Abstract
A precise balance between protein degradation and synthesis is essential to preserve skeletal muscle mass. Here, we found that TP53INP2, a homolog of the Drosophila melanogaster DOR protein that regulates autophagy in cellular models, has a direct impact on skeletal muscle mass in vivo. Using different transgenic mouse models, we demonstrated that muscle-specific overexpression of Tp53inp2 reduced muscle mass, while deletion of Tp53inp2 resulted in muscle hypertrophy. TP53INP2 activated basal autophagy in skeletal muscle and sustained p62-independent autophagic degradation of ubiquitinated proteins. Animals with muscle-specific overexpression of Tp53inp2 exhibited enhanced muscle wasting in streptozotocin-induced diabetes that was dependent on autophagy; however, TP53INP2 ablation mitigated experimental diabetes-associated muscle loss. The overexpression or absence of TP53INP2 did not affect muscle wasting in response to denervation, a condition in which autophagy is blocked, further indicating that TP53INP2 alters muscle mass by activating autophagy. Moreover, TP53INP2 expression was markedly repressed in muscle from patients with type 2 diabetes and in murine models of diabetes. Our results indicate that TP53INP2 negatively regulates skeletal muscle mass through activation of autophagy. Furthermore, we propose that TP53INP2 repression is part of an adaptive mechanism aimed at preserving muscle mass under conditions in which insulin action is deficient.
Introduction
Skeletal muscle mass is determined by a fine balance between protein synthesis and degradation (1). Alterations in any of these processes can result in muscle atrophy when there is a net increase in the protein degradation rate and in muscle hypertrophy when there is an increase in the net protein synthesis rate. Regarding protein degradation, the two main proteolytic systems in skeletal muscle are the ubiquitin-proteasome system (UPS) and macroautophagy (hereafter referred to as autophagy). Autophagy is a highly conserved pathway that degrades long-lived proteins and organelles from the cell (2, 3). Briefly, it consists of the engulfment by a double-membrane vacuole of a portion of the cytosol in which there are organelles, protein aggregates, etc., that need to be removed. Later on, this vacuole will fuse with the lysosomes, and all the cargos inside these autolysosomes will be degraded (4). A correct balance of autophagy is necessary to maintain skeletal muscle mass and its correct function. Thus, autophagy is enhanced in different situations that cause muscle atrophy favoring muscle loss (5, 6). This has been detected basically by increased formation of GFP-LC3–positive puncta in muscle fibers (7) and increased transcription of genes involved in that process (8–10). However, a certain degree of autophagy is necessary to remove damaged proteins and organelles to ensure proper mass and function of skeletal muscle (11). Thus, complete autophagy inhibition through ATG7 ablation causes sarcomere disorganization, abnormal mitochondria accumulation, distension of sarcoplasmic reticulum, accumulation of ubiquitinated proteins, and enhanced muscle wasting upon denervation or prolonged fasting (11). Actually, basal levels of autophagy have been reported to be highly selective and to constitute a quality control mechanism important for the maintenance of cellular homeostasis (3, 12).
Type 1 diabetes (13–15), cancer cachexia (16, 17), renal failure (18), and sepsis (19) are examples of pathological situations in which there is muscle wasting due to the activation of the proteolytic systems in skeletal muscle. However, whole-body protein turnover is essentially spared in type 2 diabetes (20, 21), and leg and arm muscle mass is greater in type 2 diabetic individuals compared with that in nondiabetic subjects (22). The molecular mechanism by which deficient insulin action does not reduce muscle mass has not yet been elucidated.
Diabetes and obesity regulated (DOR) gene, also named Tp53inp2, is a metazoan gene (23) highly expressed in skeletal muscle (24). Tp53inp2 encodes a bifunctional protein acting as a nuclear coactivator (24, 25) and as a key regulator of basal autophagy in cellular models and in Drosophila melanogaster (26–28). TP53INP2 interacts directly with LC3 and its family members and stimulates autophagosome formation and protein degradation (26). Here, we have studied the functional role of Tp53inp2 by gain-of-function and ablation approaches in mice. Our data indicate that TP53INP2 is a negative regulator of skeletal muscle mass that enhances basal autophagy. TP53INP2 is highly repressed in muscle from type 2 diabetic patients, and we propose that TP53INP2 repression is part of a mechanism responsible for the preservation of muscle mass in conditions characterized by reduced insulin action.
Results
TP53INP2 gain of function causes a reduction in muscle mass.
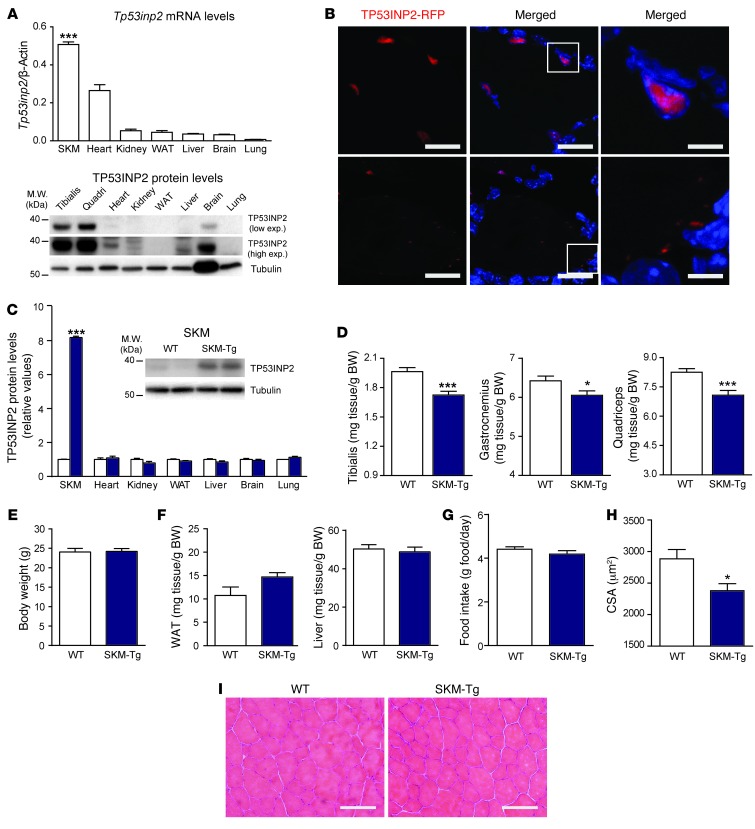
Skeletal muscle shows the highest expression of Tp53inp2 mRNA and protein compared with that in other mouse tissues (Figure 1A). This suggests that TP53INP2 must play a relevant role in skeletal muscle. TP53INP2 localization in muscle fibers was analyzed by electrotransfer of a plasmid encoding RFP-tagged TP53INP2 in mouse muscle. We have reported that TP53INP2-RFP and endogenous TP53INP2 proteins behave similarly in cultured cells (28). TP53INP2 was localized inside the nucleus and in cytoplasmic puncta (Figure 1B), in keeping with previous observations in cells (26).
Figure 1. Muscle-specific TP53INP2 gain of function reduces skeletal muscle mass in a transgenic mouse model (SKM-Tg).
(A) Tp53inp2 mRNA and protein levels in different mouse tissues. The skeletal muscle (SKM) used for mRNA analysis was tibialis anterior, and tissues from 3 different male mice were used. ***P < 0.001, skeletal muscle vs. the other tissues. (B) Confocal images showing TP53INP2 localization in transverse sections from the tibialis anterior muscle. Adult muscles were electrotransferred with TP53INP2-RFP. TP53INP2 is shown in red and nuclei are shown in blue (Hoechst33342 staining). Scale bar: 20 μm (left and middle columns); 5 μm (right column). Boxes in the middle column are shown at higher magnification to the right. (C) Quantification of TP53INP2 protein levels in tissue homogenates from WT and SKM-Tg mice (n = 4). Data are shown as relative TP53INP2 levels in WT mice within each tissue. Representative images from quadriceps homogenates are shown. (D) Weights of tibialis anterior, gastrocnemius, and quadriceps muscles from 4-month-old WT and SKM-Tg mice. (E) Body weight of WT and SKM-Tg mice. (F) Epididymal adipose tissue and liver weights from WT and SKM-Tg mice. (G) Food intake of WT and SKM-Tg mice. (H) Mean cross-sectional area (CSA) of 150 myofibers per each tibialis anterior muscle. Data in D to H were obtained from 6 WT and 6 SKM-Tg mice. (I) Representative images of hematoxylin/eosin staining from WT and SKM-Tg mice. Scale bar: 100 μm. Data represent mean ± SEM. *P < 0.05, ***P < 0.001 vs. control mice.
