Abstract
Background
Detection of specific targets by PCR is used to confirm a diagnosis of spotted fever, but serological tests are still widely used. In this prospective study, nested PCR was performed on skin biopsy specimens to confirm the diagnosis of spotted fever.
Methods
In 58 clinically suspected cases of spotted fever, nested PCR, to detect gltA, 17 kDa lipoprotein antigen gene (17 kDa), ompA and ompB, from skin biopsy of the rash was performed. Sequencing was carried on amplicons representing the four targets to confirm specificity of amplification. This was followed by phylogenetic analysis using MEGA version 4.0 software.
Results
The gltA, 17 kDa, ompA, and ompB genes were detected from skin biopsy specimens in 38, 23, 27, and 22 individuals. Sequence analysis revealed that the gltA, 17 kDa, ompA, and ompB sequences belonged to spotted fever group (SFG) rickettsia. Of the six partial ompA gene sequences, only one was dissimilar to the previously reported ‘Candidatus Rickettsia kellyi’.
Conclusion
Further evidence indicates that SFG rickettsiae resembling ‘Candidatus Rickettsia kellyi’ cause fever and rash in southern India. More detailed phylogenetic analysis following isolation of rickettsia in culture is required for providing irrefutable proof for the occurrence of novel spotted fever rickettsiae in this region.
Keywords: Spotted fever group (SFG) rickettsia, Fever with rash, Skin biopsy, Nested PCR, ompA gene, Phylogeny
Introduction
Rickettsial infections are re-emerging and a number of cases have been reported from different parts of India.1,2 These infections are caused by obligate intracellular parasites of the genus Rickettsia, which includes those responsible for spotted fever and typhus fever, having a predilection for vascular endothelium.3 The clinical features of rickettsioses consist of fever, a macular or maculopapular rash often involving the palms and soles, and sometimes an eschar. According to the CDC, the case definition of confirmed spotted fever rickettsiosis includes both clinical evidence and specific laboratory confirmation. The latter could be either detection of specific DNA by PCR, antigen by immunohistochemistry, demonstration of the organism in cell culture, or a fourfold rise in IgG antibody titres on paired samples taken 2–4 weeks apart.4 The five genes usually targeted by PCR for detection and diagnosis are citrate synthase (gltA), gene D (sca4), the 17 kDa lipoprotein precursor antigen gene (17 kDa), and genes for outer membrane proteins A and B (ompA and ompB).5
Detection of the gltA gene confirms that the amplicon belongs to the rickettsial genus which includes the typhus group (TG) and the spotted fever group rickettsia (SFGR).5–7 The molecular amplification of ompA gene is conclusive evidence for SFGR, whereas the 17 kDa gene and ompB gene found in the TG and SFGR can be used to confirm the presence of SFGR depending on the primer sequence used.5,8–11 The taxonomical position of a rickettsial sequence amplified by PCR can be ascertained up to the level of genus, group, and species using the algorithm described by Fournier et al..12 If the putative sequence is showing a homology for the 16 rRNA (rrs) and gltA of ≥98.1 and ≥86.5%, then it is said to belong to the genus Rickettsia. It is classified as a member of the SFGR if the ompA gene is amplified. In the absence of amplification of the ompA gene, the sequence should demonstrate a sequence similarity in two of the four criteria described. They are a sequence homology of ≥98.8%, ≥92.7%, ≥85.8%, and ≥82.2% for the rrs, gltA, ompB, and gene D with any one SFGR. If a similarity of <99.9%, <98.8%, <99.2%, and <99.3% for the gltA, ompA, and ompB genes and gene D is observed, then that isolate can be classified as a novel rickettsial species.12 This of course needs to be validated by subsequent isolation of the organism in culture and full elucidation of all biological properties including full gene sequences of the aforementioned genes found in this isolate.
This study was undertaken to detect spotted fever group rickettsial DNA by PCR in skin biopsies of rashes among individuals with clinically suspected spotted fever. We amplified four targets, one of which identified the isolate to genus level (gltA for genus Rickettsia) and the other three (17 kDa, ompA, and ompB) identified the PCR product as belonging to the SFGR.
