Abstract
Outbreak of a novel influenza virus is usually triggered by mutational change due to the process known as ‘antigenic shift’ or re-assortment process that allows animal-to-human or avian-to-human transmission. Birds are a natural reservoir for the influenza virus, and subtypes H5, H7, and H9 have all caused outbreaks of avian influenza in human populations. An especially notorious strain is the HPAI influenza virus H5N1, which has a mortality rate of approximately 60% and which has resulted in numerous hospitalizations, deaths, and significant economic loss. In March 2013, in Eastern China, there was an outbreak of the novel H7N9 influenza virus, which although less pathogenic in avian species, resulted in 131 confirmed cases and 36 deaths in humans over a two-month span. The rapid outbreak of this virus caused global concern but resulted in international cooperation to control the outbreak. Furthermore, cooperation led to valuable research-sharing including genome sequencing of the virus, the development of rapid and specific diagnosis, specimen sharing for future studies, and vaccine development. Although a H7N9 pandemic in the human population is possible due to its rapid transmissibility and extensive surveillance, the closure of the live-bird market will help mitigate the possibility of another H7N9 outbreak. In addition, further research into the source of the outbreak, pathogenicity of the virus, and the development of specific and sensitive detection assays will be essential for controlling and preparing for future H7N9 outbreaks.
Keywords: Global alert, Avian influenza, H5N1, H7N9
Introduction
Influenza viruses are the main cause of respiratory tract diseases in the human population and many species of animals. Influenza viruses are members of the Orthomyxoviridae family. There are three genera in this virus family, which are categorized based on their antigenic differences in the core proteins; influenza A, B, and C. To classify into subtypes, the surface hemagglutinin (HA) and neuraminidase (NA) protein combinations are used for viral characterization. For influenza A viruses, there are 16 different HA and nine different NA subtypes which result in many possible HA and NA combinations.
History of Pandemic Alert
Influenza virus type A is the most common subtype and is responsible for serious epidemics and pandemics throughout human history such as the 1918 (H1N1) ‘Spanish flu’, which was the most virulent virus in human history and caused an estimated 20–40 million deaths worldwide. Other pandemics were caused by the 1957 (H2N2) ‘Asian flu’ and 1968 (H3N2) ‘Hong Kong flu’, which resulted in approximately 2 million and 1 million deaths worldwide, respectively.1 In mid-March 2009, the World Health Organization (WHO) announced a new pandemic due to the triple-reassorted virus (H1N1) 2009, which was the first declared influenza pandemic of the 21st century.2 Although the illness caused by this recent pandemic is relatively mild, the actual number of cases and deaths are likely underestimated.3 Currently, pH1N1 has joined the repertoire of the typical seasonal influenza viruses circulating within the human population along with other seasonal viruses such as the influenza A (H3N2) subtype and influenza B viruses.4
Birds as Major Source of influenza A Virus
Many studies have concluded that wild waterfowl are the source of influenza A viruses, which currently circulate in human and animal populations. Gene segments of past human pandemic viruses also show signatures of avian viruses with the exception of the 1918 (H1N1) virus.5,6 Influenza virus can easily undergo genetic variation and result in diverse strains because it has several genome segments which allow for re-assortment between two or more different subtypes.3 Furthermore, if the re-assorted fragments are HA and NA, the resulting influenza virus can be a new subtype, which may lead to an epidemic or pandemic.7
Since 1996, there has been evidence that avian influenza viruses can be transmitted from their natural reservoir to domestic poultry and subsequently to humans. For example, the avian influenza virus subtypes H7, H5, and H9 have been reported to cause serious illness and death in humans.8,9 It has been suggested that pigs can serve as a ‘mixing vessel’ because pigs share receptors with both humans (SAα2, 6Gal) and avian species (SAα2, 3Gal).10 Thus swine may be an intermediate host that allows for the re-assortment, mutation, and adaptation of avian viruses into viruses that may be transmitted to human populations.11
The pathogenesis of avian influenza viruses has been classified into high pathogenicity (HPAI) and low pathogenicity (LPAI) according to their pathogenesis in terrestrial poultry and molecular genetic characteristics including their HA cleavage mechanism.12,13
The viruses usually are transmitted to poultry through the fecal–oral route.1 While influenza virus in its primary natural reservoir simply causes a mild intestinal tract infection, transmission of the LPAI viruses – especially H5 and H7 subtypes – from wild water fowl to poultry can potentially trigger evolution into HPAI.14 Every time the virus causes an outbreak in poultry, sporadic infections are expected to occur in people who have direct contact with sick or dead birds and their contaminated feces and/or environment.
