Abstract
We performed diagnosis and species identification of parasites in lesion samples from suspected cutaneous leishmaniasis patients in four villages, three of which are in a known Leishmania tropica endemic region in Kenya. Samples were analyzed both by microscopy and PCR for Leishmania, and typed by an assay using four ribosomal DNA-based species-identification PCRs. The lesions were demonstrated to be caused by L. tropica, which confirms the re-emergence of cutaneous leishmaniasis from this species after a period of reduced incidence in the endemic zone. Our report highlights the importance of an intervention and sustained Leishmania control program.
Keywords: Diagnosis, Kenya, Leishmania tropica, PCR, Molecular typing
Introduction
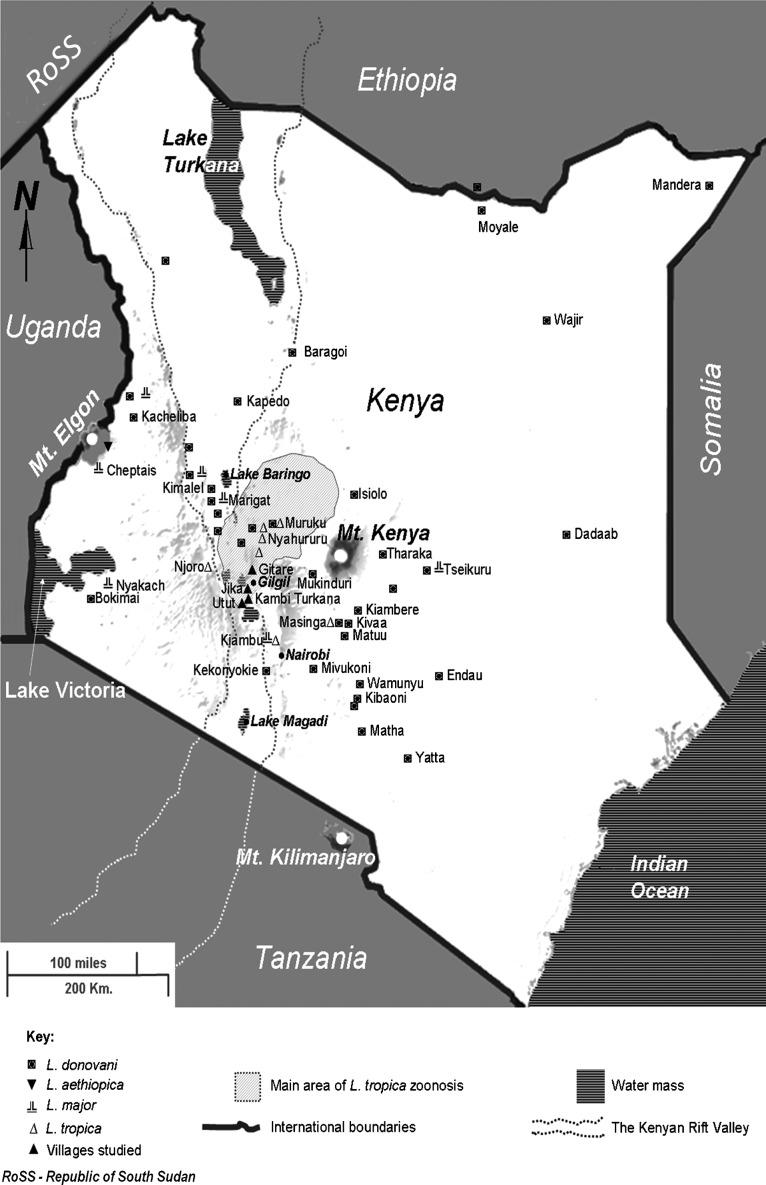
In Kenya, the etiological agent of leishmaniasis can be any of four Old World Leishmania species: visceral leishmaniasis (VL) is caused by L. donovani,1 while infection with L. major, L. tropica, or L. aethiopica is responsible for cutaneous leishmaniasis (CL).2–4 Only in one instance L. donovani was reported to cause CL. Country-wide data on the prevalence of both diseases are lacking. The species are differentially endemic (Fig. 1), as determined by multilocus enzyme electrophoresis characterization of isolates.5,6 L. donovani predominates, being distributed widely within the country. Leishmania tropica is historically reported to occur within the proximity of eastern escarpments of the central Kenyan Rift Valley and in the Laikipia plateau.2,5,7–9 This endemic zone overlaps to the west with a major L. donovani transmission area. L. tropica has also been isolated in Njoro, Kiambu, and Masinga, which are outside the main zone of endemicity as shown in Fig. 1.5
Figure 1.