Materials and Methods
Between November 2006 and April 2008, 58 individuals above the age of 6 months with fever ⩾99°F and fever duration ⩾5 and ⩽15 days) with non-confluent maculopapular, erythematous, or purpuric rash, were enrolled into the study. All the subjects were thoroughly examined by a dermatologist and were recruited after clinically ruling out drug reactions and viral exanthematous infections as described below. Drug rash was excluded mainly by history of rash with temporal relationship to a drug intake within the past 6 weeks. Those with enanthems or predominantly vesicular rash or desquamation were excluded as also those with pruritic rash and urticaria. A skin biopsy from the rash (3×3 mm, punch biopsy) was collected from all study participants after informed consent was obtained. The skin biopsies were placed in a sterile container and transported on ice immediately to the laboratory where they were stored at −70°C before DNA extraction. The study was approved by the Ethics Committee and Institutional Review Board of the Christian Medical College, Vellore.
Nested PCR was performed to detect Rickettsia genus-specific citrate synthase gene (gltA),6,7 in DNA extracted from skin rash biopsies. Spotted fever group (SFG)-specific DNA was detected by nested PCR which amplified 17 kDa,8,9 ompA,10 and ompB.11 The details of the primers used are given in Table 1. Cross-contamination was avoided as the DNA extraction, preparation of PCR master mix, addition of DNA template, thermal cycling and detection of the PCR product were performed in separate rooms using dedicated labware. All samples were considered to be PCR-positive only if the required amplicon was detected twice.
