Abstract
Coronary heart disease (CHD) is a complex and heterogeneous cardiovascular disease. There are many genome-wide association studies (GWAS) performed worldwide to extract the causative genetic factors. Moreover, each population may have some exceptional genetic characteristic. Thus, the background of our study is from the previous Lithuanian studies (the LiVicordia Project), which demonstrated the differences of the atherosclerosis process between Lithuanian and Swedish male individuals.
In this study we performed GWAS of 32 families of Lithuanian origin in search of significant candidate genetic markers [single nucleotide polymorphisms (SNPs)] of CHD in this population. After careful clinical and biochemical phenotype evaluation, the ∼770K SNPs genotyping (Illumina HumanOmniExpress-12 v1.0 array) and familial GWAS analyses were performed.
Twelve SNPs were found to be significantly associated with the CHD phenotype (p value <0.0001; the power >0.65). The odds ratio (OR) values were calculated. Two SNPs (rs17046570 in the RTN4 gene and rs11743737 in the FBXL17 gene) stood out and may prove to be important genetic factors for CHD risk. Our results correspond with the findings in other studies, and these two SNPs may be the susceptibility loci for CHD.
Keywords: Atherosclerosis, Coronary heart disease (CHD), Genome-wide association study(ies) (GWAS), Transmission disequilibrium test (TDT)
INTRODUCTION
Coronary heart disease (CHD), also called coronary artery disease, is a complex and heterogeneous cardiovascular disease (CVD). It belongs to a group of atherosclerotic CVD that is defined as a chronic disorder which develops insidiously throughout life and usually progresses to an advanced stage by the time symptoms occur [1]. The critical underlying process of pathogenesis is atherosclerosis (AS) that, in itself, is a multifactorial and peculiar condition. There are a number of known controllable and uncontrollable factors, one of the last-mentioned is genetic, named as strong family history of premature CVD [2]. There are many genome-wide association studies (GWAS) performed worldwide to determine the main genetic factors that could be used for CVD identification and creation of useful tests for effective diagnosis, prognosis and treatment. Regrettably, the genetic factors and their importance are not yet sufficiently applied in clinical practice [1]. Moreover each population may have some exceptional genetic characteristic that does not necessarily correspond with results from other studies.
The background of our study is from the previous Linkoping-Vilnius CHD risk assessment study [3], which demonstrated the differences of atherosclerotic process between Lithuanian and Swedish male individuals. Subsequently, other study aimed to identify potential genetic markers associated with AS and CHD in the Lithuanian population [4]. The results lacked significant values for strong association of single nucleotide polymorphisms (SNPs) and disease. Novel genotyping techniques and platforms provided an improved opportunity for a more precise analysis of whole genome variation associated with human complex diseases. Thus, in this study we performed the GWAS in 32 families of Lithuanian ethnicity in search of significant genetic markers (SNPs) of CHD that may elucidate the underlying specificity of AS in this population.
MATERIALS AND METHODS
All study protocols were approved by the Vilnius Regional Biomedical Research Ethics Committee (No. 158200-11-255-067LP2; 2010-11-05). Informed consent was obtained from all individuals who participated in the study.
Individuals and Phenotype Definition
According to the Department of Statistics of Lithuania, in 2011, the total number of inhabitants was 3,030,200, and 17.3% of these were of non Lithuanian ethnicity. Considering the demographics and the fact that trios with CHD patients are generally rare, in total we collected 32 relevant families (trios of premature CHD patients and their parents) from different regions throughout Lithuania, i.e., 96 individuals in total. The patient group was represented by 31 males and only one female. Thus, conclusions from our study are suitable to extrapolate only for the male population.
Patients were clinically examined at the Clinic of Cardiology and Angiology, Faculty of Medicine, Vilnius University, Vilnius, Lithuania. The patients’ clinical phenotype was evaluated after assessment of anthropometrical measurements, clinical and instrumental examination, and laboratory biochemical testing. Information about CHD risk factors, other diseases and treatment was obtained during the conventional anamnesis.
