Abstract
Aim
The origins of ecological diversity in continental species assemblages have long intrigued biogeographers. We apply phylogenetic comparative analyses to disentangle the evolutionary patterns of ecological niches in an assemblage of European birds. We compare phylogenetic patterns in trophic, habitat and climatic niche components.
Location
Europe.
Methods
From polygon range maps and handbook data we inferred the realized climatic, habitat and trophic niches of 405 species of breeding birds in Europe. We fitted Pagel's lambda and kappa statistics, and conducted analyses of disparity through time to compare temporal patterns of ecological diversification on all niche axes together. All observed patterns were compared with expectations based on neutral (Brownian) models of niche divergence.
Results
In this assemblage, patterns of phylogenetic signal (lambda) suggest that related species resemble each other less in regard to their climatic and habitat niches than they do in their trophic niche. Kappa estimates show that ecological divergence does not gradually increase with divergence time, and that this punctualism is stronger in climatic niches than in habitat and trophic niches. Observed niche disparity markedly exceeds levels expected from a Brownian model of ecological diversification, thus providing no evidence for past phylogenetic niche conservatism in these multivariate niches. Levels of multivariate disparity are greatest for the climatic niche, followed by disparity of the habitat and the trophic niches.
Main conclusions
Phylogenetic patterns in the three niche components differ within this avian assemblage. Variation in evolutionary rates (degree of gradualism, constancy through the tree) and/or non-random macroecological sampling probably lead here to differences in the phylogenetic structure of niche components. Testing hypotheses on the origin of these patterns requires more complete phylogenetic trees of the birds, and extended ecological data on different niche components for all bird species.
Keywords: Birds, Brownian motion, comparative method, disparity through time, ecological diversification, Europe, macroecological sampling, niche conservatism, phylogenetic signal
Introduction
Birds present dazzling ecological diversity, with species differing in their climatic requirements, the habitats they use for feeding and breeding and the food resources they consume (Del Hoyo et al., 1992-2010). Ecological diversity originates when these characteristics of evolving lines of species repeatedly diverge during or between speciation events (Mayr, 1963). Beyond this fundamental fact, we do not understand for any large avifauna which characteristics have diverged little from ancestral conditions, that is, which display phylogenetic niche conservatism (PNC; Harvey & Pagel, 1991) and which have diversified substantially and account for most ecological diversity. The importance of PNC for explaining patterns of morphological and ecological characteristics is increasingly recognized (Wiens et al., 2010), to the extent that it could potentially serve as a null expectation for the diversification, or lack thereof, of avian ecological characteristics. Nonetheless, several studies of the ecological diversification of birds (Böhning-Gaese et al., 2003), mammals (Cooper et al., 2011) and amphibians (Olalla-Tarraga et al., 2011) suggest that some ecological aspects of species could undergo diversification that does not conform to the expectations of PNC. It remains unclear whether the ecological characteristics of birds differ in aspects of historical patterns of diversification, and whether or not the ecological diversification that is represented in a large bird assemblage generally conforms to an expectation of PNC.
An important debate has emerged over the last years on how PNC should be quantified or diagnosed relative to neutral expectations (Losos, 2008; Wiens et al., 2010). Perhaps the most commonly used neutral evolutionary model for this purpose is Brownian motion (BM), in which the ecological characteristics of lineages diverge at a constant rate across an isotropic phenotypic space and with which many metrics can be compared (Cooper et al., 2010). Two important properties have been widely used in comparative biology to formulate tests of deviations from the patterns expected under BM. First, phylogenetic signal expresses the case in which related species tend to share similar ecological characteristics; second, the degree of evolutionary gradualism expresses whether niche divergence increases gradually over time or whether niches diverge punctually, that is, independently of time. These two properties can be quantified with the Pagel metrics lambda and kappa (Pagel, 1997; Pagel, 1999), which provide insightful descriptions of phylogenetic patterns of species niches. Nonetheless, recent simulation work suggests the limited usefulness of these metrics alone in diagnosing PNC because different rates of niche evolution (different degrees of niche conservatism) can yield a similar phylogenetic signal, while a low phylogenetic signal can also emerge under strong niche conservatism (Revell et al., 2008; Ackerly, 2009). Another line of evidence can arise through examination of the temporal course of multivariate niche diversification, in a way that is independent of constraints imposed by evolutionary models, by conducting an analysis of multivariate disparity through time (DTT; Harmon et al., 2003). This analysis compares the observed course of ecological diversification to expectations from a Brownian motion model in a multivariate niche space. Our objectives in this paper are to examine whether phylogenetic patterns of ecological niches of European birds support the hypothesis of PNC and to compare degrees of ecological niche diversification in the climatic requirements, habitat use and trophic habits of these birds.
While patterns of species diversification in birds are becoming increasingly clear (Jetz et al., 2012), comprehensive ecological data to evaluate and compare ecological diversification broadly across many ecological characteristics are generally lacking for regional avifaunas. The European avifauna provides a notable exception because sufficient observational data exist to evaluate multiple components of Hutchinson's realized environmental niche (Hutchinson, 1978). In particular, the members of this avifauna can be distinguished using existing data to describe three niche components: the habitat niche (often called the Grinnellian niche; Grinnell, 1917), the climate niche (Austin et al., 1990) and the trophic niche (often called the Eltonian niche; Elton, 1927). Although other niche concepts could be considered (Chase & Leibold, 2003), we place our work in the conceptual framework recently proposed by Devictor et al. (2010), which allows depiction of all niche realms based on the same multivariate Hutchinsonian space. We focus on the habitat and trophic niches of birds because of their historical importance and prevalence in the ecological literature, their continued persistence in ongoing discussions regarding niche and their straightforward operationalization for use in quantitative analysis of observational data. We also include the climate niche because, in the case of birds at least, the distinction between physical habitat and climate plausibly reflects the larger geographical scale of the latter, in which variation in the former may be nested. Interestingly, these three components of species niches have never been investigated jointly in a comprehensive framework.
