Abstract
Bovine lactoferrin (bLf) is a multifunctional glycoprotein that plays an important role in innate immunity against infections, including influenza. Here we have dissected bLf into its C- and N-lobes and show that inhibition of influenza virus hemagglutination and cell infection is entirely attributable to the C-lobe and that all major virus subtypes, including H1N1 and H3N2, are inhibited. By far-western blotting and sequencing studies, we demonstrate that bLf C-lobe strongly binds to the HA2 region of viral hemagglutinin, precisely the highly conserved region containing the fusion peptide. By molecular docking studies, three C-lobe fragments were identified which inhibited virus hemagglutination and infection at fentomolar concentration range. Besides contributing to explain the broad anti-influenza activity of bLf, our findings lay the foundations for exploiting bLf fragments as source of potential anti-influenza therapeutics.
Keywords: Influenza, Bovine lactoferrin, C-lobe, Peptides, Antiviral
Introduction
Influenza viruses are a serious cause of morbidity and mortality worldwide, particularly among people with immunodeficiency associated with aging or underlying predisposing conditions. While vaccines are the core measure for infection control, the immunization programs are not fully effective because of rapid antigenic drift and emergence of new viral subtypes. Antiviral chemotherapy is based on two classes of drugs: inhibitors of the M2 proton-selective ion channel protein (amantadine and its derivative rimantadine),1 and neuraminidase (NA) inhibitors (oseltamivir and zanamivir).2 Amantadine and derivatives reduce the duration of symptoms, but major side effects and the emergence of drug-resistant variants have been described.3,4 NA inhibitors remain, at present, the primary treatment against influenza. However, they have limited efficacy if administered late in infection and their widespread use is likely to result in the emergence of resistant viral strains.5–7 New therapeutic strategies are therefore a global public health priority.
On the basis of the above considerations, an ideal target for therapy should be a viral component, other than M2 and NA, whose function is essential for virus infection. In this contest, the influenza A virus hemagglutinin (HA) represents a very promising target. The HA of influenza is the major glycoprotein component of the viral envelope. There are 16 known HA subtypes, divided into two groups. HA is homotrimeric and each monomer is composed of two polypeptide segments, designated HA1 and HA2, attached to each other via a disulfide bond.8 The HA1 segments mediate HA attachment to the host cell surface by binding to sialic acid-containing cell surface glycans.9 After attachment, virions internalized by endocytoses undergo an irreversible acid-induced structural rearrangement in which the highly hydrophobic amino terminus of HA2 is exposed. This hydrophobic fusion peptide is then translocated to the endosomal membrane, thereby mediating fusion of the viral envelope with the membranes and formation of a fusion pore.10 Since this conformational change is crucial for the fusogenic activity of HA and for viral entry, this event represents an interesting target for the development of antiviral agents. Moreover, as the hydrophobic fusion peptide is the only universally conserved epitope in all influenza viruses, the identification of new antiviral drugs possibly targeting this region represents an efficacious strategy for a broad-spectrum protection against seasonal and pandemic influenza viruses.
Breast-feeding has been recognized to protect against respiratory and gastrointestinal infections in infants.11 Milk, besides secretory IgA and IgM, also contains a number of various non-antibody components with known antimicrobial activity, including lactoferrin (Lf).11,12 Lf, an 80-kDa multifunctional cationic glycoprotein belonging to the transferrin family, possesses a variety of biological functions such as: influence on iron homeostasis, immunomodulation, and inhibitory activity towards different pathogens, including influenza viruses.12,13 Bovine lactoferrin (bLf) is a potent inhibitor of different enveloped viruses14–23 and, like lactoferrin of other mammalian species, is folded in two symmetric globular lobes, each one including two domains (N-lobe: N1 and N2; C-lobe: C1 and C2).24,25 It has been shown that the principal mechanism of bLf antiviral activity follows a direct binding to viral particles20,26–28 or to host cell receptor molecules.17 An additional effect of bLf on a later step of virus infection has been described26 and, more recently, we demonstrated that bLf treatment is able to prevent influenza virus-induced apoptosis by interfering with function of caspase 3.29 This caused the block of nuclear export of viral ribonucleoproteins with consequent viral assembly prevention. However, little is known on the molecular mechanisms whereby bLf binds to the influenza virus and causes viral inhibition. Thus, in an effort to identify new antiviral therapies effective against influenza virus, we have focused on molecular dissection of bLf and interaction of its molecular fragments to precise locations on viral HA. Based on this new approach, we have retrieved molecular and structural information helping us identify peptide fragments of bLf C-lobe capable of exerting potent antiviral activity.
Materials and Methods
Virus strains
The following influenza A virus strains were used: A/Solomon/3/2006 H1N1, A/Wisconsin/67/2005 H3N2, and SW X-179A swine pandemic H1N1 (inactivated vaccine subunit preparations); A/Puerto Rico/8/34 H1N1 (PR8 virus), A/RomaISS/2/08 H1N1 oseltamivir-sensitive virus, A/Parma/24/09 H1N1 oseltamivir-resistant virus, A/Parma/05/06 H3N2, Avian 29/05/06 H5N1 (inactivated subunit vaccine), and A/Turkey/Italy/2676/99 H7N1. Virus titers were determined by a hemagglutinin titration and/or plaque assay according to the standard procedures.30,31
Cells
Madin-Darby canine kidney (MDCK, ATCC, CRL-2936) cells were grown at 37°C in minimal essential medium (MEM, Invitrogen, Paisley, UK) containing 1.2 g/l NaHCO3, and supplemented with 10% inactivated fetal calf serum (FCS, Invitrogen, Paisley, UK), 2 mM glutamine, nonessential amino acids, penicillin (100 IU/ml), and streptomycin (100 μg/ml).