Table 1. Sequences of primers used for detection of rickettsial DNA by nested PCR.
| Gene | Target detected | Primers | Primer sequence (5′–3′) | Product size (bp) |
| gltA6.7 | All rickettsiae (typhus group and SFGR) | RpCS.877p | GGGGACCTGCTCACGGCGG | 381 |
| RpCS.1258n | ATTGCAAAAAGTACAGTGAACC | |||
| RpCS.896p | GGCTAATGAAGCAGTGATAA | 338 | ||
| RpCS.1233n | GCGACGGTATACCCATAGC | |||
| 17 kDa8,9 | All SFGR | R17122 | CAGAGTGCTATGAACAAACAAGG | … |
| R17500 | CTTGCCATTGCCCATCAGGTTG | |||
| Tz 15 | TTC TCA ATT CGG TAA GGG C | 246 | ||
| Tz 16 | ATA TTG ACC AGT GCT ATT TC | |||
| ompA10 | All SFGR except Rickettsia helvetica | Rr190k.71p | TGGCCAATATTTCTCCAAAA | 650 |
| Rr190k.720n | TGCATTTGTATTACCTATTGT | |||
| Rr190 k.71p | TGGCGAATATTTCTCCAAAA | 532 | ||
| Rr190k.602n | AGTGCAGCATTCGCTCCCCCT | |||
| ompB11 | All SFGR | Rc.rompB.4362p | GTCAGCGTTACTTCTTCGATGC | 475 |
| Rc.rompB.4836n | CCGTACTCCATCTTAGCATCAG | |||
| Rc.rompB.4496p | CCAATGGCAGGACTTAGCTACT | 267 | ||
| Rc.rompB.4762n | AGGCTGGCTGATACACGGAGTAA |
Phylogenetic trees for gltA, 17 kDa (data not shown), ompA (Fig. 1), and ompB (Fig. 2) were constructed using the MEGA version 4.0 software and the neighbour-joining method to infer the evolutionary relatedness. Evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. All positions containing gaps and missing data were eliminated from the dataset.13
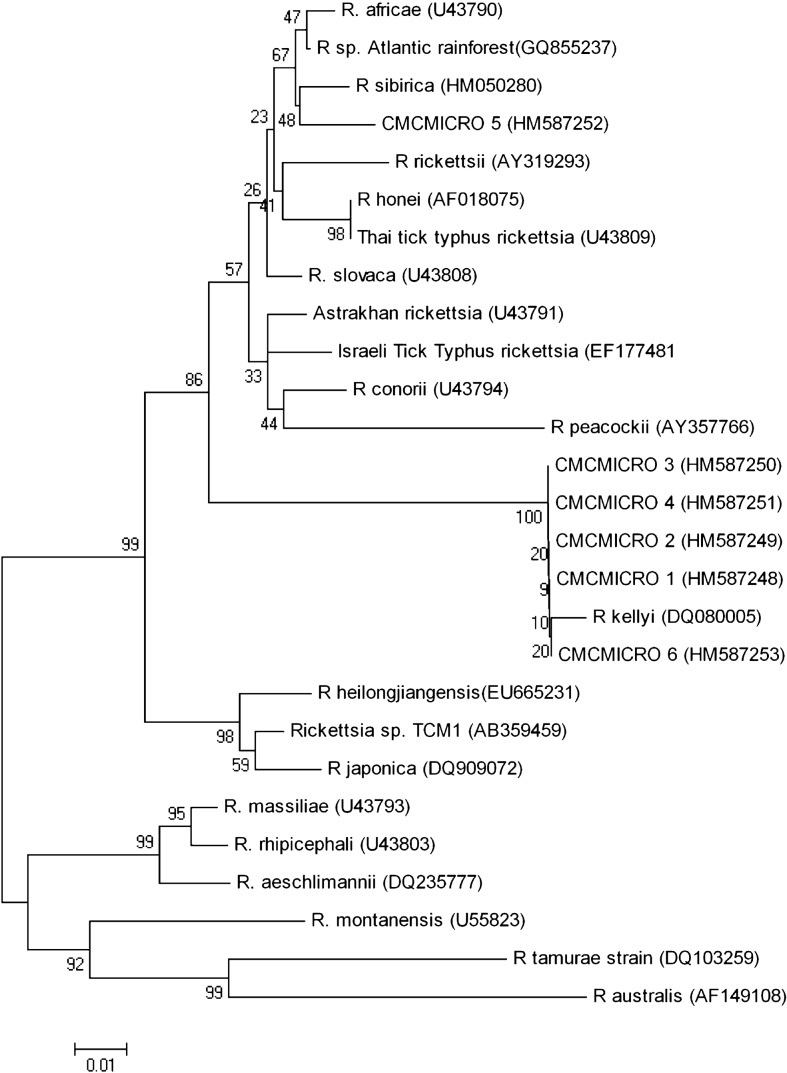
Figure 1.
Neighbour-joining dendrogram showing the relationships between six partial ompA sequences (represented by CMCMICRO1–6) from the skin biopsies of the rash from Indian patients with suspected SFG rickettsiosis compared to a spectrum of other Rickettsia species.
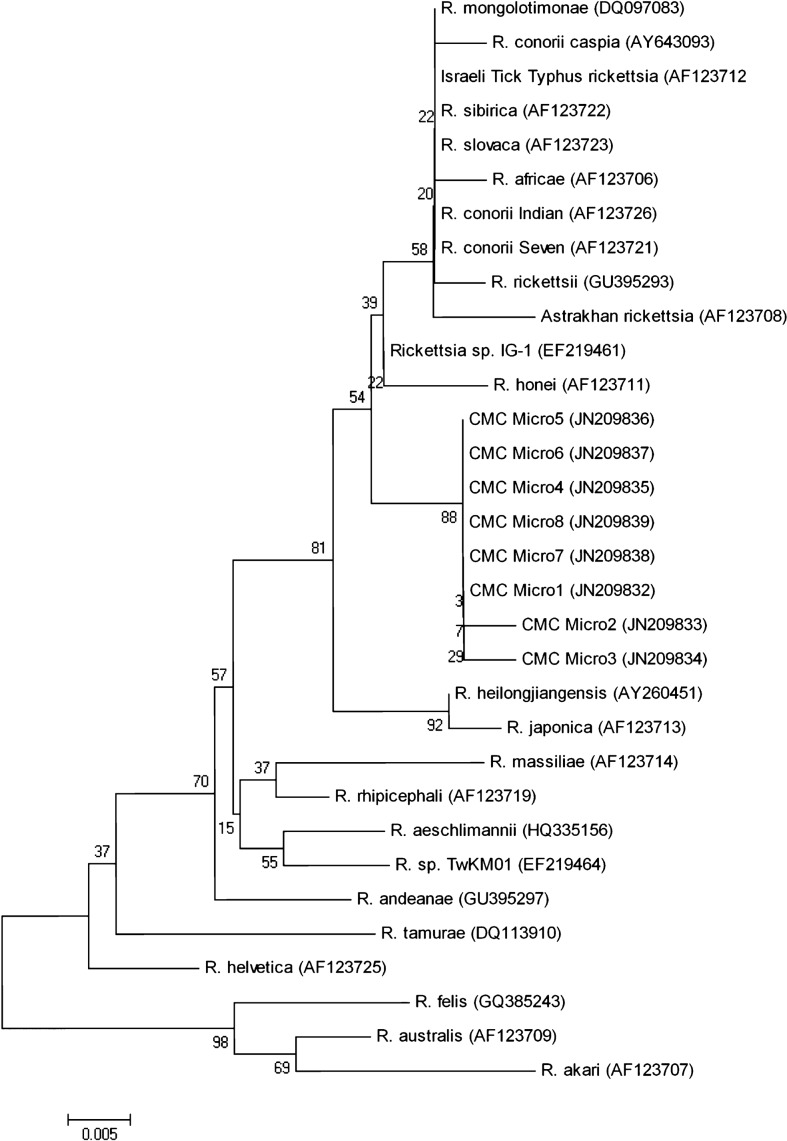
Figure 2.