Map of Kenya showing geographical endemicity of different Leishmania species based on characterized isolates.5,6 The locations of the four villages studied in this report within the larger zone of L. tropica transmission are also shown. The gray shading indicates elevation.
The Laikipia plateau and the adjacent escarpments consist mostly of alternating subsistence farmlands, private ranches and acacia bush lands with considerable wildlife population.5,10 Presence of fault scarp ranges, cragged rocks, and numerous caves provide potential habitats for both the main reservoir host of L. tropica (the rock hyrax Procavia johnstoni), and the sand fly vectors (Phlebotomus guggisbergi and P. aculeatus).11,12 Proximity or overlap of human settlements with forested areas presents a major risk factor of L. tropica CL. Other risk factors include pronounced poverty associated with poor shelter, which allows uncontrolled vector to human contact, and the apparent lack of prioritization and awareness on CL as a public health problem.
In 2009, in response to reports of an outbreak of leishmaniasis-like lesions near Gilgil on the edge of the Kenyan Rift Valley, we undertook a diagnostic survey for CL in Gitare and three villages from the known endemic zone. The areas being situated within a wider region of known L. tropica endemicity, this species was suspected as the most likely cause of the reported lesions. A clinical examination of the lesions was done, followed by parasitological diagnosis and species identification.
Materials and Methods
Study area
The study area was selected on the basis of an increase in suspected CL cases among school aged children seeking medical attention at the Nakuru district hospital, located in the Kenyan Rift Valley. As the majority of referral patients were from the larger Utut forest area, a previously known CL endemic zone,10 two villages were selected in this region: Kambi Turkana and Jika. Because the remaining patients came from Gitare, where CL had not previously been reported, this village was included in our study as well.
Sample collection
Patient screening was done in January 2009 as part of the TRYLEIDIAG study (see acknowledgements), and involved active case detection. A team from the Kenya Medical Research Institute and the Ministry of Public Health and Sanitation visited the three villages Kambi Turkana, Jika, and Gitare, with the assistance of a public health officer based at the sub-district hospital in Gilgil. Study participants included school going children as well as adults. Subjects were examined by a trained clinician for body and facial lesions, and a skin slit specimen was taken from suspected CL lesions for microscopical and molecular diagnosis. Finally, one sample was collected from Utut village, also situated in the Utut forest area.
Samples were taken from in total 25 suspected CL patients (Table 1). Lesions and the peripheral skin were sterilized with 70% ethanol. Tissue was obtained from carefully selected fresh indurated edges of the lesions, and was transferred to a microscopy slide, and preserved in 200 μl of L3TM buffer13 to stabilize the sample for transport and storage at 4°C prior to DNA extraction.
Table 1. Sample origin and assay results.
| Sample | Village* | Patient gender† | Age (years) | Microscopy results‡ | SSU-rDNA PCR | Identified species |
| 1 | Kambi Turkana | M | 35 | 1+ | + | L. tropica |
| 2 | Jika | F | 15 | 0 | + | L. donovani |
| 3 | Jika | M | 52 | 0 | + | |
| 4 | Kambi Turkana | F | 56 | 1+ | + | |
| 5 | Kambi Turkana | F | 62 | 0 | + | |
| 6 | Kambi Turkana | F | 6 | 0 | + | (L. tropica)§ |
| 7 | Jika | M | 6 | 0 | + | L. tropica |
| 8 | Kambi Turkana | F | 9 | 0 | + | |
| 9 | Kambi Turkana | F | 2.5 | 0 | + | L. tropica |
| 10 | Utut village | M | 35 | 0 | + | |
| 11 | Kambi Turkana | M | 5 | 1+ | + | L. tropica |
| 12 | Kambi Turkana | M | 2.3 | 1+ | + | L. tropica |
| 13 | Gitare | M | 10 | 0 | + | |
| 14 | Gitare | M | 5 | 1+ | + | |
| 15 | Gitare | M | 7 | 1+ | + | L. tropica |
| 16 | Gitare | M | 13 | 3+ | + | L. tropica |
| 17 | Gitare | M | 10 | ND | + | |
| 18 | Gitare | F | 13 | 0 | + | |
| 19 | Gitare | F | 13 | 0 | + | |
| 20 | Gitare | F | 9 | 0 | + | |
| 21 | Gitare | M | 7 | 1+ | + | |
| 22 | Gitare | M | 13 | ND | + | |
| 23 | Gitare | F | 7 | 1+ | + | |
| 24 | Gitare | M | 3 | 2+ | + | L. tropica |
| 25 | Gitare | F | 11 | 1+ | + | L. tropica |
Notes: *See Fig. 1 for location of villages. These are the sites where active case detection was done. Not all patients included at these sites lived in the village itself.