Lactoferrin
Lactoferrin from bovine milk (bLf) (Morinaga Milk Industries, Zama City, Japan) was deprived of endotoxin as previously described.32
Enzymatic hydrolysis of bLf and HPLC separation, purification and characterization of N- and C-lobes
These procedures were carried out as reported elsewhere.32,33 Briefly, bLf was dissolved in 50 mM ammonium bicarbonate, pH 8.5, and trypsin (from bovine pancreas TPCK-treated, Sigma Chemical Company, St.Louis, MO, USA) digestion was performed at 37°C overnight using an enzyme to substrate ratio of 1∶50 w/w. The N- and C-lobes obtained by enzymatic hydrolysis were purified by RP-HPLC chromatography and analysed by SDS-PAGE and mass spectrometry to check identity and purity.
Peptide synthesis
Synthesis was performed in Solid Phase Peptide Synthesis (SPPS) with the Fmoc-chemistry by Primm Biotech, Inc., Milan, Italy.
Cytotoxicity assay
This procedure was performed as reported elsewhere.29 Briefly, two-fold serial dilutions of each protein in culture medium were incubated at 37°C with confluent MDCK cells grown in 96-well tissue culture microplates (Nalge Nunc Europe Ltd, Neerijse, Belgium). After 24 hours, cell morphology, viability, and proliferation were evaluated. Protein dilutions that did not affect any of these parameters were considered as non-cytotoxic concentrations and utilized for antiviral assays.
Hemagglutination inhibition assay (HI)
Virus in PBS was incubated for 1 hour at 4°C with serial dilutions of bLf or peptidic fragments in PBS. An equal volume of 0.5% turkey erythrocytes was then added and allowed to agglutinate. Titers were expressed as the reciprocal of the protein dilutions giving 50% hemagglutination of erythrocytes by four virus agglutinating units.
Neutralization assay
Neutralization was carried out by incubating serial twofold bLf or peptidic fragment dilutions, starting from 12.5 μM, in culture medium with equal volumes (250 μl) of virus suspension containing 106 p.f.u. for 1 hour at 4°C. In negative controls, culture medium was used instead of bLf or peptidic fragments in the same volume. MDCK cells, grown in 96-well tissue culture microplates (Nalge Nunc Europe Ltd, Neerijse, Belgium), were infected with 100 μl/well (10 p.f.u./cell; in quadruplicate) of the virus-protein mixtures. After adsorption, cells were rinsed thoroughly and incubated at 37°C for 24 hours. The viral cytopathic effect (c.p.e.) was measured by neutral red staining as reported elsewhere by our laboratory.17
Far-western-blot
Purified viral subunits preparations of Solomon (H1N1) and Wisconsin (H3N2) strains were resolved by polyacrylamide-SDS gel electrophoresis (SDS-PAGE), as described by Laemmli34 on 15% acrylamide gel. Separated proteins were then transferred from gel to nitrocellulose membranes and ligand-blot assays were carried out as reported elsewhere by our laboratory.28
Sequencing and alignment
For identification of HA region recognized by bLf in far-western blot, A/Solomon 3/2006 H1N1 subunits, separated on 15% acrylamide gel in denaturing conditions, were sequenced by Primm Biotech, Inc., Milan, Italy with Edman degradation method. Sequences of HAs under study were aligned to those of the HAs whose X-ray structures were available in the Protein Data Bank. FASTA sequences alignment was carried out applying the default parameters. The same procedure was applied to align the N- and C-lobe of bLf.
Protein–protein docking
The protein–protein docking calculations were performed using the C-lobe of bLf (PDB code 3IB0) and the crystal structure of HA with PDB code 2WRG, because it showed the highest sequence similarity with the HAs isolated from the H1N1viral strains used in the assays. Both X-ray structures were submitted to the Protein Preparation routine in Maestro (Maestro, version 9.2, Schrödinger, LLC, New York, NY, 2011). 3IB0 missing residues were built using the homology modeling routine Prime (Prime, version 3.0, Schrödinger, LLC, New York, NY, 2011). The structures were subjected to four different docking approaches using the software: Autodock-MacroModel,35 ZDock-RDock,36,37 ClusPro,38 PatchDock-FireDock.39–41 All procedures comprised a preliminary docking calculation followed by a refinement step.
Resulting complexes from the four docking calculations were collected and those showing the bLf interacting with the fusion peptide were selected for further analysis. One hundred sixty one resulting complexes were clustered in order to find a consensus between poses generated with different approaches. The clustering was carried out using R (R, The R foundation for statistical computing) and applying the Ward clustering metric.