Neighbour-joining dendrogram showing the relationships between eight partial ompB sequences (represented by CMCMicro1–8) from the skin biopsies of the rash from Indian patients with suspected SFG rickettsiosis compared to a spectrum of other Rickettsia species.
Serum collected from the patients enrolled was subjected to an ELISA for detection of IgM antibodies to spotted fever ((PanBio Ltd, Brisbane, Australia), and a value of ⩾16 units was considered as positive.
Results
None of our patients had eschars and 34 subjects were children under the age of 6 years and constituted the largest group (58.6%). The gltA, 17 kDa, ompA, and ompB genes were detected in skin biopsies from 38, 23, 27, and 22 patients, respectively (Table 1). SFG-specific DNA was detected in 34 (58.6%) cases, confirming these as cases of spotted fever rickettsiosis in accordance with the CDC case definition. The results of the four nested PCR assays are shown in Table 2. In 27 of these 34 patients, spotted fever IgM ELISA was positive, with concordance of 79.4%.
Table 2. Results of nested PCR assays among 58 patients with suspected SFG rickettsiosis.
| PCR result | Total | Spotted fever |
| All four PCR assays negative | 20 | Negative |
| All four PCR assays positive | 15 | Confirmed |
| Only gltA and ompA PCR positive | 4 | Confirmed |
| Only gltA and 17 kDa PCR positive | 5 | Confirmed |
| Only gltA, 17 kDa, and ompA PCR positive | 3 | Confirmed |
| Only gltA, ompA, and ompB PCR positive | 5 | Confirmed |
| Only gltA and ompB PCR positive | 2 | Confirmed |
| Only gltA PCR positive | 4 | Negative |
| Total | 58 |
Although three amplicons each for the gltA and 17 kDa antigen genes were sequenced, only one for each gene was submitted to GenBank (GQ260637 and GQ260636), as the three sequences for these genes were found to be identical by ClustalW multiple sequence alignment. As all the six ompA and the eight ompB sequences were different, they were deposited in the GenBank (ompA: HM587248-53, ompB: JN209832-39).
The gltA gene sequence showed 99% similarity to Rickettsia parkeri, Rickettsia africae, Rickettsia sibirica and Rickettsia mongolotimonae, while the 17 kDa gene amplified in this study demonstrated a 99% similarity to Rickettsia japonica, Rickettsia honei, Rickettsia rickettsii, and Rickettsia conorii. A 99% similarity to Rickettsia spp. IG-1 and 98% similarity to R. honei, R. conori, and R. mongolotimonae were observed with ompB sequences. In contrast, five of the six ompA sequences showed 98% similarity to Candidatus Rickettsia kellyi, whereas one ompA (HM587252) showed 100% similarity to R. parkeri, 98% R. sibirica, R. africae, and R. slovaca, but only 91% for Candidatus Rickettsia kellyi.
Our gltA sequence and the six ompA and eight ompB sequences were closely related to the R. rickettsii cluster of the SFG. The previously published ‘Candidatus Rickettsia kellyi’ sequence13 showed close homology to five out of the six ompA sequences elucidated in this study. The lone sequence which was divergent was closely related to R. parkeri, R. sibirica, and R. africae. These two clusters were supported by high bootstrap values (Fig. 1), whereas the gltA, 17 kDa (data not shown) and ompB phylogenetic trees had lower bootstrap values (Fig. 2) for the R. rickettsii cluster. The eight ompB sequences all clustered together and are closely related to SFG rickettsial strain IG-1 and R. honei.
Discussion
This is the first prospective study in which PCR has been performed on a group of patients clinically suspected to have spotted fever at a tertiary care centre in India. Various reports in the recent past point to the occurrence of spotted fever rickettsiosis in India,1,2,14–16 suggesting that this disease needs to be considered among the differential diagnosis in all patients presenting with fever and rash.