†M: male; F: female.
‡Parasite load grading. ND: not done.
§In Fig. 3, the suspected L. tropica fragment appears slightly higher, and could represent an aspecific amplicon. To be on the safe side, this was not considered positive for L. tropica.
Ethical considerations
Permission for the study was obtained from the respective community leaders, and an informed consent was given by individual persons participating. Minors were represented by their parents or guardians. The outcome of microscopy was communicated to the respective study participants a day after the screening exercise, and the result of the molecular diagnosis was given several weeks later. Confirmed CL patients were referred to the Ministry of Health sub-district hospital for treatment. Health education was performed at each village to increase awareness of the disease. Ethical guidelines of the Kenya Medical Research Institute ethical review board were followed (reference SSC/1084).
Microscopical diagnosis
Microscopical examination was done at the sub-district hospital in Gilgil. Microscopy slides were stained for 20 minutes with 10% Giemsa stain diluted in phosphate-buffered saline pH 7, and examined for amastigotes under a microscope with a ×100 oil immersion objective and a ×10 ocular lens. Observed amastigotes were counted, and samples were graded according to Chulay et al.,14 ranging from 0 (indicating no observed amastigotes) to 6+ (⩾100 parasites/field).
Molecular analyses
Nucleic acid extraction was done as described by Boom et al.,15 and eluted from the silica particles in 30 μl of TE buffer. DNA samples were assayed by a small ribosomal subunit (SSU) rDNA-based PCR16 in a 25-μl reaction volume to confirm the presence of Leishmania DNA. They were scored positive when a 115-bp amplicon was detected. Reactions were spiked with an internal positive control template (plasmid pICR1Leish28) to check for sample inhibition, consisting of a cloned phage lambda DNA fragment flanked by Leishmania-specific annealing sites, resulting in an amplicon of 230 bp (Fig. 2). Failure to amplify the internal positive control indicates PCR inhibition or suboptimal PCR reaction conditions. In such case, negative PCR results cannot be trusted, as they can be caused by either absence of template in the sample, or alternatively failure to amplify the template when present. All PCRs were checked for contamination by running a negative non-template control.
Figure 2.

Internal control used to spike the SSU-rDNA PCR. Vector pCR4-TOPO® (tools.invitrogen.com/content/sfs/vectors/pcr4topo_map.pdf) was used to clone a PCR product using the T:A overhang strategy. The cloning site of the vector is shown in the outer shaded areas. The central shaded box describes the internal phage lambda fragment of the cloned PCR amplicon, with positions referring to accession J02459 (www.ebi.ac.uk, accession number J02459, release 106). The regions corresponding to the SSU-rDNA Leishmania specific primers in the flanking sequences are indicated in the outlined boxes, with the arrows in the 5′–3′ direction. The orientation of the PCR amplicon in plasmid pICR1Leish28 is as shown.
Subsequently, the DNA was tested with four species-specific PCRs as described by Ogado Ceasar Odiwuor et al.:17 LT2 (L. tropica), LM (L. major), LDI (L. donovani/L. infantum), and LA (L. aethiopica), whereby 2 μl of DNA was used. All assays included internal positive controls, at twice the amount described by Ogado Ceasar Odiwuor et al.17 A sample was scored positive for a certain species if the species-specific fragment (Fig. 3) was detected. The PCR was considered invalid if neither a species-specific nor the internal control amplicon was detected. A species was finally assigned to the sample if all four PCRs were valid, and only one of them showed a species-specific amplicon, as described in the combined assay of Ogado Ceasar Odiwuor et al.17 All PCRs were checked for contamination by running a negative non-template control.