Results
Interaction of bLf and its lobes with viral hemagglutinin and neutralization of influenza virus
The interaction between influenza virus and bLf was initially tested by HI. As shown in Table 1, concentrations of bLf ranging from about 0.05 pM to 6 nM were able to prevent HA activity of all tested viruses, inclusive of H1N1, H3N2, H5N1 and H7N1 subtypes. Table 1 also shows that for all viruses HI activity was exclusively expressed by bLf C-lobe. For inhibition of virus replication, representatives of the two influenza virus groups 1 and 2 were used. As shown in Table 2, virus replication was markedly inhibited by bLf with SI values of about 106 and 105 for influenza A/H1N1 and A/H3N2 virus subtypes, respectively. In keeping with HI studies, virus replication was strongly inhibited by C-lobe with SI values 2.5–5-fold higher than those obtained with bLf, while the N-lobe was ineffective (data not shown).
Table 1. Interaction of bLf, N-lobe, and C-lobe with viral HA.
| HI titre | ||||
| Viral strain | Subtype | bLf | N-lobe | C-lobe |
| A/Roma-ISS/2/08 (Brisbane-like) | H1N1 | 6.0 nM | – | 10.0 nM |
| A/Parma/24/09 (Brisbane-like) | H1N1 | 6.0 nM | – | 10.0 nM |
| A/PR/8/34 | H1N1 | 3.0 nM | – | 12.0 nM |
| A/Solomon (3/06) | H1N1 | 5.0 pM | – | 5.0 pM |
| A/Parma/5/06 (Wisconsin-like) | H3N2 | 3.0 nM | – | 6.0 nM |
| A/Wisconsin (67/05) | H3N2 | 0.3 pM | – | 1.2 pM |
| SW X-179A 1089/09 | H1N1 | 46.5 fM | – | 46.5 fM |
| Avian 29/05/06 | H5N1 | 2.9 pM | – | 2.4 pM |
| A/Turkey/Italy/2676/99 | H7N1 | 93.1 fM | – | 93.1 fM |
Table 2. In vitro antiviral activity of bLf and C-lobe towards influenza virus infection.
| Viral strain | Subtype | CC50* | EC50° | SÎ | |
| Lactoferrin | A/Roma-ISS/2/08 | H1N1 | >25 μM | 25±0.95 pM | >106 |
| A/Parma/24/09 | H1N1 | >25 μM | 25±1.01 pM | >106 | |
| A/Parma/05/06 | H3N2 | >25 μM | 0.25±0.01 nM | >105 | |
| C-lobe | A/Roma-ISS/2/08 | H1N1 | >25 μM | 10±0.62 pM | >2.5×106 |
| A/Parma/24/09 | H1N1 | >25 μM | 10±0.81 pM | >2.5×106 | |
| A/Parma/05/06 | H3N2 | >25 μM | 50±1.96 pM | >5.0×105 |
Note: *CC50 the reciprocal substance dilution at which 50% of cells were protected from substance toxicity; °EC50 the reciprocal substance dilution at which 50% of cells were protected from the virus induced killing; ˆSI (selectivity index) the ratio between CC50 and EC50.
The mean values of three independent experiments with standard deviations are shown.
Far-western-blot and protein identification
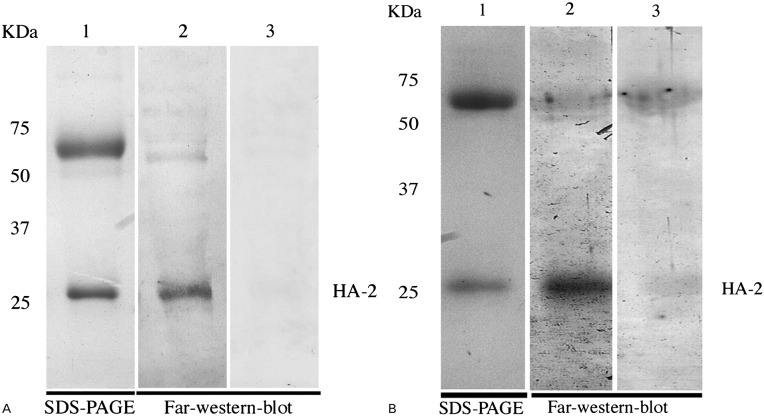
We attempted to identify the bLf-virus binding components, by first examining bLf interaction with viral envelope proteins. Thus, envelope proteins present in Solomon (H1N1) and Wisconsin (H3N2) subunit vaccines were separated by electrophoresis and probed with bLf by a far-western blot assay. In both viral strains bLf more consistently bound to a viral protein of apparent molecular mass of 28 kDa, tentatively corresponding to the HA2 subunit (Fig. 1, panels A and B). N-terminus protein sequencing of H1N1 Solomon subunit confirmed that the virus component bound by bLf was indeed the HA2 subunit as shown by the exact correspondence of the first 18 amino acids of the sequenced peptide GLFGAIAGFIEGGWTGMV with the highly conserved fusion peptide of HA2 subunit.42
Figure 1.
