Abstract
Arthropod borne diseases cause significant human morbidity and mortality and, therefore, efficient measures to control transmission of the disease agents would have great impact on human health. One strategy to achieve this goal is based on the manipulation of bacterial symbionts of vectors. Bacteria of the Gram-negative, acetic acid bacterium genus Asaia have been found to be stably associated with larvae and adults of the Southeast Asian malaria vector Anopheles stephensi, dominating the microbiota of the mosquito. We show here that after the infection of Anopheles gambiae larvae with Asaia the bacteria were stably associated with the mosquitoes, becoming part of the microflora of the midgut and remaining there for the duration of the life cycle. Moreover they were passed on to the next generation through vertical transmission. Additionally, we show that there is an increase in the developmental rate when additional bacteria are introduced into the organism which leads us to the conclusion that Asaia plays a yet undetermined crucial role during the larval stages. Our microarray analysis showed that the larval genes that are mostly affected are involved in cuticle formation, and include mainly members of the CPR gene family.
Keywords: Asaia, Anopheles gambiae, Larval development, Cuticle proteins, Paratransgenesis, Symbiont
Introduction
Arthropod borne diseases cause significant human morbidity and mortality. In the absence of effective vaccines, with the exception of the one directed against yellow fever, current preventive methods rely heavily on vector control. Since the discovery of DDT's insecticidal properties in the 1930s, insecticides have played a major role in this approach. However, the emergence of insecticide resistance in many vector species and the worldwide non-acceptance of extensive spraying of chemicals necessitate the development of new strategies for the control of arthropod vectors.1
The last few years have witnessed an increased use of mosquito bednets, whose efficacy may be enhanced tremendously if soaked, before use, in insecticidal formulations.2 Moreover, in addition to an intensified search for new and environment-friendly insecticides, novel strategies being developed include the use of genetically modified organisms in different combinations of approaches. Such proposed strategies include the replacement of vector populations by engineered ones that cannot transmit the pathogens,3 and the creation of modified vectors that promote the ‘emergence’ of only one sex, rendering the application of the sterile insect technique simpler.4 For the latter, field experiments are already underway.5
More recently a great deal of attention has been devoted to the development of another approach, based on the manipulation of bacterial symbionts of vectors. For example, the idea is to generate symbiotic bacteria that can be modified to produce anti-plasmodial effector molecules inside mosquitoes,6 thus preventing malaria transmission. In the case of malaria the mosquito midgut is of special interest, since this is where the Plasmodium parasite first enters the vector. Here, the first steps of the parasite development take place. Ookinete development and sporogony represent bottlenecks in the malarial parasite life cycle;7 in the wild usually only a few oocysts develop inside a single mosquito.8
Genetic manipulation of bacteria is simpler and faster than genetic manipulation of mosquitoes. Bacteria are easy to introduce into mosquito populations and can be produced cheaply and efficiently in large quantities. Furthermore, the introduction of modified bacteria into a vector population will bypass any genetic barriers of reproductively isolated mosquito populations, which often occur in regions of disease endemicity.9 There are, of course, requirements that must be met for this approach to work for a given vector/parasite combination.10 Several characteristics have been identified that are necessary for a successful paratransgenic control strategy.11 This was first demonstrated through the transformation of the bacteria Rhodococcus rhodnii, a symbiont of the kissing bug Rhodnius prolixus, which is a major vector of Chagas disease.12
Bacteria of the Gram-negative, acetic acid bacterium genus Asaia have been found to be stably associated with larvae and adults of the Southeast Asian malaria vector Anopheles stephensi, dominating the microbiota of the mosquito. PCR analysis showed that Asaia spp. DNA is present in eggs, pupae, and different larval stages, as well as in various mosquito organs, including gut, salivary glands, ovaries, and testes.13
Asaia bacteria, which had been transformed with green fluorescent protein (GFP)-expressing plasmids, were used to re-infect adult mosquitoes by adding the bacteria to sugar or blood meals. Transformed Asaia were found in different mosquito tissues, which are sites for pathogen development, such as midgut and salivary glands. They were also found in male and female reproductive tracts, although at lower numbers. Vertical and venereal transmission of Asaia was demonstrated by crossing males fed with the GFP-tagged Asaia with normal females of A. stephensi in the laboratory. After mating, fluorescent bacteria could be detected in the spermatheca and in the terminal portions of the gastrointestinal tract, thus indicating the transmission of the bacterium along with sperm. Furthermore, the vertical transmission of the bacterium to the progeny was also observed.13
Here we report the infection of mosquito larvae with the GFP-expressing Asaia strain in order to explore its effects on the mosquito, its suitability as a paratransgenic vehicle, and its role in the organism.
