Abstract
Microscopic examination of eggs of parasitic helminths in stool samples has been the most widely used classical diagnostic method for infections, but tiny and low numbers of eggs in stool samples often hamper diagnosis of helminthic infections with classical microscopic examination. Moreover, it is also difficult to differentiate parasite eggs by the classical method, if they have similar morphological characteristics. In this study, we developed a rapid and sensitive polymerase chain reaction (PCR)-based molecular diagnostic method for detection of Clonorchis sinensis eggs in stool samples. Nine primers were designed based on the long-terminal repeat (LTR) of C. sinensis retrotransposon1 (CsRn1) gene, and seven PCR primer sets were paired. Polymerase chain reaction with each primer pair produced specific amplicons for C. sinensis, but not for other trematodes including Metagonimus yokogawai and Paragonimus westermani. Particularly, three primer sets were able to detect 10 C. sinensis eggs and were applicable to amplify specific amplicons from DNA samples purified from stool of C. sinensis-infected patients. This PCR method could be useful for diagnosis of C. sinensis infections in human stool samples with a high level of specificity and sensitivity.
Keywords: Clonorchis sinensis, Egg, PCR diagnosis, Retrotransposon
Introduction
Human clonorchiasis is an infection caused by the Chinese liver fluke, Clonorchis sinensis, which is endemic in China, Korea, Taiwan, northern Vietnam, and eastern Russia. It is estimated to infect about 35 million people in the endemic areas, particularly 15 millions in China.1,2 Humans are usually infected by ingesting raw or inadequately cooked freshwater fish infected with C. sinensis metacercariae. Metacercariae excyst in the duodenum, migrate into the intrahepatic biliary duct and then grow to adult worms in the bile duct. The adult worms chronically inhabit the bile duct and can cause periductal inflammation, fibrosis, cholangitis, cholelithiasis, cholangiectasis, and even by cholangiocarcinoma.3,4
Diagnosis of C. sinensis infection has been classically performed by microscopic detection of the parasite’s eggs in stool samples. However, microscopic examination of C. sinensis eggs in stool samples is usually cumbersome and time consuming, and requires well-trained experts. Moreover, the eggs are sometimes difficult to identify with microscopic examination method, especially in patients with low worm burdens, and it is also difficult to differentiate C. sinensis eggs from other parasitic trematode eggs with similar morphological characteristics, such as Opisthorchis spp. and Metagonimus yokogawai. And therefore, various efforts based on immunodiagnostic assays for C. sinensis infection using crude or recombinant antigens have been developed to overcome these limitations,5–9 but these assays also have their own limitations including low sensitivity and cross-reactivity with proteins from other closely related helminth parasites. These tests also yield false-positive results in individuals who are cured from the infection but still have antibodies against C. sinensis antigens. Therefore, development of a new method that can improve specificity and sensitivity is required.
Accumulation of large amounts of genetic information for various parasitic organisms has made it possible to identify species-specific sequences for each parasite. Molecular approaches based on genetic differences between parasitic helminthes have been attempted to differentiate morphologically similar parasites.10–13 These lead to the development of polymerase chain reaction (PCR)-based molecular diagnostic methods for C. sinensis,14–19 hookworm,20 O. viverrini,21,22 Schistosoma mansoni and S. haematobium,23,24 and Taenia spp.25
In this study, we developed a single-step rapid and sensitive PCR detection method for C. sinensis, using multiple pairs of species-specific primers based on the long-terminal repeat (LTR) of C. sinensis retrotransposon1 (CsRn1) gene, and evaluated its diagnostic potential for C. sinensis eggs in stool samples. Our results suggested the PCR method developed in this study showed high specificity and sensitivity for C. sinensis and could be applicable to diagnose C. sinensis infection by detecting parasite eggs in human stool samples.