Patient recruitment criteria was as follows: men aged 35–55, women aged 35–65; individuals who were experiencing acute coronary syndrome for the first time in their lives, who were hospitalized at an intensive cardiology unit for myocardial infarction (MI) with or without Q wave or unstable angina pectoris (confirmed by examination of common electrocardiographic and/or coronographic changes, assessment of cardio-specific markers); any previous or current evidence of significant atherosclerotic CHD (MI, percutaneous coronary angioplasty, coronary artery bypass graft or coronary angiography with hemo-dynamically significant stenosis). Anatomical vascular changes were confirmed by non invasive methods, evaluating the presence of the atherosclerotic plaques as well as examining the arterial stiffness and endothelial function. Patient exclusion criteria was as follows: diabetes treated with insulin, kidney function deficiency, III–IV functional class of heart deficiency, tumors (except skin basalioma), alcoholism and other social factors that may influence the study results.
Metabolic syndrome was diagnosed by determining three or more criteria described in the NCEP ATP III program [5]: waist circumference in men >102 cm, in women >88 cm, triglycerides ≥1.7 mmol/L, high-density lipoprotein cholesterol (HDL-C) <1.0 in men and <1.2 in women, blood pressure ≥130/85 mm Hg, fasting glucose ≥5.6 mmol/L or type 2 diabetes.
Evaluation of biochemical phenotype included inflammatory and metabolic markers, participating in the patho-genesis of atherosclerosis: C-reactive protein (CRP), Hb A1c, lipoprotein [Lp(a)], apolipoprotein A1 (ApoA1), apolipoprotein B (ApoB), ratio ApoB/A1, lipids, oxidized low-density lipoprotein (oxLDL) homocysteine, interleukin-6 (IL-6), fasting glucose in plasma, potassium (K), sodium (Na), urea, creatinin.
Genotyping
The DNA samples were extracted from the peripheral venous blood using phenol-chloroform DNA isolation or TECAN Freedom EVO® platform (TECAN Group Ltd., Männedorf, Switzerland) protocols. All DNA samples were genotyped according to the Illumina® provided protocols on Illumina HiScanSQ™ genetic analysis platform using an Illumina HumanOmni Express-12 v1.0 array comprised of ∼770K SNP markers (Illumina Inc., San Diego, CA, USA).
The primary genotyping results were visualized, inspected and prepared for further analysis by using the GenomeStudio software (Illumina Inc.). Subsequent data quality control, filtering and analysis were performed by using the PLINK v1.07 software [6] integrated in to the BC|Gene platform (Biocomputing Platforms Ltd., Espoo, Finland).
Statistical Analysis
The transmission-disequilibrium test (TDT) was used to determine the association between SNPs and CHD phenotype using the McNemar test [7]. Single nucleotide polymorphisms included in the TDT analysis met the following criteria: 1) minor allele frequency (MAF) greater than 0.01 (MAF >0.01); 2) missingness rate smaller than 0.1 (GENO <0.1); 3) the p value (<0.0005) of the Hardy-Weinberg equilibrium test; also if the subjects’ missing genotype rate was lower than 0.05 (MIND <0.05). For the computation of the empirical power, the frequency of the informative transmission of disease alleles and the number of informative transmissions of marker alleles was used. The binomial distribution was used for the approximation of the TDT power [8]. The 106 random variables distributed according to the TDT statistics under alternative hypothesis were generated. Odds ratio (OR) with 95% confidence interval (CI 95%) was calculated for selected SNPs. The level of significance was set at a = 10−4. For multiple testing, the correction adaptive permutation procedure was used. Maximum number of permutations was 106. The statistical analysis was performed with the program R (v2.15.3) (R Foundation for Statistical Computing, Vienna, Austria).
RESULTS
According to the Illumina Inc. protocol guidelines, all of the samples except one, were of good quality and had been properly processed (call rate >98; LogRDev <0.3; coincidental sex list file created). At the beginning of the analysis there were 731,412 SNPs genotyped in the group of 96 individuals. After the data filtering procedure, two individuals were removed from further analysis for low genotyping (MIND >0.05); 25,293 heterozygous genotypes were excluded from analysis because the second allele of the genotype was missing; 298 SNPs were excluded based on the Hardy-Weinberg equilibrium test (p >0.0005); 2528 SNPs failed missingness test (GENO >0.1); 82,552 SNPs failed frequency test (MAF <0.01); 591 SNP were not used because of homogeneity over all individuals. After the final frequency and genotyping pruning, 646,445 SNPs in 31 patient and 63 parents were included for further association analysis.
Twelve SNPs were found to be significantly associated with CHD phenotype with p values smaller than 0.0001. The SNPs annotation (transmitted allele, chromosomal position, gene, gene function) along with the χ2, p value, OR and empirical power based on the sample size calculations, are presented in Table 1. The SNPs are annotated according to the National Center for Biotechnology Information (NCBI) dbSNP and Gene databases [9].