Materials and Methods
Mosquitoes
Anopheles gambiae mosquitoes from the Ngousso strain14 were obtained from the insectary of Imperial College in London. The mosquito stocks were reared under standard conditions (16/8 light/dark cycle, RT = 28°C, humidity = 80%) while the experiments on the treated and non-treated specimens were conducted under slightly different conditions (16/8 light/dark cycle, RT = 27°C, humidity = 60%), after initially making sure that these changes had no effect on the results obtained.
Colonization of larvae and adult mosquitoes with Asaia sp. SF2.1(Gfp)
Asaia sp. SF2.1(Gfp)13 was grown in 3 ml cultures for 48 hours at 30°C in GLY medium (glycerol 25 g/l, yeast extract 10 g/l, pH = 5.0) to which 50 μg/ml kanamycin was added. Cells were harvested by centrifugation and washed with PBS three times. Finally 50 μl of the bacterial pellet was resuspended in 1 ml PBS. Fifty 1st instar A. gambiae larvae were transferred to containers filled with 500 ml H2O. To half of the containers the Asaia re-suspension was added. For the experiments where the mosquitoes were fed with dead bacteria two different methods were used. The bacteria were killed either by freezing in liquid nitrogen for 10 minutes or by heat treatment (20 minutes at 80°C). Development of the mosquitoes was monitored by stereoscopic observation at 24-hour intervals (20 larvae for each time point). At 72 hours larvae were dissected and the guts and peritrophic membranes were mounted in Vectashield (Vector Laboratories, Inc., Burlingame, CA, USA), followed by observation with a Bio-Rad confocal microscope (Bio-Rad Laboratories, Hercules, CA, USA) attached to a Zeiss Axioskop 2 plus microscope (Carl Zeiss, Oberkochen, Germany).
Midguts were also dissected from adult mosquitoes and observed with a Zeiss Axioskop 2 plus fluorescent microscope. In each experiment samples from non-infected mosquitoes were also viewed in parallel using the same settings; in no case was a fluorescence signal detected.
RNA extraction
After initial infection with Asaia, samples of 10 larvae were removed at 24-hour intervals until the pupal stage was reached; they were then homogenized using a mortar-and-pestle. RNA was extracted by following the protocol for whole organisms provided with the Qiagen RNeasy Mini kit (Qiagen, Valencia, CA, USA; cat. no. 74104), together with the Qiagen RNase-Free DNase set (cat no.79254). RNA concentration was measured with a Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and the samples were kept at −80°C.
Microarray study
The samples originating from the RNA extraction were pooled into two groups. One group contained samples from first and second instar larvae and the other the samples from the third and the fourth instars. The RNA was amplified and labeled using the Agilent Low Input Quick Amp Labeling Kit, two-color (Agilent Technologies, Santa Clara, CA, USA; Agilent #5190-2306). The RNA was then hybridized to Agilent arrays (name: Pfalcip_Agamb_2009, design ID: 026247) using the Agilent hybridization kit according to the manufacturer’s instructions. Following hybridization and washing, the slides were scanned using a GenePix scanner (Molecular Devices, LLC, Sunnyvale, CA, USA; GenePix 4000B).
Microarray data analysis
The raw gene expression data were extracted using the GenePix Pro Software v7 and imported into the Agilent GeneSpring GX software for further analysis. The data sets were normalized and all of the data were interpreted using the log-ratio setting. The affected genes were identified using the parameters fold-change>2 and p-value<0.05. Gene ontology (GO) analysis was used to identify the GO categories to which the affected genes belonged.
Results
Colonization of mosquito larvae with Asaia sp.