Until now, only LPAI avian influenza viruses H9N2, H7N2, H7N3, H7N7, H10N7 along with the recently reported H7N9 infection and HPAI avian influenza viruses H5N1, H7N3, and H7N7 have crossed the species barrier into humans.15 To date, there has been no or little evidence of sustained human-to-human transmission of avian influenza viruses because tissue tropism limits virus transmission from birds to humans and similarly, from human to human.15,16
Time Line of Avian Influenza in Humans
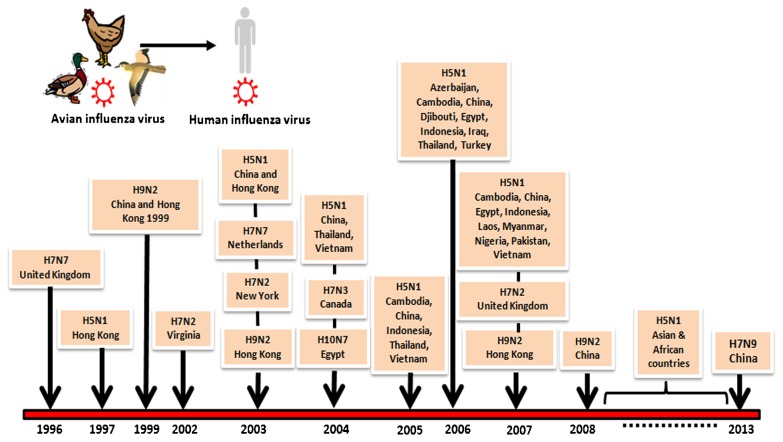
Since 1996, there have been several recorded LPAI and HPAI outbreaks that spread to humans (Fig. 1). The clinical symptoms are usually mild (e.g. flu-like symptoms and conjunctivitis) with very low fatality rates, HPAI H5N1 being the major exception.
Figure 1.
Time line of avian influenza in a human.
H5N1 Outbreak in Asia
HPAI H5N1 virus has been a public health concern due to their ability to cause severe respiratory illness. The first HPAI H5N1 viruses in human descended from the 1996 viral detection in geese from Guangdong Province in southern China, which the first cases occurred in Hong Kong in 1997 and out of the 18 recorded cases, six of them died.17 In 2003, HPAI H5N1 re-appeared in east and south-east Asia including Korea, Vietnam, Hong Kong, Japan, and Thailand, and has had devastating economic consequences due to its effect on the poultry industry and high mortality rate in reported human cases (approximately 60%).18 By 2005, the virus had spread to Indonesia, China, and Malaysia as well as Africa, Europe, the Pacific, and the Middle East. Currently these HPAI H5N1 viruses continue to circulate in poultry and humans with their genetic evolution aimed at adaptation to their hosts over time.19
HPAI H5N1 can be transmitted from wild birds to poultry by a fecal–oral route, oral–oral route, and also indirectly by respiratory droplets and aerosols.20,21 Most human cases, however, were exposed through unprotected direct or indirect contact with contaminated environments.22
In addition, mammalian carnivores (e.g. tigers, cats, and dogs) can be infected with HPAI H5N1 after feeding on infected birds. To date, the number of H5N1 WHO-confirmed human cases amounts to 628 with 374 deaths from 15 countries.23 Human infections tend to occur during December–March and are associated with bird migratory paths and seasonal outbreaks in poultry.24
Other HPAI and LPAI infections of humans have also been reported albeit less frequently. For example, humans have occasionally been shown to be infected with H7, H9, and H10 subtypes. However, when infected with these subtypes, the associated symptoms tend to be mild flu-like-illness, conjunctivitis or gastrointestinal symptoms25–27 except for one fatal case during an outbreak of HPAI H7N7 in the Netherlands.28
Avian influenza H7N9 Outbreak in China
