotrypsin-inhibitor from soybeans. Int. J. Pept. Protein Res. 25: 113–131. [DOI] [PubMed] [Google Scholar]
- Bock R. (2010). The give-and-take of DNA: Horizontal gene transfer in plants. Trends Plant Sci. 15: 11–22. [DOI] [PubMed] [Google Scholar]
- Botella J.R., Arteca J.M., Schlagnhaufer C.D., Arteca R.N., Phillips A.T. (1992). Identification and characterization of a full-length cDNA encoding for an auxin-induced 1-aminocyclopropane-1-carboxylate synthase from etiolated mung bean hypocotyl segments and expression of its mRNA in response to indole-3-acetic acid. Plant Mol. Biol. 20: 425–436. [DOI] [PubMed] [Google Scholar]
- Bouzidi M.F., Franchel J., Tao Q., Stormo K., Mraz A., Nicolas P., Mouzeyar S. (2006). A sunflower BAC library suitable for PCR screening and physical mapping of targeted genomic regions. Theor. Appl. Genet. 113: 81–89. [DOI] [PubMed] [Google Scholar]
- Cai J., Zhao R., Jiang H., Wang W. (2008). De novo origination of a new protein-coding gene in Saccharomyces cerevisiae. Genetics 179: 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvunis A.-R., et al. (2012). Proto-genes and de novo gene birth. Nature 487: 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., III, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66: 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley R.R., Russell R.B., Ponting C.P. (2001). Sialidase-like Asp-boxes: Sequence-similar structures within different protein folds. Protein Sci. 10: 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly N.L., Clark R.J., Plan M.R., Craik D.J. (2006). Kalata B8, a novel antiviral circular protein, exhibits conformational flexibility in the cystine knot motif. Biochem. J. 393: 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C.C., Wurdack K.J. (2004). Host-to-parasite gene transfer in flowering plants: Phylogenetic evidence from Malpighiales. Science 305: 676–678. [DOI] [PubMed] [Google Scholar]
- Donoghue M.T., Keshavaiah C., Swamidatta S.H., Spillane C. (2011). Evolutionary origins of Brassicaceae specific genes in Arabidopsis thaliana. BMC Evol. Biol. 11: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J., Rambaut A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunse K.M., Stevens J.A., Lay F.T., Gaspar Y.M., Heath R.L., Anderson M.A. (2010). Coexpression of potato type I and II proteinase inhibitors gives cotton plants protection against insect damage in the field. Proc. Natl. Acad. Sci. USA 107: 15011–15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlanger B.F., Kokowsky N., Cohen W. (1961). The preparation and properties of two new chromogenic substrates of trypsin. Arch. Biochem. Biophys. 95: 271–278. [DOI] [PubMed] [Google Scholar]
- Feeney R.E., Means G.E., Bigler J.C. (1969). Inhibition of human trypsin, plasmin, and thrombin by naturally occurring inhibitors of proteolytic enzymes. J. Biol. Chem. 244: 1957–1960. [PubMed] [Google Scholar]
- Forslund, K., and Sonnhammer, E.L. (2012). Evolution of protein domain architectures. In Evolutionary Genomics, M. Anisimova, ed (Totowa, NJ: Humana Press), pp. 187–216. [DOI] [PubMed] [Google Scholar]
- Gough J. (2005). Convergent evolution of domain architectures (is rare). Bioinformatics 21: 1464–1471. [DOI] [PubMed] [Google Scholar]
- Grishin N.V. (2001). Fold change in evolution of protein structures. J. Struct. Biol. 134: 167–185. [DOI] [PubMed] [Google Scholar]
- Gruis D., Schulze J., Jung R. (2004). Storage protein accumulation in the absence of the vacuolar processing enzyme family of cysteine proteases. Plant Cell 16: 270–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güntert, P. (2004). Automated NMR structure calculation with CYANA. In Protein NMR Techniques, A.K. Downing, ed (Totowa, NJ: Humana Press), pp. 353–378. [DOI] [PubMed] [Google Scholar]
- Hara-Hishimura I., Takeuchi Y., Inoue K., Nishimura M. (1993). Vesicle transport and processing of the precursor to 2S albumin in pumpkin. Plant J. 4: 793–800. [DOI] [PubMed] [Google Scholar]
- Hartl M., Giri A.P., Kaur H., Baldwin I.T. (2010). Serine protease inhibitors specifically defend Solanum nigrum against generalist herbivores but do not influence plant growth and development. Plant Cell 22: 4158–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson E.G., Thornton J.M. (1996). PROMOTIF—A program to identify and analyze structural motifs in proteins. Protein Sci. 5: 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., et al. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P.J., Palmer J.D. (2008). Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 9: 605–618. [DOI] [PubMed] [Google Scholar]
- Kim K.-J., Choi K.-S., Jansen R.K. (2005). Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae). Mol. Biol. Evol. 22: 1783–1792. [DOI] [PubMed] [Google Scholar]