The bacterium used to infect the mosquito larvae was Asaia sp., strain SF2.1, which is kanamycin resistant and expresses GFP.13 This particular strain of Asaia has been shown previously to colonize the mosquitoes Aedes aegypti and A. stephensi as well as the leafhopper Scaphoideus titanus,15 and it is considered to be a good candidate microorganism for potential paratransgenesis uses.
We first wanted to test whether this strain of Asaia would establish symbiosis in the A. gambiae Ngousso strain, an M strain recently isolated in Cameroon and now widely used for genomic experiments.14 Newly hatched 1st stage larvae were added to larval pans (50 per container) including the bacteria. Bacteria were not added to the pans after this initial feed during the development of the mosquitoes. After 72 and 96 hours the larvae were dissected and observed under a fluorescent microscope. Figure 1 shows a dissected midgut that heavily fluoresces. Further analysis demonstrated that the bacteria were present only on the peritrophic membrane; no bacteria were detected inside the actual epithelial cells of the midgut. Infection was maintained throughout the whole duration of the four larval stages.
Figure 1.
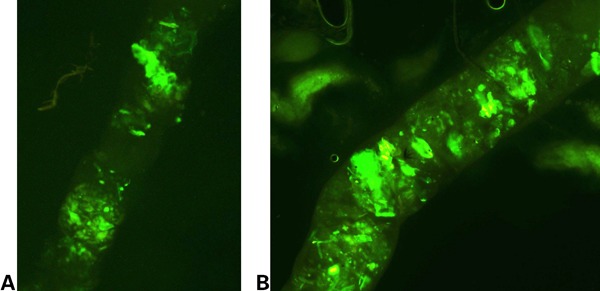
Section of the peritrophic matrix of Anopheles gambiae larvae infected with Asaia. (A) Peritrophic matrix clearly showing green fluorescent protein (GFP)-expressing bacteria throughout its length, at 72 hours post-infection (larval stage 3). (B) Peritrophic matrix showing the growth of GFP-expressing bacteria, at 96 hours post-infection (larval stage 4).
Developmental effects of Asaia sp. on A. gambiae larvae
During the infection of mosquito larvae with the bacterium we observed a very significant change in the developmental rate of the insects. Infected larvae progressed through the four larval stages at an accelerated rate, reaching the pupal stage almost 48 hours faster than their non-infected counterparts. The very marked change in the developmental rate seemed to be occurring on the threshold between stage 2 and stage 3 (Table 1). There was a clearly visible size difference 72 hours post-infection, which was maintained throughout the larval stages (Fig. 2). We determined that this developmental effect was dependent upon the presence of viable bacteria; when larvae were fed with dead bacteria there was no change in the duration of larval development (data not shown), irrespective of the procedure used to inactivate the bacteria (high or ultra-low temperature). The length of the pupal stage was unaffected by the presence of Asaia bacteria in the food.
Table 1. Differences in developmental rate between infected and non-infected larvae.
| Infected | Uninfected | |
| 24 hours | 1st instar | 1st instar |
| 48 hours | 2nd instar | 1st instar |
| 72 hours | 3rd instar | 2nd instar |
| 96 hours | 4th instar | 2nd instar |
| 5 days | 4th instar/pupae | 3rd instar |
| 6 days | Pupae/adults | 3rd instar |
| 7 days | Pupae/adults | 4th instar |
| 8 days | Adults | Pupae |
| 9 days | Adults | Pupae/adults |
Figure 2.
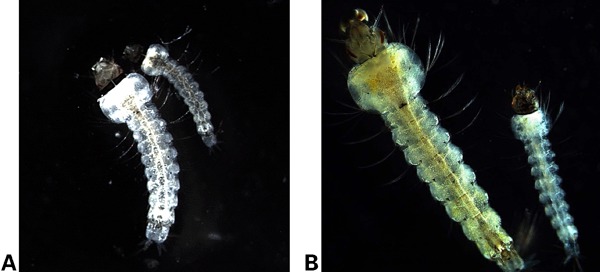
Accelerated development of Asaia-infected mosquito larvae. (A) Two larvae can be seen at 72 hours after hatching. The bigger larva, on the left, was infected with Asaia 72 hours prior to the capture. The smaller larva, to the right, is the non-infected control. The difference in developmental advancement is clearly visible. (B) At 96 hours post-infection, the infected larva on the left has clearly reached the 4th stage, while the non-infected control remains in the 2nd instar.