Materials and Methods
Parasites
Metacercariae of C. sinensis were collected from the second intermediated host, Pseudorasbora parva, caught at Shenyang, Liaoning Province, China. The fish was digested with artificial gastric juice for 1 hour at 37°C.26 Metacercariae were collected and then orally administered to New Zealand White rabbits. Adult worms were collected from the bile duct 2 months after infection and washed with cold physiological saline several times to remove host contaminants. Metacercariae of M. yokogawai were collected from sweetfish Plecoglossus altivelis, caught at Gwangyang-si, Jeollanam-do, Korea, and digested with the same artificial gastric juice described above. Dogs were orally infected with the metacercariae and adult worms were obtained from the small intestine 2 weeks after infection. Metacercariae of Paragonimus westermani were collected from naturally infected crayfish Cambaroides similes caught at Haenam-gun, Jeollanam-do, Korea. Dogs were orally infected with 200 metacercariae and were sacrificed 3 months after infection. The adult worms were recovered from the lungs. The eggs of each parasite were produced by incubating the adult worms overnight in physiological saline at 37°C. The eggs were collected under a dissection microscope and washed several times with cold physiological saline. All experimental animals used in study were cared and handled in the Korea Food and Drug Administration (KFDA) animal facility followed by guidelines of AAALAC International Animal Care policies (Accredited Unit, KFDA; Unit Number 000996). Approval for animal experiments was obtained from KFDA animal facilities (NIH-08-19).
Extraction of genomic DNA from adult worms
Genomic DNA from adult C. sinensis, M. yokogawai, and P. westermani worms was extracted using NucleoSpin Tissue Kit (Macherey-Nagel, Duren, Germany) according to the manufacturer’s instructions. Five adult worms were homogenized using a glass-Teflon unit (Glas-Col, Terre Haute, IN, USA) followed by the manufacturer’s instructions. The purified genomic DNA of each parasite was diluted in distilled water to 25 ng/μl prior to use.
Design of oligonucleotide primers and optimization of polymerase chain reaction
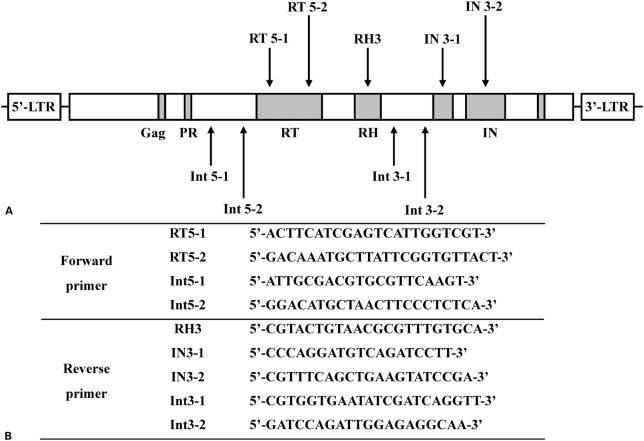
Specific oligonucleotide primers were designed based on the C. sinensis retrotransposon LTR (CsRn1; GenBank Accession No. AY013558-AY013571). The positions and sequences of the primers are shown in Fig. 1. Seven primer sets were combined as follows (expected amplicon size in parentheses): RT5-1 and RH3 (929 bp), RT5-2 and IN3-1 (999 bp), RT5-2 and IN3-2 (1,215 bp), Int5-1 and Int3-1 (1,466 bp), Int5-2 and Int3-2 (1,589 bp), Int5-1 and Int3-2 (1,706 bp), Int5-2 and Int3-1 (1,349 bp). Polymerase chain reaction was performed with Accupower PCR Premix (Bioneer, Daejeon, Korea), genomic DNA of C. sinensis (25 ng), 50 pM each primer, 1 U Taq DNA polymerase, 250 μM dNTPs, 10 mM Tris-HCl; pH 9.0, 40 mM KCl, 1.5 mM MgCl2, in a final volume of 20 μl. Thermal cycling was performed with denaturation at 98°C for 10 minutes followed by 30 cycles of 95°C for 30 seconds, 54°C for 1 minute, 72°C for 1 minute, and final extension at 72°C for 5 minutes. Polymerase chain reaction amplicons were electrophoresed on 1% agarose gel and stained with ethidium bromide. To confirm the sequences of the amplified products, each PCR product was purified with the Qiaquick Purification Kit (Qiagen, Hilden, Germany), ligated into a pCR2.1-TOPO TA-cloning vector (Invitrogen, Carlsbad, CA, USA), and sequenced with an automatic sequencer (Applied Biosystems, Foster City, CA, USA).