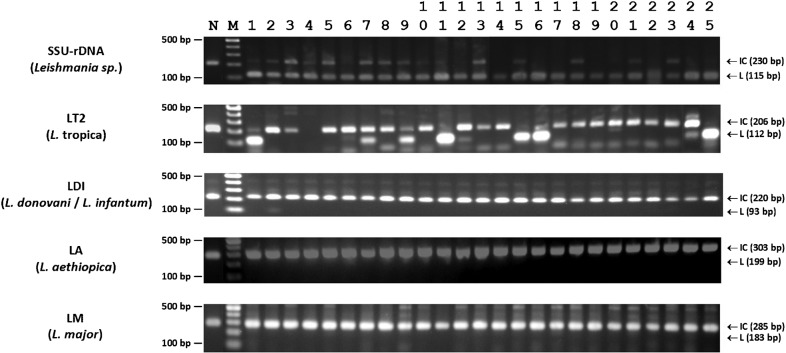
Figure 3.
Results of Leishmania diagnostic and four species-specific PCR tests17 for the 25 samples listed in Table 1. PCRs are listed on the left, with their specificity between brackets. The size of the internal controls (IC) and the Leishmania specific (L) fragments are indicated by arrows on the right. The GeneRuler 100-bp molecular weight marker (Fermentas, St Leon-Rot, Germany) is used as size reference (M), of which the region between 100 and 500 bp is depicted. The negative no-template controls (N) are shown in the first lane of the gels, where only the IC fragment is amplified.
Results
Results are shown in Fig. 3 and Table 1. Of the 25 patients with suspected CL, 11 were positive in microscopy, and all 25 in SSU-rDNA PCR, indicating the presence of Leishmania DNA in all lesions. In nine samples, L. tropica parasites were detected (4 from Kambi Turkana, 4 from Gitare, and 1 from Jika), while L. donovani was found in one patient from Jika. From the remaining 15 samples, no species-specific PCR amplicons were obtained. One of the samples proved invalid in the LT2 PCR, as neither an L. tropica fragment nor an internal control was amplified. However, as all other species-specific PCRs were valid for this sample, we could exclude the possibility of another Leishmania species being present. The variation observed in the LT2 PCR fragment intensities is likely caused by the fluctuations in parasite load in the samples, as lesions were of different age and size, and samples were taken with a scalpel, which is hard to standardize.
Discussion
The identification of L. tropica from the three villages Kambi Turkana, Gitare, and Jika confirms the known epidemiology of CL lesions caused by this species in the region. The fact that the species could not be identified from all lesions is not a surprise, as the species-specific PCRs are less sensitive than the diagnostic SSU-rDNA PCR. The species in the only sample originating from Utut village could not be confirmed.
In one of the CL samples from Jika, an L. donovani amplicon was obtained. Although this species is generally associated with VL, a CL case from Nyahururu,18 situated within the larger L. tropica endemic zone (Fig. 1), and several from Sudan19 have been documented. In addition, the study area is close to previously identified VL foci (Fig. 1), indicating a possible L. donovani transmission in the area. As we identified only one L. donovani sample, we cannot estimate the frequency of the event, and the result should be confirmed by additional lesions diagnosed with the same species.
Despite the increase in both research and clinical interventions for leishmaniasis in recent years,20 much of the effort has been directed towards VL, with cutaneous disease being ignored largely because of its lesser morbidity. For this reason, much of the indexed data on L. tropica transmission are from studies done in the late 1980s and the 1990s. Despite the lack of systematic follow-up and monitoring, irregular reports of outbreaks indicate that L. tropica continues to be endemic in Kenya. One of the villages (Utut) is a known historical focus of leishmaniasis,10 and it is likely that leishmaniasis spread from there to the surrounding regions, including the other villages studied. Incidences could have risen over time as the human population increased, resulting in expansion of settlements into virgin bush lands. As demonstrated by the outbreaks documented in this report, with the lack of any medical intervention and the perennial presence of the major risk factors, CL is expected to persist in the study area. Apart from causing unsightly lesions, L. tropica is capable of more serious disease in synergy with other infections such as HIV,21–23 for which the study area is at risk. Therefore, it cannot be ignored as a public health problem.