The literature on avian ecology and evolution provides alternative empirical hypotheses regarding the relative contribution of the three niche components to the evolution of avian ecological diversity. Potentially, phylogenetic lability of any one of the three niche components, climate, habitat or trophic, could be primarily responsible for the ecological diversity of European birds, resulting in four testable hypothesis (Table 1). Vicariant speciation and climate cycling have frequently exposed forming avian species to different climate regimes and may play a large role in diversification (Lovette, 2005), suggesting (1) a principal role for climate niche lability in ecological diversification. However, feeding generalization has been linked to avian diversification rates (Phillimore et al., 2006), which suggests (2) a leading role for variation in the trophic niche in ecological diversification. Similarly, divergent habitat use for breeding and feeding between closely related avian species has long been recognized (Lack, 1944), implying (3) an important role for the diversification of habitat use in overall ecological diversification. A fourth hypothesis arises directly from the claim of the pervasiveness of PNC (Wiens et al., 2010). In this case, all three niche components should diversify in accordance with (or slower than; Losos, 2008) a BM model of trait diversification (Table 1).
Table 1.
Hypotheses for the primary phylogenetic origin of ecological diversity in Western Palaearctic birds
| Hypothesis | Potential processes | Potential supporting evidence |
|---|---|---|
| Climate occupation divergence | Global climate cycling and geological processes separate breeding populations geographically (vicariance); speciation occurs with adaptation to regional climate | Climate niche has the greatest disparity, smallest phylogenetic signal and greatest divergence from a Brownian evolutionary model |
| Habitat divergence | Interspecific competition occurs as species occupy habitat used for feeding and reproduction, causing habitat use to diverge as in ecological character displacement | Habitat niche has the greatest disparity, smallest phylogenetic signal and greatest divergence from a Brownian model |
| Trophic divergence | Interspecific competition occurs for food resources, resulting in character and food preference displacement between closely related species | Trophic niche has the greatest disparity, smallest phylogenetic signal and greatest divergence from a Brownian model |
| Niche conservatism | Lineages gradually diverge due to responses of ecological traits to selection that is random in direction, and uncorrelated temporally and among species | All three niches show similar levels of disparity and phylogenetic signal, both of which are consistent with a Brownian model of evolutionary divergence; trait divergence is consistent with evolutionary gradualism |
In this paper, we compare phylogenetic patterns of trophic, habitat and climatic niches expressed during the breeding season in an assemblage of 405 birds that breed in Europe, west of the Urals. First, we test for phylogenetic signal and gradualism in each of the three niche components (Pagel, 1997, 1999). Second, we reconstruct past diversification of niches based on the computation of multivariate disparity over time (see Fig. S1 in Appendix S1 in the Supporting Information). We then compare the results for each niche component with the diversification that would be expected under BM. These two approaches allow us to finally examine the four alternative hypotheses for the primary origin of ecological diversity in this avian assemblage.
Methods
We selected all European breeding bird species for which global breeding distribution data were available, a total of 405 species. We obtained data on the global breeding distribution of these species from a global database of avian species distributions (Birdlife International and Natureserve, 2012). These data represent the best available information on the global breeding distribution of this avian assemblage. We converted polygons to species presence and absence on a 10-arcmin resolution grid. We used the entire global distribution of each species in order to avoid missing portions of the species realized climate niche. We focused on ‘The Complete Birds of the Western Palearctic’ CD-ROM (Perrins & Ogilvie, 1998) as the data source for the habitat and trophic niches of these avian species because of its extensive treatment and literature review of habitat use, foraging behaviour, foraging location, food items and nesting characteristics. We addressed niche components only during the breeding season because distribution and ecological data regarding this season are much more robust than wintering data, especially for tropical migrants. We excluded from consideration all species feeding exclusively in the pelagic zone during the breeding season because data are lacking to meaningfully distinguish trophic differences among species.
We decomposed patterns of diversification for all niche axes separately for each niche component (climatic, habitat and trophic). We applied multivariate analyses to continuous and discrete ecological characters to produce species-specific values on orthogonal niche axes. We selected ordination methods appropriate for analysis of categorical or continuous variables, or a mixture of the two, and which produced continuous numeric values that were compatible with the phylogenetic comparative methods we employed (detailed below). We included in the analysis a number of axes from each niche ordination sufficient to describe a uniform threshold percentage (90%) of the total ecological variation in each niche. Thus, the number of ordination axes varied among niches, but we needed to ensure that any differences among niches during phylogenetic comparative analysis were not due to consideration of arbitrary and variable percentages of ecological variation among niches. Use of a standard proportion of ecological variation for each set of variables enabled us to make consistent and comparable estimates of variability for the analysis of ecological divergence of species in terms of the three niche components. All ecological ordinations were conducted in R (R Development Core Team, 2013) with the package ade4 (Dray & Dufour, 2007).
Trophic niche
The 35 variables of the trophic niche (see Table S1 in Appendix S1; Elton, 1927) were non-exclusive, focused on the breeding season and included variables that characterized food type (14 variables), behaviours used in acquiring food (9), substrate from which food is taken (9) and the period of day during which a species forages actively (3). The variables were scored as either 0 or 1, with the exception of body weight, which was scored as an average of values provided for individuals weighed during the breeding season. Body weight was included as a trophic variable because body weight is likely to correlate with total energy needs and prey size (Price et al., 2000). A food type variable (e.g. small birds, seeds, invertebrates, etc.) was scored as 1 when the substance reportedly formed more than 10% of stomach contents, 10% or more of observed foraging successes or when the species was described as principally foraging on a particular item based on qualitative observation. Items described exclusively as being taken only ‘rarely’, ‘occasionally’, or presented as a unique observation for a species, led to a score of 0 being assigned to the type of food item under consideration (see the Table of Avian Niche Traits in Appendix S2 for species scores).
Species received a score of 1 on each foraging behaviour attributed to the species, unless the species was described as ‘rarely’ performing a behaviour. The nine substrate variables received a score of 1 if specified in the description of foraging behaviours for species. Three variables (nocturnal, crepuscular and diurnal) described the daily period of foraging activity and were scored as 1 if the species was reported as being active (or ‘occasionally active’) during that period. When information existed regarding variation in trophic variables between the breeding and non-breeding seasons, only data concerning the breeding season (e.g. food type) were scored as being used. We scored behaviours and food types as unused (0) when they were reported for the species but exclusively from a geographical area outside Europe. Body weight was centred and normalized to unit standard deviation before ordination. Hill–Smith ordination (Hill & Smith, 1976) was used to summarize the matrix of binary and mixed variables, and provided scores for each species on orthogonal axes.