Figure 1.
(A) Schematic for each primer. The primers were designed based on the sequence of long-terminal repeat (LTR) of C. sinensis retrotransposon (CsRn1). Gag, protease (PR), reverse transcriptase (RT), RNase H (RH), and three subdomains of integrase (IN) regions are represented in gray. (B) Nucleotide sequences of each olionucleotide primer.
Oligonucleotide primers.
Specificity of PCR method
To evaluate the specificity of the PCR method, PCR was performed with genomic DNA (25 ng) purified from M. yokogawai and P. westermani adult worms as a template, respectively. The amplification condition was the same as described above. The amplification products were analyzed by agarose gel electrophoresis followed by ethidium bromide staining.
Stool samples
The stool samples used in this study were collected from patients with clonochiasis (n = 15) or normal healthy individuals (n = 10) residing in C. sinensis endemic areas in Korea. All participants were informed about this study and signed an informed consent according to the ethical guideline of the Korea National Institute of Health (KNIH). Stool samples were collected followed by an ethical guideline of the KNIH and were diagnosed by formalin ether sedimentation method and microscopic detection of eggs in the samples. The normal stool samples were confirmed to be not infected with any kind of parasite. No helminth eggs other than C. sinensis were identified in patients’ stool samples.
Detection of C. sinensis eggs in stool samples
To investigate the applicability of the PCR methods for direct detection of C. sinensis eggs in stool samples, the genomic DNA of C. sinensis eggs in the stool (200 mg) of patients with clonorchiasis were purified using QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer instructions. For negative control, genomic DNA extracted from stool samples of normal healthy individuals was used. Polymerase chain reaction was performed with the purified genomic DNA (50 ng/μl) as described above. In order to analyze the minimum number of C. sinensis eggs in stool sample, which can be detected by the PCR method, we performed seeding experiments. Briefly, different numbers of C. sinensis eggs (1,000, 100, and 10) were artificially seeded into stool of a normal healthy person, who has proved not to be infected with any helminth parasite by microscopic stool examination. The genomic DNA was extracted from each stool sample and applied to PCR analysis with the same method described above. All experiments were performed triplicate. To assess the detection limit of the PCR, we also performed PCR with different numbers of C. sinensis eggs (1,000, 100, 10, and 1) with the same amplification condition described above.
Results and Discussion
Several PCR-based diagnostic methods for C. sinensis have been developed to overcome the drawbacks of traditional microscopic stool examination method for detecting C. sinensis eggs in stool samples and serodiagnostic methods using crude or recombinant antigens of C. sinensis.14–19 These studies suggest applicability of PCR methods as reliable molecular diagnostic tools for rapid and sensitive diagnosis of C. sinensis infection in humans and animals. Previously developed PCR methods are basically based on real-time PCR or nested PCR in order to enhance their sensitivity and specificity.14–19 Real-time PCR is becoming more widely used for routine diagnosis of various pathogens due to its flexibility and speed, providing an obvious experimental advantage for routine laboratory diagnosis. However, real-time quantitative PCR methods require high cost to maintain compared to conventional PCR methods and the instrument is still not widely distributed to all small local laboratories, which carry out first-line diagnosis. Nested PCR methods have advantages of high sensitivity and specificity, but these methods essentially require two-steps of amplification processes that can be time-consuming. Therefore, development of a reliable single-step conventional PCR method that can be easily applicable in all laboratories is highly required for routine diagnosis of C. sinensis infection. In this study, we developed a new single-step conventional PCR method, which can be applied to direct detection of C. sinensis eggs in stool samples. Our PCR method targets the LTR of CsRn1, a class of transposable elements.27 LTR retrotransposons are distributed in nearly all eukaryotes, including nematodes28 and echinoderms.29 An advantage of using the LTR of C. sinensis retrotransposon1 (CsRn1) gene as a target for amplification is its presence in more than 100 copies per haploid genome of the parasite,27 which enables to maximize the amplification efficacy.