In conclusion, we have successfully applied species-specific PCRs to confirm that the etiological agent of a CL epidemic of 2009 is indeed Leishmania tropica. As the latest reports on this species in the region date back to the late 1990s, we strongly recommend that species typing should be performed systematically for surveillance of the leishmaniases in Kenya.
Acknowledgments
We thank Simonne De Doncker for technical assistance. The study was supported by funds from the European Union’s sixth Framework Program INCO-CT-2005-015379 (TRYLEIDIAG project) and the Belgian Directorate General for Development Cooperation (DGDC) through the third framework agreement with the Institute of Tropical Medicine Antwerp. TRYLEIDIAG funded the research while DGDC provided salary (GVDA) and scholarship (SO) support.
REVIEWER/AUTHOR UNRESOLVED DISCUSSIONS
The editor has decided to publish the referee’s and author’s comments in order for the reader to make their own opinion. To comment on this article, please visit the dedicated blog for this issue: http://www.pathogensandglobalhealth.com/2011/12/issue2/
10/01/2012 — Reviewer Comment Thread 1: The authors for reasons that are not quite clear added to each PCR reaction an internal control.
09/02/2012 — Author’s reply: The reason behind including an internal control is extensively discussed in Ogado Ceasar Odiwuor et al.17 The internal control is used to verify that the PCR reaction is properly set up, and that no enzyme inhibitors are present in the reaction. Because a minimal quantity of the internal control template is added to the PCR mix, a suboptimal reaction set-up or thermal cycling profile, or sample inhibition, will prevent amplification. This is clarified in Materials and Methods, section Molecular analyses: ‘Failure to amplify the internal positive control indicates PCR inhibition or suboptimal PCR reaction conditions. In such case negative PCR results cannot be trusted, as they can be caused by either absence of template in the sample, or alternatively failure to amplify the template when present.’
15/02/2012 — The authors contend that their species identification results were validated in a previous paper and therefore no external controls are necessary. However as it is almost impossible in biological studies to repeat an experiment exactly as performed previously, relying on data acquired prior to the present study is a limitation.
18/02/2012 — Author’s reply: It is true that repeating PCR conditions exactly is not easy to do. Nevertheless, (1) in the paper that describes these assays,17 we report the development of the PCRs at the Institute of Tropical Medicine Antwerp, and their validation at the Kenya Medical Research Institute in Nairobi, using other reagents and cyclers. We performed this validation precisely to ensure these PCRs are robust enough to withstand changing conditions, as our aim was to develop assays that are solid and easily transferable to other laboratories and settings. As we were hence able to reproduce our results in a limited-resource setting, we feel very confident we can also repeat these in the same lab where they were developed, using the same thermocycler. (2) In addition, and this is even more important, the internal controls included in the PCRs were developed exactly to ensure optimal PCR set-up and thermal cycling (see previous reply). As the internal controls in our study were successfully amplified, there was no problem with the reactions. (3) Third, if there was a problem with reduced specificity in our PCRs, this would have been evident from combining the 4 species-specific reactions, as more than one species would be detected in a particular sample, which is not the case. (4) Finally, we think that including one reference strain in each PCR is inferior to our method of using internal controls and a combination of the 4 species-specific PCRs. Relying on one reference strain can never capture the intra-species variability that we took into account during assay development.17 In addition, to ensure PCR specificity we would have to include not only a reference strain of the amplified species, but also of all species that should not amplify in a particular assay.
10/01/2012 — Reviewer Comment Thread 2: It is a deficit of the work that no attempt was made to isolate cultures of the samples. The whole paper depends on the interpretation of the Fig. 3.
09/02/2012 — Author’s reply: The methods we used in the paper were designed exactly to circumvent the need for parasite isolation from clinical samples, as this is laborious work that can be avoided, and the success rate of culturing is generally low. As described in Ogado Ceasar Odiwuor et al.,17 these PCRs were tested and validated on a large representative panel of Old World Leishmania reference strains, and were shown to be 100% specific.