Table 1.
Statistically significant single nucleotide polymorphisms associated with coronary hearth disease.
| Chromosome | SNP | Transmitted Allele | Position | Gene | Function | χ2 | p Value | OR (95% CI) | Power |
|---|---|---|---|---|---|---|---|---|---|
| 2p16.1 | rs17046570 | A | 55220584 | RTN4 | neuroendocrinous secretion; apoptosis processes | 15.21 | 9.62×10−5 | 18 (2.4–134.8) | 0.74 |
| 3p23 | rs294314 | A | 31067452 | – | – | 15.38 | 8.77×10−5 | 0.13 (0.04–0.43) | 0.65 |
| 5q31.1 | rs1346440 | G | 134573007 | LOC340073; LOC100996485 | uncharacterized | 18.24 | 1.946×10−5 | 0.12 (0.03–0.38) | 0.82 |
| 5q31.1 | rs2019973 | G | 134573992 | LOC340073; LOC100996485 | uncharacterized | 16.33 | 5.312×10−5 | 0.13 (0.04–0.42) | 0.68 |
| 5q21.3 | rs11743737 | A | 107263580 | FBXL17 | SCF complex; protein ubiquitination | 15.38 | 8.77×10−5 | 7.67 (2.30–25.53) | 0.65 |
| 9q22.23 | rs10819695 | G | 98762901 | – | – | 15.70 | 7.439×10−5 | 0.10 (0.02–0.41 | 0.69 |
| 12p11.23 | rs11048567 | A | 26689621 | ITPR2 | intracellular Ca2+ relaxation | 17.19 | 3.38×10−5 | 0.05 (0.01–0.37 | 0.81 |
| 20q11.21 | rs6141273 | A | 29904377 | – | – | 16.20 | 5.70×10−5 | 0.05 (0.01–0.39) | 0.77 |
| 20p12 | rs1321936 | G | 12826999 | – | – | 16.03 | 6.23×10−5 | 0.18 (0.07–0.46 | 0.64 |
| Possible Artefacts | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1q32.1 | rs12734338 | G | 202469723 | PPP1R12B | regulation myosin phosphatase activity; augments Ca2+ sensitivity of the contractile apparatus | 26.13 | 3.19×10−7 | 29.0 (3.95–212.9) | 0.997 |
| 4q25 | rs3853444 | G | 111734136 | – | – | 20.83 | 5.023×10−6 | 0.13 (0.05–0.37) | 0.880 |
| 15q25.2 | rs3883013 | G | 85088657 | UBE2Q2P1 | psuedogene | 26.13 | 3.19×10−7 | 29.0 (3.95–212.9) | 0.997 |
Ca2+: calcium ion. Out of 12 significant SNPs, at least two (italicized and bold rows) had promising OR values in addition to power and p values.
The acceptable power values were greater than or equal to 0.65, and thus fell partly into the desired range between 0.8 and 0.95 [10]. Only the power value of the significant SNP rs1321936 diverged and was excluded from further evaluation.
The OR values in Table 1 show the size of the effect. The greater the deviation of OR is from the value of 1, the more significant the test is.
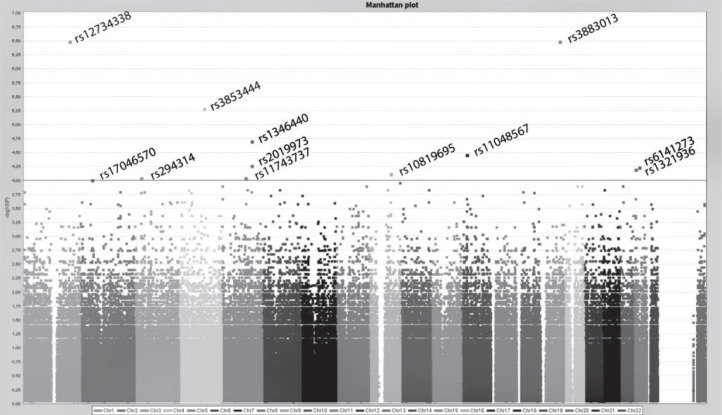
As can be seen from the Manhattan plot (Figure 1), there are three significant markers (rs12734338, rs3883013, rs3853444) that do not have the significant adjacent SNPs (according to the nucleotide’s position), i.e., the correlation is absent. Thus, these markers could be artefacts. We could also suspect that not all of the adjacent SNPs were genotyped. This is more likely to happen with rs3853444, as there were two adjacent SNPs that were excluded from the analysis.