Habitat niche
A total of 38 habitat variables (Table S1 in Appendix S1) were distinguished from climatic variables because the former are central to the original definition of the habitat niche (Grinnell, 1917). We developed 18 variables to describe habitat preferred for nesting (breeding habitat niche). Three mutually exclusive variables were used to describe nest position (elevated > 1 m in a tree or bush; tree-hole; ground or other substrate or surface). Other nesting habitat categories were not mutually exclusive and a score of 1 was assigned to additional breeding habitat variables to describe the nesting preferences of the species. An additional 20 variables described the habitat used for foraging (the foraging habitat niche). In both types of habitat niche, all types of wet grassland (wet tundra, fen, sedge meadow, seasonally-flooded meadow, etc.) were pooled. Dry grassland, steppe and agricultural fields were pooled and shrub and bush were also pooled. These grassland variables were presented as both foraging and breeding habitats.
Sand, beach and gravel were also pooled into one category that was present as both breeding and foraging niche variables. Mud flats and wind- or water-deposited silt habitat were pooled but only as considered foraging habitat. Near-shore marine habitat was a foraging variable that was scored for species feeding in marine waters, within 500 m of the shore during the breeding season. Forest edge was scored when the species description specified that this habitat was used for foraging. We pooled naturally open forest, open forest caused by disturbance and early stage successional forest, and included the category in both the breeding and foraging habitat niches. We distinguished garden habitats and urban environment to accommodate European species with populations that breed and/or forage in one or both of these anthropogenic habitat types. Variation in habitat niche among species was quantified using multiple correspondence analysis, which is appropriate for categorical variables (Tenenhaus & Young, 1985).
Climatic niche
We estimated the breeding distribution of each species by determining both resident and breeding areas from digital maps from the Birdlife International database of bird distributions and gridding them to 10-arcmin resolution (Birdlife International and Natureserve, 2012). Climate variables at 10-arcmin resolution were obtained from the WorldClim database (Hijmans et al., 2005; http://www.worldclim.com). We used climate variables for the months of April to September as a period when European species would be on the nest and/or fledging chicks. The variables captured range, central tendency and variability of temperature and precipitation during the 6-month period (Table S1 in Appendix S1). This span of months should capture much intra- and interspecific variation in timing of reproduction throughout the European range of the focal species, but excludes the mid-winter period that is irrelevant to the breeding conditions of almost all species except some large, northern owls (Perrins & Ogilvie, 1998). We judged the alternative of directly taking only climate variation during nesting as being of too fine a scale for our purposes, likely to obscure broad geographical differences in climate and needing comprehensive observational data across large areas, elevations and times of year for all the species. These latter data were unavailable at the comprehensive level necessary to cover an analysis of all 405 species. The monthly averages of the climate variables we used were determined over a base period of the years 1951 to 2000. We estimated species climatic niches (Austin et al., 1990) using the outlying mean index (OMI) analysis (Dolédec et al., 2000). Climatic niche position for each species along the two selected axes was extracted by projecting species centroids orthogonally to the ordination axes (Thuiller et al., 2004). We used these values as measures of preferred climate during the breeding season.
Phylogenetic reconstruction
We used the phylogenetic tree of European birds constructed for a previous study (Thuiller et al., 2011), following a published pipeline (Roquet et al., 2013). To do so we retrieved from GenBank: 10 mitochondrial gene regions (12S, ATP6, ATP8, COII, COIII, ND1, ND3, ND4, ND5, ND6) plus six nuclear ones (28S, c-mos, c-myc, RAG1, RAG2, ZENK). The percentage of genetic sampling of species varied among regions (Table S2 in Appendix S1). The sequences were aligned with several algorithms, including ClustalW (Larkin et al., 2007), Kalign (Lassmann & Sonnhammer, 2006), mafft (Katoh & Toh, 2010) and muscle (Edgar, 2004), and checked by eye. The best alignment for each region was selected with mumsa (Lassmann & Sonnhammer, 2006) and depurated with TrimAl (Capella-Gutierrez et al., 2009). Then, DNA matrices were concatenated to obtain a supermatrix. We conducted phylogenetic analyses using maximum likelihood within RaxML (Stamatakis et al., 2008), applying the GTRCAT model with partitions defined for each region, and a prior tree constraint at the ordinal level for the birds dataset (based on Hackett et al., 2008). The inferred tree generally had a satisfying level of node support (for details on bootstrap and support analyses, refer to the supplementary materials of Thuiller et al., 2011). We kept 100 most likely trees for our analysis to account for the uncertainty associated with phylogenetic inference. We dated these trees by applying penalized likelihood with r8s (Sanderson, 2003) and fossils relevant for 14 clades (Table S3 in Appendix S1). The best ML tree can be found in Treebase (http://treebase.org/treebase-web/home.html; study number 10770).
Phylogenetic signal and gradualism of niche traits
For each niche component we evaluated the degree to which evolutionary relatedness of species was correlated to ecological similarity (i.e. shows phylogenetic signal) by examining values of Pagel's lambda and kappa (Pagel, 1997), which quantify phylogenetic signal and gradualism respectively. A lambda value of 1 indicates a magnitude of phylogenetic signal that is consistent with that produced by a Brownian evolutionary model, while a value of 0 indicates the absence of phylogenetic signal. A kappa value of 1 indicates evolutionary gradualism, with character change proportional to branch length. Kappa values near 0 indicate that character change is independent of divergence time, and suggest rapid change at or immediately following speciation. The estimation of these parameters and the test of their significance relative to 0 and 1 were performed based on likelihood optimization, using the motmot package in R (Thomas & Freckleton, 2012). For phylogenetic signal, lambda was preferred to another metric, Blomberg's K, because a recent simulation study showed that lambda estimated the expected strength of the phylogenetic signal better than other metrics, and was insensitive to tree size and tree uncertainty (Münkemüller et al., 2012). We chose not to make inferences based on the third Pagel index, delta, which captures the timing (late versus recent) of ecological diversification. Calculation of this parameter often causes computation problems as it does not increase asymptotically with increasingly late character divergence, and hence becomes impossible to estimate when diversification occurs very late on the tree. Nonetheless, late diversification is well captured by lambda, which is negatively correlated with delta estimates in simulated data (data not shown).