15/02/2012 — The authors argue that their method supersedes the culture method as ‘this is laborious work that can be avoided, and the success rate of culturing is generally low’. Before the advent of molecular techniques, culturing parasites was the method of choice for almost 100 years. Its advantage is that the parasites can be characterized further by more sophisticated methods. For future reference, if one can do a slide for microscopy, one could add a sample for culture almost as easily, if one has the technical ability.
18/02/2012 — Author’s reply: We are quite surprised by this argument. The advance of PCR has been a great addition to typing and diagnosis, and we fail to see why we should stick to ancient, expensive, and time-consuming methods, even though these are indispensable for many purposes. It is true that culturing allows more analyses on these strains, but this was not the goal of our study.
References
- 1.Perkins PV, Githure JI, Mebrahtu Y, Kiilu G, Anjili C, Ngumbi PS, et al. Isolation of Leishmania donovani from Phlebotomus martini in Baringo district, Kenya. Trans R Soc Trop Med Hyg. 1988;82:695–700. doi: 10.1016/0035-9203(88)90204-0. [DOI] [PubMed] [Google Scholar]
- 2.Mebrahtu Y, Oster CN, Shatry AM, Hendricks LD, Githure JI, Rees PH, et al. Cutaneous leishmaniasis caused by Leishmania tropica in Kenya. Trans R Soc Trop Med Hyg. 1987;81:923–4. doi: 10.1016/0035-9203(87)90352-x. [DOI] [PubMed] [Google Scholar]
- 3.Mebrahtu Y, Lawyer P, Githure J, Kager P, Leeuwenburg J, Perkins P, et al. Indigenous human cutaneous leishmaniasis caused by Leishmania tropica in Kenya. Am J Trop Med Hyg. 1988;39:267–73. doi: 10.4269/ajtmh.1988.39.267. [DOI] [PubMed] [Google Scholar]
- 4.Sang DK, Okelo GB, Chance ML. Cutaneous leishmaniasis due to Leishmania aethiopica, on Mount Elgon, Kenya. Ann Trop Med Parasitol. 1993;87:349–57. doi: 10.1080/00034983.1993.11812778. [DOI] [PubMed] [Google Scholar]
- 5.Mebrahtu YB, Lawyer PG, Pamba H, Koech D, Perkins PV, Roberts CR, et al. Biochemical characterization and zymodeme classification of Leishmania isolates from patients, vectors, and reservoir hosts in Kenya. Am J Trop Med Hyg. 1992;47:852–92. doi: 10.4269/ajtmh.1992.47.852. [DOI] [PubMed] [Google Scholar]
- 6.Tonui WK. Situational analysis of leishmaniases research in Kenya. Afr J Health Sci. 2006;13:7–21. [PubMed] [Google Scholar]
- 7.Mebrahtu YB, Lawyer PG, Ngumbi PM, Kirigi G, Mbugua J, Gachihi G, et al. A new rural focus of cutaneous leishmaniasis caused by Leishmania tropica in Kenya. Trans R Soc Trop Med Hyg. 1992;86:381–7. doi: 10.1016/0035-9203(92)90230-a. [DOI] [PubMed] [Google Scholar]
- 8.Sang DK, Okelo GB, Ndegwa CW, Ashford RW. New foci of cutaneous leishmaniasis in central Kenya and the Rift Valley. Trans R Soc Trop Med Hyg. 1993;87:629–32. doi: 10.1016/0035-9203(93)90265-r. [DOI] [PubMed] [Google Scholar]
- 9.Massamba NN, Mutinga MJ, Kamau CC. Characterisation of Leishmania isolates from Laikipia District, Kenya. Acta Trop. 1998;71:293–303. doi: 10.1016/s0001-706x(98)00071-0. [DOI] [PubMed] [Google Scholar]
- 10.Sang DK, Njeru WK, Ashford RW. A zoonotic focus of cutaneous leishmaniasis due to Leishmania tropica at Utut, Rift Valley Province, Kenya. Trans R Soc Trop Med Hyg. 1994;88:35–7. doi: 10.1016/0035-9203(94)90486-3. [DOI] [PubMed] [Google Scholar]