Since March 2013, 130 people in China’s eastern region and one confirmed case in Taiwan, who had visited China, have been infected with avian influenza H7N9 virus, a subgroup of H7 viruses, leading to 36 deaths.29 However, the total number of cases has been suspected to be much higher than those reported since mild infections frequently remain undiagnosed. Similar to seasonal human influenza, these viruses tends to infect the elderly rather than children and many patients did not have any recorded contact with birds. Although these LPAI H7 viruses have been commonly isolated from birds, this is the first reported instance of human infection. Symptoms include fever and shortness of breath and may progress to severe pneumonia and respiratory failure after 5–7 days.30 So far, there has been no indication that the virus can be transmitted between humans and currently the origin of H7N9 is still unknown, the virus has been found in chickens, ducks, and pigeons at live-bird markets, although the level of infection in poultry farms is relatively low without causing any visible sign of illness in birds. Hence, there is concern about the virus’s potential to spread among populations and threaten to create a pandemic. Yet, the actual risks are still unknown.
Characteristics of the H5N1 and Novel H7N9 Avian Influenza Viruses
According to phylogenic analysis, the novel H7N9 avian influenza virus might derive from a triple re-assorted virus. The HA gene is closely related to the HA from A/duck/Zhejiang/12/2011 (H7N3) whereas the NA gene shares the highest similarity with the NA from A/wild bird/Korea/A14/2011 (H7N9). The remaining six internal genes comprising PB2, PB1, PA, NP, M, and NS might have originated from A/brambling/Beijing/16/2012-like viruses (H9N2).30 Molecular characterization of the novel H7N9 avian influenza virus cause LPAI in poultry compared to HPAI H5N1. First, the novel H7N9 virus lacks polybasic amino acid insertions within the cleavage site of HA. In addition, a four amino acid deletion has been established within the stalk region of neuraminidase (NA) and was established in the novel H7N9 virus. In comparison, a 20-amino-acid deletion indicative of high pathogenicity has been found in the H5N1 virus. Moreover, no PDZ domain (ESEV) has been observed at the C-terminal of NS1 of the novel H7N9 virus whereas the H5N1 virus contains ESEV residues responsible for increased virulence in animals.30
However, some characteristics of the novel H7N9 avian influenza virus might be taken into consideration with regard to infection of humans. Amino acid change Q226L in the HA have been identified in some strains of the novel H7N9 viruses implying a possible shift from avian to human receptor binding.31 In addition, the E627K substitution has been found in PB2 which is associated with an increased ability for replication and transmission in mammals.17 Moreover, the S31N substitution has been observed in M2 of the novel H7N9 avian influenza virus which is the substitution responsible for amantadine resistance.32 Lastly, the R293K mutation has been found in NA of some strains of the novel H7N9 avian influenza virus which indicates a potential resistance to NA inhibitors.33
Comparison of Epidemiological and Clinical Disease Patterns between Avian Influenza H5N1 and H7N9
Comparison of epidemiology, clinical characteristics, risk factors, and transmission between H5N1 and H7N9 influenza virus infection are shown in Table 1.
Table 1. Epidemiology and clinical pattern of diseases comparison between avian influenza H5N1 and H7N934,35.