Colonization of adult mosquitoes with Asaia sp.
In order to determine if there were any effects on the development from larvae to adult, we allowed the larvae to reach the full adult stage. No additional Asaia was given after molting. There were no visible differences in the size of the infected compared to the non-infected insects. Midgut dissections were performed on 10 mosquitoes, male and female, at different time-points of their adult lives. Bacteria were present in all samples (Fig. 3).
Figure 3.
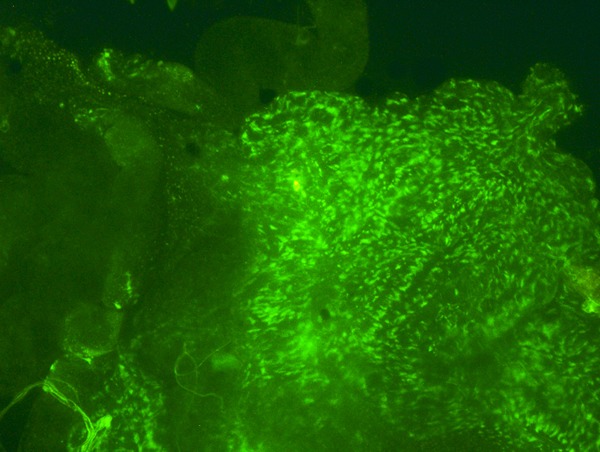
Section of the midgut of an infected, 24-day-old adult mosquito. Green fluorescent protein (GFP)-expressing Asaia bacteria are present in the gut.
Infected adult mosquitoes were blood-fed and allowed to produce eggs to investigate if the faster developmental rate persisted in the next generation. However, no similar acceleration effect was visible on the F1 larvae of the infected cohort. Dissections of third and fourth instar larvae showed that the genetically modified Asaia sp. was still present in the endo-peritrophic space (Fig. 4).
Figure 4.
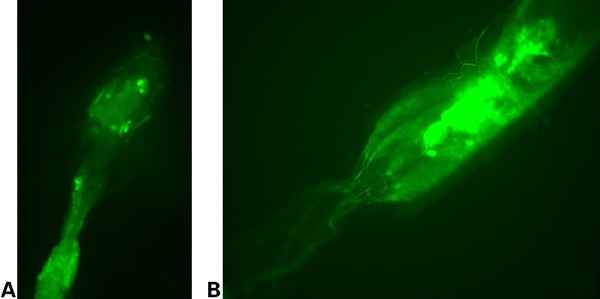
Dissected guts of larvae derived from parents infected with Asaia during their larval stage. (A and B) The continuous growth and spread of the genetically modified bacteria is shown here. The bacteria reside along the length of the gut.
RNA profiling of larval samples
Larval RNA was extracted every 24 hours from 10 infected and 10 non-infected individuals until the pupal stage was reached. A microarray analysis of the larval RNA followed. Two groups of samples were created, Group A containing the combined RNA of first and second instar larvae, and Group B containing the combined RNA of third and fourth instar larvae. The results can be summarized as follows. A comparison between infected and non-infected larvae of Group A identified a total of 205 genes, which were significantly up- or down-regulated. The threshold chosen was either above or below a two-fold change with a p-value<0.05. One hundred and thirty genes were down-regulated, while 75 were up-regulated. In Group B, 925 genes fit the significance parameters, with 416 being down-regulated and the remaining 409 being up-regulated. The smaller number of genes affected in Group A can be explained by the observation that the actual change in the developmental rate was initiated between larval stages 2 and 3.
Gene ontology analyses were performed to obtain more detailed functional information regarding the genes whose expression level was affected by the presence of Asaia. In Group A, additional information was obtained for 104 genes, while in Group B, for 249 genes (see Fig. 5 for details).
Figure 5.
