Abstract
Aim:
Cytomegalovirus (CMV) infection is one of the most common complications in patients submitted to hematopoietic stem cell transplant (HSCT). Pre-emptive therapy has been indicated in patients with laboratory evidence of CMV replication. The aims of this study were to compare real-time PCR or pp65 antigen assay methodologies to detect CMV replication in HSCT patients, define a viral load threshold for initiation of pre-emptive therapy, and assess the feasibility of its implementation in hospitals of countries with low and middle income.
Material and methods:
Human CMV detection by real-time PCR and pp65 antigen assay was carried out in blood and plasma samples of HSCT patients collected weekly during 3 months. Pre-emptive therapy was based on CMV antigenemia results.
Results:
Twenty-one patients were monitored with a total of 227 samples collected; 13 (62%) patients were children. A poor correlation was observed between qualitative results, though quantitative results showed statistically significant difference, with higher viral loads detected in patients with positive antigenemia. Compared to a positive antigenemia, a cutoff value of 1067.5 copies/ml, 3.03 log10/ml, for viral load was obtained with 100% sensitivity and 71% specificity.
Conclusion:
CMV real-time PCR in whole blood was suitable for monitoring HSCT patients. However, its high cost is a limiting factor, and it could be used to monitor selected patients, those with prolonged leukopenia and underweight children, and subsequently switched to pp65 antigen test. Further studies involving larger numbers of patients should be performed to confirm this statement.
Keywords: CMV, Hematopoietic stem cell transplant, Real time PCR, pp65 antigenemia
Introduction
Human infection by cytomegalovirus (CMV) is common in the general population, with no significant morbidity in immune competent patients. Nevertheless, primary infection or reactivation of latent CMV in the immunocompromised patients can cause life-threatening diseases with higher mortality rates.1–4
Usually, CMV reactivation occurs within the first 3 months after transplantation. The use of steroids for prophylaxis and treatment of graft-versus-host disease (GvHD), which impair the recovery of CD4+ and CD8+ T cells including those specific for CMV, can expose the patients to the risk of CMV disease for a longer period.5
Early detection of reactivated CMV replication in hematopoietic stem cell transplant (HSCT) is essential to the management of the disease. In this context, detection of pp65 antigen, a marker of active viral replication, has considerably improved the clinical management of this group of patients; thus, pre-emptive therapy could replace universal prophylaxis, reducing CMV disease risk, with better results. Nevertheless, pp65 antigenemia assay has several limitations regarding its use in the clinical practice: it is a manual test, blood samples should be processed within 6–8 hours of collection, and high level of technical expertise is required to read the slides. Leukopenia is another significant limitation to this test, especially in patient submitted to HSCT.6–9
In this setting, quantitative PCR (qPCR) is an alternative method for early detection of viral replication and indicates pre-emptive therapy of CMV infection as it is not affected by leukopenia. Other advantages of CMV qPCR are the possibility of processing a higher number of samples, viral load quantification with higher sensitivity; it is a faster method, has a low risk of carryover contamination and has a broader linear range.6,9,10
However, the threshold value of viral load to indicate pre-emptive therapy has not yet been well defined, varying on the sample used to quantify viral DNA (whole blood or plasma) and the group of patients studied (solid organ transplant or HSCT patients). Besides, the impact of the high cost of this new methodology can be a limiting factor for its implementation in many public health services in Brazil.
Based on that, we conducted a study to compare the results of pp65 antigenemia assay and qPCR for detection of early active CMV infection in patients submitted to HSCT, comparing the results with clinical outcome.
Methodology
Samples and patients
This was a prospective study that compared pp65 assay and PCR tests on plasma and whole blood samples from 21 patients undergoing HSCT at the Bone Marrow Transplant Unit, Hospital de Clínicas, Universidade Federal do Paraná, Southern Brazil, from January to April 2011. The study was approved by the Committee on the Ethics of Research on Human Beings of the Hospital de Clínicas, Universidade Federal do Paraná, under registration number 2377.271/2010-11.
Patients were monitored for CMV infection with simultaneous determination of pp65-antigen test on polymorphonuclear cells and CMV DNA viral load on plasma and whole blood at weekly intervals, from day +15 to day +98 after transplantation. A total of 227 samples were analyzed, with a median number of samples per patient of 10.6 (range: 5–13). Pre-emptive therapy with intravenous ganciclovir was administered based on pp65-antigen and was started when more than 1 pp65-positive cells/200.000 leukocytes were detected. Patients’ characteristics are summarized in Table 1.