| Category | H5N1 | H7N9 |
| Start date and countries | 1997 (Hong Kong)2003 (SE Asia) | March 2013 (China) |
| Duration (to present) | 10 years | 2 months (so far) |
| Global distribution | 1997 (Hong Kong) | China (East China) |
| 2003 (South-east Asia, European countries (birds), African countries) | Taiwan (ex mainland China) | |
| Source | Live-bird poultry and wild birds | Live-bird poultry |
| Total cases (24 May 2013) | 682 | 130+1 (ex mainland China) |
| Death (24 May 2013) (%) | 384 (59) | 36 (at least 27) |
| Age (Mean), years | 19.8 | 60.9 |
| Age (Median), years | 18.0 (range 0.3–75) | 63 (range 4–87) |
| Sex ratio (M∶F) | 1∶1.19 | 1∶0.45 |
| Coexisting condition | Uncommon | Common (Elderly, hypertension, diabetes, coronary heart disease, immunosuppression etc.) |
| Risk factors | Direct contact with poultry (Most of them) | Direct contact with poultry (Three quarters) |
| Incubation period | 7 days or less | 2–8 days |
| Clinical symptoms | Influenza-like symptoms: Pneumonia, diarrhoea, vomiting, myalgia±Conjunctivitis | Fever, cough, pneumonia |
| No conjunctivitis and encephalopathy | ||
| Acute kidney injury, Rhabdomyolysis | ||
| Respiratory failure | Rapidly progressive | Rapidly progressive |
| Lab findings | Leukopenia, lymphopenia, thrombocytopenia | Leukopenia, lymphopenia, thrombocytopenia |
| Elevated aminotransferase | Elevated aminotransferase | |
| Person-to-person transmission | No | Unclear |
| Sensitive to neuraminidase inhibitors | Yes | Yes |
Duration, Mortality and Velocity of Outbreak
Although the mortality rate in H7N9 infection (20%) is lower than that attributed to H5N1 virus (approximately 60%), H7N9 virus spreads faster than H5N1. While HPAI H5N1 has resulted in pandemics in 1993 and 2003, only sporadic cases have been reported since 2003. In addition, while the cumulative number of patients infected with H5N1 is 628 over the last 10 years,23 in only 2 months H7N9 has resulted in 131 confirmed cases.29 This suggests that the novel influenza virus can spread more rapidly in a shorter period of time than H5N1 avian influenza virus. This further implies that subsequent to a future mutation potentially altering its characteristics to become highly pathogenic, this virus would be more severe and have a higher potential to trigger a pandemic.
The mortality rate of H7N9 infected patients deserves further consideration. In fact, the exact number of people infected with H7N9 virus has not been determined due to some asymptomatic infections and mild cases that did not require hospitalization. Thus, this number might either be much larger or lower than that of patients whose disease was aggravated by specific complications. Prevalence of infection and severity of this virus can be determined by testing the healthy population for seroprevalence using microneutralization test (MN) or hemagglutination inhibition test (HI), which needs to be validated in the respective assay protocol.
Sources of Virus And Risk Factors
Previous studies have established that both wild birds and poultry were responsible for the spread of H5N1,39,40 currently there has been no report of H7N9 in migratory birds. Some study has reported H7N9 virus in live-bird markets in Anhui, Jiangsu, and Zhejiang, which are also places that reported H7N9 human cases suggesting that direct contact with poultry is one of the key factors contributing to H7N9 infection in humans.41 So far, there has been no report of H7N9 virus in poultry farms in China. If the spread of the virus is solely due to migratory birds, then the virus will be more difficult to eradicate because it is difficult to control the migration of wild birds. Thus, due to a current lack of understanding of the source of H7N9, it is important that governments remain alert and prepared for a possible reemergence of the virus.
Age Uncertainties and Sex Imbalance
Moreover, there are some areas of uncertainty that should be clarified. First, the sexual imbalance of patients with two out of three H7N9 patients being male should be mentioned. In contrast, H5N1 patients showed a balanced sex profile. Second, is the difference of age pattern between H5N1 and H7N9 patients. Patients were infected with H5N1 at an earlier age when compared to those infected with H7N9 (the median age of H5N1 patients is 18 years while that of H7N9 patients is 63 years).34,35 This age uncertainty may be ascribed to the pathogenicity of this virus. There has been no report of H7N9 virus infection in humans until the recent one from China. Therefore, this virus may be able to infect people of all age groups due to a lack of cross-reactivity antibodies but can cause symptomatic infection only in vulnerable populations; e.g. people with underlying complications, which are more prevalent among elderly people while H7N9 infection in children caused mild fever and asymptomatic infection. Unlike H7N9 infection, H5N1 infection can be more severe in infants and school-aged children, which may result from cross-reactivity from previous outbreaks of H5 or virus with the same epitope. The sex imbalance in H7N9 infection is harder to explain. It may be due to the socio-cultural behavior patterns in the areas of outbreak, but any host–virus interaction responsible for the observed sex imbalance would require further investigation.