Figure 1.
Manhattan plot. Single nucleotide polymorphism distribution with logarithmic transformation of the p value. The horizontal line depicts the selected significance level (α = 0.0001). The x axis represents the SNPs’ positions according to each chromosome, the y axis shows −log10(P) values of the SNPs.
It was previously mentioned that the study group included only male patients and their parents. A male could possess the SNP allele on either his X or Y chromosome and this affects the analysis algorithm. Each transmission from a heterozygous mother to a male offspring should be given twice the weight of a transmission to a female offspring [11]. Thus, the standard TDT appears to be unsuitable for the analysis of SNPs in sex chromosomes and eventually sex chromosomes were excluded from the analysis.
DISCUSSION
Our aim was to identify the particular genetic factors for Lihuanian CHD patients. In order to avoid population stratification we planned the familial GWAS and subsequently performed TDT analysis. Out of 12 significant SNPs, at least two (italicized and bold rows in Table 1) had promising OR values in addition to power and p values. Despite relatively wide OR intervals, caused by a modest sample size, these SNPs may indicate the potential genes that could be involved in CHD pathogenesis.
The SNP rs17046570 is located in an intron of the reticulon 4 coding gene (RTN4) on chromosome 2. Retic-ulons are associated with endoplasmic reticulum, and are involved in neuroendocrine secretion or membrane trafficking and apoptotic processes. In particular, reticulon 4 has been identified as a potential inhibitor of central nervous system regeneration by means of the inhibition of neuron outgrowth [11,12]. Common RTN4 variants that are associated with schizophrenia in the Japanese population [13] and also blood lipid phenotypes [14], are cited. RTN4 is a candidate gene associated with vascular cell apoptosis and AS modulation [12]. It is thought to participate in vascular remodeling and is a considerable new factor for atherogenesis process [15–17]. It was also stated that reticulons may be factors that mediate between the apoptosis and AS processes [12]. Thus, our results are consistent with these findings.
Another SNP on chromosome 5, rs11743737, is located in an intron of the F-box and leucine-rich repeat protein 17 coding gene FBXL17. The FBXL17 protein has an F-box that is a 40 amino acid motif typical for F-box containing proteins. F-box containing proteins together with culin and SKP1 (S-phase kinase-associated protein 1) make up the SCF complex (SKP1, cullin, F-box containing complex) that is a protein ubiquitine ligase [18]. The SCF is a key complex in the ubiquitine-proteosome system (UPS) that is involved in 70.0–90.0% of protein degradation processes including the degradation of a number of proteins important for the cardiovascular system. The UPS is also important in the regulation of endothelial cell cycle. The effect of oxidative stress on the SCF complex may disrupt the function of UPS and in turn the function of the endothelium that is regulated the by UPS [19]. According to the NCBI Gene database review of association results [from National Human Genome Research Institute (NHGRI) Catalogue and association results submitted to the database of Genotypes and Phenotypes (dbGaP)] there are many SNPs in the FBXL17 gene region associated with the various phenotypes including cholesterol, high-density lipoproteins, body mass index. These findings do not compromise our findings either. Moreover, these summarized results might show us the complexity and universality of the FBXL17 protein function in the pathogenesis of different diseases. It is possible that other SNPs that are in linkage disequilibrium with the identified CHD associated SNPs were not identified during this TDT analysis but may also be involved in the development of the disease.
Conclusions
Our results suggest that the RTN4 and FBXL17 genes may be the susceptibility loci for the CHD in the Lithuanian male population. In addition, the genotypes of the significantly associated SNPs with OR >7 may prove to be informative and specific for the genetic risk of CHD evaluation in the Lithuanian population and could be taken under consideration in further hypothesis validation.
Acknowledgments
We thank Professor A. Metspalu and his colleagues from Tartu University Institute of Molecular and Cell Biology, Tartu, Estonia, for their assistance.
Footnotes
Declaration of Interest: This study was partly supported by the Research Council of Lithuania (LIG-21/2010). The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
REFERENCES
- 1.Perk J, De Backer G, Gohlke H, Graham I, Reiner Z, Verschuren WM, et al. European guidelines on cardiovascular disease prevention in clinical practice (version 2012). The Fifth Joint Task Force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice (constituted by representatives of nine societies and by invited experts) Eur Heart J. 2012;33(13):1635–1701. doi: 10.1093/eurheartj/ehs092. [DOI] [PubMed] [Google Scholar]
- 2. National Heart Lung and Blood Institute ( http://www.nhlbi.nih.gov/health/health-topics/topics/atherosclerosis/) (accessed 3 April 2013).