We calculated lambda and kappa indices using the species values on each of the selected ordination axes from analyses of niches. We repeated the calculations across 100 nearly maximum likelihood trees to account for topological uncertainty in the species phylogeny, then averaged estimates across these trees for all ecological axes. The axes of ecological variation pinpointed by the multivariate ordinations of climatic, habitat and trophic niches had different associated values of inertia. We weighted the lambda and kappa estimates for each axis of the climatic, habitat and trophic niches by the associated inertia, in order to retrieve separate weighted summary estimates of phylogenetic signal and gradualism for each niche component.
Disparity through time
The analyses of disparity through time were based on the calculation of Euclidian interspecific ecological distances for each niche component (R package cluster; Maechler et al., 2013). These distances were computed in the multivariate space defined by species scores on the selected axes of the corresponding ordination. We used these distance matrices to compute the disparity (mean ecological distance) within all extant subclades at each time step, marked by node ages (Fig. S1 in Appendix S1). After standardizing disparity values between 1 and 0, we plotted the values for all subclades against the age of the parent node subtending these subclades as originally done by Harmon et al. (2003). The temporal profile of niche disparity was compared with that obtained from 1000 multivariate Brownian simulations, which were based on the inferred variance–covariance matrix of the niche component (Revell & Collar, 2009). We accounted for phylogenetic uncertainty by repeating the calculations using the 100 most-likely phylogenetic trees. The deviation between observed disparity and Brownian simulations was quantified using the minimum discrimination index (MDI) statistic, which estimated the area between the curves of observed disparity and mean Brownian simulated disparity (as in Harmon et al., 2003). This analysis was repeated identically for the climatic, habitat and trophic niches. All these steps were performed using available functions in the package geiger (Harmon et al., 2003, script available upon request). The comparisons of empirical and modelled disparity are robust to incomplete taxon sampling that generally occurs on assemblage trees because empirical disparity curves were compared with corresponding neutral (Brownian) expectations based on phylogenetic divergences among the species. All disparity analyses were performed using both weighted and unweighted species scores on each axis of ecological variation to determine whether our results were robust to the unequal inertia among axes in niche ordinations.
Results
The first 22 of 35 axes of the Hill–Smith ordination of the trophic niche capture 90% of trophic niche differentiation among all species. The first 24 of 38 axes of the multiple correspondence analysis of the habitat niche capture 90% of differentiation among all species. In both these ordinations, the variation ascribed to the successive axes is fairly evenly distributed (Fig. S2 in Appendix S1). The first two of the eight axes of the OMI analysis of climatic niche values capture 96% of climatic niche differentiation between species, with the first axis explaining 86.5% of total inertia. Correlations among ordination axes of different niches are generally low, indicating that the three niches can be considered independent (Fig. S3 in Appendix S1). A high proportion of nodes of the maximum likelihood phylogenetic tree are supported by RaxML bootstrap analyses (BS): 71.1% of the nodes had BS equal or higher than 70%; 8.2% of the nodes received low support (BS of 50–70%); and 20.8% of the nodes receive no support (BS < 50%).
Phylogenetic signal and gradualism
Estimated lambda and kappa values, across combinations of niche axes and phylogenetic trees, are significantly lower than 1, which is less than expected under a scenario of Brownian evolution. Non-zero lambda estimates show that the three niche components exhibit moderate and significant phylogenetic signal (Fig. 1a) for most combinations of ecological axes and trees except for the habitat niche, which exhibits non-zero phylogenetic signal for 83% of the cases. The weighted average of lambda indicates that phylogenetic signal is greater for trophic niches than for climatic and habitat niches, which are similar. These trends demonstrate that related species of European birds resemble one another less in the climates they prefer during breeding, and in the habitats they use, than in their food preferences (Fig. S4 in Appendix S1).
Figure 1.
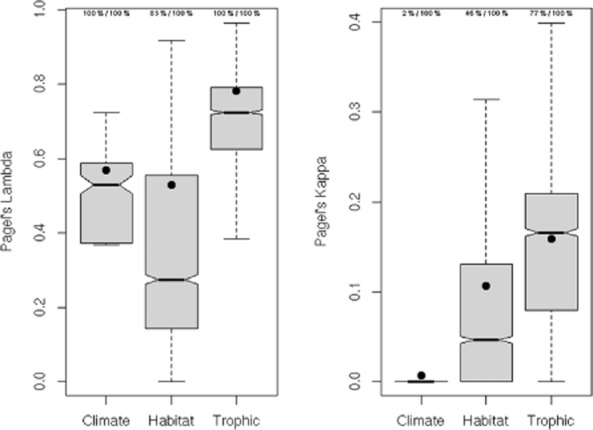
Boxplots of Pagel's lambda and kappa as measures of phylogenetic signal and gradualism, respectively, estimated for all ordination axes of ecological variation relevant to climatic, habitat and trophic niches. Replicate estimates of lambda and kappa for each axis were made using 100 nearly maximum likelihood phylogenetic trees. The values at the top of each box give the percentage of tests (across trees and axis combinations) that yielded lambda or kappa estimates that were significantly greater than 0 and lower than 1, respectively, based on likelihood ratio tests. To account for the importance of each ordination axis, lambda and kappa values were weighted according to the variances (eigenvalues) of the corresponding ordination axis. The box waist indicates the median score. Filled black points indicate the weighted average estimates of lambda and kappa for climatic, habitat and trophic multivariate niches, across trees and axes.
Estimates of kappa are generally low for the three niche components, ranging from 0 to 0.4 across all niche axes and phylogenetic trees, and are always significantly lower than 1 and, thus, lower than the BM expectation (Fig. 1b). However, kappa values estimated on certain combinations of ecological axes and phylogenetic trees are not significantly different from 0, and the proportion of non-zero kappa values increases from climatic to habitat and then trophic niches (2, 46 and 77%, respectively). Accordingly, the average estimates of kappa are lowest (very close to 0) for the climatic niche, with kappa values for the habitat niche and the trophic niche being successively greater. These trends indicate that evidence for punctual evolution is strongest for the climatic niches and somewhat less for the habitat and trophic niches.