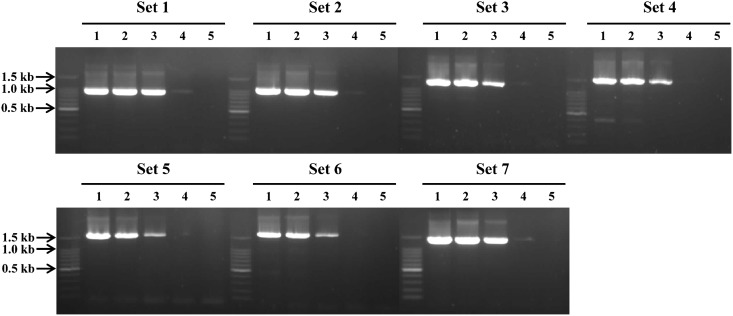
Polymerase chain reaction analyses using seven different primer sets and genomic DNA of C. sinensis adult worms resulted in amplicons with expected sizes, respectively: RT5-1 and RH3 (929 bp), RT5-2 and IN3-1 (999 bp), RT5-2 and IN3-2 (1,215 bp), Int5-1 and Int3-1 (1,466 bp), Int5-2 and Int3-2 (1,58 = 9 bp), Int5-1 and Int3-2 (1,706 bp), Int5-2 and Int3-1 (1,349 bp) (Fig. 2A). The sequence of each amplicon was confirmed to be identical with each target sequence (data not shown). Polymerase chain reaction analyses with each primer set with genomic DNA of P. westermani or M. yokogawai did not show any corresponding size of amplicon, even though non-specific amplified products with different sizes were identified for P. westermani and M. yokogawai with primer set 4 and 5, respectively (Fig. 2B). To evaluate the sensitivity of the PCR methods, we also performed PCR analyses with each primer set and different quantity of genomic DNA of C. sinensis (50 ng, 5 ng, 500 pg, 50 pg, and 5 pg) as a template. The detection limit of each PCR with different set of primers slightly differed, but all PCR methods were able to detect at least 500 pg of C. sinensis genomic DNA (Fig. 3). In particular, PCRs with primer set 1, 2, 3, and 7 could detect 50 pg of genomic DNA. These results collectively suggest that our PCR method is highly specific for C. sinensis and is sensitive to detect as little as 50–500 pg of genomic DNA of the parasite.
Figure 2.
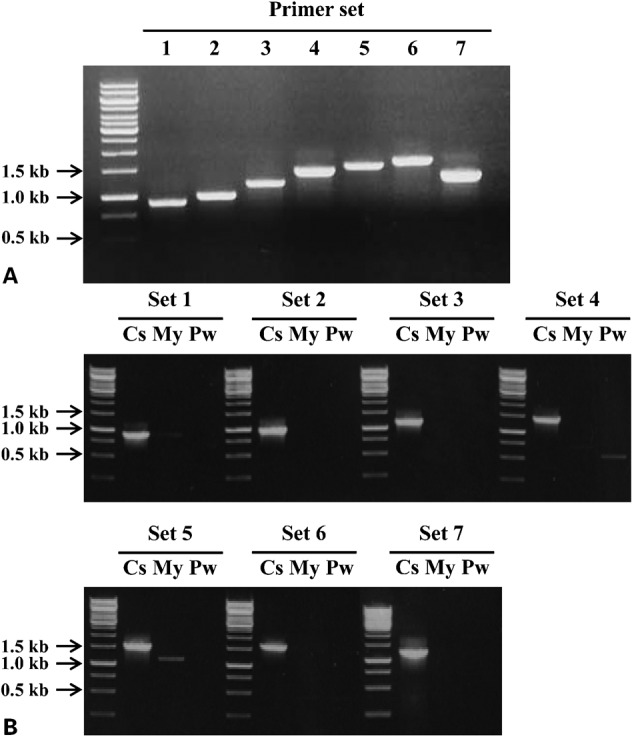
(A) Polymerase chain reaction (PCR) analyses using each set of primers and genomic DNA of C. sinensis (25 ng).The primer sets were as follows: Set 1, RT5-1 and RH3 (929 bp); Set 2, RT5-2 and IN3-1 (999 bp); Set 3, RT5-2 and IN3-2 (1,215 bp); Set 4, Int5-1 and Int3-1 (1,466 bp); Set 5, Int5-2 and Int3-2 (1,589 bp); Set 6, Int5-1 and Int3-2 (1,706 bp); Set 7, Int5-2 and Int3-1 (1,349 bp). Each amplicon had an expected size and the sequence was confirmed by automatic sequencing. (B) Specificity of PCR. PCR analyses with each primer set with genomic DNA (25 ng) of C. sinensis, M. yokogawai, or P. westermani were performed in the same amplification condition. Cs, C. sinensis; My, M. yokogawai; Pw, P. westermani.