Table 1. HSCT and CMV monitoring: demographic and clinical findings.
| Characteristics | N |
| Patients | 21 |
| Age (years) | |
| Median (range) | 10 (2–54) |
| Gender | |
| Male/female | 14/7 |
| Diagnosis | |
| Acute leukemia | 9 |
| Severe aplastic anemia | 5 |
| Fanconi anemia | 5 |
| Myelodisplastic syndrome | 1 |
| Congenital dyserythropoietic anemia type 1 | 1 |
| Type of transplant | |
| Related | 12 |
| Unrelated | 9 |
| Source of stem cells | |
| Bone marrow | 15 |
| Cord blood | 4 |
| Peripheral blood | 2 |
| Total body irradiation | |
| No | 18 |
| Yes | 3 |
| Prophylaxis of aGvHD | |
| CyA | 3 |
| CyA+MTX | 17 |
| CyA+PDN | 1 |
| CMV status (donor/recipient) | |
| Positive/positive | 13 |
| Negative/positive | 2 |
| Positive/negative | 6 |
| Deaths | 3 |
| CMV-related | 2 |
Active CMV infection was defined by the detection of pp65 antigen-positive and/or CMV DNA in blood. CMV disease was defined by demonstration of viral inclusion in immunohistochemical analysis of biopsy specimens in association with clinical symptoms, such as unexplained fever (> 38°C), leukopenia, hepatitis, enteritis, retinitis, and pneumonitis.3,7 CMV syndrome was considered when clinical symptoms were present, with positive pp65 antigen detection, but without a confirmation by biopsy; and other virus infection as human herpesvirus 6 and adenovirus were excluded.11
pp65 antigenemia assay
CMV antigenemia was carried out within 4 hours of specimen collection by indirect immunofluorescence method using CMV Brite™ Turbo kit (IQ Products, Groningen, The Netherlands) according to manufacturer’s instruction. Results are expressed as the number of positive cells per 2×105 leukocytes.
CMV DNA quantification on plasma and whole blood
DNA extraction
Total nucleic acids were extracted at the same time from 200 μl of whole blood and plasma using Biopur™ Kit and AxyPrep™ Body Fluid Viral DNA/RNA Miniprep Kit, respectively, following the manufacturer’s protocols.
Quantitative real-time PCR
CMV DNA was quantified using the commercial kit Nanogen Q-CMV Real Time Complete Kit (Nanogen Advanced Diagnostics, Torino, Italy) that has the UL123 gene as target region. The method was performed according to manufacturer’s protocols.
Sample results are referred to 1 ml of sample multiplying the value obtained per reaction with a factor that takes in consideration the starting specimen volume for the extraction, the final elution volume, and the volume of DNA used for PCR amplification and reported as copies/ml. The assay’s analytical sensitivity as assessed by the manufacturer was between 20 and 106 HCMV DNA equivalent genomes per reaction.
Statistical analysis
CMV DNA values were reported as log10 copies/ml and pp65 antigen detection was expressed as the number of positive cells/200 000 leukocytes. Descriptive analysis was performed for all the variables. Nonparametric Wilcoxon test was used for comparing the results of the two methods. Relationship between the results of pp65 antigen detection and qPCR was estimated by Spearman’s correlation coefficient. Positive rates and levels of agreement between these methods were assessed using Kappa coefficients of agreement. Kappa values above 0.75 indicate strong agreement; values between 0.40 and 0.75 represent fair to good agreement and values below 0.40 reflect poor agreement. Non-parametric Mann–Whitney test was used in the comparison of groups with positive antigenemia regarding quantitative PCR results in whole blood. Receiver-operating characteristic (ROC) curve was adjusted for the determination of the threshold value of CMV DNA in whole blood to start pre-emptive therapy considering a level ⋛ 1 pp65-positive cells as the reference standard. P values < 0.05 were considered statistically significant. Data were compiled using JMP Software Version 5.2.1 and analyzed with the software Statistica v.8.0.
Results
Comparison of qualitative results between whole blood/plasma qPCR and pp65 antigen detection
Two hundred and twenty-seven samples were analyzed in the study; invalid results were observed in 10%, 7% ,and 5% for pp65 antigenemia assay, qPCR from plasma and whole blood samples, respectively (P = 0.15).
Comparison of qualitative results showed a poor agreement between the number of positive and negative samples obtained by pp65 antigenemia and qPCR assays (whole blood and plasma) (kappa: 0.31; 95% CI: 0.10–0.52 and kappa: 0.20; 95% CI: 0.01 – 0.38, respectively). It was observed a larger number of positive specimens in the qPCR methodology (Table 2). Only one case of positive pp65 antigenic with negative PCR result was detected in an asymptomatic patient. Thirteen patients had negative antigenemia in all collected samples, and four patients had all PCR tests negative.