Different gene targets have been used with varying degrees of success for confirming the diagnosis of spotted fever by PCR from blood clots,17,18 serum,7,11,19 skin biopsies,20 and eschars.21 Fournier and Raoult20 reported that among 103 skin biopsies from patients proven to have rickettsial infection, ‘suicide PCR’ was positive in 70 (68%), whereas regular nested PCR and culture detected rickettsia in 43 (45.6%) and 32 (31%), respectively. In addition, ‘suicide PCR’ was also positive in skin biopsies from 17 of the 104 individuals with a probable diagnosis of rickettsiosis. In our study, 34 (58.6%) of the 58 clinically suspected cases of spotted fever rickettsiosis were confirmed by nested PCR.
According to the criteria proposed by Fournier et al.,12 a novel SFGR should ideally show a sequence similarity of <99.8% and <99.9%, for the 16S rRNA (rrs) and gltA gene, and <98.8%, <99.2%, and <99.3% for ompA, ompB, and gene D sequences, with the most closely related validated species described. Our gltA and partial ompA, and ompB sequences are less similar to the most homologous species, but sequence data are unavailable for other commonly targeted genes such as rrs (16S rRNA gene) and sca4 (gene D). In spite of this drawback, the current sequence data further strengthen the earlier observation that novel Rickettsia species may be a cause of disease in this region.14 Further studies to detect these agents from vector hosts, isolation of the organism by culture both from humans and vectors, and also determination of animal reservoirs, especially potential rodent hosts, are required to validate and extend these preliminary findings.
The current study provides further evidence for the occurrence of SFG rickettsiae as important causes of acute febrile illness with rash in southern India. The available sequence data strengthen the assumption that SFGR resembling ‘Candidatus Rickettsia kellyi’ is responsible for spotted fever in these patients.
In the future, paired serum samples will be required to serologically confirm rickettsial infection using micro-immunofluorescence. Owing to shortcomings of nested PCR, we will explore the diagnostic utility of the highly sensitive and specific quantitative real-time PCR assay as we previously described.22
In conclusion, this is the first prospective study where specific nested PCRs to diagnose spotted fever were performed in clinically suspected cases in a tertiary care centre in India. Isolation of the organism in culture followed by molecular enumeration of the necessary genes is essential to provide irrefutable taxonomic proof that a novel rickettsial species is the causative agent of spotted fever rickettsiosis in this part of India. However, these data strengthen the concept of an expanding role for established and novel SFG rickettsiae in the occurrence of febrile disease in India and around the world.
Acknowledgments
This work is supported by the Institutional Review Board, Christian Medical College, Vellore 632004, India (Fluid Research Grant Nos. 22X138 and 22X245).
References
- 1.Somashekar HR, Moses PD, Pavithran S, Mathew LG, Agarwal I, Rolain JM, et al. Magnitude and features of scrub typhus and spotted fever in children in India. J Trop Pediatr. 2006;52:228–9. doi: 10.1093/tropej/fmi096. [DOI] [PubMed] [Google Scholar]
- 2.Mahajan SK, Kashyap R, Sankhyan N, Sharma V, Rolain JM, Prasher BS, et al. Spotted fever group rickettsioses in Himachal Pradesh. J Assoc Physicians India. 2007;55:868–70. [PubMed] [Google Scholar]
- 3.Walker DH. Rickettsiae and rickettsial infections: the current state of knowledge. Clin Infect Dis. 2007;45:S39–44. doi: 10.1086/518145. [DOI] [PubMed] [Google Scholar]
- 4.Spotted Fever Rickettsiosis: 2010 Case Definition. CSTE Position Statement Number: 09-ID-16. Available from: http://www.cdc.gov/ncphi/disss/nndss/casedef/spottedfever current.htm, accessed 31 Dec 2011. [Google Scholar]