Optimization and specificity of polymerase chain reaction (PCR).
Figure 3.
Polymerase chain reaction (PCR) analyses with each primer set and different amount of genomic DNA of C. sinensis (5 pg to 50 ng) were performed. The primer sets used were the same as previously described. Lane 1, 50 ng; lane 2, 5 ng; lane 3, 500 pg; lane 4, 50 pg; lane 5, 5 pg.
Sensitivity assay.
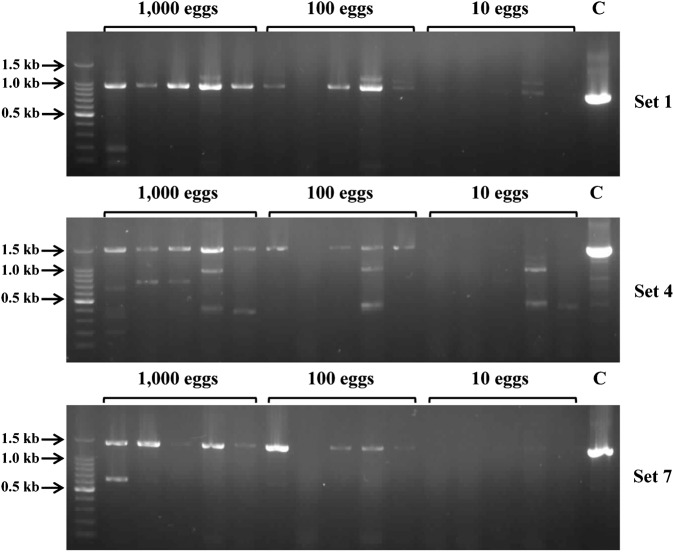
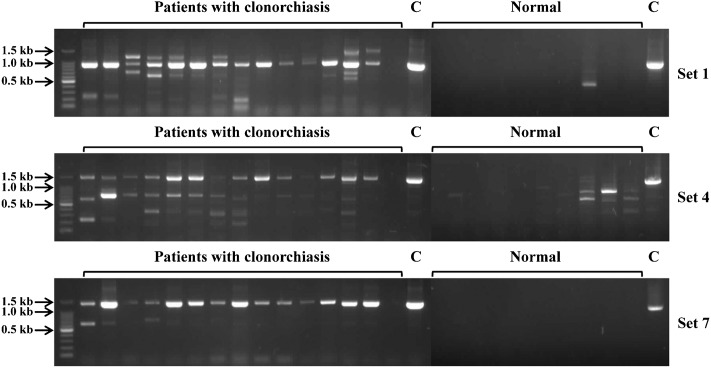
To further investigate the applicability of the PCR method for stool samples, we selected three sets of PCR primers (set 1, 4, and 7), which showed good performances in our system, and analyzed their potential as diagnostic tools. PCR analyses using genomic DNA purified from stool samples, which had different numbers of C. sinensis eggs (1,000, 100, and 10) that were experimentally seeded in normal human stools, showed that all three primer sets amplified their corresponding amplicons with expected sizes in all five stool samples with 1,000 C. sinensis eggs (Fig. 4). Although non-specific amplified products were observed in the cases of primer set 4 and 7, each amplified product with expected size were clearly identified. All three primer sets also successfully amplified their amplicons in four stool samples with 100 eggs. Meanwhile, no amplification was observed in all stool samples seeding 10 eggs. PCR amplifications of three primer sets were able to detect a single C. sinensis egg (Supplementary Material 1 http://dx.doi.org/10.1179/2047773213Y.0000000099.S1), but negative results were obtained in the stool samples with 10 eggs. This suggests that a large amount of genomic DNA of the parasite probably was lost during the genomic DNA purification process, and therefore, further effort to enhance the quality of genomic DNA from stool samples would be necessary. However, considering the number of eggs produced by a single C. sinensis adult worm is estimated to about 100 per day per gram of feces (EPG),1,30,31 the PCR methods developed here would be reliable methods to apply stool samples, if the diagnosis would be carried out with at least 1 g of stool sample. Indeed, our further investigation with the stool samples obtained from 15 clonorchiasis patients with low egg burden (EPG range: 200–999) revealed that positive results were obtained in fourteen stool samples (Fig. 5). We also tried direct PCR analysis with stool samples without prior genomic DNA preparation process, but we failed to obtain reliable results, which was probably because stool contains various substances that can inhibit the PCR reaction.32 And therefore, further effort to develop a new method, which can apply stool samples for direct PCR would be necessary to simplify the PCR process.