Ligand-blot assay of bLf binding to influenza A virus subunits: (A) Lane 1: SDS-PAGE of reduced A/Solomon 3/2006 H1N1 subunits. Lane 2: lactoferrin blot overlay. Lane 3: specificity control (rabbit anti-bLf antibodies+HRP-conjugated anti-rabbit antibodies). (B) Lane 1: SDS-PAGE of reduced A/Wisconsin (67/05) H3N2 subunits. Lane 2: lactoferrin blot overlay. Lane 3: specificity control (rabbit anti-bLf antibodies+HRP-conjugated anti-rabbit antibodies).
Protein–protein docking
To gain more insight into the binding mode between bLf C-lobe and HA, a protein–protein docking protocol was set up. Resulting complexes were visually inspected to eliminate all the unsuitable binding poses: i.e. those involving not accessible regions of C-lobe. Also complexes where C-lobe binds just the HA1 of HA were ignored in following analysis.
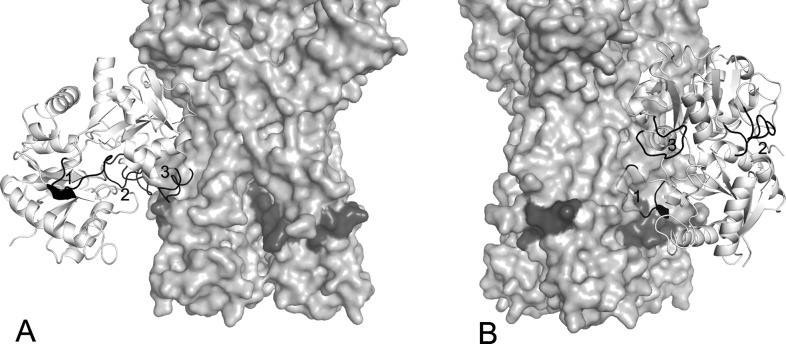
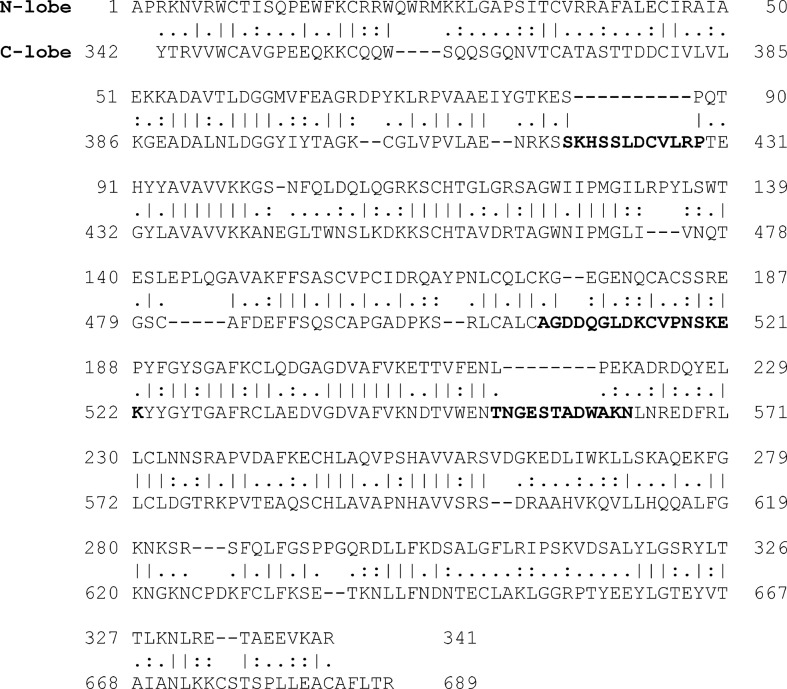
As shown in Fig. 2, most of the clustered binding poses gathered in two regions of the HA stem: the first one in the cleft between two monomers (Fig. 2A) and the second one very close to the fusion peptide (Fig. 2B). The bLf C-lobe binding to HA was mediated, in most cases, by three surface-exposed loops characterized by the following amino acid sequences: SKHSSLDCVLRP (aa 418–429), TNGESTADWAKN, (aa 552–563) and AGDDQGLDKCVPNSKEK (aa 506–522). Interestingly sequences 418–429 and 552–563 correspond to loops that are not present in the N-lobe, as demonstrated by aligning the sequences of the N- and C-lobe (Fig. 3).
Figure 2.
Putative binding mode of bLf C-lobe (white ribbon) with the HA stm (light grey solid surface): (A) close to the fusion peptide (dark grey surface); (B) in the cleft between two monomers. Selected bLf sequences correspond to the numbered black loops of bLf C-lobe: (1) SKHSSLDCVLRP; (2) AGDDQGLDKCVPNSKEK; (3) TNGESTADWAKN.
Figure 3.
Sequence alignment of the N- and C-lobe of bLf. The bold sequences correspond to C-lobe loops supposed to be involved in binding.
Interaction of bLf derived peptides with viral hemagglutinin and neutralization of influenza virus
The docking results suggested the possibility of obtaining inhibitory peptides from bLf regions interacting with HA. Thus, the three peptides whose sequence was mentioned above were synthesized and assessed for HI activity. Because of poor stability of TNGESTADWAKN peptide, a modified sequence (NGESSADWAKN) has been designed where the first threonine residue was removed and the other one was replaced by serine. As shown in Table 3, each peptide was able to inhibit HA activity of all tested virus strains at concentrations much lower than those shown by the C-lobe (compare with Table 1).