- 11.Lawyer PG, Mebrahtu YB, Ngumbi PM, Mwanyumba P, Mbugua J, Kiilu G, et al. Phlebotomus guggisbergi (Diptera: Psychodidae), a vector of Leishmania tropica in Kenya. Am J Trop Med Hyg. 1991;44:290–8. doi: 10.4269/ajtmh.1991.44.290. [DOI] [PubMed] [Google Scholar]
- 12.Johnson RN, Ngumbi PM, Mwanyumba JP, Roberts CR. Host feeding preference of Phlebotomus guggisbergi, a vector of Leishmania tropica in Kenya. Med Vet Entomol. 1993;7:216–8. doi: 10.1111/j.1365-2915.1993.tb00679.x. [DOI] [PubMed] [Google Scholar]
- 13.Basiye FL, Schoone GJ, Beld M, Minnaar R, Ngeranwa JN, Wasunna MK, et al. Comparison of short-term and long-term protocols for stabilization and preservation of RNA and DNA of Leishmania, Trypanosoma, and Plasmodium. Diagn Microbiol Infect Dis. 2011;69:66–73. doi: 10.1016/j.diagmicrobio.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 14.Chulay JD, Anzeze EM, Koech DK, Bryceson AD. High-dose sodium stibogluconate treatment of cutaneous leishmaniasis in Kenya. Trans R Soc Trop Med Hyg. 1983;77:717–21. doi: 10.1016/0035-9203(83)90213-4. [DOI] [PubMed] [Google Scholar]
- 15.Boom R, Sol CJ, Salimans MM, Jansen CL, Dillen Wertheim-vanPM, Noordaa van derJ. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deborggraeve S, Boelaert M, Rijal S, Doncker DeS, Dujardin JC, Herdewijn P, et al. Diagnostic accuracy of a new Leishmania PCR for clinical visceral leishmaniasis in Nepal and its role in diagnosis of disease. Trop Med Int Health. 2008;13:1378–83. doi: 10.1111/j.1365-3156.2008.02154.x. [DOI] [PubMed] [Google Scholar]
- 17.Odiwuor Ogado CeasarS, Saad AgeedA, Doncker DeS, Maes I, Laurent T, Safi ElS, et al. Universal PCR assays for the differential detection of all Old World Leishmania species. Eur J Clin Microbiol Infect Dis. 2011;30:209–18. doi: 10.1007/s10096-010-1071-3. [DOI] [PubMed] [Google Scholar]
- 18.Mebrahtu YB, Eys VanG, Guizani I, Lawyer PG, Pamba H, Koech D, et al. Human cutaneous leishmaniasis caused by Leishmania donovani s. l. in Kenya. Trans R Soc Trop Med Hyg. 1993;87:598–601. doi: 10.1016/0035-9203(93)90101-u. [DOI] [PubMed] [Google Scholar]
- 19.Elamin EM, Guerbouj S, Musa AM, Guizani I, Khalil EA, Mukhtar MM, et al. Uncommon clinical presentations of cutaneous leishmaniasis in Sudan. Trans R Soc Trop Med Hyg. 2005;99:803–8. doi: 10.1016/j.trstmh.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Kolaczinski JH, Reithinger R, Worku DT, Ocheng A, Kasimiro J, Kabatereine N, et al. Risk factors of visceral leishmaniasis in East Africa: a case-control study in Pokot territory of Kenya and Uganda. Int J Epidemiol. 2008;37:344–52. doi: 10.1093/ije/dym275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jafari S, Hajiabdolbaghi M, Mohebali M, Hajjaran H, Hashemian H. Disseminated leishmaniasis caused by Leishmania tropica in HIV-positive patients in the Islamic Republic of Iran. East Mediterr Health J. 2010;16:340–3. [PubMed] [Google Scholar]
- 22.Khandelwal K, Bumb RA, Mehta RD, Kaushal H, Lezama-Davila C, Salotra P, et al. A patient presenting with diffuse cutaneous leishmaniasis (DCL) as a first indicator of HIV infection in India. Am J Trop Med Hyg. 2011;85:64–5. doi: 10.4269/ajtmh.2011.10-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soni P, Prasad N, Khandelwal K, Ghiya BC, Mehta RD, Bumb RA, et al. Unresponsive cutaneous leishmaniasis and HIV co-infection: report of three cases. Indian J Dermatol Venereol Leprol. 2011;77:251. doi: 10.4103/0378-6323.77484. [DOI] [PubMed] [Google Scholar]