To determine the functional role of TP53INP2 in skeletal muscle, we generated transgenic mice overexpressing TP53INP2 specifically in skeletal muscle [Tg(Mlc1-Tp53inp2) (SKM-Tg) mice]. Transgenic line SKM-Tg showed higher TP53INP2 protein levels in skeletal muscle compared with that in controls (WT), whereas TP53INP2 expression was unaltered in other tissues (Figure 1C). SKM-Tg male mice showed a reduction in the weight of muscles compared with that in their control littermates (Figure 1D), which were without changes in body weight (Figure 1E). This effect was specific to skeletal muscle, and it was not detected in other tissues, such as liver or epididymal adipose tissue (Figure 1F). No differences in food intake were detected between WT and transgenic animals (Figure 1G). Transgenic mice showed a marked decrease in myofiber cross-sectional area compared with that in controls (Figure 1H and Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI72327DS1), which occurred in the absence of other histological abnormalities in muscle (Figure 1I).
In order to assess whether the reduction in muscle mass was specific to a certain period during the life of the mouse, we used a microcomputed tomography approach in which the lean tissue volume along the fibula from the right hind limb was measured (Supplemental Figure 1B). This allowed the monitoring of the same group of animals over time. In keeping with the decreased muscle weight and fiber size, we found a reduction in the lean tissue volume in SKM-Tg mice compared with that in controls from age 3 to 12 months (Supplemental Figure 1C).
A second transgenic line, SKM-Tg’ (Supplemental Figure 1D), confirmed the muscle phenotype. Thus, SKM-Tg’ mice also showed reduced muscle weight compared with that of controls (Supplemental Figure 1E), which were without changes in body (Supplemental Figure 1F), adipose tissue, or liver weights (Supplemental Figure 1G). In accordance, lean tissue volume was reduced in 4-month-old SKM-Tg’ mice (Supplemental Figure 1H). This indicates that the phenotype observed is specific to TP53INP2 gain of function and is not dependent on the insertion site of the transgene.
The reduction in muscle mass was sex-independent, and age-matched transgenic female mice also showed reduced muscle weight (Supplemental Figure 2A), without differences in perigonadal adipose tissue, liver (Supplemental Figure 2B), or body weights (Supplemental Figure 2C) compared with those of WT female mice. Lean tissue volume of transgenic female mice was also reduced from age 3 to 12 months (Supplemental Figure 2D). Moreover, myofiber cross-sectional area of transgenic females was markedly reduced (Supplemental Figure 2E).
The alterations detected in SKM-Tg mice were not a consequence of changes in muscle fiber-type composition. Thus, no differences were found in the distribution of type IIb, IIx, or IIa myofibers in tibialis anterior muscles from WT and SKM-Tg mice (Supplemental Figure 3, A and B). TP53INP2 gain of function caused a reduction in the cross-sectional area from type IIb, IIx, and IIa myofibers (Supplemental Figure 3, C and D). Moreover, TP53INP2 gain of function did not alter mitochondrial content or functionality (Supplemental Figure 4).
Because TP53INP2 loss of function has been reported to cause a delay in the myogenic differentiation of C2C12 myoblasts (24), we analyzed whether the alterations detected in mice upon TP53INP2 overexpression were secondary to changes in muscle formation. To this end, tibialis anterior muscle damage was induced with cardiotoxin injection in WT and SKM-Tg mice, and muscle regeneration was monitored 7 days after injury by quantifying the cross-sectional area of muscle fibers. No differences were detected between transgenic and control mice (Supplemental Figure 5A). TP53INP2 overexpression did not alter the rate of apoptosis in muscle (Supplemental Figure 5B).
Muscle TP53INP2 gain of function did not alter glucose blood levels either in fed or fasting conditions (Supplemental Figure 6A) and did not alter glucose utilization upon a glucose tolerance test (Supplemental Figure 6B). Moreover, there were no differences in circulating insulin, cholesterol, or triglyceride levels between fasted WT and SKM-Tg animals (Supplemental Figure 6C).
TP53INP2 ablation causes muscle hypertrophy.
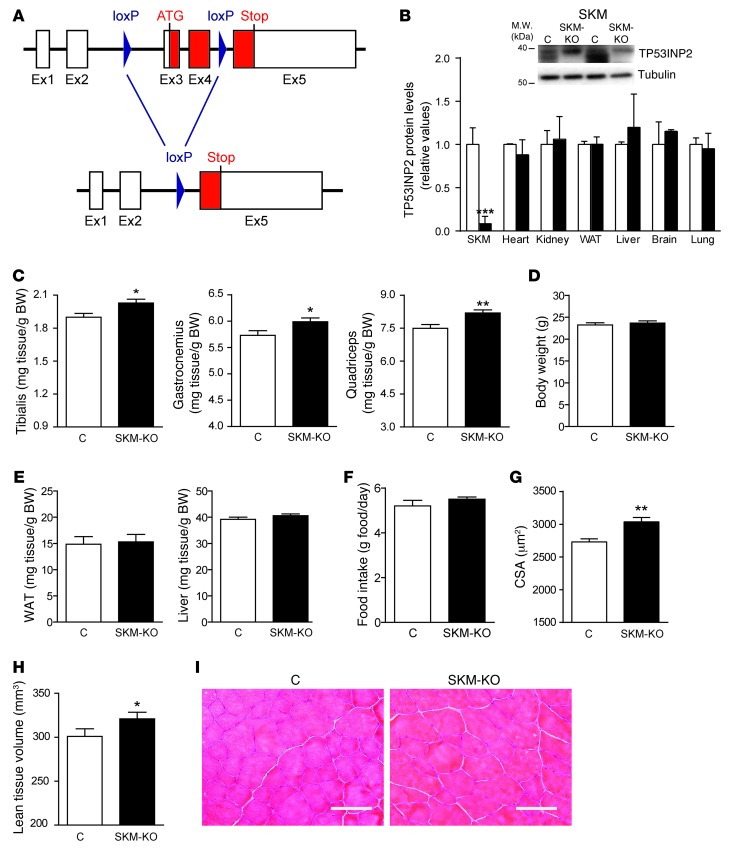
Next, we generated a muscle-specific Tp53inp2 knockout mouse line (SKM-KO) by crossing homozygous Tp53inp2loxP/loxP mice with a mouse strain expressing Cre recombinase under the control of the myosin light chain 1 promoter (29). Deletion of exons 3 and 4 driven by Cre recombinase caused the ablation of Tp53inp2 expression (Figure 2A), and, indeed, TP53INP2 protein was undetectable in skeletal muscle from SKM-KO mice (Figure 2B). However, other tissues showed a normal TP53INP2 expression in SKM-KO mice (Figure 2B).
Figure 2. TP53INP2-specific ablation in skeletal muscle causes muscle hypertrophy in a knockout mouse model (SKM-KO).
(A) Genomic structure of Tp53inp2 gene showing 5 exons and the corresponding 4 introns. LoxP sequences were inserted into introns 2 and 4. After Cre recombinase action, exons 3 and 4 were excised, eliminating the initiation codon. (B) Quantification of TP53INP2 protein levels in tissue homogenates from control (C) (nonexpressing Cre Tp53inp2loxP/loxP mice) and SKM-KO mice (expressing Cre Tp53inp2loxP/loxP mice). Representative images from quadriceps homogenates are shown. Data are shown as relative TP53INP2 levels in control mice within each tissue. (C) Weights of tibialis anterior, gastrocnemius, and quadriceps muscles from 4-month-old control and SKM-KO mice. (D) Body weight of control and SKM-KO mice. (E) Epididymal adipose tissue and liver weights from control and SKM-KO mice. (F) Food intake of control and SKM-KO mice. (G) Mean cross-sectional area of 150 myofibers per each tibialis anterior muscle. (H) Lean tissue volume of the right hind limb of 4-month-old control and SKM-KO mice. Data in C to H were obtained from 6 control and 10 SKM-KO mice. (I) Representative images of hematoxylin/eosin staining from control and SKM-KO mice Scale bar: 100 μm. Data represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control mice.
In contrast to what was observed upon Tp53inp2 gain of function, SKM-KO mice showed muscle hypertrophy compared with control mice (nonexpressing Cre Tp53inp2loxP/loxP littermates), as assessed by increased tibialis anterior, gastrocnemius, or quadriceps muscle weights (Figure 2C), without changes in body weight (Figure 2D). Again, the effects were specific for muscle and no alterations in epididymal adipose tissue or liver weights were detected (Figure 2E). SKM-KO mice showed normal food intake (Figure 2F). In accordance with the increased muscle weight, we documented increased cross-sectional area of muscle fibers (Figure 2G and Supplemental Figure 7) and increased lean tissue volume in SKM-KO mice compared with that in controls (Figure 2H). SKM-KO mice did not display histological abnormalities (Figure 2I) or changes in fiber-type composition (Supplemental Figure 8, A and B). However, TP53INP2 ablation caused an increase in the cross-sectional area of type IIb, IIx, and IIa myofibers from tibialis anterior muscles (Supplemental Figure 8, C and D).
Muscle regeneration capacity was analyzed in the same way as performed in the SKM-Tg mice. Cardiotoxin was injected into tibialis anterior muscles from SKM-KO and control mice, and cross-sectional area was quantified 7 days after injury. No differences were found between groups (Supplemental Figure 9A). TP53INP2 ablation in muscle did not alter the number of apoptotic nuclei (Supplemental Figure 9B) and the mitochondrial content or functionality (Supplemental Figure 10). Moreover, there was no difference in glycemia between 4-month-old control and SKM-KO mice (Supplemental Figure 11A), and they responded in a similar way to a glucose tolerance test (Supplemental Figure 11B).