Table 3. Interaction of SKHSSLDCVLRP, AGDDQGLDKCVPNSKEK, and NGESSADWAKN peptides with viral HA.
| HI titre | ||||
| Viral strain | Subtype | SKHSSL DCVLRP | AGDDQGLDK CVPNSKEK | NGESSA DWAKN |
| A/Roma-ISS/2/08 (Brisbane-like) | H1N1 | 1.4 pM | 1.4 pM | 0.7 fM |
| A/Parma/24/09 (Brisbane-like) | H1N1 | 1.4 pM | 1.4 pM | 0.3 fM |
| A/PR/8/34 | H1N1 | 0.7 nM | 0.35 nM | 0.7 fM |
| A/Solomon (3/06) | H1N1 | 46.5 fM | 46.5 fM | 93.0 fM |
| A/Parma/5/06 (Wisconsin-like) | H3N2 | 0.7 pM | 0.7 pM | 0.3 pM |
| A/Wisconsin (67/05) | H3N2 | 23.2 fM | 11.6 fM | 5.8 fM |
| SW X-179A 1089/09 | H1N1 | 5.8 fM | 46.0 fM | 0.3 fM |
| Avian 29/05/06 | H5N1 | 18.0 fM | 70.0 fM | 93.1 fM |
| A/Turkey/Italy/2676/99 | H7N1 | 18.0 fM | 46.5 fM | 5.8 fM |
We also examined whether, and to what extent, bLf-derived peptides were able to affect virus replication. Two strains of the influenza A/H1N1 virus subtype (A/Roma-ISS/2/08 and A/Parma/24/09), and one strain of A/H3N2 virus subtype (A/Parma/05/06) were used in these experiments. Similarly to HI data, bLf-derived peptides were better inhibitors than the entire protein, their selectivity index being about one or two order of magnitude higher, depending on virus strain. In particular, the SI of NGESSADWAKN tested against H1N1-infected cells reached values about 500 times higher than the SI of bLf against the same virus-cell system (Table 4). Notably, the H1N1 used in these experiments is an oseltamivir-resistant virus strain.
Table 4. In vitro antiviral activity of SKHSSLDCVLRP, AGDDQGLDKCVPNSKEK, and NGESSADWAKN peptides towards influenza virus infection.
| Viral strain | Subtype | CC50* | EC50° | SÎ | |
| SKHSSLDCVLRP | A/Roma-ISS/2/08 | H1N1 | >25 μM | 4±0.37 pM | >6.25×106 |
| A/Parma/24/09 | H1N1 | >25 μM | 3.1±0.12 pM | >8×106 | |
| A/Parma/05/06 | H3N2 | >25 μM | 5.8±0.7 pM | >4.3×106 | |
| AGDDQGLDKCVPNSKEK | A/Roma-ISS/2/08 | H1N1 | >25 μM | 3.7±0.35 pM | >6.75×106 |
| A/Parma/24/09 | H1N1 | >25 μM | 3.4±0.14 pM | >7.35×106 | |
| A/Parma/05/06 | H3N2 | >25 μM | 7.3±0.65 pM | >3.42×106 | |
| NGESSADWAKN | A/Roma-ISS/2/08 | H1N1 | >25 μM | 225±5.8 fM | >1.11×108 |
| A/Parma/24/09 | H1N1 | >25 μM | 50±1.37 fM | >5×108 | |
| A/arma/05/06 | H3N2 | >25 μM | 22.5±1.16 pM | >1.11×106 |
Note: *CC50 the reciprocal substance dilution at which 50% of cells were protected from substance toxicity; °EC50 the reciprocal substance dilution at which 50% of cells were protected from the virus induced killing; ˆSI (selectivity index) the ratio between CC50 and EC50.
The mean values of three independent experiments with standard errors are shown.
Discussion
Influenza remains one of the leading causes of morbidity and death worldwide. Efforts to prevent influenza by vaccination are complicated by the virus ability to rapidly mutate and recombine into antigenically new virions, sometimes leading to the emergence of a totally new virus.43 The availability of broad-spectrum antiviral drugs is an important asset in the fight against influenza. Particularly, a combination of different anti-influenza drugs, each directed against a different viral target or endowed with different mechanism of action, would be expected to be more active in influenza treatment and minimize the emergence of drug resistance.
It is well known that breast-feeding protects against both respiratory and enteric infections in infants.11 We have previously demonstrated that lactoferrin, a main antimicrobial component of milk, is able to inhibit influenza-induced apoptosis by interfering with caspase 3 function and by inhibiting the export of viral ribonucleoproteins from the nucleus to the cytoplasm.29
In the present study, we demonstrate that bLf is able to bind viral HA and inhibit hemagglutination and infection of influenza A viruses belonging to both group 1 and group 2 subtypes. Of note, bLf was shown to bind to the HA2 subunit, an HA region which is known to contain the universally conserved HA epitope, thus explaining the broad specificity of the observed anti-influenza activity. Our data also demonstrate that bLf binding to HA stem region results in the inhibition of virus attachment to target cells. The stem region of HA is mainly formed by the HA2 domain, with a minimal but important contribution of several residues in the N- and C-terminal segments of HA1.44,45 Molecular modeling studies46 have shown a strong electrostatic attraction between the HA1 subunits (positively charged) and the HA2 subunits (negatively charged) at neutral pH. Thus, at neutral pH, the electrostatic repulsion between the three HA1 subunits on the one hand and that between the three HA2 subunits on the other is overcome by electrostatic attraction between the HA1 and HA2 domains, preserving a stable trimeric association of the monomers. It can be hypothesized that bLf interaction with HA2 stem region of different HA monomers can lead to a trimer conformational change, so hindering HA binding to sialic acid receptors.