Disparity through time analysis
Results for weighted and unweighted disparity are virtually the same, and we report only those results obtained with weighted scores. Observed disparity for all three niche components exceeds the corresponding levels expected under a Brownian model of diversification (Fig. 2). Multivariate disparity of the climatic niche of European breeding birds exceeds that of the other two niche components through most of the time span (Fig. 2). Similarly, disparity values for the climatic and habitat niches are also consistently greater than expected under the Brownian model, beginning early on the trees (Fig. 2). Further, the observed disparity of the trophic niche, while similar to the disparity for the other two niche components, is of lower magnitude. MDI scores estimated for each curve of the three niches (Fig. S5 in Appendix S1) further support highest overall disparity for the climate niche and lower deviation of disparity from the Brownian expectation for the trophic niche.
Figure 2.
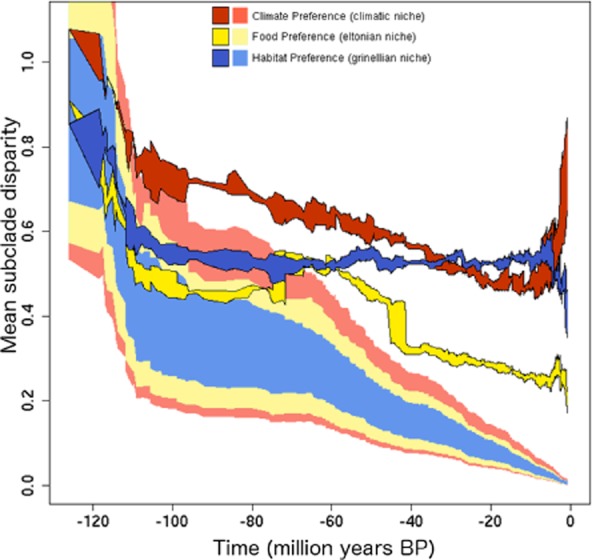
Plots of observed disparity through time (DTT) for observed data and results from Brownian evolution models for three components of the environmental niches of 405 species of European birds. The span of solid colors in the vertical direction indicates the range of disparity values that arises from topological uncertainty across 100 nearly maximum likelihood phylogenetic trees. Values of disparity for all three niches (saturated colours) differ from neutral expectations derived from 1000 Brownian evolution simulations of each niche (pale colours).
Discussion
A lack of support for phylogenetic niche conservatism
Together, our analyses consistently show that patterns of ecological niches of European birds diverge from those expected under PNC. Although patterns tend to vary among the three niche components, estimated values of phylogenetic signal and gradualism are systematically lower than expected under a Brownian model. Operative niche conservatism should have generated values indistinguishable from or greater than those expected under the BM assumption. In particular, we find a recurrent tendency towards punctualism for the three niches, suggesting that niche shifts have been largely decoupled from evolutionary time. Consistent with the low values of these indices, within-clade disparity for each of the niche components exceeds expectations under neutral (Brownian) niche divergence. Our results suggest that PNC, though potentially widespread (Wiens et al., 2010), is not an operative principle in the ecological diversification of European birds. This has an important implication for understanding the evolution and adaptation of species to changing environments. The gradualist view of evolution suggests that natural selection is often very weak, causing little change to accumulate over time, and that evolution ends up being uncoupled from ecology (for a recent review see Bell, 2013). Our results suggest that (1) niche evolution deviates from gradualism in birds and can sometimes be rapid and independent of time and thus (2) emphasize the potential for detectable relationships between macroevolutionary patterns of niche evolution and ongoing diversification of ecological traits (Salamin et al., 2010).
Scenarios of ecological diversification in different niche components
Four potentially conflicting hypotheses attribute the primary origin of avian ecological diversity to: (1) greater divergence of the climatic niche arising from global climate cycling and geographical barriers (Lovette, 2005; Outlaw & Voelker, 2008); (2) greater divergence of habitat use (Lack, 1944; Richman & Price, 1992; Cicero & Johnson, 1998); (3) greater divergence of trophic preferences (Richman & Price, 1992; Grant & Grant, 2006); or (4) a level of divergence consistent with neutral evolutionary drift (Table 1; Felsenstein, 1985; Wiens et al., 2010). Our results suggest that neutral (BM) evolution (4) alone is not adequate to explain the variety of among-niche patterns of disparity we observe. However, each of the other hypotheses has some support in the literature and differences in degree of diversification of the three niche components could exist.
Ecological diversification of the climatic niche, as shown in the DTT analysis, is somewhat greater than that of the other two niches. This suggests the hypothesis that the primary evolutionary driver of ecological diversity held in these birds arises from greater divergence in species climatic requirements, which have sometimes occurred quite abruptly (hence the evidence for strong punctualism). One potential mechanism for this involves global climate cycling, which has probably driven species repeatedly in and out of geographically isolated refugia in the Northern Hemisphere and promoted vicariant speciation (Lovette, 2005). Such processes could have operated since the Pliocene (Outlaw & Voelker, 2008) and could be common compared with mechanisms of sympatric speciation (Phillimore et al., 2008). Nonetheless, ecological diversity in European birds is the product of ecological diversification in many different clades, many members of which are not present in Europe. Knowledge of these species would be necessary for an unbiased comparison of rates of niche evolution. Relatively rapid diversification of the climatic niche, if so confirmed, would contrast with rates of niche evolution in tropical avian assemblages, where the effects of climate cycling on niche diversification could be less important (Lovette, 2005). In contrast, our results suggest that evolutionary divergence of habitat preferences could contribute to ecological diversity, secondarily to diversification of the climatic niche. This finding contrasts with previous findings that divergent preferences for feeding and breeding habitat exist among many closely related avian species (Lack, 1944), and may be the dominant contributor to ecological diversification in certain avian groups (Richman & Price, 1992; Cicero & Johnson, 1998).
The analyses suggest that there is greater phylogenetic signal in the trophic characteristics of the species in this assemblage than in the other two niche components, and that trophic niches tend to diversify in a somewhat less punctuated (or more gradual) manner than other niches. The limited diversification of trophic habits in comparison with the other two niche components could be due to relatively slow evolutionarily rates. For example, constraints might occur from slow or infrequent divergence of specialized morphology that is associated with resource capture and feeding, and which may have diversified relatively early in the history of different lineages. Our results are partially consistent with earlier studies that report evidence for phylogenetic signal in the trophic niche of the assemblage of European birds. Böhning-Gaese and Oberrath (1999) use Mantel tests on an ecological dissimilarity matrix and a matrix representing phylogenetic distance among 151 species to find phylogenetic signal in both diet and breeding habitat. Similarly, Brandle et al. (2002) examines taxonomic levels to find that significant phylogenetic conservatism exists in the dietary niche breadth of 142 European bird species at the level of family and genus. Nevertheless, one potentially testable hypothesis is that low levels of disparity of trophic preferences (compared with the other niches) arise because macroecological sampling preferentially ‘selects’ lineages having particular trophic characteristics. Such sampling, and not variation in evolutionary rates, could be the cause of this pattern.