TP53INP2 mainly activates basal autophagy in skeletal muscle.
TP53INP2 has been described as a protein with two different functions, i.e., a nuclear coactivator and an autophagy regulator (24–27). This is why we decided to analyze which of these two functions could explain the phenotype observed in our mice models. In this regard, we performed a transcriptomic analysis using microarrays looking for genes differentially expressed in the quadriceps muscles of WT and SKM-Tg mice (Supplemental Table 1) as well as in control and SKM-KO animals (Supplemental Table 2). Only a reduced number of genes was dysregulated upon TP53INP2 manipulation (13 genes upregulated in SKM-Tg mice and 6 genes downregulated in SKM-KO mice) (Supplemental Tables 1 and 2). In addition, most of the genes underwent mild changes in expression (1.3- to 2.9-fold). These data strongly suggest that TP53INP2 does not operate as a nuclear cofactor in mouse skeletal muscle, at least, not under the conditions we have studied.
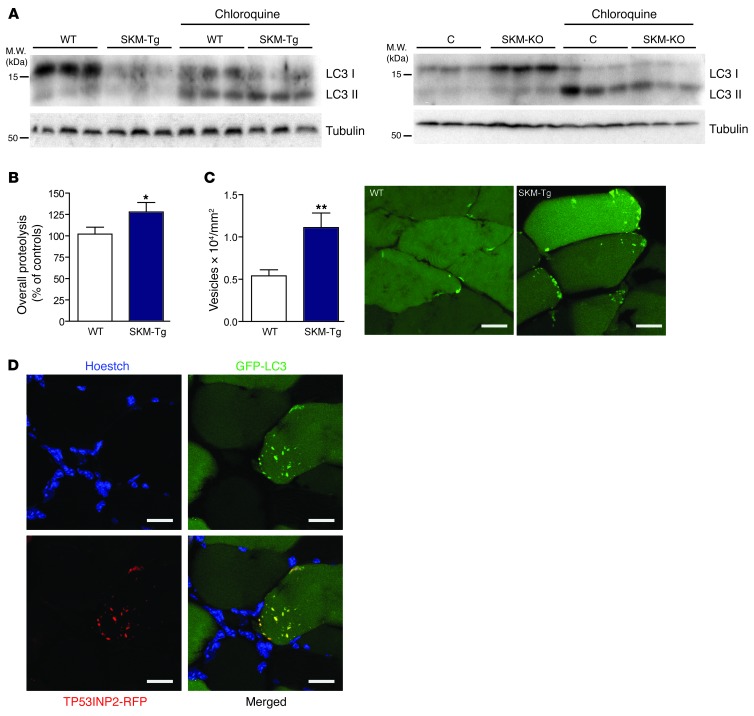
Next, we studied whether TP53INP2 regulates autophagy in muscle. Autophagic activity was analyzed by measuring the abundance of LC3I and LC3II in muscles from fed mice (basal conditions). Male SKM-Tg mice showed a marked reduction of LC3I protein and an increased LC3II/LC3I ratio (Figure 3A and Supplemental Figure 12A). Similar data were obtained in female SKM-Tg mice (Supplemental Figure 12B) and male SKM-Tg’ mice (Supplemental Figure 12C). In contrast, SKM-KO mice showed increased levels of both LC3I and LC3II compared with the control group in basal conditions (Figure 3A and Supplemental Figure 12D).
Figure 3. TP53INP2 increases basal autophagy in skeletal muscle by inducing the formation of autophagosomes.
(A) Western blot analysis of LC3I and LC3II content in total homogenates of gastrocnemius muscles from WT and SKM-Tg mice or from control and SKM-KO mice. Representative images are shown. Mice were treated with chloroquine as indicated. (B) Overall proteolysis assessed as the rate of tyrosine released to the media in incubated extensor digitorum longus muscles (n = 14). (C) Autophagosomes were quantified by counting EGFP-LC3–positive dots normalized for cross-sectional area from tibialis anterior muscles transfected with EGFP-LC3 from WT and SKM-Tg mice fasted for 16 hours (n = 5). Representative images of transverse sections from adult tibialis anterior muscles transfected with EGFP-LC3 of WT and SKM-Tg mice are shown. Scale bar: 15 μm. (D) Confocal images of tibialis anterior muscle transverse sections showing colocalization between TP53INP2 and LC3. Adult muscles were transfected with TP53INP2-RFP and EGFP-LC3. TP53INP2 is stained in red, LC3 is stained in green, and nuclei are stained in blue (Hoechst33342 staining). Scale bar: 30 μm. Data represent mean ± SEM. *P < 0.05, **P < 0.01 vs. control values.
These changes in LC3 protein levels cannot be explained by changes in the mRNA levels (Supplemental Figure 12E). We further analyzed autophagic flux to evaluate whether changes in LC3 content were due to altered degradation rate of LC3. To this end, SKM-Tg mice were treated with chloroquine to inhibit lysosomal protein degradation. Under these conditions, SKM-Tg animals showed higher accumulation of LC3II protein than control mice (Figure 3A and Supplemental Figure 12F), indicating an enhanced autophagy flux. On the other hand, SKM-KO mice treated with chloroquine showed a lower accumulation of LC3II than controls (Figure 3A and Supplemental Figure 12G), indicating a reduced autophagy flux in muscle upon TP53INP2 ablation.
Based on the greater changes detected in autophagy, we selected SKM-Tg mice to analyze whether the increased basal autophagy activity shown in these muscles had an impact on protein degradation. The protein degradation rate was 28% greater in incubated extensor digitorum longus muscles from fasted SKM-Tg mice compared with that in controls (Figure 3B). However, no changes were found in proteasome activity in total homogenates from control and transgenic animals (Supplemental Figure 13A). Given that the UPS and autophagy are the major proteolytic systems in skeletal muscle, these data suggest that the increased total proteolysis observed in TP53INP2-overexpressing muscles is due to increased autophagy.
TP53INP2 increases basal autophagy in cells by increasing the formation of autophagosomes through the interaction with different members of the ATG8 family proteins, including LC3 (26, 27). To assess whether TP53INP2 functions in a similar manner in skeletal muscle, tibialis anterior muscles from SKM-Tg and WT mice were electrotransferred with EGFP-LC3 construct, and the number of EGFP-LC3–positive dots was counted. Fasted SKM-Tg mice showed a marked increase in the number of EGFP-LC3–positive vesicles compared with that in the control group (Figure 3C). In addition, muscles transfected with both EGFP-LC3 and TP53INP2-RFP showed a high colocalization between these 2 proteins in muscle fibers (Figure 3D). Specifically, 77% ± 4% of EGFP-LC3 dots colocalized with TP53INP2-RFP and, vice versa, 83% ± 4% of TP53INP2-RFP dots colocalized with EGFP-LC3. This is consistent with an interaction between TP53INP2 and LC3 occurring in skeletal muscle. In keeping with results observed in cellular models, TP53INP2 was localized almost exclusively in cytosolic puncta upon EGFP-LC3 overexpression (23, 26).
Electron microscopy of muscle sections from SKM-Tg mice did not show major ultrastructural changes (Supplemental Figure 13B). However, the abundance of autophagosome-related structures was higher in muscles from SKM-Tg mice compared with that in controls (Supplemental Figure 13C).
TP53INP2 is involved in the autophagic degradation of ubiquitinated proteins.
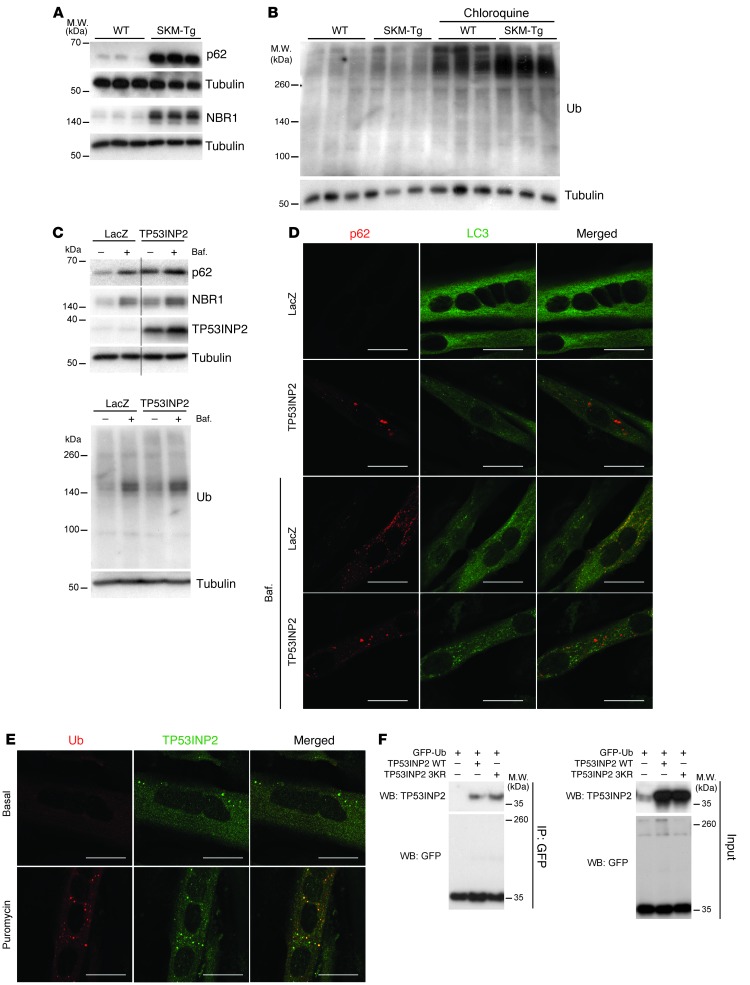
To further characterize TP53INP2 action on basal autophagy, we decided to analyze the levels of autophagy receptors p62 and NBR1 in total homogenates from WT and SKM-Tg mice (30–32). TP53INP2 overexpression increased p62 and NBR1 protein levels both in muscle (Figure 4A and Supplemental Figure 14A) and in C2C12 myotubes (Supplemental Figure 14B). This accumulation was not explained by changes in mRNA levels (Supplemental Figure 14C).