- Konarev A.V., Anisimova I.N., Gavrilova V.A., Vachrusheva T.E., Konechnaya G.Y., Lewis M., Shewry P.R. (2002). Serine proteinase inhibitors in the Compositae: Distribution, polymorphism and properties. Phytochemistry 59: 279–291. [DOI] [PubMed] [Google Scholar]
- Korsinczky M.L., Schirra H.J., Rosengren K.J., West J., Condie B.A., Otvos L., Anderson M.A., Craik D.J. (2001). Solution structures by 1H NMR of the novel cyclic trypsin inhibitor SFTI-1 from sunflower seeds and an acyclic permutant. J. Mol. Biol. 311: 579–591. [DOI] [PubMed] [Google Scholar]
- Kortt A.A., Caldwell J.B., Lilley G.G., Higgins T.J.V. (1991). Amino acid and cDNA sequences of a methionine-rich 2S protein from sunflower seed (Helianthus annuus L.). Eur. J. Biochem. 195: 329–334. [DOI] [PubMed] [Google Scholar]
- Kreis M., Shewry P.R. (1989). Unusual features of cereal seed protein structure and evolution. Bioessays 10: 201–207. [DOI] [PubMed] [Google Scholar]
- Lawn R.M., Schwartz K., Patthy L. (1997). Convergent evolution of apolipoprotein(a) in primates and hedgehog. Proc. Natl. Acad. Sci. USA 94: 11992–11997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M.T., Jones C.D., Kern A.D., Lindfors H.A., Begun D.J. (2006). Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc. Natl. Acad. Sci. USA 103: 9935–9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang C., Xu X., Wang J., Yu H., Lai R., Gong W. (2007). Trypsin inhibitory loop is an excellent lead structure to design serine protease inhibitors and antimicrobial peptides. FASEB J. 21: 2466–2473. [DOI] [PubMed] [Google Scholar]
- Luckett S., Garcia R.S., Barker J.J., Konarev A.V., Shewry P.R., Clarke A.R., Brady R.L. (1999). High-resolution structure of a potent, cyclic proteinase inhibitor from sunflower seeds. J. Mol. Biol. 290: 525–533. [DOI] [PubMed] [Google Scholar]
- Lupas A.N., Ponting C.P., Russell R.B. (2001). On the evolution of protein folds: Are similar motifs in different protein folds the result of convergence, insertion, or relics of an ancient peptide world? J. Struct. Biol. 134: 191–203. [DOI] [PubMed] [Google Scholar]
- Miller, M.A., Pfeiffer, W., and Schwartz, T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In 2010 Gateway Computing Environments Workshop (GCE 2010), (Institute of Electrical and Electronics Engineers), pp. 1–8. [Google Scholar]
- Mylne J.S., Chan L.Y., Chanson A.H., Daly N.L., Schaefer H., Bailey T.L., Nguyencong P., Cascales L., Craik D.J. (2012). Cyclic peptides arising by evolutionary parallelism via asparaginyl-endopeptidase-mediated biosynthesis. Plant Cell 24: 2765–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylne J.S., Colgrave M.L., Daly N.L., Chanson A.H., Elliott A.G., McCallum E.J., Jones A., Craik D.J. (2011). Albumins and their processing machinery are hijacked for cyclic peptides in sunflower. Nat. Chem. Biol. 7: 257–259. [DOI] [PubMed] [Google Scholar]
- Neme R., Tautz D. (2013). Phylogenetic patterns of emergence of new genes support a model of frequent de novo evolution. BMC Genomics 14: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Yang Z. (1998). Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148: 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. (1984). Birth of a unique enzyme from an alternative reading frame of the preexisted, internally repetitious coding sequence. Proc. Natl. Acad. Sci. USA 81: 2421–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panero, J.L. (2007). Compositae: Key to the tribes of the Heliantheae alliance. In Flowering Plants, Eudicots, Asterales, J.W. Kadereit and C. Jeffrey, eds (Berlin: Springer-Verlag), pp. 391–395. [Google Scholar]
- Pavesi A., Magiorkinis G., Karlin D.G. (2013). Viral proteins originated de novo by overprinting can be identified by codon usage: application to the “gene nursery” of Deltaretroviruses. PLOS Comput. Biol. 9: e1003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C.P., Russell R.R. (2002). The natural history of protein domains. Annu. Rev. Biophys. Biomol. Struct. 31: 45–71. [DOI] [PubMed] [Google Scholar]
- Puillandre N., Watkins M., Olivera B.M. (2010). Evolution of Conus peptide genes: Duplication and positive selection in the A-superfamily. J. Mol. Evol. 70: 190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey M., Ohno S., Pintor-Toro J.A., Llobell A., Benitez T. (1998). Unexpected homology between inducible cell wall protein QID74 of filamentous fungi and BR3 salivary protein of the insect Chironomus. Proc. Natl. Acad. Sci. USA 95: 6212–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice D.W., Alverson A.J., Richardson A.O., Young G.J., Sanchez-Puerta M.V., Munzinger J., Barry K., Boore J.L., Zhang Y., dePamphilis C.W., Knox E.B., Palmer J.D. (2013). Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 342: 1468–1473. [DOI] [PubMed] [Google Scholar]