- 5.Parola P, Paddock CD, Raoult R. Tick-borne rickettsioses around the world: emerging diseases challenging old concepts. Clin Microbiol Rev. 2005;18:719–56. doi: 10.1128/CMR.18.4.719-756.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roux V, Rydkina E, Eremeeva M, Raoult D. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int J Syst Bacteriol. 1997;47:252–61. doi: 10.1099/00207713-47-2-252. [DOI] [PubMed] [Google Scholar]
- 7.Choi YJ, Jang WJ, Ryu JS, Lee SH, Park KH, Paik HS, et al. Spotted fever group and typhus group rickettsioses in humans, South Korea. Emerg Infect Dis. 2005;11:237–44. doi: 10.3201/eid1102.040603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massung RF, Davis LE, Slater K, McKechnie DB, Puerzer M. Epidemic typhus meningitis in the southwestern United States. Clin Infect Dis. 2001;32:979–82. doi: 10.1086/319351. [DOI] [PubMed] [Google Scholar]
- 9.Tzianabos T, Anderson BE, McDade JE. Detection of Rickettsia rickettsii DNA in clinical specimens by using polymerase chain reaction technology. J Clin Microbiol. 1989;7:2866–8. doi: 10.1128/jcm.27.12.2866-2868.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishikura M, Ando S, Shinagawa Y, Matsuura K, Hasegawa S, Nakayama T, et al. Phylogenetic analysis of spotted fever group rickettsiae based on gltA, 17-kDa, and rompA genes amplified by nested PCR from ticks in Japan. Microbiol Immunol. 2003;47:823–32. doi: 10.1111/j.1348-0421.2003.tb03448.x. [DOI] [PubMed] [Google Scholar]
- 11.Choi YJ, Lee SH, Park KH, Koh YS, Lee KH, Baik HS, et al. Evaluation of PCR-based assay for diagnosis of spotted fever group rickettsiosis in human serum samples. Clin Diagn Lab Immunol. 2005;12:759–63. doi: 10.1128/CDLI.12.6.759-763.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fournier PE, Dumler JS, Greub G, Zhang J, Wu Y, Raoult D. Gene sequence-based criteria for identification of new rickettsia isolates and description of Rickettsia heilongjiangensis sp. nov. J Clin Microbiol. 2003;41:5456–65. doi: 10.1128/JCM.41.12.5456-5465.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura K, Dudley J, Nei M, Kumar S. Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 14.Rolain JM, Mathai E, Lepidi H, Somashekar HR, Mathew LG, Prakash JA, et al. ‘Candidatus Rickettsia kellyi’, India. Emerg Infect Dis. 2006;12:483–5. doi: 10.3201/eid1203.050853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murali N, Pillai S, Cherian T, Raghupathy P, Padmini V, Mathai E. Rickettsial infections in South India — how to spot the spotted fever. Indian Pediatr. 2001;38:1393–6. [PubMed] [Google Scholar]
- 16.Sundhindra BK, Vijayakumar S, Kutty KA, Tholpadi SR, Rajan RS, Mathai E, et al. Rickettsial spotted fever in Kerala. Natl Med J India. 2004;17:51–2. [PubMed] [Google Scholar]
- 17.Furuya Y, Katayama T, Yoshida Y, Kaiho I. Specific amplification of Rickettsia japonica DNA from clinical specimens by PCR. J Clin Microbiol. 1995;33:487–9. doi: 10.1128/jcm.33.2.487-489.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanaoka N, Matsutani M, Kawabata H, Yamamoto S, Fujita H, Sakata A, et al. Diagnostic assay for Rickettsia japonica. Emerg Infect Dis. 2009;15:1994–7. doi: 10.3201/eid1512.090252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leitner M, Yitzhaki S, Rzotkiewicz S, Keysary A. Polymerase chain reaction-based diagnosis of Mediterranean spotted fever in serum and tissue samples. Am J Trop Med Hyg. 2002;67:166–9. doi: 10.4269/ajtmh.2002.67.166. [DOI] [PubMed] [Google Scholar]
- 20.Fournier PE, Raoult D. Suicide PCR on skin biopsy specimens for diagnosis of rickettsioses. J Clin Microbiol. 2004;42:3428–34. doi: 10.1128/JCM.42.8.3428-3434.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paddock CD, Koss T, Eremeeva ME, Dasch GA, Zaki SR, Sumner JW. Isolation of Rickettsia akari from eschars of patients with rickettsialpox. Am J Trop Med Hyg. 2006;75:732–8. [PubMed] [Google Scholar]
- 22.Prakash JA, Reller ME, Barat N, Dumler JS. Assessment of a quantitative multiplex 5' Nuclease real-time PCR for spotted fever and typhus group rickettsioses and Orientia tsutsugamushi. Clin Microbiol Infect. 2009;15(Suppl. 2):292–3. doi: 10.1111/j.1469-0691.2008.02242.x. [DOI] [PubMed] [Google Scholar]