Figure 4. TP53INP2 promotes degradation of ubiquitinated proteins in skeletal muscle.
(A) p62 and NBR1 protein levels in total homogenates of gastrocnemius muscles from WT and SKM-Tg mice. Representative images are shown. (B) Western blot analysis of ubiquitinated protein content in total homogenates of gastrocnemius muscles from WT and SKM-Tg mice with or without chloroquine treatment. Representative images are shown. (C) Western blot analysis of p62, NBR1, TP53INP2, and ubiquitinated protein (FK2) content in protein extracts from day 5 C2C12 myotubes overexpressing TP53INP2 or LacZ (control). Cells were treated with bafilomycin (Baf.) as indicated. Thin black lines indicate that lanes were run on the same gel but were noncontiguous. Representative images are shown. (D) Immunofluorescence against p62 and LC3 in control (LacZ) or TP53INP2-overexpressing C2C12 day 5 myotubes. Cells were treated with bafilomycin at 200 nM during 3 hours as indicated. p62 is stained in red and LC3 is stained in green. Scale bar: 20 μm. (E) Immunofluorescence against ubiquitin (FK2) and TP53INP2 in C2C12 day 5 myotubes. Cells were treated with puromycin at 50 μg/ml during 4 hours as indicated. Ub (FK2) is stained in red and TP53INP2 is stained in green. Scale bar: 20 μm. (F) HEK293T cells were transfected with plasmids coding for GFP-Ub and/or mouse TP53INP2 or mouse TP53INP2 3KR mutant. Pull-down was performed using GFP-Trap beads and inputs and pellets were probed in Western blot assays with anti-GFP and anti-TP53INP2 antibodies.
To further confirm that p62 was accumulated, we performed immunofluorescence in transversal sections of tibialis anterior muscles from WT and SMK-Tg animals (Supplemental Figure 14D). We did not detect p62-positive dots in 4-month-old WT animals (Supplemental Figure 14D). SKM-Tg mice showed a moderate number of small p62-positive dots (Supplemental Figure 14D). p62 immunofluorescence was also performed in C2C12 myotubes, and we observed that TP53INP2 overexpression caused p62 accumulation (Supplemental Figure 14E).
This was an unexpected result since TP53INP2 overexpression enhances autophagy in skeletal muscle, and normally accumulation of p62 and NBR1 has been associated with autophagy inhibition (30, 31). p62 and NBR1 are autophagy receptors of ubiquitinated proteins that interact simultaneously with LC3 and ubiquitin chains and promote the degradation of ubiquitinated protein aggregates (30–32). By doing so, p62 and NBR1 are selectively degraded through autophagy together with their cargo (30, 31). Thus, we decided to analyze whether TP53INP2-induced accumulation of those proteins implied an impairment in the degradation of ubiquitinated proteins through autophagy. We monitored the content of ubiquitinated proteins in homogenates from WT and SKM-Tg mice treated or not with chloroquine (Figure 4B and Supplemental Figure 14F). No statistically significant changes in the content of ubiquitinated proteins were found between SKM-Tg and WT mice in basal conditions (Figure 4B and Supplemental Figure 14F). However, chloroquine treatment caused a greater accumulation of ubiquitinated proteins in transgenic animals compared with that in controls (Figure 4B). These results suggest that TP53INP2 gain of function in skeletal muscle, not only does not impair, but also accelerates the degradation of ubiquitinated proteins, despite of the accumulation of p62 and NBR1.
We hypothesized that TP53INP2 also acts as an autophagy receptor for ubiquitinated proteins and, when overexpressed, displaces p62 and NBR1 from their normal function. Given that p62 and NBR1 are specifically degraded through autophagy (30, 31), the displacement from their function should cause their accumulation. Thus, we monitored p62 and NBR1 accumulation in TP53INP2-overexpressing or control C2C12 myotubes upon autophagy inhibition (Figure 4C and Supplemental Figure 14, G and H). As previously shown, TP53INP2 overexpression caused an increase in p62 and NBR1 protein levels in basal conditions (Figure 4C and Supplemental Figure 14, G and H). However, meanwhile control muscle cells showed a large accumulation of p62 upon autophagy blockage with bafilomycin A1 (Figure 4C and Supplemental Figure 14G), and TP53INP2-overexpressing myotubes displayed reduced changes in p62 upon autophagy inhibition (Figure 4C and Supplemental Figure 14G). NBR1 was accumulated in both control and TP53INP2-overexpressing myotubes upon bafilomycin A1 treatment, although the accumulation was larger in LacZ-expressing muscle cells (Figure 4C and Supplemental Figure 14H). In accordance with data obtained in muscle, the accumulation of p62 and NBR1 occurred without any impairment in the autophagic degradation of ubiquitinated proteins (Figure 4C and Supplemental Figure 14I). Our results indicate that TP53INP2 gain of function displaces p62 and NBR1 from their function as autophagy receptors.
To complement Western blot data, we performed immunofluorescence in C2C12 myotubes, and we analyzed the colocalization between LC3 and p62. Almost no LC3-positive dots were detected either in control or in TP53INP2-overexpressing myotubes (Figure 4D). In order to promote autophagosome accumulation, muscle cells were treated with bafilomycin A1. Under these conditions, control myotubes presented a large colocalization between p62 and LC3 (Figure 4D). In fact, 82% ± 2% of LC3-positive dots also contained p62. TP53INP2 overexpression reduced the extent of colocalization between LC3 and p62 to 21% ± 3% (Figure 4D). These results indicate that p62 is displaced from autophagosomes upon TP53INP2 gain of function. Moreover, in LacZ-expressing myotubes, only 7.5% ± 0.9% of LC3-positive dots colocalized with TP53INP2 upon bafilomycin A1 treatment (Supplemental Figure 15A). However, in TP53INP2-overexpressing myotubes, the 94% ± 1% of LC3-positive dots also contained TP53INP2 (Supplemental Figure 15A). This increase in the colocalization between TP53INP2 and LC3 correlates with the reduction in the colocalization between p62 and LC3.
To further characterize the putative role of TP53INP2 as a novel autophagy receptor for ubiquitinated proteins, we performed again immunofluorescence in C2C12 myotubes, analyzing whether TP53INP2 colocalized with ubiquitinated protein aggregates. This analysis was performed under basal conditions or upon treatment with puromycin, which stimulates the formation of protein aggregates (ref. 30 and Figure 4E). Ubiquitin-positive dots were barely detected in C2C12 myotubes under basal conditions (Figure 4E). In the presence of puromycin, endogenous TP53INP2 showed a partial colocalization with ubiquitin-positive dots (17.1% ± 4.5% of TP53INP2-positive dots colocalized with ubiquitin dots) (Figure 4E) in accordance with its role as an autophagy receptor for ubiquitinated proteins.
Finally, we analyzed whether TP53INP2 was able to interact with ubiquitin. HEK293T cells were transfected with GFP-Ub and TP53INP2, and cell lysates were pulled down with GFP-Trap beads. Data shown in Figure 4F indicate that TP53INP2 coimmunoprecipitates with GFP-ubiquitin. To confirm that TP53INP2 was pulled down, because it interacted with ubiquitin and not because it was ubiquitinated, HEK293T cells were transfected with GFP-Ub and TP53INP2 3KR mutant. This mutant form of TP53INP2 has all 3 lysine residues mutated to arginine and, therefore, cannot be ubiquitinated. TP53INP2 3KR mutant was still pulled down together with GFP-Ub (Figure 4F), confirming that TP53INP2 is able to interact with GFP-Ub. To further confirm this interaction, we performed the reverse pull-down (Supplemental Figure 15B). Thus, HEK293T cells were transfected with Flag-TP53INP2 and HA-Ub, and whole cell lysates were pulled down with Flag resin. When both constructs were transfected, HA-Ub was pulled down together with Flag-TP53INP2, both in basal conditions and when incubated with HBSS (which stimulates protein degradation) (Supplemental Figure 15B).