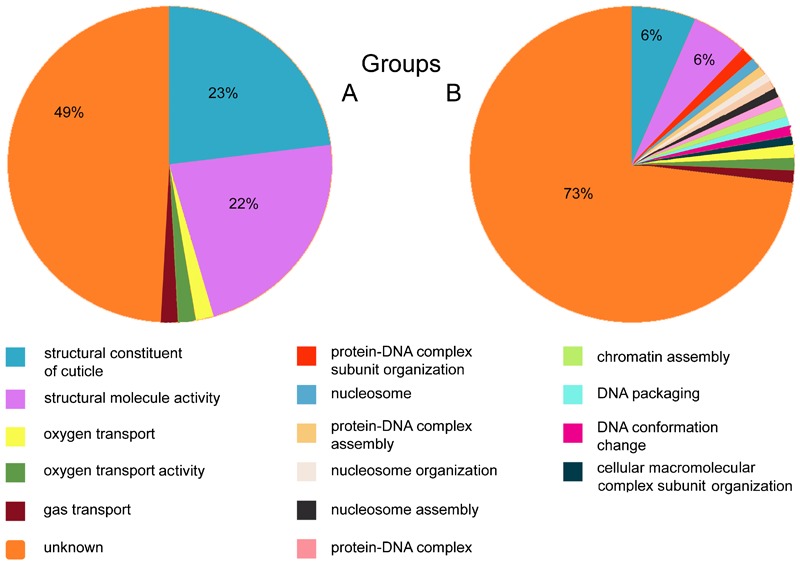
Gene ontology (GO) analysis of the affected genes of Group A (left pie) and Group B (right pie). In Group A 49% of the genes were annotated with GO terms referring to unknown functions and processes. The rest belonged to two large categories (23% each) that had GO annotations related to the cuticle, as well as three small groups (about 2% of the genes each) with GO annotations referring to oxygen transport, oxygen transporter activity, and gas transport. For 73% of the affected genes in Group B no GO terms could be identified. Of the remaining, 6% each belonged to the same two main categories described for Group A (cuticle-related). The remaining genes belonged to small clusters (about 1% each) with GO annotations referring to protein–DNA complex subunit organization, protein–DNA complex assembly, nucleosome assembly, gas transport, chromatin assembly, oxygen transport, nucleosome, nucleosome organization, protein–DNA complex, DNA packaging, oxygen transporter activity, and DNA conformation change. The GO terms that belong to the individual clusters are color-coded (shown below the pies).
The most common category in both groups was that of ‘structural constituent of the cuticle’ (GO:0042302), containing the CPR genes, a family of structural cuticular proteins (CPs).16 This is a large CP gene family that is characterized by the presence of the Rebers and Riddiford (1988) domain.17 In Group A, of the 45 genes that belonged to this family 42 were down-regulated and 3 were up-regulated, while in Group B, 24 were down-regulated and 24 were up-regulated. Nineteen CPR genes were shown to be affected in both Groups A and B. Of the three up-regulated CPR genes in Group A two were found to be up-regulated in Group B as well and in total 12 genes were down-regulated in both groups as well (Table 2) while five were both up- and down-regulated. Using VectorBase’s biomart tool (http://www.vectorbase.org/) we were able to determine that of the CPR genes that were affected by the Asaia infections, there were 33 genes containing the RR-1 motif, 38 genes containing the RR-2 motif, and one gene containing an RR-3 motif (see Table 3).18,19 Further analysis with the Pfam database (http://pfam.sanger.ac.uk) showed that they had the same chitin_bind_4 domain (PF00379).
Table 2. Up- and down-regulated CPR genes of Group A and Group B individually and those affected in both groups.