Case Definitions of Avian Influenza
To control outbreaks of emerging disease, accurate definition of the diseases is crucial for diagnosis and treatment so that the spread of the pathogens can be curbed and control measures put in place. The WHO has provided definitions for suspected, probable, and confirmed cases of emerging infectious diseases. The definitions have been mentioned elsewhere.42
Clinical Symptoms, Lab Findings, and Transmissibility of Avian Influenza
Avian influenza symptoms can manifest as typical influenza illnesses and may include acute respiratory distress, fever, cough, diarrhea, malaise, and muscle aches. In severe cases and in most cases of H5N1, patients may have other complications such as pneumonia, respiratory failure, multiple organ failure, and death.43
There are several uncommon features in H7N9 infection that should be reviewed. Compared to H5N1 infection – whose initial symptoms started with influenza-like illness, such as high fever, sore throat, cough, nasal congestion, rhinorrhea, followed by vomiting and diarrhea – H7N9 infection causes some uncommon clinical features such as conjunctivitis and encephalopathy while some common clinical symptoms such as nasal congestion and rhinorrhea are not apparent. A study found that, similar to H5N1, H7N9 infection can develop rapidly and attack the lower respiratory tract. This lead to hypoxemia, bilateral pulmonary infiltrates, and multiple organ failure which is the major cause of death. However, the incubation period of H7N9 infection (5–7 days) was less than that of H5N1 (2–8 days). The laboratory findings of patients infected with avian influenza virus suggest many abnormalities, such as leucopenia, thrombocytopenia, and elevated liver enzyme (aminotransferase). While some cases infected with H7N9 have normal white cells count, H5N1 infection were more likely to have lower white blood cell in fatal H5N1 cases.41 In addition, unlike with the H5N1 outbreak, the ability of the virus to be transmitted from person-to-person is still unclear. Infected patients represented some family clusters and although this might indicate human-to-human transmission capability of the virus, they might also have been infected with the virus because they live in the same environment and have the same contact with poultry. Unlike with H5N1 infection, the presence of comorbidities such as coronary heart disease, chronic obstructive pulmonary disease, hypertension, and diabetes is common in H7N9 virus infection.41
Severity of H7N9 Infection
During the large scale surveillance of influenza-like illness patients in China, more than 46,807 specimens were obtained both in the area affected by the H7N9 outbreak and the non-affected area. Of those, two children (2 and 4 years of age) had developed mild diseases, compared with four adults with more severe disease. The data suggests that severity of H7N9 virus infection severity increases with increasing age.44The risk factors associated with moderate-to-severe ARDS tended to manifest at the age of 65 years or older in the form of coexisting medical conditions, lymphocyte count<1000 cells/mm3, aspartate aminotransferase level>40 U/l, creatine kinase level>200 U/l and>3 days time from symptom onset to initiation of antiviral therapy.35
Global Planning and Alert to H7N9 Outbreak
This recent outbreak of avian influenza in China has demonstrated the effective preparedness measures the Chinese government employed by providing early detection and prompt treatment of patients with respiratory infections and their close contacts, who are being followed-up. This outbreak also proved the success of international collaboration between organizations, including human and animal health sectors and emphasized the importance of surveying LPAI influenza viruses. The timely sharing of surveillance data and whole genome sequences of isolated virus have enabled scientists around the world to develop virus detection protocols, diagnostic tests, vaccine development, and further virus characterization. In addition, the Chinese national surveillance system was able to effectively trace the source back to poultry farms. Despite the substantial progress made, however, many aspects of transmission and possible sources of infection are still under investigation.