Next, to characterize the interaction between TP53INP2 and ubiquitin, recombinant His-tagged mono-Ub, K48-Ub, and K63-Ub were coupled to a cobalt resin and incubated with Flag-TP53INP2 previously pulled down with Flag resin (Supplemental Figure 15C). Under these conditions, TP53INP2 showed preferential binding to mono- and K63-linked ubiquitin (Supplemental Figure 15C). These data further suggest that TP53INP2 interacts directly with ubiquitin.
TP53INP2 enhances muscle wasting in streptozotocin-induced diabetes.
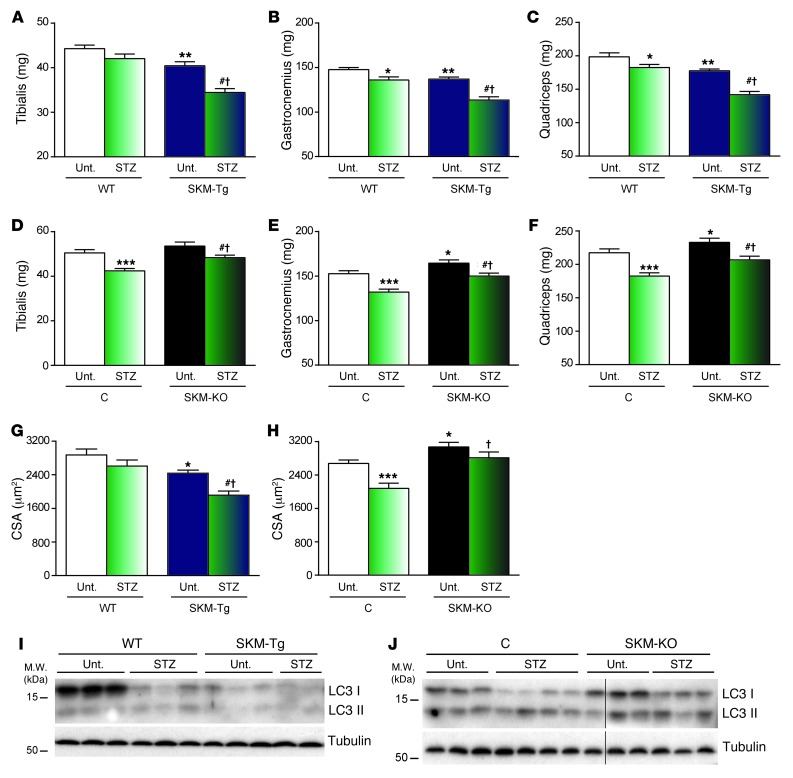
Based on prior data suggesting that TP53INP2 is a negative regulator of muscle mass, we analyzed whether changes in TP53INP2 expression could affect the muscle loss caused by major catabolic stimuli. Thus, we induced diabetes in our mouse models by injecting streptozotocin on 2 consecutive days, and, 4 days later, we monitored the effects of the treatment on muscle weight and cross-sectional area of muscle fibers. Streptozotocin-induced diabetes is an optimal model to study muscle wasting, which is detectable only a few days after diabetes induction. Muscles underwent a weight reduction in WT mice upon induction of diabetes (Figure 5, A–C). However, SKM-Tg mice showed a much more dramatic muscle loss (Figure 5, A–C). In contrast to what was observed in SKM-Tg mice, the reduction in muscle weights from SKM-KO mice upon streptozotocin treatment was less than the one displayed by their control littermates (Figure 5, D–F). Cross-sectional area quantification of muscle fibers from tibialis anterior muscles corroborated the prior data on muscle weight (Figure 5, G and H). Thus, the reduction in cross-sectional area induced by streptozotocin was larger in SKM-Tg mice when compared with that in controls (WT) (Figure 5G). On the other hand, the reduction in the tibialis anterior cross-sectional area was larger in control mice than in SKM-KO mice (Figure 5H). In all, our data indicate that TP53INP2 promotes muscle wasting in streptozotocin-induced diabetes, and TP53INP2 ablation partially prevents muscle wasting induced by diabetes.
Figure 5. TP53INP2 enhances diabetes-induced muscle loss while it is ameliorated by TP53INP2 ablation.
(A–F) Weights of (A and D) tibialis anterior, (B and E) gastrocnemius, or (C and F) quadriceps muscles from WT and SKM-Tg mice or control and SKM-KO mice with or without streptozotocin (STZ) treatment. (G and H) Mean cross-sectional area of 150 myofibers per tibialis anterior muscle from (G) WT and SKM-Tg mice or (H) control and SKM-KO mice with or without streptozotocin treatment. (I and J) Western blot analysis of LC3I and LC3II content in total homogenates of gastrocnemius muscles from WT and SKM-Tg mice or control and SKM-KO mice treated or not with streptozotocin. Thin black lines indicate that lanes were run on the same gel but were noncontiguous. Data represent mean ± SEM. (A–C and G) Data were obtained from 8 untreated WT, 7 untreated SKM-Tg, 8 streptozotocin-treated WT, and 7 streptozotocin-treated SKM-Tg mice. (D–F and H) Data were obtained from 7 untreated control, 7 untreated SKM-KO, 10 streptozotocin-treated control, and 12 streptozotocin-treated SKM-KO mice. *P < 0.05, **P < 0.01, ***P < 0.001 vs. untreated WT or control mice; #P < 0.01 vs. untreated SKM-Tg or SKM-KO mice; †P < 0.01 vs. treated WT or control mice.
To analyze whether changes in autophagy may be responsible for these effects, we measured LC3I and LC3II content in muscles from untreated and streptozotocin-treated WT and SKM-Tg mice. Diabetes in WT mice caused a marked reduction in LC3I protein content in skeletal muscle, which indicates an activation of autophagy (Figure 5I and Supplemental Figure 16A). As shown in Figure 3, genetic manipulation of Tp53inp2 in muscle altered autophagy activity, and reduced levels of LC3I were detected in SKM-Tg mice (Figure 5I and Supplemental Figure 16A). Interestingly, the induction of diabetes in SKM-KO mice caused a blunted reduction in LC3I levels (Figure 5J and Supplemental Figure 16B). These results suggest that autophagy in SKM-KO diabetic mice is not as active as that in diabetic control mice, which explains the partial protection in muscle loss observed in streptozotocin-treated SKM-KO mice.
However, to corroborate that TP53INP2 promotion of muscle wasting depends on autophagy, we analyzed whether TP53INP2 gain of function was still able to enhance streptozotocin-induced muscle loss upon autophagy blockage. Thus, we induced diabetes in WT and SKM-Tg animals, and mice were treated daily with chloroquine, starting the day of the first streptozotocin injection. Under these conditions, control and TP53INP2-overexpressing mice lost muscle mass to a similar extent (Supplemental Figure 17A). In keeping with this, cross-sectional area was also reduced to a similar extent in both genotypes when mice were treated with chloroquine (Supplemental Figure 17B). To analyze autophagy behavior under these conditions, we monitored LC3 content in muscle from nondiabetic and streptozotocin-diabetic WT and SKM-Tg mice treated with chloroquine. An increase in LC3II protein levels was detectable in both WT and SKM-Tg diabetic mice (Supplemental Figure 17C), which is in accordance with autophagy blockage caused by chloroquine and an increased LC3 expression caused by diabetes induction (Supplemental Figure 17D).
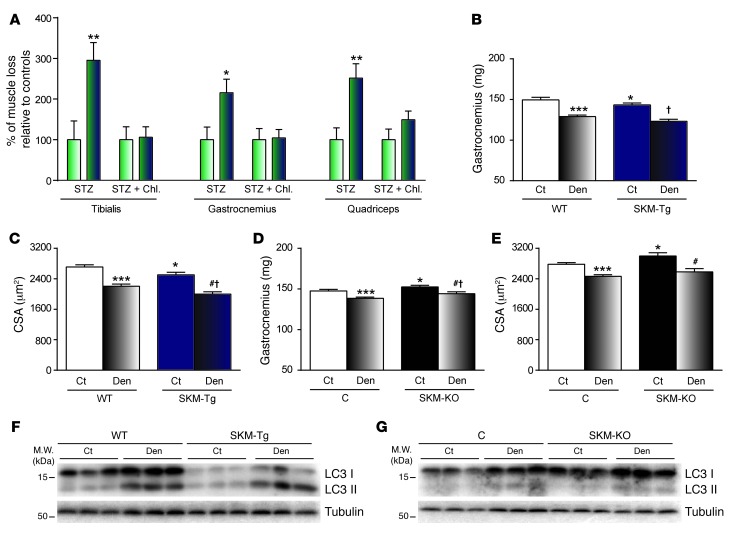
Overall, these data suggest that TP53INP2 favors autophagy-dependent muscle wasting upon induction of diabetes. Thus, SKM-Tg mice lost between 2 to 3 times more skeletal muscle (depending on the muscle analyzed) than controls (Figure 6A). However, when autophagy was blocked by chloroquine treatment, WT and SKM-Tg mice lost the same amount of muscle (Figure 6A).