- 3.Kristenson M, Olsson AG, Kucinskiene Z. Good self-rated health is related to psychosocial resources and a strong cortisol response to acute stress: the LiVicordia study of middle-aged men. Int J Behav Med. 2005;12(3):153–160. doi: 10.1207/s15327558ijbm1203_4. [DOI] [PubMed] [Google Scholar]
- 4.Pepalyte I, Kucinskiene Z, Grigalionienė K, Petrulionienė Z, Dzenkeviciute V, Bagdonaite L, et al. Genetic variants that participate in oxidation processes and/or oxidative stress and are associated with atherosclerosis. Eur Med Health Pharmaceut J. 2012;3:13–16. [Google Scholar]
- 5.Lorenzo C, Serrano-Rios M, Martinez-Larrad MT, Gonzalez-Sanchez JL, Seclen S, Villena A, et al. Geographic variations of the International Diabetes Federation and the National Cholesterol Education Program-Adult Treatment Panel III definitions of the metabolic syndrome in non-diabetic subjects. Diabetes Care. 2006;29(3):685–691. doi: 10.2337/diacare.29.03.06.dc05-1796. [DOI] [PubMed] [Google Scholar]
- 6.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: The insulin gene region and insulin-dependent diabetes mellitus (iddm) Am J Hum Genet. 1993;52(3):506–516. [PMC free article] [PubMed] [Google Scholar]
- 8.Germanas Š, Jakaitiene A. Power approximation of the transmission disequilibrium test. Lithuanian Mathemat J. 2012:56–61. Series B, 53(Series B) (in Lithuanian) [Google Scholar]
- 9. National Center for Biotechnology Information (NCBI) Gene database ( http://www.ncbi.nlm.nih.gov/gene/57142) (accessed 3 April 2013).
- 10.Whitley E, Ball J. Statistics review 4: sample size calculations. Crit Care. 2002;6(4):335–341. doi: 10.1186/cc1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrett J, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, et al. A genome-wide association study and metaanalysis indicate that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41(6):703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Zhao S, Xiang R. RTN3 and RTN4: candidate modulators in vascular cell apoptosis and atherosclerosis. J Cell Biochem. 2010;111(4):797–800. doi: 10.1002/jcb.22838. [DOI] [PubMed] [Google Scholar]
- 13.Jitoku D, Hattori E, Iwayama Y, Yamada K, Toyota T, Kikuchi M, et al. Association study of Nogo-related genes with schizophrenia in a Japanese case-control sample. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(5):581–592. doi: 10.1002/ajmg.b.31199. [DOI] [PubMed] [Google Scholar]
- 14.Kathiresan S, Manning AK, Demissie S, D’Agostino RB, Surti A, Guiducci C, et al. A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S17. doi: 10.1186/1471-2350-8-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Acevedo L, Yu J, Erdjument-Bromage H, Miao RQ, Kim JE, Fulton D, et al. A new role for Nogo as a regulator of vascular remodelling. Nat Med. 2004;10(4):382–388. doi: 10.1038/nm1020. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez-Feo JA, Hellings WE, Verhoeven BAN, Moll FL, de Kleijn DPV, Prendergast J, et al. Low levels of Nogo-B in human carotid atherosclerotic plaques are associated with an atheromatous phenotype, restenosis, and stenosis severity. Arterioscler Thromb Vasc Biol. 2007;27:1354–1360. doi: 10.1161/ATVBAHA.107.140913. [DOI] [PubMed] [Google Scholar]
- 17.Harrison KD, Miao RQ, Fernandez-Hernándo C, Suárez Y, Dávalos A, Sessa WC. Nogo-B receptor stabilizes Niemann-Pick type C2 protein and regulates intra-cellular cholesterol trafficking. Cell Metab. 2009;10(3):208–218. doi: 10.1016/j.cmet.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18(21):2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Depre C, Powell SR, Wang X. The role of the ubiquitin-proteasome pathway in cardiovascular disease. Cardiovasc Res. 2010;85(2):251–252. doi: 10.1093/cvr/cvp362. [DOI] [PubMed] [Google Scholar]