| Group A | Group B | Group A & B | |||
| Up-regulated | Down-regulated | Up-regulated | Down-regulated | Up-regulated | Down-regulated |
| CPR150 | CPR101 | CPR100 | CPR101 | CPR150 | CPR101 |
| CPR24 | CPR106 | CPR109 | CPR106 | CPR98 | CPR106 |
| CPR98 | CPR121 | CPR110 | CPR117 | CPR121 | |
| CPR135 | CPR111 | CPR119 | CPR136 | ||
| CPR136 | CPR131 | CPR120 | CPR33 | ||
| CPR148 | CPR142 | CPR121 | CPR42 | ||
| CPR149 | CPR150 | CPR123 | CPR43 | ||
| CPR32 | CPR34 | CPR125 | CPR44 | ||
| CPR33 | CPR35 | CPR129 | CPR45 | ||
| CPR35* | CPR38 | CPR136 | CPR47 | ||
| CPR36 | CPR40 | CPR2 | CPR49 | ||
| CPR37 | CPR66 | CPR24* | CPR70 | ||
| CPR38* | CPR73 | CPR32 | |||
| CPR39 | CPR77 | CPR33 | |||
| CPR41 | CPR78 | CPR4 | |||
| CPR42 | CPR80 | CPR42 | |||
| CPR43 | CPR92 | CPR43 | |||
| CPR44 | CPR93 | CPR44 | |||
| CPR45 | CPR94 | CPR45 | |||
| CPR46 | CPR95 | CPR47 | |||
| CPR47 | CPR96 | CPR49 | |||
| CPR48 | CPR97 | CPR67 | |||
| CPR49 | CPR98 | CPR70 | |||
| CPR50 | CPR99 | CPR71 | |||
| CPR51 | |||||
| CPR52 | |||||
| CPR53 | |||||
| CPR54 | |||||
| CPR60 | |||||
| CPR61 | |||||
| CPR62 | |||||
| CPR66* | |||||
| CPR67 | |||||
| CPR70 | |||||
| CPR73 | |||||
| CPR85 | |||||
| CPR86 | |||||
| CPR87 | |||||
| CPR88 | |||||
| CPR89 | |||||
| CPR90 | |||||
| CPR91 | |||||
Genes with an asterisk are both up- and down-regulated.
Table 3. Genes containing the RR-1, RR-2, and RR-3 motifs.
| RR-1 motif | RR-2 motif | RR-3 motif |
| CPR106 | CPR100 | CPR111 |
| CPR125 | CPR101 | |
| CPR129 | CPR109 | |
| CPR24 | CPR110 | |
| CPR32 | CPR117 | |
| CPR33 | CPR119 | |
| CPR34 | CPR120 | |
| CPR35 | CPR121 | |
| CPR36 | CPR123 | |
| CPR37 | CPR131 | |
| CPR38 | CPR136 | |
| CPR39 | CPR142 | |
| CPR40 | CPR2 | |
| CPR41 | CPR4 | |
| CPR42 | CPR42 | |
| CPR43 | CPR43 | |
| CPR44 | CPR44 | |
| CPR45 | CPR45 | |
| CPR46 | CPR47 | |
| CPR47 | CPR49 | |
| CPR48 | CPR67 | |
| CPR49 | CPR70 | |
| CPR50 | CPR71 | |
| CPR51 | CPR85 | |
| CPR52 | CPR86 | |
| CPR61 | CPR87 | |
| CPR62 | CPR88 | |
| CPR66 | CPR89 | |
| CPR66 | CPR90 | |
| CPR73 | CPR91 | |
| CPR77 | CPR92 | |
| CPR78 | CPR93 | |
| CPR80 | CPR94 | |
| CPR95 | ||
| CPR96 | ||
| CPR97 | ||
| CPR98 | ||
| CPR99 |
Discussion
Symbiotic bacteria residing inside arthropods have been proposed as paratransgenic vehicles.6 The bacteria can either, as such, negatively affect disease agents or be engineered to produce toxic factors. This way, strains of arthropod vectors could be produced that can be employed in strategies aimed at reducing the disease burden. It is however important to understand the role these symbionts play in the host insect under ‘natural’, i.e. non-engineered conditions. We show here that after the infection of A. gambiae larvae with Asaia, the bacteria were stably associated with the mosquitoes, becoming part of the microflora of the midgut and remaining there for the duration of the life cycle. Moreover they were passed on to the next generation through vertical transmission. This suggests that bacteria of the Asaia genus could be used to express an effector gene; in addition, the bacteria could possibly be transmitted to subsequent generations. This would render the need for repeated introductions unnecessary, which would reduce the cost substantially for such a strategy. As recently shown in another study,20 when mosquitoes are deprived of Asaia symbionts they experience a delay in larval development. This, combined with our observations that there is an increase in the developmental rate when additional bacteria are introduced into the organism, leads us to the conclusion that Asaia plays a yet undetermined crucial role during the larval stages. This role is most likely dependent upon live bacteria, as we saw that the addition of killed bacteria did not result in a similar acceleration. This suggests that acceleration is not just the result of a ‘better’ food source for larvae, but that live Asaia provides specific nutrients or other molecules which are limiting under normal larval development using the conditions in the laboratory. Possible explanations for this finding are that the bacteria in the gut assist in the digestion of nutrients in the mosquito’s gut and/or that the bacteria produce molecule(s) that allow faster development. Since adult development was neither impaired nor aided by the presence of the bacteria, it follows that Asaia does not play a pivotal role in the adult mosquito.