Figure 6. TP53INP2 does not affect muscle wasting when autophagy is blocked.
(A) Muscle loss caused by streptozotocin-induced diabetes in WT (green and white bars) and SKM-Tg mice (green and blue bars) with or without chloroquine treatment. Data are presented as percentage relative to control values. (B and D) Weights of gastrocnemius muscle and (C and E) mean cross-sectional area of 150 myofibers per tibialis anterior muscle from WT and SKM-Tg mice or control and SKM-KO mice. Sciatic nerve from right hind limb was transected (Den) and left hind limb was used as control (Ct). (F and G) Western blot analysis of LC3I and LC3II content in total homogenates of gastrocnemius muscles from denervated or nondenervated WT and SKM-Tg mice or from control and SKM-KO mice. Data represent mean ± SEM. Data in A and B were obtained from 14 WT and 11 SKM-Tg mice. Data in C and D were obtained in 9 control and 11 SKM-KO mice. *P < 0.05, **P < 0.01, ***P < 0.001 vs. nondenervated WT or control mice; #P < 0.05 vs. untreated SKM-Tg or SKM-KO mice; †P < 0.05 vs. treated WT or control mice.
In addition, to further confirm that TP53INP2 effects on muscle wasting were autophagy-dependent, we performed denervation in our mice models. Denervation is a condition that induces an accelerated muscle wasting, characterized by a high activation of the UPS (6, 33, 34). However, autophagy in denervated muscles is suppressed and does not contribute to denervation-induced muscle wasting (35). In fact, autophagy blockage by muscle-specific ATG7 ablation not only does not prevent denervation induced muscle loss but also enhances muscle atrophy (11).
Right sciatic nerves from WT and SKM-Tg mice were transected, and, 5 days later, effects were analyzed by comparing the right hind limb muscles with the left nondenervated hind limb muscles. SKM-Tg mice displayed a similar reduction in gastrocnemius muscle weight and tibialis anterior cross-sectional area compared to that in WT mice (Figure 6, B and C). In accordance, the reduction in gastrocnemius weight and tibialis anterior cross-sectional area was also similar in SKM-KO mice compared with that in their control littermates (Figure 6, D and E). These results suggest that TP53INP2 does not participate in the muscle atrophy induced by denervation.
Next, to analyze whether this effect was explained by the suppression of autophagy in denervated muscles, we monitored LC3I and LC3II protein levels in homogenates of gastrocnemii from WT, SKM-Tg, control, and SKM-KO mice. All groups displayed an increase in both LC3I and LC3II levels (Figure 6, F and G, and Supplemental Figure 18), which is consistent with a blockage in muscle autophagy upon denervation.
Diabetes is characterized by TP53INP2 repression in skeletal muscle.
Murine models of diabetes, such as streptozotocin-induced diabetes and db/db mice, showed reduced muscle TP53INP2 expression (Supplemental Figure 19, A and B). Both models are also characterized by a large reduction in skeletal muscle mass (refs. 36, 37; Figure 5; and Supplemental Figure 19C). Considering that TP53INP2 is a negative regulator of muscle mass, these data suggest that TP53INP2 repression may be a mechanism to prevent muscle loss under pathological conditions.
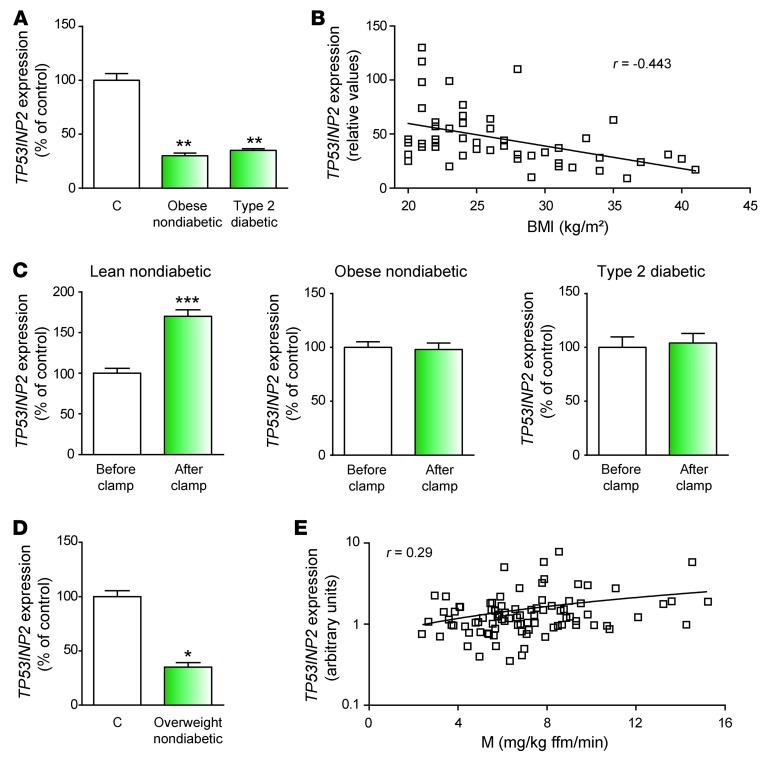
In humans, type 2 diabetes is characterized by greater muscle mass and by an enhanced risk of muscle loss in aged individuals (20, 22, 38). This is in contrast to the muscle atrophy reported in type 1 diabetic patients (14, 15). The situation in type 2 diabetes can only be explained if a robust adaptive mechanism aimed at preventing muscle loss over the years is in place. Based on the functional role of TP53INP2 in controlling muscle mass, we analyzed whether the expression of TP53INP2 was regulated in muscles during insulin resistance in 2 different human studies. Type 2 diabetic and obese nondiabetic subjects were analyzed in a first study (Lyon study; see Supplemental Table 3). Muscle TP53INP2 expression was 70% lower in both type 2 diabetic patients and in obese nondiabetic subjects compared with that in controls (Figure 7A). In agreement with these data, a negative correlation (r = –0.443, P = 0.0014) between TP53INP2 mRNA levels and BMI was detected in control, obese, or type 2 diabetic patients (Figure 7B). In keeping with these data, TP53INP2 gene expression was markedly enhanced in muscle from lean nondiabetic subjects (see Supplemental Table 4) in response to 3 hours of euglycemic-hyperinsulinemic clamp (Figure 7C). However, this response was blunted in obese and type 2 diabetic subjects (Figure 7C) (see Supplemental Tables 5 and 6).
Figure 7. TP53INP2 is repressed in skeletal muscle from overweight, obese, and type 2 diabetic patients.
(A) TP53INP2 mRNA levels in skeletal muscle from type 2 diabetic and obese subjects (Lyon study). (B) TP53INP2 mRNA levels in skeletal muscle are inversely proportional to BMI. Pearson correlation analysis yielded r = –0.443 and P = 0.0014 (n = 49) (Lyon study). (C) TP53INP2 mRNA levels in skeletal muscle from nondiabetic, obese, and type 2 diabetic subjects before and after 3 hours of euglycemic-hyperinsulinemic clamp (Lyon study). (D) TP53INP2 mRNA levels in skeletal muscle from overweight subjects (Bialystock study). (E) TP53INP2 mRNA levels in skeletal muscle positively correlate with in vivo insulin action determined by the euglycemic clamp (M value; ref. 50) in nondiabetic subjects (Bialystock study). Data represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 vs. control group.
To further confirm these results, we analyzed TP53INP2 expression in overweight subjects (Bialystok study; see Supplemental Table 7). TP53INP2 expression was 65% lower in overweight insulin-resistant subjects compared with that in lean aged-matched subjects (Figure 7D), suggesting that insulin resistance is characterized by TP53INP2 repression in human muscle. In keeping with this view, a moderate positive correlation (r = 0.29, P = 0.006) between TP53INP2 gene expression and insulin sensitivity was detected in nondiabetic subjects (Figure 7E and see Supplemental Table 8).
In all, our data indicate that muscles from obese and type 2 diabetic patients show TP53INP2 gene repression, which may be relevant in the sparing of muscle mass that takes place under this pathological state.
Discussion
A major conclusion of this study is that TP53INP2 protein is a novel negative regulator of skeletal muscle mass. Thus, TP53INP2 gain of function in skeletal muscle causes a reduction in muscle mass, whereas TP53INP2 ablation causes muscle hypertrophy. Furthermore, TP53INP2 exacerbates muscle wasting during insulinopenic diabetes. Previous studies carried out in cellular models had defined TP53INP2 as a regulator of basal autophagy (26–28). Here, we have extended these observations to skeletal muscle, given that TP53INP2 stimulates autophagy in muscles from fed mice (basal conditions). The stimulatory effects of TP53INP2 on autophagy explains its role as a negative regulator of muscle mass.