Niche disparity, conservatism and species sampling
Several processes other than simple heterogeneity in evolutionary rates and degree of niche diversification may influence the patterns we report. For instance, we focus on disparity as arising from variation in niche position and ignore possible effects of changes in niche breadth. The development of methods to address niche breadth in a comparative analysis (e.g. Kostikova et al., 2013) may shed additional light on differences among niches in phylogenetic patterns and underlying processes. Observed differences in DTT among niches could also involve heterogeneity in rates of niche evolution within the complete clade from which the European avifauna is drawn (Diniz-Filho et al., 2010). Such non-stationarity in rates of niche diversification, in combination with incomplete and biased macroecological sampling from within the encompassing clade, might create the appearance of heterogeneity among niche components in their DTT values. However, this heterogeneity might not be a good representation of any rate heterogeneity in the full tree of birds. Another possibility is that effects of macroecological sampling bias could arise when the birds of the European assemblage are not a random sample from the inclusive clade of birds of which they are members. As macroecological sampling would affect which species are found in Europe, the climatic niche characteristics of the assemblage might be notably influenced.
Macroecological sampling of the lineages in this assemblage could involve additional evolutionary processes that are not identified as niche evolution per se. These might include range evolution during vicariance events and the development of migratory behaviour (Lundberg, 1988; Perez-Tris et al., 2004; Mila et al., 2006), which may eventually precipitate evolutionary shifts in resource use or habitat selection. One effect of the evolution of migratory behaviour on niche characteristics is that the environmental niche of migratory birds during the breeding season may be less conserved than in the winter range (Martinez-Meyer et al., 2004). This suggests that species niche characteristics during residency in the breeding range may become labile during the evolution of migratory behaviour. Change in migratory patterns could also contribute to geographical variation within species in their niche characteristics (Martinez-Meyer et al., 2004), as in the case of partial migration (Lundberg, 1988). Such potential effects of changes in migration strategies and geographical range on niche evolution have been virtually unexplored from a macroevolutionary perspective.
Differences in methodologies used for quantification of the three different niche components could conceivably contribute to variability among species and translate into differences in diversification among the three niche components. While there is certainly error in all the empirical data reported here, we see no reason to suspect that niche variability among species, and resulting patterns of diversification, should be biased toward one niche component or another. We made every effort to carefully evaluate and include all ecological data from our sources that were pertinent to the European avifauna. Preliminary analyses conducted previously use only climate niche values based solely on the European (not global) distribution of the study species. The current analyses derive climate occupancy from polygons that express global species ranges, and which may contain areas of false presence. Preliminary analyses with restricted atlas data on species distributions show even greater diversification in the climate niche than in the other components (results not presented). Thus, our use of polygon data on species global ranges probably results in conservative differences in disparity between the climate niche and the other two niche components.
Conclusions
Over the history and diversity represented by the European avifauna, we find no support for phylogenetic conservatism in any of the three multivariate niche components we investigated. While PNC has been proposed as an evolutionary principle, this perspective may arise from focus on a small number of ecological characteristics or a biased sample thereof, while our ecological description of species niches is likely to capture most aspects of climatic requirements, habitat selection and trophic habits. Alternatively, conservatism of some avian niche components may be greater in other regions of the world, some of which (e.g. tropical areas) are much more species rich than Europe or have been diversifying under stronger environmental stability. Several processes may contribute to the patterns we observe here, including the processes of macroecological sampling and relative rates of ecological diversification of niche components. Hypotheses regarding the importance of variation in underlying evolutionary rates for variation in ecological disparity, phylogenetic signal and gradualism may be testable with greater ecological and phylogenetic information. Finally, the application of additional methods of comparative analysis to large assemblages is likely to reveal new insights that can drive hypothesis generation and testing.
Acknowledgments
We thank two anonymous referees whose comments contributed to the improvement of earlier manuscript versions. This research received funding from the European Research Council under the European Community's Seven Framework Programme FP7/2007–2013 Grant Agreement no. 281422 (TEEMBIO). This work was also funded by a French ANR project called EVORANGE (ANR-09-PEXT-011). P.B.P. and N.E.Z. acknowledge support from EU grant ENVCT-2009-226544 ‘MOTIVE’. P.B.P. acknowledges support of Swiss National Fund grant 31003A_122433 ‘ENNIS’. R.W. was supported by Swiss National Fund grant CRSII3_125240 ‘SPEED’. C.R. was supported by a Fundación Ramón Areces postdoctoral grant when she conducted the phylogenetic inference.
Biosketch
Peter B. Pearman is a broadly trained ecologist who is interested in the relationships between organisms and their biotic and abiotic environments and how these relationships influence species distributions. His recent work has addressed potential effects of climate change on species richness patterns in the Swiss Alps and on intraspecific genetic diversity, including in large African mammals. He has additional interests in the use of phylogeographical information to inform and constrain distribution models, and the application of these constrained models to questions in palaeoecology. All co-authors share interests in the evolution of ecological diversity, in the context of the niche concept and functional traits of plants and animals. They apply phylogenetic comparative methods to understand the development and distribution of diversity at large geographical scales, and to identify threats to the persistence of biodiversity.
Author contributions: All authors contributed to conception of the project. P.B.P. led the writing, constructed the matrix of trophic and habitat niche values and contributed to ecological ordinations and comparative analyses; S.L. led comparative analyses and provided R scripts; C.R. constructed the dated phylogenetic trees; R.W. contributed to constructing the ecological niche matrix; W.T. constructed matrix of species occurrence and environmental data, and provided scripts for ecological ordinations. All authors contributed substantially to the writing and revision.
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher's web-site.
Supporting figures and tables.
Table of avian niche traits.