- Robson P., Wright G.M., Youson J.H., Keeley F.W. (2000). The structure and organization of lamprin genes: Multiple-copy genes with alternative splicing and convergent evolution with insect structural proteins. Mol. Biol. Evol. 17: 1739–1752. [DOI] [PubMed] [Google Scholar]
- Rokyta D.R., Wray K.P., Lemmon A.R., Lemmon E.M., Caudle S.B. (2011). A high-throughput venom-gland transcriptome for the Eastern Diamondback Rattlesnake (Crotalus adamanteus) and evidence for pervasive positive selection across toxin classes. Toxicon 57: 657–671. [DOI] [PubMed] [Google Scholar]
- Russell R.B. (1998). Detection of protein three-dimensional side-chain patterns: New examples of convergent evolution. J. Mol. Biol. 279: 1211–1227. [DOI] [PubMed] [Google Scholar]
- Shen Y., Delaglio F., Cornilescu G., Bax A. (2009). TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44: 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry, P. (1999). Enzyme inhibitors of seeds: Types and properties. In Seed Proteins, P. Shewry and R. Casey, eds (Dordrecht, The Netherlands: Kluwer), pp. 587–615. [Google Scholar]
- Shewry, P., and Pandya, M. (1999). The 2S albumin storage proteins. In Seed Proteins, P. Shewry and R. Casey, eds (Dordrecht, The Netherlands: Kluwer), pp. 563–586. [Google Scholar]
- Shimada T., et al. (2003). Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J. Biol. Chem. 278: 32292–32299. [DOI] [PubMed] [Google Scholar]
- Stevens J.A., Dunse K.M., Guarino R.F., Barbeta B.L., Evans S.C., West J.A., Anderson M.A. (2013). The impact of ingested potato type II inhibitors on the production of the major serine proteases in the gut of Helicoverpa armigera. Insect Biochem. Mol. Biol. 43: 197–208. [DOI] [PubMed] [Google Scholar]
- Tautz D., Domazet-Lošo T. (2011). The evolutionary origin of orphan genes. Nat. Rev. Genet. 12: 692–702. [DOI] [PubMed] [Google Scholar]
- Tavaré S. (1986). Some probabilistic and statistical problems on the analysis of DNA sequences. Lect. Math Life Sci. 17: 57–86. [Google Scholar]
- Tekaia F., Yeramian E. (2006). Evolution of proteomes: fundamental signatures and global trends in amino acid compositions. BMC Genomics 7: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoyts P.J.E., Napier J.A., Millichip M., Stobart A.K., Griffiths W.T., Tatham A.S., Shewry P.R. (1996). Characterization of a sunflower seed albumin which associates with oil bodies. Plant Sci. 118: 119–125. [Google Scholar]
- Tyndall J.D.A., Nall T., Fairlie D.P. (2005). Proteases universally recognize beta strands in their active sites. Chem. Rev. 105: 973–999. [DOI] [PubMed] [Google Scholar]
- Wagner G. (1990). NMR investigations of protein structure. Prog. Nucl. Magn. Reson. Spectrosc. 22: 101–139. [Google Scholar]
- Westesson O., Lunter G., Paten B., Holmes I. (2012). Accurate reconstruction of insertion-deletion histories by statistical phylogenetics. PLoS ONE 7: e34572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S., Bigam C.G., Holm A., Hodges R.S., Sykes B.D. (1995). 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR 5: 67–81. [DOI] [PubMed] [Google Scholar]
- Won H., Renner S.S. (2003). Horizontal gene transfer from flowering plants to Gnetum. Proc. Natl. Acad. Sci. USA 100: 10824–10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D.-D., Irwin D.M., Zhang Y.-P. (2011). De novo origin of human protein-coding genes. PLoS Genet. 7: e1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich, K. (1986). NMR of Proteins and Nucleic Acids. (New York: Wiley-Interscience). [Google Scholar]
- Yamada K., Shimada T., Kondo M., Nishimura M., Hara-Nishimura I. (1999). Multiple functional proteins are produced by cleaving Asn-Gln bonds of a single precursor by vacuolar processing enzyme. J. Biol. Chem. 274: 2563–2570. [DOI] [PubMed] [Google Scholar]
- Yang S., Bourne P.E. (2009). The evolutionary history of protein domains viewed by species phylogeny. PLoS ONE 4: e8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (1998). Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15: 568–573. [DOI] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z., Huang J. (2011). De novo origin of new genes with introns in Plasmodium vivax. FEBS Lett. 585: 641–644. [DOI] [PubMed] [Google Scholar]
- Yang Z., Wong W.S.W., Nielsen R. (2005). Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22: 1107–1118. [DOI] [PubMed] [Google Scholar]
- Yoshida S., Maruyama S., Nozaki H., Shirasu K. (2010). Horizontal gene transfer by the parasitic plant Striga hermonthica. Science 328: 1128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.