Continuous evaluation of the provided diagnostic kits will improve their specificity and sensitivity of detection in animals and human cases. Also, successful treatment strategies should be distributed to all health care facilities along with disease information and good management practices according to national and WHO guideline.
As a requirement for influenza virus prevention, global and local health authorities should employ intervention methods, which have been proven to reduce transmission and decrease the number of cases. Examples include the temporal closure of live poultry markets, distribution of health information and situation updates to the general population that will assist in understanding the virus-related risks and promote good personal hygiene practices. Educational campaigns should provide guidance on proper equipment and operations as well as cleaning and sanitation of facilities to poultry sellers and slaughterers. Similarly, all staff on poultry farms should be made aware of the most suitable management and biosafety practices. Along with appropriate hygiene, proper preparation and cooking of poultry products are advisable to reduce the risk of avian influenza virus infection and a range of other diseases. Importantly, strengthened surveillance could enhance preparedness for global epidemics and pandemics to better monitoring the spread of a new disease and thus, provide early warnings of any unusual respiratory disease or mortality due to unknown causes. Even though the authorities have already announced the end of the emergency response, people should remain alert to influenza virus prevention.
Conflict of Interests
The authors have declared that no conflicts of interests exist.
Author Contributions
YP, SP, SP and JK reviewed, wrote the manuscript. All authors approved the manuscript.
Acknowledgments
This study was supported by The Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission (HR1155A), the Center of Excellence in Clinical Virology, Chulalongkorn University and Hospital, Centenary Academic Development Project, the RGJ PhD program of the Thailand Research Fund, Thailand Research Fund (DPG5480002), Integrated Innovation Academic Center; Chulalongkorn University Centenary Academic Development Project (CU56-HR01);the Outstanding Professor of the Thailand Research Fund (DPG5480002); and King Chulalongkorn Memorial Hospital, National Research Council of Thailand. We would like to thank Ms Petra Hirsch for reviewing the manuscript.
References
- 1.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–79. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC. Center for disease control and prevention, outbreak of swine-origin influenza A (H1N1) virus infection -Mexico, March–April 2009. Morb Mortal Wkly Rep. 2009;58:467–470 Available from: http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5817a5.htm. [PubMed] [Google Scholar]
- 3.CDC. Center for disease control and prevention, flu news & spotlights, first global estimates of 2009 h1n1 pandemic mortality released by cdc-led collaboration. Atlanta (GA): CDC. Available from: http://www.cdc.gov/flu/spotlights/pandemic-global-estimates.htm (cited 2013 May 10) [Google Scholar]
- 4.WHO. Influenza: Influenza Update N° 185 [internet]. Geneva: WHO. Available from: http://www.who.int/influenza/surveillance_monitoring/updates/2013_05_10_surveillance_update_185.pdf (cited 2013 May 10) [Google Scholar]
- 5.Shrestha SS, Swerdlow DL, Borse RH, Prabhu VS, Finelli L, Atkins CY, et al. Estimating the burden of 2009 pandemic influenza A (H1N1) in the United States (April 2009–April 2010). Clin Infect Dis. 2011;52:S75–82. doi: 10.1093/cid/ciq012. [DOI] [PubMed] [Google Scholar]