Figure 4.
Polymerase chain reaction (PCR) analyses were performed with genomic DNA purified from human stool samples experimentally seeded with 1,000, 100, or 10 C. sinensis eggs. C, control with genomic DNA purified from C. sinensis adult worm (25 ng).
Detection of C. sinensis eggs in stool samples.
Figure 5.
Polymerase chain reaction (PCR) analyses were performed with gnomic DNA purified from stool samples of 15 C. sinensis- infected patients with light worm burden (EPG<999) and 10 normal humans. The patients were confirmed to be infected with C. sinensis by general microscopic method. C, control with genomic DNA purified from C. sinensis adult worm (25 ng).
Detection of C. sinensis eggs in stool samples from patients with clonorchiasis.
In conclusion, in this study, we developed a single-step conventional PCR method that could detect C. sinensis eggs and might be applicable for stool samples. Although the overall sensitivity of the PCR method is slightly lower than other previously developed real-time or nested PCR methods,14–19 its diagnostic value could be greater than other PCR methods previously developed in the consideration of low cost, time-saving, and easy application with simple instrument. The PCR method developed in the present study can be a useful alternative approach to detect C. sinensis eggs in human stool samples for clinical and epidemiological purposes in endemic areas, which in turn will contribute to the effective control of clonorchiasis.
Acknowledgments
This work was supported by Inha University research grant.
References
- 1.Rim HJ. Clonorchiasis: an update. J Helminthol. 2005;79:269–81. doi: 10.1079/joh2005300. [DOI] [PubMed] [Google Scholar]
- 2.Lun ZR, Gasser RB, Lai DH, Li AX, Zhu XQ, Yu XB, et al. Clonorchiasis: a key foodborne zoonosis in China. Lancet Infect Dis. 2005;5:31–41. doi: 10.1016/S1473-3099(04)01252-6. [DOI] [PubMed] [Google Scholar]
- 3.Choi BI, Han JK, Hong ST, Lee KH. Clonorchiasis and cholangiocarcinoma: etiologic relationship and imaging diagnosis. Clin Microbiol Rev. 2004;17:540–52. doi: 10.1128/CMR.17.3.540-552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, et al. WHO International Agency for Research on Cancer Monograph Working Group. A review of human carcinogens-Part B: biological agents. Lancet Oncol. 2009;10:321–2. doi: 10.1016/s1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- 5.Yong TS, Yang HJ, Park SJ, Kim YK, Lee DH, Lee SM. Immunodiagnosis of clonorchiasis using a recombinant antigen. Korean J Parasitol. 1998;36:183–90. doi: 10.3347/kjp.1998.36.3.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Na BK, Lee HJ, Cho SH, Lee HW, Cho JH, Kho WG, et al. Expression of cysteine proteinase of Clonorchis sinensis and its use in serodiagnosis of clonorchiasis. J Parasitol. 2002;88:1000–6. doi: 10.1645/0022-3395(2002)088[1000:EOCPOC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 7.Zhao QP, Moon SU, Lee HW, Na BK, Cho SY, Kong Y, et al. Evaluation of Clonorchis sinensis recombinant 7-kilodalton antigen for serodiagnosis of clonorchiasis. Clin Diagn Lab Immunol. 2004;11:814–7. doi: 10.1128/CDLI.11.4.814-817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma C, Hu X, Hu F, Li Y, Chen X, Zhou Z, et al. Molecular characterization and serodiagnosis analysis of a novel lysophospholipase from Clonorchis sinensis. Parasitol Res. 2007;101:419–25. doi: 10.1007/s00436-007-0481-3. [DOI] [PubMed] [Google Scholar]