We also observed that although the genetically modified bacteria were passed on to the next generation, the effects on larval development were no longer observed. This is possibly due to the fact that during the first generation the number of bacteria was much higher in the environment and/or midgut than in the F1 generation.
Our microarray analysis showed that the genes that are mostly affected are involved in cuticle formation. Currently, 12 different families of CPs have been recognized, in which each family shares common features.16 In our analysis the main category affected included mainly members of the CPR gene family. These genes are characterized by the presence of a conserved 35–36 amino acid motif (R&R consensus) first identified by Rebers and Riddiford (1988).17 This finding is consistent with the fact that larval molting was accelerated due to the effect that overexposure to Asaia has on the larval development. One explanation for this could be that the accelerated development induced by the bacteria goes along with an increased necessity to build ‘new’ integument, at rates that are higher than the ones usually seen. The biomart and Pfam analysis showed that all of the affected genes contained the chitin_bind_4 domain and that those genes had both subgroups of the R&R consensus: RR-1, which is found in proteins that form soft cuticles, and RR-2, which is present in proteins that form hard cuticles.21,22 One protein, finally, contains the RR-3 domain, which is a variant of the R&R consensus.
In this study we showed that genetically modified Asaia can be easily introduced into A. gambiae and maintained through at least one generation, without any negative effects on the adult mosquito. Furthermore, we have uncovered evidence that bacterial symbiotic organisms may play a significant role in larval development as the addition of Asaia to the larval environment showed a significant boost in developmental rate. Therefore, these bacteria can be considered excellent candidates for use as agents of paratransgenesis in a vector-centered fight against malaria. Taken together with the observation that Asaia is reduced using rifampicin treatment,20 we tend to conclude that the bacteria are needed to maintain the developmental rate of the larvae. The molecular nature of the developmental accelerator, though, remains to be identified.
Acknowledgments
We thank Prof. John Vontas for hosting EM for the mosquito rearing experiments and Dr. Dina Vlachou for sharing the DNA microarrays and analysis tools prior to publication. This work was supported by grants 223736 (Transmalariabloc) and 201588 (Evimalar) of the FP7 (HEALTH) Programme of the European Commission. CL was partly supported by the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 267232.
References
- 1.Enayati A, Hemingway J. Malaria management: past, present, and future. Annu Rev Entomol. 2010;55:569–91 [DOI] [PubMed] [Google Scholar]
- 2.Curtis CF, Jana-Kara B, Maxwell CA. Insecticide treated nets: impact on vector populations and relevance of initial intensity of transmission and pyrethroid resistance. J Vector Borne Dis. 2003;40(1–2):1–8 [PubMed] [Google Scholar]
- 3.Marshall JM, Taylor CE. Malaria control with transgenic mosquitoes. PLoS Med. 2009;6(2):e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alphey L, Nimmo D, O'Connell S, Alphey N. Insect population suppression using engineered insects. Adv Exp Med Biol. 2008;627:93–103 [DOI] [PubMed] [Google Scholar]
- 5.Lacroix R, McKemey AR, Raduan N, Kwee Wee L, Hong Ming W, Guat Ney T, et al. Open field release of genetically engineered sterile male Aedes aegypti in Malaysia. PloS one. 2012;7(8):e42771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riehle MA, Jacobs-Lorena M. Using bacteria to express and display anti-parasite molecules in mosquitoes: current and future strategies. Insect Biochem Mol Biol. 2005;35(7):699–707 [DOI] [PubMed] [Google Scholar]