Currently available data indicate that autophagy is necessary for the maintenance of skeletal muscle mass and quality (11). Thus, complete abrogation of autophagy by ablation of ATG7 in skeletal muscle leads to muscle atrophy as well as to the accumulation of abnormal organelles, altering the structure and the function of this tissue (11). In this study, we have analyzed the affect of gain of function or loss of function of TP53INP2 protein that modulates basal autophagy, although it has been shown that it is not an essential player as ATG7 (there is still autophagic activity in the absence of TP53INP2). In this connection, our data permit us to conclude that moderate changes in basal autophagy do not cause major alterations in skeletal muscle morphology or integrity but have a substantial affect on muscle mass. This supports the concept that a certain level of autophagy is required for the preservation of muscle structure and function, but modulation above a certain threshold will just represent enhanced protein degradation and a reduction of muscle fiber size.
Muscle mass and quality are key factors that determine muscle performance, although these 2 parameters do not perfectly correlate in humans (39–41). In this connection, the reduction in muscle strength observed in older adults is faster than muscle mass loss occurring with age (39). This suggests that the reduction in muscle quality is also a contributor to the decline in muscle performance. On the other hand, conditions such as type 2 diabetes mellitus are characterized by reduced muscle strength and quality in spite of an increase in muscle mass (41). In this report, we show that genetic manipulation of Tp53inp2 in skeletal muscle leads to changes in muscle mass in mice. Under these conditions, we found no evidence for ultrastructural alterations in muscle fibers, suggesting that muscle quality remains unchanged.
Autophagy was initially considered an unselective process to degrade long-lived proteins and organelles in situations of nutrient deprivation. However, it is becoming clear that autophagy can be a very selective process to degrade protein aggregates, mitochondria, and other organelles (42, 43). This selectivity is an important feature of basal autophagy, described as a quality-control mechanism important for the maintenance of the homeostasis of the cell and the tissue. In this regard, recent reports have identified and characterized different proteins involved in the degradation of selective substrates through autophagy (42, 43). For example, p62 and NBR1 are autophagy receptors for the degradation of ubiquitinated proteins (30, 31). Our results suggest that TP53INP2 not only activates basal autophagy in skeletal muscle but is also involved in the autophagic degradation of ubiquitinated proteins. TP53INP2 colocalizes with ubiquitinated protein aggregates, interacts with ubiquitin, and promotes the degradation of ubiquitinated proteins in spite of the p62 and NBR1 accumulation. In fact, TP53INP2 overexpression displaces p62 from autophagosomes. Accumulation of p62 has also been reported upon overexpression of the TP53INP2 homologous protein TP53INP1 (also referred to as SIP) (44). TP53INP1 overexpression enhances autophagy and causes p62 accumulation due to the fact that TP53INP1 interacts with LC3 with higher affinity than p62 (44). So, the authors reasoned that TP53INP1 is able to displace p62 from LC3 and from its function in autophagy (44). It is feasible that TP53INP2 is acting in a similar way as TP53INP1.
Another major conclusion of our study is the fact that TP53INP2 favors muscle wasting specifically in those catabolic conditions in which autophagy is activated. We showed that streptozotocin-induced diabetes activates autophagy, and, therefore, this is an additional contributor to muscle wasting under these conditions. Furthermore, TP53INP2 gain of function accelerates muscle atrophy in diabetic mice, suggesting that enhanced basal autophagy drives further muscle loss during diabetes. These data complement prior observations indicating that experimental diabetes causes increased UPS activity through atrogin 1 and MuRF1 induction (36, 37, 45). On the other hand, TP53INP2 ablation mitigates muscle loss during streptozotocin-induced diabetes together with reduced autophagy activation in those muscles. In fact, under conditions in which autophagy is not a contributor to muscle loss (as it occurs in denervation or by chloroquine treatment), TP53INP2 does not have an affect on muscle atrophy. In all, our data strongly support the view that TP53INP2 effects on muscle wasting depend solely on its role as a regulator of autophagy.
We have documented that the expression of TP53INP2 protein is reduced in muscle from streptozotocin-induced diabetes as well as in obese diabetic db/db mice. In parallel, muscle atrophy develops in these 2 conditions. We propose that TP53INP2 repression is part of a mechanism that reduces the extent of muscle wasting. In this regard, db/db mice represent a mouse model of sarcopenic obesity, a condition that affects near 15% of type 2 diabetic patients (46).
There is clear evidence for altered protein metabolism and muscle atrophy in type 1 diabetes mellitus (14, 15, 47). In contrast, type 2 diabetes is characterized by greater muscle mass (22, 38, 41). Body composition studies in humans also indicate the existence of a positive correlation between body fat and muscle mass (48, 49), indicating the existence of greater muscle mass in human obesity. An enhanced rate of muscle loss has been reported in aged type 2 diabetic patients (22, 38). Overall, currently available data strongly indicate the existence of robust adaptive mechanisms that prevent muscle loss in spite of deficient insulin action in type 2 diabetic patients. In this regard, we documented that TP53INP2 gene expression is downregulated in muscles from type 2 diabetic patients as well as in nondiabetic obese or in young overweight subjects. Our data are consistent with the view that in humans the progression to obesity and/or insulin resistance shuts off TP53INP2 expression in an attempt to minimize muscle loss. In fact, TP53INP2 mRNA levels in skeletal muscle were upregulated by hyperinsulinemia in lean nondiabetic subjects but not in obese or in type 2 diabetic patients.
We propose that muscle TP53INP2 expression is modulated as part of an adaptive mechanism to preserve homeostasis in highly anabolic or catabolic conditions. Thus, the repression of TP53INP2 that occurs during the development of obesity or type 2 diabetes prevents muscle loss in these pathological conditions. The repression of TP53INP2 leads to reduced basal autophagy, which mitigates the danger of accelerated protein loss under catabolic conditions. On the other hand, high insulin action that deeply inhibits catabolism may be compensated by an increased TP53INP2 expression and enhanced basal autophagy.
Our data also suggest that TP53INP2 may be a target to prevent muscle wasting under specific pathological conditions. Because TP53INP2 repression reduces autophagy flux without blocking it completely, TP53INP2 targeting may permit the sparing of muscle mass, avoiding the negative side effects of a complete blockage of autophagy, such as reduced muscle quality (11). In those conditions in which TP53INP2 is partially repressed (as it occurs in obesity or in type 2 diabetes), further reduction in TP53INP2 expression may be used as a preventive intervention in people at risk of muscle loss.
Methods
Supplemental methods.
A list of antibodies and plasmids used in this work as well as a detailed description of subjects and mouse strain characteristics and protocols used is found in the Supplemental Methods.
Transcriptomic analysis.
Microarray data is available at GEO (accession no. GSE54917). See the Supplemental Methods for more information.
Statistics.
The data presented here were analyzed using 2-tailed Student’s t test or analysis of variance. Statistical significance was established at P < 0.05.
Study approval.
Animal studies were approved by the University of Barcelona Committee on Animal Care. Subject studies were approved by the Ethics Committee of the Comité consultatif de protection des personnes dans la recherche biomédicale Lyon A (RG/FL-2003-039-125 A) and by the Bioethics Committee of the Medical University of Bialystok and were performed according to the principles of the Helsinki Declaration and the French legislation (for the Lyon studies). All subjects gave written informed consent before entering the study.
Supplementary Material
Acknowledgments
We thank the Advanced Digital Microscopy Facility (IRB Barcelona), the Biostatistics/Bioinformatics Unit (IRB Barcelona), the Functional Genomics Facility (IRB Barcelona), the Unit of Electron Cryo-Microscopy (Scientific and Technological Centers, Universitat de Barcelona), V. Lukesova, J.M. Seco, I. Castrillón, and J.C. Monasterio for technological assistance. D. Sala was the recipient of a FPU fellowship from the “Ministerio de Educación y Cultura,” Spain. V. Ribas was supported by a postdoctoral fellowship from the Instituto de Salud Carlos III (Ministerio de Economía y Competitividad, Spain). This study was supported by research grants from the MINECO (SAF2008-03803), grant 2009SGR915 from the “Generalitat de Catalunya,” CIBERDEM (“Instituto de Salud Carlos III”), FIS-PS09/01267 and FIS-PI13/025 from “Instituto de Salud Carlos III,” Spain, SB/CP2013-0167/16642 from Association Française contre les Myopathies (AFM), Interreg IV-B-Sudoe-Feder (DIOMED, SOE1/P1/E178), and UDA-POIG.01.03.01-00-128/08 from the Innovative Economy Program 2007-2013, partially financed by the European Union within the European Regional Development Fund. A. Zorzano was the recipient of a Science Intensification Award from the University of Barcelona.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article:J Clin Invest. 2014;124(5):1914–1927. doi:10.1172/JCI72327.