- 9.Shen C, Lee JA, Allam SR, Bae YM, Han ET, Takeo S, et al. Serodiagnostic applicability of recombinant antigens of Clonorchis sinensis expressed by wheat germ cell-free protein synthesis system. Diagn Microbiol Infect Dis. 2009;64:334–9. doi: 10.1016/j.diagmicrobio.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Kane RA, Rollinson D. Comparison of the intergenic spacers and 3’ end regions of the large subunit (28S) ribosomal RNA gene from three species of Schistosoma. Parasitol. 1998;117:235–42. doi: 10.1017/s0031182098003059. [DOI] [PubMed] [Google Scholar]
- 11.Lee SU, Huh S, Sohn WM, Chai JY. Sequence comparisons of 28S ribosomal DNA and mitochondrial cytochrome c oxidase subunit I of Metagonimus yokogawai, M. takahashii and M. miyatai. Korean J Parasitol. 2004;42:129–35. doi: 10.3347/kjp.2004.42.3.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le TH, De NV, Blair D, Sithithaworm P, McManus DP. Clonorchis sinensis and Opisthorchis viverrini: development of a mitochondrial-based multiplex PCR for their identification and discrimination. Exp Parasitol. 2006;112:109–14. doi: 10.1016/j.exppara.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Park KM. Genetic comparison of liver flukes, Clonorchis sinensis and Opisthorchis viverrini, based on rDNA and mtDNA gene sequences. Parasitol Res. 2007;100:351–7. doi: 10.1007/s00436-006-0269-x. [DOI] [PubMed] [Google Scholar]
- 14.Parvathi A, Kumer HA, Prakasha BK, Lu J, Xu X, Hu W, et al. Clonorchis sinensis: development and evaluation of a nested polymerase chain reaction (PCR) assay. Exp Parasitol. 2007;115:291–5. doi: 10.1016/j.exppara.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Kim EM, Verweij JJ, Jalili A, van Lieshout L, Choi MH, Bae YM, et al. Detection of Clonorchis sinensis in stool samples using real-time PCR. Ann Trop Med Parasitol. 2009;103:513–8. doi: 10.1179/136485909X451834. [DOI] [PubMed] [Google Scholar]
- 16.Rahman SM, Bae YM, Hong ST, Choi MH. Early detection and estimation of infection burden by real-time PCR in rats experimentally infected with Clonorchis sinensis. Parasitol Res. 2011;109:297–303. doi: 10.1007/s00436-011-2253-3. [DOI] [PubMed] [Google Scholar]
- 17.Cai XQ, Yu HQ, Bai JS, Tang JD, Hu XC, Chen DH, et al. Development of a TaqMan based real-time PCR assay for detection of Clonorchis sinensis DNA in human stool samples and fishes. Parasitol Int. 2012;61:183–6. doi: 10.1016/j.parint.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Sanpool O, Intapan PM, Thanchomnang T, Janwan P, Lulitanond V, Doanh PN, et al. Rapid detection and differentiation of Clonorchis sinensis and Opisthorchis viverrini eggs in human fecal samples using a duplex real-time fluorescence resonance energy transfer PCR and melting curve analysis. Parasitol Res. 2012;111:89–96. doi: 10.1007/s00436-011-2804-7. [DOI] [PubMed] [Google Scholar]