References
- Ackerly D. Conservatism and diversification of plant functional traits: evolutionary rates versus phylogenetic signal. Proceedings of the National Academy of Sciences USA. 2009;106:19699–19706. doi: 10.1073/pnas.0901635106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin MP, Nicholls AO. Margules CR. Measurement of the realized qualitative niche: environmental niches of 5 Eucalyptus species. Ecological Monographs. 1990;60:161–177. [Google Scholar]
- Bell G. Evolutionary rescue and the limits of adaptation. Philosophical Transactions of the Royal Society B: Biological Sciences. 2013;368:20120080. doi: 10.1098/rstb.2012.0080. doi: 10.1098/rstb.2012.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdlife International and Natureserve. Bird species distribution maps of the world. Cambridge, UK and Arlington, USA: BirdLife International and NatureServe; 2012. [Google Scholar]
- Böhning-Gaese K. Oberrath R. Phylogenetic effects on morphological, life-history, behavioural and ecological traits of birds. Evolutionary Ecology Research. 1999;1:347–364. [Google Scholar]
- Böhning-Gaese K, Schuda MD. Helbig AJ. Weak phylogenetic effects on ecological niches of Sylvia warblers. Journal of Evolutionary Biology. 2003;16:956–965. doi: 10.1046/j.1420-9101.2003.00605.x. [DOI] [PubMed] [Google Scholar]
- Brandle M, Prinzing A, Pfeifer R. Brandl R. Dietary niche breadth for Central European birds: correlations with species-specific traits. Evolutionary Ecology Research. 2002;4:643–657. [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM. Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase JM. Leibold MA. Ecological niches: linking classical and contemporary approaches. Chicago, IL: University of Chicago Press; 2003. [Google Scholar]
- Cicero C. Johnson NK. Molecular phylogeny and ecological diversification in a clade of New World songbirds (genus Vireo. Molecular Ecology. 1998;7:1359–1370. doi: 10.1046/j.1365-294x.1998.00483.x. [DOI] [PubMed] [Google Scholar]
- Cooper N, Jetz W. Freckleton RP. Phylogenetic comparative approaches for studying niche conservatism. Journal of Evolutionary Biology. 2010;23:2529–2539. doi: 10.1111/j.1420-9101.2010.02144.x. [DOI] [PubMed] [Google Scholar]
- Cooper N, Freckleton RP. Jetz W. Phylogenetic conservatism of environmental niches in mammals. Proceedings of the Royal Society B: Biological Sciences. 2011;278:2384–2391. doi: 10.1098/rspb.2010.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Hoyo J, Elliott A. Sargatal J. Handbook of birds of the world, vols 1–15. Barcelona: Lynx Edicions; 1992–2010. [Google Scholar]
- Devictor V, Clavel J, Julliard R, Lavergne S, Mouillot D, Thuiller W, Venail P, Villeger S. Mouquet N. Defining and measuring ecological specialization. Journal of Applied Ecology. 2010;47:15–25. [Google Scholar]
- Diniz-Filho JAF, Terribile LC, Da Cruz MJR. Vieira LCG. Hidden patterns of phylogenetic non-stationarity overwhelm comparative analyses of niche conservatism and divergence. Global Ecology and Biogeography. 2010;19:916–926. [Google Scholar]
- Dolédec S, Chessel D. Gimaret-Carpentier C. Niche separation in community analysis: a new method. Ecology. 2000;81:2914–2927. [Google Scholar]
- Dray S. Dufour AB. The ade4 package: implementing the duality diagram for ecologists. Journal of Statistical Software. 2007;22:1–20. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton C. Animal ecology. London: Sidgwick and Jackson; 1927. [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. The American Naturalist. 1985;125:1–15. doi: 10.1086/703055. [DOI] [PubMed] [Google Scholar]
- Grant PR. Grant BR. Evolution of character displacement in Darwin's finches. Science. 2006;313:224–226. doi: 10.1126/science.1128374. [DOI] [PubMed] [Google Scholar]
- Grinnell J. The niche-relationships of the California thrasher. Auk. 1917;34:427–433. [Google Scholar]
- Hackett SJ, Kimball RT, Reddy S, Bowie RCK, Braun EL, Braun MJ, Chojnowski JL, Cox WA, Han KL, Harshman J, Huddleston CJ, Marks BD, Miglia KJ, Moore WS, Sheldon FH, Steadman DW, Witt CC. Yuri T. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- Harmon LJ, Schulte JA, Larson A. Losos JB. Tempo and mode of evolutionary radiation in iguanian lizards. Science. 2003;301:961–964. doi: 10.1126/science.1084786. [DOI] [PubMed] [Google Scholar]
- Harmon LJ, Weir JT, Brock CD, Glor RE. Challenger W. GEIGER: investigating evolutionary radiations. Bioinformatics. 2008;24:129–131. doi: 10.1093/bioinformatics/btm538. [DOI] [PubMed] [Google Scholar]
- Harvey PH. Pagel M. The comparative method in evolutionary biology. Oxford: Oxford University Press; 1991. [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG. Jarvis A. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology. 2005;25:1965–1978. [Google Scholar]
- Hill MO. Smith AJE. Principal component analysis of taxonomic data with multi-state discrete characters. Taxon. 1976;25:249–255. [Google Scholar]
- Hutchinson GE. An introduction to population ecology. New Haven, CT: Yale University Press; 1978. [Google Scholar]
- Jetz W, Thomas GH, Joy JB, Hartmann K. Mooers AO. The global diversity of birds in space and time. Nature. 2012;491:444–448. doi: 10.1038/nature11631. [DOI] [PubMed] [Google Scholar]
- Katoh K. Toh H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 2010;26:1899–1900. doi: 10.1093/bioinformatics/btq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostikova A, Litsios G, Salamin N. Pearman PB. Linking life-history traits, ecology and niche breadth evolution in North American eriogonoids (Polygonaceae) The American Naturalist. 2013;182 doi: 10.1086/673527. doi: 10.1086/673527. [DOI] [PubMed] [Google Scholar]
- Lack D. Ecological aspects of species-formation in passerine birds. Ibis. 1944;86:260–286. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ. Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lassmann T. Sonnhammer ELL. Kalign, Kalignvu and Mumsa: web servers for multiple sequence alignment. Nucleic Acids Research. 2006;34:W596–W599. doi: 10.1093/nar/gkl191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losos JB. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecology Letters. 2008;11:995–1003. doi: 10.1111/j.1461-0248.2008.01229.x. [DOI] [PubMed] [Google Scholar]
- Lovette IJ. Glacial cycles and the tempo of avian speciation. Trends in Ecology and Evolution. 2005;20:57–59. doi: 10.1016/j.tree.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Lundberg P. The evolution of partial migration in birds. Trends in Ecology and Evolution. 1988;3:172–175. doi: 10.1016/0169-5347(88)90035-3. [DOI] [PubMed] [Google Scholar]
- Maechler M, Rousseeuw P, Struyf A, Hubert M. Hornik K. cluster: cluster analysis basics and extensions. 2013. R package version 1.14.2. Available at: http://www.cran.r-project.org/web/packages/cluster/index.html.