We also show here that the antiviral activity of bLf is entirely mediated by the C-lobe, with no apparent role of the N-lobe. This helped us in further dissection of the bLf molecule in an attempt to unveil the smallest most active anti-influenza virus regions of the C-lobe. Based on sequencing and protein–protein docking calculations, three C-lobe peptides were identified, synthesized and tested for their antiviral activity. Both hemagglutination and virus infection were inhibited by these peptides in a concentration range (fento- to picomolar) that suggests particularly high affinity for the HA2 target. Besides strengthening the evidence of an involvement of specific sequences of the C-lobe as mediators of antiviral activity of bLf, the elevated activity of these peptides, together with the highly conserved nature of their HA2 target, would suggest that bLf peptides and their mimics are worthy being further studied as anti-influenza therapeutics. It is of some interest making a comparison between the inhibitory activity of bLf, C-lobe and C-lobe peptides and that shown by neutralizing antibodies, also in view of the surging interest in the immunotherapy of influenza.47 Overall, neutralizing antibodies against influenza virus have been found to act by two different mechanisms, mirroring the dual functions of HA: (i) prevention of attachment to target cells, (ii) inhibition of entry (membrane fusion). The mechanism of neutralization depends on the HA site recognized by the HA-specific molecule. Usually, antibodies against HA act by inhibiting attachment. This occurs as they bind HA1 and physically hinder the interaction with sialic acid receptors on target cells.48 Some antibodies have been found to prevent membrane fusion; most of them recognize sites in the HA2 region, far away from the receptor-binding site.42,49 Quite recently, some so-called ‘universal antibodies’ have been generated which bind to highly conserved epitopes of the HA2 stem region, and one of them (F16) has been shown to target part of the fusion peptide.50 Differently from previously described heterosubtypic antibodies, F16 binds and neutralizes both group 1 and group 2 influenza A viruses. However, at variance with bLf and C-lobe peptides, the antibody described by Corti et al.50 does not inhibit hemagglutination, suggesting that bLf and the neutralizing mAb reported by Corti et al.50 target differently the F subdomain of HA. At any rate it is of interest that bLf and some antibodies, typical representatives of innate and adaptive immunity, respectively, can inhibit influenza virus by targeting the same or overlapping antigenic regions of the virus. In conclusion, we have identified the bLf regions most likely interacting with influenza virus, and expressing the broad anti-influenza activity of bLf. In addition, by exploiting the molecular information retrieved from bLf and C-lobe. interaction with the HA2 virus subunit, we have generated particularly potent peptide inhibitors of both hemagglutination and virus replication in cell cultures. These peptides would constitute a new class of antivirals since the commercially existing ones target the neuraminidase or the M2 protein, not the HA, of the influenza virus. These neutralizing peptides, targeting HA of viral subtypes belonging to the two major phylogenetic groups of influenza virus, may represent a useful tool in neutralizing viral infection, clearing virus, and suppressing viral spread as well as a valuable resource for therapeutic possibilities. For all these reasons we plan to test bLf, the C-lobe and the peptides themselves for their anti-influenza activity in suitable in vivo models.
Acknowledgments
We are grateful to Dr Isabella Donatelli (ISS, Italy) for viral isolates.
This work was supported by a grant from Ministero della Salute-Istituto Superiore di Sanità- within the Research Project: ‘Studio e sviluppo di nuovi farmaci antivirali contro infezioni A-H1N1’.