- 7.Alavi Y, Arai M, Mendoza J, Tufet-Bayona M, Sinha R, Fowler K, et al. The dynamics of interactions between Plasmodium and the mosquito: a study of the infectivity of Plasmodium berghei and Plasmodium gallinaceum, and their transmission by Anopheles stephensi, Anopheles gambiae and Aedes aegypti. Int J Parasitol. 2003;33(9):933–43 [DOI] [PubMed] [Google Scholar]
- 8.Ghosh A, Edwards MJ, Jacobs-Lorena M. The journey of the malaria parasite in the mosquito: hopes for the new century. Parasitol Today. 2000;16(5):196–201 [DOI] [PubMed] [Google Scholar]
- 9.Fontenille D, Cohuet A, Awono-Ambene P, Kengne P, Antonio-Nkondjio C, Wondji C, et al. Vecteurs de paludisme: du terrain a la genetique moleculaire. Recherches en Afrique [Malaria vectors: from the field to genetics. Research in Africa]. Revue d'epidemiologie et de sante publique. 2005;53(3):283–90 [DOI] [PubMed] [Google Scholar]
- 10.Damiani C, Ricci I, Crotti E, Rossi P, Rizzi A, Scuppa P, et al. Mosquito-bacteria symbiosis: the case of Anopheles gambiae and Asaia. Microb Ecol. 2010;60(3):644–54 [DOI] [PubMed] [Google Scholar]
- 11.Beard CB, Cordon-Rosales C, Durvasula RV. Bacterial symbionts of the triatominae and their potential use in control of Chagas disease transmission. Annu Rev Entomol. 2002;47:123–41 [DOI] [PubMed] [Google Scholar]
- 12.Durvasula RV, Gumbs A, Panackal A, Kruglov O, Aksoy S, Merrifield RB, et al. Prevention of insect-borne disease: an approach using transgenic symbiotic bacteria. Proc Natl Acad Sci U S A. 1997;94(7):3274–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Favia G, Ricci I, Damiani C, Raddadi N, Crotti E, Marzorati M, et al. Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proc Natl Acad Sci U S A. 2007;104(21):9047–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris C, Lambrechts L, Rousset F, Abate L, Nsango SE, Fontenille D, et al. Polymorphisms in Anopheles gambiae immune genes associated with natural resistance to Plasmodium falciparum. PLoS Pathog. 2010;6(9):e1001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonella E, Crotti E, Rizzi A, Mandrioli M, Favia G, Daffonchio D, et al. Horizontal transmission of the symbiotic bacterium Asaia sp. in the leafhopper Scaphoideus titanus Ball (Hemiptera: Cicadellidae). BMC Microbiol. 2012;12(Suppl 1):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willis JH. Structural cuticular proteins from arthropods: annotation, nomenclature, and sequence characteristics in the genomics era. Insect Biochem Mol Biol. 2010;40(3):189–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rebers JE, Riddiford LM. Structure and expression of a Manduca sexta larval cuticle gene homologous to Drosophila cuticle genes. J Mol Biol. 1988;203(2):411–23 [DOI] [PubMed] [Google Scholar]
- 18.Cornman RS, Togawa T, Dunn WA, He N, Emmons AC, Willis JH. Annotation and analysis of a large cuticular protein family with the R&R Consensus in Anopheles gambiae. BMC genomics. 2008;9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersen SO. Studies on proteins in post-ecdysial nymphal cuticle of locust, Locusta migratoria, and cockroach, Blaberus craniifer. Insect Biochem Mol Biol. 2000;30(7):569–77 [DOI] [PubMed] [Google Scholar]
- 20.Chouaia B, Rossi P, Epis S, Mosca M, Ricci I, Damiani C, et al. Delayed larval development in Anopheles mosquitoes deprived of Asaia bacterial symbionts. BMC Microbiol. 2012;12(Suppl 1):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen SO. Exoskeletal proteins from the crab, Cancer pagurus. Comp Biochem Physiol, Part A: Mol Integr Physiol. 1999;123(2):203–11 [DOI] [PubMed] [Google Scholar]
- 22.Iconomidou VA, Willis JH, Hamodrakas SJ. Is beta-pleated sheet the molecular conformation which dictates formation of helicoidal cuticle? Insect Biochem Mol Biol. 1999;29(3):285–92 [DOI] [PubMed] [Google Scholar]