- Martinez-Meyer E, Peterson AT. Navarro-Siguenza AG. Evolution of seasonal ecological niches in the Passerina buntings (Aves: Cardinalidae) Proceedings of the Royal Society B: Biological Sciences. 2004;271:1151–1157. doi: 10.1098/rspb.2003.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. Animal species and evolution. Cambridge, MA: Belknap; 1963. [Google Scholar]
- Mila B, Smith TB. Wayne RK. Postglacial population expansion drives the evolution of long-distance migration in a songbird. Evolution. 2006;60:2403–2409. [PubMed] [Google Scholar]
- Münkemüller T, Lavergne S, Bzeznik B, Dray S, Jombart T, Schiffers K. Thuiller W. How to measure and test phylogenetic signal. Methods in Ecology and Evolution. 2012;3:743–756. [Google Scholar]
- Olalla-Tarraga MA, McInnes L, Bini LM, Diniz-Filho JAF, Fritz SA, Hawkins BA, Hortal J, Orme CDL, Rahbek C, Rodriguez MA. Purvis A. Climatic niche conservatism and the evolutionary dynamics in species range boundaries: global congruence across mammals and amphibians. Journal of Biogeography. 2011;38:2237–2247. [Google Scholar]
- Outlaw DC. Voelker G. Pliocene climatic change in insular Southeast Asia as an engine of diversification in Ficedula flycatchers. Journal of Biogeography. 2008;35:739–752. [Google Scholar]
- Pagel M. Inferring evolutionary processes from phylogenies. Zoologica Scripta. 1997;26:331–348. [Google Scholar]
- Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- Perez-Tris J, Bensch S, Carbonell R, Helbig AJ. Telleria JL. Historical diversification of migration patterns in a passerine bird. Evolution. 2004;58:1819–1832. doi: 10.1554/03-731. [DOI] [PubMed] [Google Scholar]
- Perrins CM. Ogilvie MA. The complete birds of the Western Palearctic. Oxford, UK: Oxford University Press and Optimedia; 1998. CD-ROM Version 1.0. [Google Scholar]
- Phillimore AB, Freckleton RP, Orme CDL. Owens IPF. Ecology predicts large-scale patterns of phylogenetic diversification in birds. The American Naturalist. 2006;168:220–229. doi: 10.1086/505763. [DOI] [PubMed] [Google Scholar]
- Phillimore AB, Orme CDL, Thomas GH, Blackburn TM, Bennett PM, Gaston KJ. Owens IPF. Sympatric speciation in birds is rare: insights from range data and simulations. The American Naturalist. 2008;171:646–657. doi: 10.1086/587074. [DOI] [PubMed] [Google Scholar]
- Price T, Lovette IJ, Bermingham E, Gibbs HL. Richman AD. The imprint of history on communities of North American and Asian warblers. The American Naturalist. 2000;156:354–367. doi: 10.1086/303397. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- Revell LJ. Collar DC. Phylogenetic analysis of the evolutionary correlation using likelihood. Evolution. 2009;63:1090–1100. doi: 10.1111/j.1558-5646.2009.00616.x. [DOI] [PubMed] [Google Scholar]
- Revell LJ, Harmon LJ. Collar DC. Phylogenetic signal, evolutionary process, and rate. Systematic Biology. 2008;57:591–601. doi: 10.1080/10635150802302427. [DOI] [PubMed] [Google Scholar]
- Richman AD. Price T. Evolution of ecological differences in the Old-World leaf warblers. Nature. 1992;355:817–821. doi: 10.1038/355817a0. [DOI] [PubMed] [Google Scholar]
- Roquet C, Thuiller W. Lavergne S. Building megaphylogenies for macroecology: taking up the challenge. Ecography. 2013;36:13–26. doi: 10.1111/j.1600-0587.2012.07773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamin N, Wuest RO, Lavergne S, Thuiller W. Pearman PB. Assessing rapid evolution in a changing environment. Trends in Ecology and Evolution. 2010;25:692–698. doi: 10.1016/j.tree.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P. Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Tenenhaus M. Young FW. An analysis and synthesis of multiple correspondence analysis, optimal scaling, dual scaling, homogeneity analysis and other methods for quantifying categorical multivariate data. Psychometrika. 1985;50:91–119. [Google Scholar]
- Thomas GH. Freckleton RP. MOTMOT: models of trait macroevolution on trees. Methods in Ecology and Evolution. 2012;3:145–151. [Google Scholar]
- Thuiller W, Lavorel S, Midgley G, Lavergne S. Rebelo T. Relating plant traits and species distributions along bioclimatic gradients for 88 Leucadendron taxa. Ecology. 2004;85:1688–1699. [Google Scholar]
- Thuiller W, Lavergne S, Roquet C, Boulangeat I, Lafourcade B. Araujo MB. Consequences of climate change on the tree of life in Europe. Nature. 2011;470:531–534. doi: 10.1038/nature09705. [DOI] [PubMed] [Google Scholar]
- Wiens JJ, Ackerly DD, Allen AP, Anacker BL, Buckley LB, Cornell HV, Damschen EI, Davies TJ, Grytnes JA, Harrison SP, Hawkins BA, Holt RD, McCain CM. Stephens PR. Niche conservatism as an emerging principle in ecology and conservation biology. Ecology Letters. 2010;13:1310–1324. doi: 10.1111/j.1461-0248.2010.01515.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting figures and tables.
Table of avian niche traits.