- 6.Kawaoka Y, Krauss S, Webster RG.Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol 1989:634603–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webby RJ, Webster RG. Emergence of influenza A viruses. Phil Trans R Soc Lond B Biol Sci. 2001;356:1817–28. doi: 10.1098/rstb.2001.0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, de Jong MD, et al. Avian influenza A (H5N1) infection in humans. N Engl J Med. 2005;353:1374–85. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 9.Butt KM, Smith GJD, Chen HL, Zhang LJ, Leung YHC, Xu KM, et al. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J Clin Microbiol. 2005;43:5760–7. doi: 10.1128/JCM.43.11.5760-5767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma W, Kahn RE, Richt JA. The pig as a mixing vessel for influenza viruses: human and veterinary implications. J Mol Genet Med. 2008;3:158–66. [PMC free article] [PubMed] [Google Scholar]
- 11.Taubenberger JK, Morens DM. Influenza: the once and future pandemic. Public Health Rep. 2010;125:16–26. [PMC free article] [PubMed] [Google Scholar]
- 12.Webster RG, Rott R. Influenza virus A pathogenicity: the pivotal role of hemagglutinin. Cell. 1987;50(5):665–6. doi: 10.1016/0092-8674(87)90321-7. [DOI] [PubMed] [Google Scholar]
- 13.Klenk HD, Garten W. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 1994;2:39–43. doi: 10.1016/0966-842x(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 14.Peiris JS, de Jong MD, Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev. 2007;20:243–67. doi: 10.1128/CMR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan H, Sorrell EM, Song H, Hossain MJ, Ramirez-Nieto G, Monne I, et al. Replication and transmission of H9N2 influenza viruses in ferrets: evaluation of pandemic potential. PLoS One. 2008;3:e2923. doi: 10.1371/journal.pone.0002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Hoeven N, Pappas C, Belser JA, Maines TR, Zeng H, García-Sastre A, et al. Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proc Natl Acad Sci U S A. 2009;106:3366–71. doi: 10.1073/pnas.0813172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steel J, Lowen AC, Mubareka S, Palese P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 2009;5:e1000252. doi: 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Claas ECJ, Osterhaus ADME, van Beek R, De Jong JC, Rimmelzwaan GF, Senne DA, et al. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–7. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 19.WHO. Influenza: FAQs: H5N1 influenza Geneva: WHO; Available from: http://www.who.int/influenza/human_animal_interface/avian_ influenza/h5n1_research/faqs/en/ (cited 2013 May 14) [Google Scholar]
- 20.Sturm-Ramirez KM, Ellis T, Bousfield B, Bissett L, Dyrting K, Rehg JE, et al. Reemerging H5N1 influenza viruses in Hong Kong in 2002 are highly pathogenic to ducks. J Virol. 2004;78:4892–901. doi: 10.1128/JVI.78.9.4892-4901.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeong OM, Kim MC, Kim MJ, Kang HM, Kim HR, Kim YJ, et al. Experimental infection of chickens, ducks and quails with the highly pathogenic H5N1 avian influenza virus. J Vet Sci. 2009;10:53–60. doi: 10.4142/jvs.2009.10.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CDC. Center for disease control and prevention, avian influenza (flu). Transmission of avian influenza A viruses between animals and people. Atlanta (GA): CDC. Available from: http://www.cdc.gov/flu/avianflu/virus-transmission.htm. [Google Scholar]
- 23.WHO. Influenza: Cumulative number of confirmed human cases of avian influenza A(H5N1) reported to WHO, 2003–2013, 26 April 2013 [internet]. Geneva: WHO. Available from: http://www.who.int/influenza/human_animal_interface/EN_GIP_20130426CumulativeNumberH5N1cases.pdf (cited 2013 May 22) [Google Scholar]
- 24.WHO. Weekly epidemiological record (WER): Update on human cases of influenza at the human– animal interface, 2012. Weekly epidemiological record 29 March 2013, 88(13): 137–44 [internet]. Geneva: WHO. Available from: http://www.who.int/wer/2013/wer8813.pdf (cited 2013 May 16) [Google Scholar]
- 25.Pepin KM, Wang J, Webb CT, Hoeting JA, Poss M, Hudson PJ, et al. Anticipating the prevalence of avian influenza subtypes H9 and H5 in live-bird markets. PLoS One. 2013;8:e56157. doi: 10.1371/journal.pone.0056157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chmielewski R, Swayne DE. Avian influenza: public health and food safety concerns. Annu Rev Food Sci Technol. 2011;2:37–57. doi: 10.1146/annurev-food-022510-133710. [DOI] [PubMed] [Google Scholar]