Table 2. Comparison of qualitative qPCR and antigenemia for detection of CMV.
| pp65 antigen detection | qPCR results | ||
| Positive (69) | Negative (146) | Inconclusive (12) | |
| Positive (12) | 10 | 01 | 01 |
| Negative (192) | 52 | 133 | 07 |
| Inconclusive (23) | 07 | 12 | 04 |
Comparison of quantitative results of whole blood/plasma qPCR and pp65 antigen detection
CMV pp65 antigen-positive results ranged from 1 to 20 positive cells/2×105 leukocytes (median: 1); for qPCR of whole blood samples, positive results ranged from 38 to 50 000 copies/ml (median: 750); for slightly higher than plasma samples, which varied from 28 to 23 650 copies/ml (median: 434).
Analysis of antigen detection and plasma qPCR results in the group of patients with positive pp65 antigenemia showed a statistically significant difference between the values, with the median viral load of 2512 copies/ml, while in the negative group, the viral load was considerably lower, with a median of 293 copies/ml. In the whole blood samples, the median viral load in the group of patients with positive pp65 antigen was 3058 copies/ml, while in the negative group, it was 462 copies/ml (Fig. 1 and Table 3). Considering qPCR from whole blood results in logarithmic values and antigenemia results, the median value obtained was 2.67 (range: 2.05–4.7) and 3.47 (range: 3.05–4.31) for negative and positive antigenemia, respectively (P < 0.001).
Figure 1.
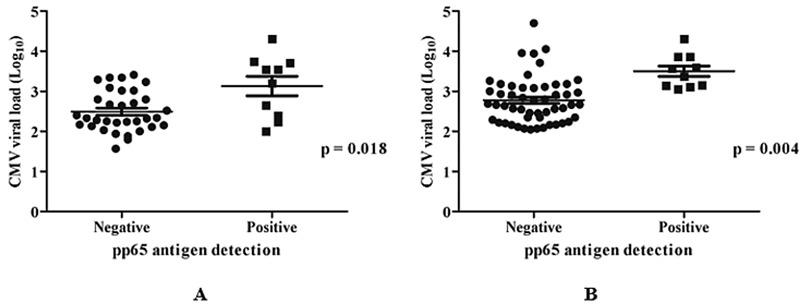
CMV pp65 antigen and qPCR results in plasma (A) and whole blood (B)
Table 3. Comparison of quantitative results of whole blood/plasma qPCR and antigenemia for CMV detection.
| qPCR results | qPCR results | |
| Whole blood (copies/ml)* | Plasma (copies/ml)* | |
| Positive CMV pp65 antigen | 3058 (1390–20 275) | 2512 (100–20 550) |
| Negative CMV pp65 antigen | 462 (112–11 337) | 293 (37–2570) |
Note: *Median (min. and max.) values.
Comparison of qualitative and quantitative results of plasma and whole blood qPCR
Real-time PCR qualitative results from plasma and whole blood samples showed a moderate agreement (kappa: 0.55; 95% CI: 0.42–0.68). Comparing the results of CMV viral load in plasma and whole blood samples in a logarithm scale, the average values obtained were 2.77 (± 0.67) and 2.96 (± 0.69), respectively (P = 0.053), and Spearman correlation coefficient was estimated to be r = 0.53 (P < 0.001) (Fig. 2). All the subsequent analyses are based on whole blood qPCR results.
Figure 2.
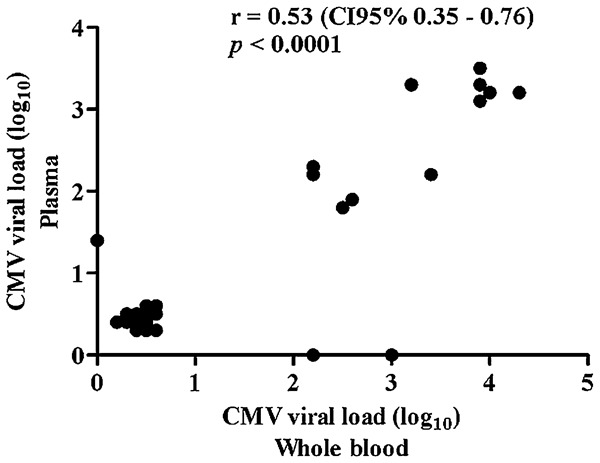
Comparison of quantitative results of whole blood and plasma qPCR to detect CMV
Comparison of qualitative results of whole blood qPCR and pp65 antigen detection by day post-transplant
Comparison of CMV pp65 antigen detection and qPCR carried out in different periods showed a better correlation between the results obtained in the day +15 and after day +70. The disagreement in the results from day +22 to +63 occurred because CMV DNAemia was detected earlier than pp65 antigen (Fig. 3).