References
- 1.Dolin R, Reichman RC, Madore HP, Maynard R, Linton PN, Webber-Jones J. A controlled trial of amantadine and rimantadine in the prophylaxis of influenza A infection. N Engl J Med. 1982;307:580–4. doi: 10.1056/NEJM198209023071002. [DOI] [PubMed] [Google Scholar]
- 2.Palese P, Shaw ML, Knipe DM, Howley PM.(editors)Orthomyxoviridae: the viruses and their replication. In: Fields virology. Philadelphia, PA: Lippincott Williams & Wilkins; 2007 [Google Scholar]
- 3.Belshe RB, Burk B, Newman F, Cerruti RL, Sim IS. Resistance of influenza A virus to amantadine and rimantadine: results of one decade of surveillance. J Infect Dis. 1989;159:430–5. doi: 10.1093/infdis/159.3.430. [DOI] [PubMed] [Google Scholar]
- 4.Hayden FG. Antiviral resistance in influenza viruses – implications for management and pandemic response. N Engl J Med. 2006;354:785–8. doi: 10.1056/NEJMp068030. [DOI] [PubMed] [Google Scholar]
- 5.Kiso M, Mitamura K, Sakai-Tagawa Y, Shiraishi K, Kawakami C, Kimura K, et al. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet. 2004;364:759–65. doi: 10.1016/S0140-6736(04)16934-1. [DOI] [PubMed] [Google Scholar]
- 6.Meijer A, Lackenby A, Hungnes O, Lina B, van-der-Werf S, Schweiger B, et al. European influenza surveillance scheme Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007–08 season. Emerg Infect Dis. 2009;15:552–60. doi: 10.3201/eid1504.081280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dharan NJ, Gubareva LV, Meyer JJ, Okomo-Adhiambo M, McClinton RC, Marshall SA, et al. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA. 2009;30:1034–41. doi: 10.1001/jama.2009.294. [DOI] [PubMed] [Google Scholar]
- 8.Wilson IA, Skehel JJ, Wiley DC. Structure of the hemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–73. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 9.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–69. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 10.Wiley DC, Skehel JJ. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–94. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 11.May JT. Microbial contaminants and antimicrobial properties of human milk. Microbiol Sci. 1988;5:42–6. [PubMed] [Google Scholar]
- 12.Levay PF, Viljoen M. Lactoferrin: a general review. Haematologica. 1995;80:252–67. [PubMed] [Google Scholar]
- 13.Palma C, Cassone A, Serbousek D, Pearson CA, Djeu JY. Lactoferrin release and interleukin-1, interleukin-6, and tumor necrosis factor production by human polymorphonuclear cells stimulated by various lipopolysaccharides: relationship to growth inhibition of Candida albicans. Infect Immun. 1992;60:4604–11. doi: 10.1128/iai.60.11.4604-4611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harmsen MC, Swart PJ, de B’ethune MP, Pawels R, De Clercq E, Th’e TH, et al. Antiviral effects of plasma and milk proteins: lactoferrin shows a potent activity against both human immunodeficiencyvirus and human cytomegalovirus replication in vitro. J Infect Dis. 1995;172:380–8. doi: 10.1093/infdis/172.2.380. [DOI] [PubMed] [Google Scholar]
- 15.Beljaars L, van der Strate BW, Bakker HI, Reker-Smit C, van Loenen-Weemaes AM, Wiegmans FC, et al. Inhibition of cytomegalovirus infection by lactoferrin in vitro and in vivo. Antivir Res. 2004;63:197–208. doi: 10.1016/j.antiviral.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Hasegawa K, Motsuchi W, Tanaka S, Dosako S. Inhibition with lactoferrin of in vitro infection with human herpes virus. Jpn J Med Sci Biol. 1994;47:73–85. doi: 10.7883/yoken1952.47.73. [DOI] [PubMed] [Google Scholar]
- 17.Marchetti M, Trybala E, Superti F, Johansson M, Bergström T. Inhibition of herpes simplex virus infection by lactoferrin is dependent on interference with the virus binding to glycosaminoglycans. Virology. 2004;18:405–13. doi: 10.1016/j.virol.2003.09.029. [DOI] [PubMed] [Google Scholar]
- 18.Marchetti M, Ammendolia MG, Superti F. Glycosaminoglycans are not indispensable for the anti-herpes simplex virus type 2 activity of lactoferrin. Biochimie. 2009;91:155–9. doi: 10.1016/j.biochi.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 19.Ammendolia MG, Marchetti M, Superti F. Bovine lactoferrin prevents the entry and intercellular spread of herpes simplex virus type 1 in Green Monkey Kidney cells. Antiviral Res. 2007;76:252–62. doi: 10.1016/j.antiviral.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Yi M, Kaneko S, Yu DY, Murakami S. Hepatitis C virus envelope proteins bind lactoferrin. J Virol. 1997;71:5997–6002. doi: 10.1128/jvi.71.8.5997-6002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy ME, Kariwa H, Mizutani T, Yoshimatsu K, Arikawa J, Takashima I. In vitro antiviral activity of lactoferrin and ribavirin upon hantavirus. Arch Virol. 2000;145:1571–82. doi: 10.1007/s007050070077. [DOI] [PubMed] [Google Scholar]
- 22.Hara K, Ikeda M, Saito S, Matsumoto S, Numata K, Kato N, et al. Lactoferrin inhibits hepatitis B virus infection in cultured human hepatocytes. Hepatol Res. 2002;24:228–35. doi: 10.1016/s1386-6346(02)00088-8. [DOI] [PubMed] [Google Scholar]
- 23.Sano H, Nagai K, Tsutsumi H, Kuroki Y. Lactoferrin and surfactant protein A exhibit distinct binding specificity to F protein and differently modulate respiratory syncytial virus infection. Eur J Immunol. 2003;33:2894–902. doi: 10.1002/eji.200324218. [DOI] [PubMed] [Google Scholar]