- 27.Arzey GG, Kirkland PD, Arzey KE, Frost M, Maywood P, Conaty S, et al. Influenza virus A (H10N7) in chickens and poultry abattoir workers, Australia. Emerg Infect Dis. 2012;18:814–6. doi: 10.3201/eid1805.111852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A. 2004;101:1356–61. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bao CJ, Cui LB, Zhou MH, Hong L, Gao GF, Wang H. Live-animal markets and influenza A (H7N9) virus infection. N Engl J Med. 2013;368:2337–9. doi: 10.1056/NEJMc1306100. [DOI] [PubMed] [Google Scholar]
- 30.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–97. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 31.Connor RJ, Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 32.Cheng PK, Leung TW, Ho EC, Leung PC, Ng AY, Lai MY, et al. Oseltamivir- and amantadine-resistant influenza viruses A (H1N1). Emerg. Infect Dis. 2009;15:966–8. doi: 10.3201/eid1506.081357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gubareva LV. Gubareva LV. Molecular mechanisms of influenza virus resistance to neuraminidase inhibitors. Virus Res. 2004;103:199–203. doi: 10.1016/j.virusres.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 34.Abdel-Ghafar AN, Chotpitayasunondh T, Gao Z, Hayden FG, Nguyen DH, de Jong MD, et al. Update on avian influenza A (H5N1) virus infection in humans. N Engl J Med. 2008;358:261–73. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- 35.Gao HN, Lu HZ, Cao B, Du B, Shang H, Gan JH, et al. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med. 2013;368:2277–85. doi: 10.1056/NEJMoa1305584. [DOI] [PubMed] [Google Scholar]
- 36.WHO . Influenza: avian influenza A(H7N9) virus. Geneva: WHO. Available from: http://www.who.int/influenza/human_animal_interface/influenza_h7n9/en/index.html (cited 2013 Apr 23) [Google Scholar]
- 37.Li Q, Zhou L, Zhou M, Chen Z, Li F, Wu H, et al. Preliminary report: epidemiology of the avian influenza A (H7N9) outbreak in China. N Engl J Med. 2013 doi: 10.1056/NEJMoa1304617. [Google Scholar]
- 38.CDC . Center for disease control and prevention, avian influenza: current H5N1 situation. Atlanta (GA): CDC. Available from: http://www.cdc.gov/flu/avian/outbreaks/current.htm (cited 2013 May 20) [Google Scholar]
- 39.Webster RG, Govorkova EA. H5N1 influenza-continuing evolution and spread. N Engl J Med. 2006;355:2174–7. doi: 10.1056/NEJMp068205. [DOI] [PubMed] [Google Scholar]
- 40.Van Kerkhove MD, Mumford E, Mounts AW, Bresee J, Ly S, Bridges CB, et al. Highly pathogenic avian influenza (H5N1): pathways of exposure at the animal-human interface, a systematic review. PLoS One. 2011;6:e14582. doi: 10.1371/journal.pone.0014582. doi: 10.1371/journal.pone.0014582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.WHO. Influenza: Avian influenza A(H7N9) virus, China—WHO joint mission on human infection with avian influenza A(H7N9) virus [internet]. Geneva: WHO. Available from: http://www.who.int/influenza/human_animal_interface/influenza_h7n9/ChinaH7N9JointMissionReport2013.pdf (cited 2013 Apr 20) [Google Scholar]
- 42.WHO. Influenza: WHO case definitions for human infections with influenza A(H5N1) virus. Geneva: WHO. Available from: http://www.who.int/influenza/resources/documents/case_definition2006_08_29/en/ (cited 2013 May 26) [Google Scholar]
- 43.WHO. Medicines publications: WHO rapid advice guidelines on pharmacological management of humans infected with avian influenza A (H5N1) virus [internet]. Geneva: WHO. Available from: http://www.who.int/medicines/publications/WHO_PSM_PAR_2006.6.pdf (cited 2013 May 22) [Google Scholar]
- 44.Xu C, Havers F, Wang L, Chen T, Shi J, Wang D, et al. Monitoring avian influenza A(H7N9) virus through national influenza-like illness surveillance, China. Emerg Infect Dis. doi: 10.3201/eid1908.130662. Available from: http://wwwnc.cdc.gov/eid/article/19/8/13-0662_article.htm (cited 23 May 2013) [DOI] [PMC free article] [PubMed] [Google Scholar]