References
- 1.Mitch WE, Goldberg AL. Mechanisms of muscle wasting. The role of the ubiquitin-proteasome pathway. N Engl J Med. 1996;335(25):1897–1905. doi: 10.1056/NEJM199612193352507. [DOI] [PubMed] [Google Scholar]
- 2.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8(11):931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 5.Bechet D, Tassa A, Taillandier D, Combaret L, Attaix D. Lysosomal proteolysis in skeletal muscle. Int J Biochem Cell Biol. 2005;37(10):2098–2114. doi: 10.1016/j.biocel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 6.Furuno K, Goodman MN, Goldberg AL. Role of different proteolytic systems in the degradation of muscle proteins during denervation atrophy. J Biol Chem. 1990;265(15):8550–8557. [PubMed] [Google Scholar]
- 7.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15(3):1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lecker SH, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18(1):39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 9.Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6(6):458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Zhao J, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6(6):472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Masiero E, et al. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10(6):507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto A, Simonsen A. The elimination of accumulated and aggregated proteins: a role for aggrephagy in neurodegeneration. Neurobiol Dis. 2011;43(1):17–28. doi: 10.1016/j.nbd.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price SR, et al. Muscle wasting in insulinopenic rats results from activation of the ATP-dependent, ubiquitin-proteasome proteolytic pathway by a mechanism including gene transcription. J Clin Invest. 1996;98(8):1703–1708. doi: 10.1172/JCI118968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krause MP, Riddell MC, Hawke TJ. Effects of type 1 diabetes mellitus on skeletal muscle: clinical observations and physiological mechanisms. Pediatr Diabetes. 2011;12(4 pt 1):345–364. doi: 10.1111/j.1399-5448.2010.00699.x. [DOI] [PubMed] [Google Scholar]
- 15.Jakobsen J, Reske-Nielsen E. Diffuse muscle fiber atrophy in newly diagnosed diabetes. Clin Neuropathol. 1986;5(2):73–77. [PubMed] [Google Scholar]
- 16.Baracos VE, DeVivo C, Hoyle DH, Goldberg AL. Activation of the ATP-ubiquitin-proteasome pathway in skeletal muscle of cachectic rats bearing a hepatoma. Am J Physiol. 1995;268(5 pt 1):E996–E1006. doi: 10.1152/ajpendo.1995.268.5.E996. [DOI] [PubMed] [Google Scholar]
- 17.Penna F, et al. Autophagic degradation contributes to muscle wasting in cancer cachexia. Am J Pathol. 2013;182(4):1367–1378. doi: 10.1016/j.ajpath.2012.12.023. [DOI] [PubMed] [Google Scholar]
- 18.Bailey JL, Zheng B, Hu Z, Price SR, Mitch WE. Chronic kidney disease causes defects in signaling through the insulin receptor substrate/phosphatidylinositol 3-kinase/Akt pathway: implications for muscle atrophy. J Am Soc Nephrol. 2006;17(5):1388–1394. doi: 10.1681/ASN.2004100842. [DOI] [PubMed] [Google Scholar]
- 19.Voisin L, et al. Muscle wasting in a rat model of long-lasting sepsis results from the activation of lysosomal, Ca2+-activated, and ubiquitin-proteasome proteolytic pathways. . J Clin Invest. 1996;97(7):1610–1617. doi: 10.1172/JCI118586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pereira S, Marliss EB, Morais JA, Chevalier S, Gougeon R. Insulin resistance of protein metabolism in type 2 diabetes. Diabetes. 2008;57(1):56–63. doi: 10.2337/db07-0887. [DOI] [PubMed] [Google Scholar]
- 21.Halvatsiotis P, Short KR, Bigelow M, Nair KS. Synthesis rate of muscle proteins, muscle functions, and amino acid kinetics in type 2 diabetes. Diabetes. 2002;51(8):2395–2404. doi: 10.2337/diabetes.51.8.2395. [DOI] [PubMed] [Google Scholar]
- 22.Park SW, et al. Accelerated loss of skeletal muscle strength in older adults with type 2 diabetes: the health, aging, and body composition study. Diabetes Care. 2007;30(6):1507–1512. doi: 10.2337/dc06-2537. [DOI] [PubMed] [Google Scholar]
- 23.Sancho A, et al. DOR/Tp53inp2 and Tp53inp1 constitute a metazoan gene family encoding dual regulators of autophagy and transcription. PLoS One. 2012;7(3):e34034. doi: 10.1371/journal.pone.0034034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baumgartner BG, et al. Identification of a novel modulator of thyroid hormone receptor-mediated action. PLoS One. 2007;2(11):e1183. doi: 10.1371/journal.pone.0001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Francis VA, Zorzano A, Teleman AA. dDOR is an EcR coactivator that forms a feed-forward loop connecting insulin and ecdysone signaling. Curr Biol. 2010;20(20):1799–1808. doi: 10.1016/j.cub.2010.08.055. [DOI] [PubMed] [Google Scholar]
- 26.Mauvezin C, et al. The nuclear cofactor DOR regulates autophagy in mammalian and Drosophila cells. EMBO Rep. 2010;11(1):37–44. doi: 10.1038/embor.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nowak J, et al. The TP53INP2 protein is required for autophagy in mammalian cells. Mol Biol Cell. 2009;20(3):870–881. doi: 10.1091/mbc.E08-07-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mauvezin C, Sancho A, Ivanova S, Palacin M, Zorzano A. DOR undergoes nucleo-cytoplasmic shuttling, which involves passage through the nucleolus. FEBS Lett. 2012;586(19):3179–3186. doi: 10.1016/j.febslet.2012.06.032. [DOI] [PubMed] [Google Scholar]
- 29.Bothe GW, Haspel JA, Smith CL, Wiener HH, Burden SJ. Selective expression of Cre recombinase in skeletal muscle fibers. Genesis. 2000;26(2):165–166. doi: 10.1002/(SICI)1526-968X(200002)26:2<165::AID-GENE22>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 30.Kirkin V, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33(4):505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 31.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 32.Bjorkoy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bodine SC, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294(5547):1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 34.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci U S A. 2001;98(25):14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quy PN, Kuma A, Pierre P, Mizushima N. Proteasome-dependent activation of mammalian target of rapamycin complex 1 (mTORC1) is essential for autophagy suppression and muscle remodeling following denervation. J Biol Chem. 2013;288(2):1125–1134. doi: 10.1074/jbc.M112.399949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Z, et al. PTEN expression contributes to the regulation of muscle protein degradation in diabetes. Diabetes. 2007;56(10):2449–2456. doi: 10.2337/db06-1731. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Hu Z, Hu J, Du J, Mitch WE. Insulin resistance accelerates muscle protein degradation: Activation of the ubiquitin-proteasome pathway by defects in muscle cell signaling. Endocrinology. 2006;147(9):4160–4168. doi: 10.1210/en.2006-0251. [DOI] [PubMed] [Google Scholar]
- 38.Park SW, et al. Excessive loss of skeletal muscle mass in older adults with type 2 diabetes. Diabetes Care. 2009;32(11):1993–1997. doi: 10.2337/dc09-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodpaster BH, et al. The loss of skeletal muscle strength, mass, and quality in older adults: the health, aging and body composition study. J Gerontol A Biol Sci Med Sci. 2006;61(10):1059–1064. doi: 10.1093/gerona/61.10.1059. [DOI] [PubMed] [Google Scholar]
- 40.Hairi NN, et al. Loss of muscle strength, mass (sarcopenia), and quality (specific force) and its relationship with functional limitation and physical disability: the Concord Health and Ageing in Men Project. J Am Geriatr Soc. 2010;58(11):2055–2062. doi: 10.1111/j.1532-5415.2010.03145.x. [DOI] [PubMed] [Google Scholar]
- 41.Park SW, et al. Decreased muscle strength and quality in older adults with type 2 diabetes: the health, aging, and body composition study. Diabetes. 2006;55(6):1813–1818. doi: 10.2337/db05-1183. [DOI] [PubMed] [Google Scholar]
- 42.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A. 2008;105(52):20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12(9):836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 44.Seillier M, et al. TP53INP1, a tumor suppressor, interacts with LC3 and ATG8-family proteins through the LC3-interacting region (LIR) and promotes autophagy-dependent cell death. Cell Death Differ. 2012;19(9):1525–1535. doi: 10.1038/cdd.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandri M, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117(3):399–412. doi: 10.1016/S0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim TN, et al. Prevalence and determinant factors of sarcopenia in patients with type 2 diabetes: the Korean Sarcopenic Obesity Study (KSOS). Diabetes Care. 2010;33(7):1497–1499. doi: 10.2337/dc09-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nair KS, Ford GC, Ekberg K, Fernqvist-Forbes E, Wahren J. Protein dynamics in whole body and in splanchnic and leg tissues in type I diabetic patients. J Clin Invest. 1995;95(6):2926–2937. doi: 10.1172/JCI118000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Micozzi MS, Harris TM. Age variations in the relation of body mass indices to estimates of body fat and muscle mass. Am J Phys Anthropol. 1990;81(3):375–379. doi: 10.1002/ajpa.1330810307. [DOI] [PubMed] [Google Scholar]
- 49.Kanehisa H, Fukunaga T. Association between body mass index and muscularity in healthy older Japanese women and men. J Physiol Anthropol. 2013;32(1):4. doi: 10.1186/1880-6805-32-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lillioja S, et al. Skeletal muscle capillary density and fiber type are possible determinants of in vivo insulin resistance in man. J Clin Invest. 1987;80(2):415–424. doi: 10.1172/JCI113088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.