- 24.Norris GE, Gartner AL, Anderson BF, Ward J, Baker EN, Rumball SV, et al. Preliminary crystallographic studies on bovine lactoferrin. J Mol Biol. 1986;191:143–5. doi: 10.1016/0022-2836(86)90432-8. [DOI] [PubMed] [Google Scholar]
- 25.Moore SA, Anderson BF, Groom CR, Haridas M, Baker EN. Three-dimensional structure of diferric bovine lactoferrin at 2.8 °A resolution. J Mol Biol. 1997;274:222–36. doi: 10.1006/jmbi.1997.1386. [DOI] [PubMed] [Google Scholar]
- 26.Superti F, Ammendolia MG, Valenti P, Seganti L. Antirotaviral activity of milk proteins: lactoferrin prevents rotavirus infection in the enterocyte-like cell line HT-29. Med Microbiol Immunol. 1997;186:83–91. doi: 10.1007/s004300050049. [DOI] [PubMed] [Google Scholar]
- 27.Pietrantoni A, Di Biase AM, Tinari A, Marchetti M, Valenti P, Seganti L, et al. Bovine lactoferrin inhibits adenovirus infection by interacting with viral structural polypeptides. Antimicrob Agents Chemother. 2003;47:2688–91. doi: 10.1128/AAC.47.8.2688-2691.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ammendolia MG, Pietrantoni A, Tinari A, Valenti P, Superti F. Bovine lactoferrin inhibits echovirus endocytic pathway by interacting with viral structural polypeptides. Antiviral Res. 2007;73:151–60. doi: 10.1016/j.antiviral.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Pietrantoni A, Dofrelli E, Tinari A, Ammendolia MG, Puzelli S, Fabiani C, et al. Bovine lactoferrin inhibits influenza A virus induced programmed cell death in vitro. Biometals. 2010;23:465–75. doi: 10.1007/s10534-010-9323-3. [DOI] [PubMed] [Google Scholar]
- 30.Gaush CR, Smith TF. Replication and plaque assay of influenza virus in an established line of canine kidney cells. Appl Microbiol. 1968;16:588–94. doi: 10.1128/am.16.4.588-594.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rimmelzwaan GF, Baars M, Claas EC, Osterhaus AD. Comparison of RNA hybridization, hemagglutination assay, titration of infectious virus and immunofluorescence as methods for monitoring influenza virus replication in vitro. J Virol Methods. 1998;74:57–66. doi: 10.1016/s0166-0934(98)00071-8. [DOI] [PubMed] [Google Scholar]
- 32.Pietrantoni A, Ammendolia MG, Tinari A, Siciliano R, Valenti P, Superti F. Bovine lactoferrin peptidic fragments involved in inhibition of Echovirus 6 in vitro infection. Antiviral Res. 2006;69:98–106. doi: 10.1016/j.antiviral.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Superti F, Siciliano R, Rega B, Giansanti F, Valenti P, Antonini G. Involvement of bovine lactoferrin metal saturation, sialic acid and protein fragments in the inhibition of rotavirus infection. Biochim Biophys Acta. 2001;1528:107–15. doi: 10.1016/s0304-4165(01)00178-7. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Morris GM, Goodsell DS, Halliday RS, Huey R, William E, Hart WE, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–62. [Google Scholar]
- 36.Chen R, Weng Z. Docking unbound proteins using shape complementarity, desolvation, and electrostatics. Proteins. 2002;47:281–94. doi: 10.1002/prot.10092. [DOI] [PubMed] [Google Scholar]
- 37.Li L, Chen R, Weng Z. RDOCK: refinement of rigid-body protein docking predictions. Proteins. 2003;53:693–707. doi: 10.1002/prot.10460. [DOI] [PubMed] [Google Scholar]
- 38.Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins. 2006;65:392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 39.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33(Web Server issue):W363–7. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andrusier N, Nussinov R, Wolfson HJ. FireDock: fast interaction refinement in molecular docking. Proteins. 2007;69:139–59. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- 41.Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008;36(Web Server issue):W229–32. doi: 10.1093/nar/gkn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–51. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kasowski EJ, Garten RJ, Bridges CB. Influenza pandemic epidemiologic and virologic diversity: reminding ourselves of the possibilities. Clin Infect Dis. 2011;52(Suppl 1):S44–9. doi: 10.1093/cid/ciq010. [DOI] [PubMed] [Google Scholar]
- 44.Russell RJ, Gamblin SJ, Haire LF, Stevens DJ, Xiao B, Ha Y, et al. H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology. 2004;325:287–96. doi: 10.1016/j.virol.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 45.Gamblin SJ, Skehel JJ. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem. 2010;285:28403–9. doi: 10.1074/jbc.R110.129809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang Q, Opitz R, Knapp EW, Herrmann A. Protonation and stability of the globular domain of influenza virus hemagglutinin. Biophys J. 2002;82:1050–8. doi: 10.1016/S0006-3495(02)75464-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mancini N, Solforosi L, Clementi N, De Marco D, Clementi M, Burioni R. A potential role for monoclonal antibodies in prophylactic and therapeutic treatment of influenza. Antiviral Res. 2011;92:15–26. doi: 10.1016/j.antiviral.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 48.Edwards MJ, Dimmock NJ. Hemagglutinin 1-specific immunoglobulin G and Fab molecules mediate postattachment neutralization of influenza A virus by inhibition of an early fusion event. J Virol. 2001;75:10208–18. doi: 10.1128/JVI.75.21.10208-10218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–73. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–6. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]