- 19.Huang SY, Tang JD, Song HQ, Fu BQ, Xu MJ, Hu XC, et al. A specific PCR assay for the diagnosis of Clonorchis sinensis infection in humans, cats and fishes. Parasitol Int. 2012;61:187–90. doi: 10.1016/j.parint.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 20.Yong TS, Lee JH, Sim SB, Lee JW, Min DY, Chai JY, et al. Differential diagnosis of Trichostrongylus and hookworm eggs via PCR using ITS-1 sequence. Korean J Parasitol. 2007;45:69–74. doi: 10.3347/kjp.2007.45.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wongratanacheewin S, Pumidonming W, Sermswan RW, Pipitgool V, Maleewong W. Detection of Opisthorchis viverrini in human stool specimens by PCR. J Clin Microbiol. 2002;40:3879–80. doi: 10.1128/JCM.40.10.3879-3880.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller B, Schmidt J, Mehlhorn H. PCR diagnosis of infections with different species of Opisthorchiidae using a rapid clean-up procedure for stool samples and specific primers. Parasitol Res. 2007;100:905–9. doi: 10.1007/s00436-006-0321-x. [DOI] [PubMed] [Google Scholar]
- 23.Hamburger J, Xu YX, Ramzy RM, Jourdane J, Ruppel A. Development and laboratory evaluation of a polymerase chain reaction for monitoring Schistosoma mansoni infestation of water. Am J Trop Med Hyg. 1998;59:468–73. doi: 10.4269/ajtmh.1998.59.468. [DOI] [PubMed] [Google Scholar]
- 24.Hamburger J, He-Na, Abbasi I, Ramzy RM, Jourdane J, Ruppel A. Polymerase chain reaction assay based on a highly repeated sequence of Schistosoma haematobium: a potential tool for monitoring schistosome-infested water. Am J Trop Med Hyg. 2001;65:907–11. doi: 10.4269/ajtmh.2001.65.907. [DOI] [PubMed] [Google Scholar]
- 25.Yamasaki H, Allan JC, Sato MO, Nakao M, Sako Y, Nakaya K, et al. DNA differential diagnosis of taeniasis and cysticercosis by multiplex PCR. J Clin Microbiol. 2004;42:548–53. doi: 10.1128/JCM.42.2.548-553.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong SJ, Seong KY, Sohn WM, Song KY. Molecular cloning and immunological characterization of phosphoglycerate kinase from Clonorchis sinensis. Mol Biochem Parasitol. 2000;108:207–16. doi: 10.1016/s0166-6851(00)00220-6. [DOI] [PubMed] [Google Scholar]
- 27.Bae YA, Moon SY, Kong Y, Cho SY, Rhyu MG. CsRn1, a novel active retrotransposon in a parasitic trematode, Clonorchis sinensis, discloses a new phylogenetic clade of Ty3/gypsy-like LTR retrotransposons. Mol Biol Evol. 2001;18:1474–83. doi: 10.1093/oxfordjournals.molbev.a003933. [DOI] [PubMed] [Google Scholar]
- 28.Bowen NJ, McDonald JF. Genomic analysis of Caenorhabditis elegans reveals ancient families of retroviral-like elements. Genome Res. 1999;9:924–35. doi: 10.1101/gr.9.10.924. [DOI] [PubMed] [Google Scholar]
- 29.Britten RJ, McCormack TJ, Mears TL, Davidson EH. Gypsy/Ty3-class retrotransposons integrated in the DNA of herring, tunicate, and echinoderms. J Mol Evol. 1995;40:13–24. doi: 10.1007/BF00166592. [DOI] [PubMed] [Google Scholar]
- 30.Wykoff DE. Studies on Clonorchis sinensis IV. Production of eggs in experimentally infected rabbits. J Parasitol. 1959;45:91–4. [PubMed] [Google Scholar]
- 31.Song IC, Lee JS, Rim HJ. Epidemiological studies on the distribution of Clonorchis sinensis infection in Korea (in Korean). Korea Univ Med J. 1983;20:165–90. [Google Scholar]
- 32.Wilde J, Eiden J, Yolken R. Removal of inhibitory substances from human fecal specimens for detection of group A rotaviruses by transcriptase and polymerase chain reaction. J Clin Microbiol. 1990;28:1300–7. doi: 10.1128/jcm.28.6.1300-1307.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]