Figure 3.
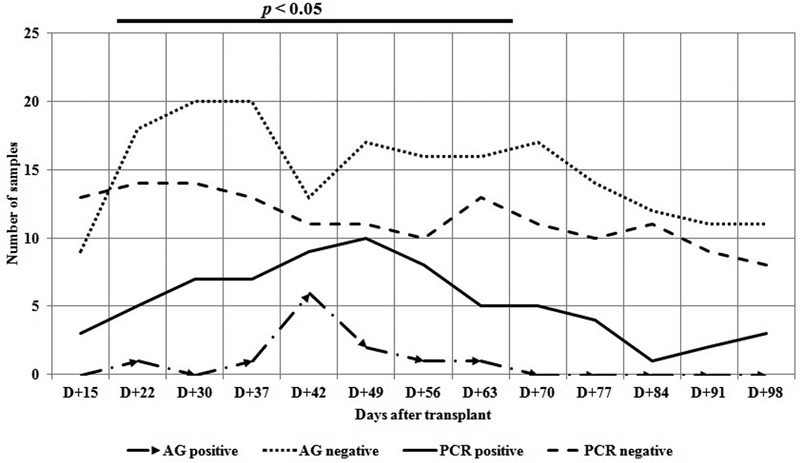
CMV pp65 antigen detection and PCR: qualitative results by day of transplant
Clinical outcome
Eight patients received pre-emptive therapy with ganciclovir and three (14%) patients died; two of them probably related with CMV disease associated with GvHD and bacterial infection. One patient (day +63) was 5 years old, with the diagnostic of Fanconi anemia, CMV IgG negative and received a non-related transplant from a CMV IgG positive donor. The second patient (day +56) was 54 years old, diagnosed with acute myeloid leukemia, CMV IgG-positive and received related transplant from a CMV IgG = negative donor. Both patients had high viral load and a positive qPCR earlier (1 week) than the antigen detection. The medians of viral load of both patients were 3050 and 5950 copies/ml. Detection of pp65 antigen could be performed in only one of these patients, with a median of 2 positive cells/200.000 leukocytes. The other patient had an insufficient number of cells to perform antigen detection.
Determination of cutoff value for qPCR
Considering all cases of positive qPCR from whole blood and antigenemia valid results, an ROC curve was adjusted (Fig. 4). The estimated area under the curve adjusted for whole blood qPCR was 0.86 with statistical significance (P < 0.001) indicating proper adjustment. The cutoff point was 1067.5 copies/ml (3.03 log10/ml); this viral load was the best value to discriminate between patients with positive and negative CMV antigenemia, with sensitivity and specificity equal to 100% and 71.1%, respectively.
Figure 4.
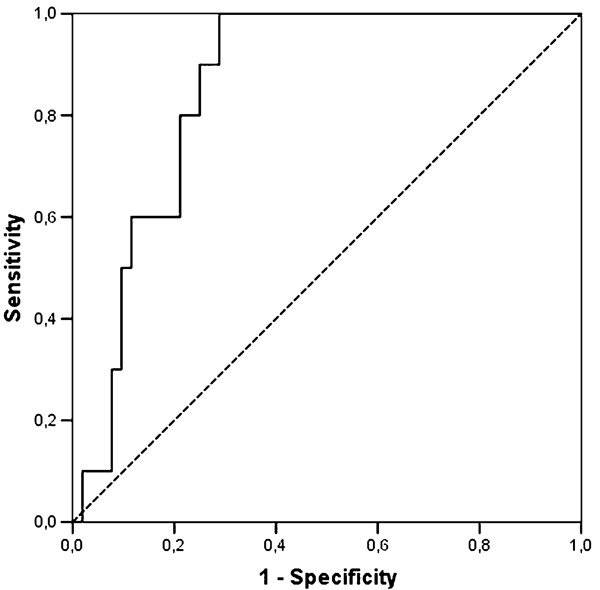
Receiver-operating characteristic (ROC) curve for qPCR as a diagnostic test for CMV reactivation in HSCT patients
Discussion
Early diagnosis of CMV infection could optimize pre-emptive therapy and improve outcome. CMV quantification methodologies, such as pp65 antigen detection and real-time PCR, are of high value in monitoring and detecting active infection. Even though qPCR threshold values were suggested in many reports, there is a consensus that these values have to be determined within the centers where the patients are monitored.
Although primary CMV infection has been regarded as a consequence of transmission through transplanted organ or administration of blood products,5 most transplanted recipients in this study were previously CMV IgG-positive and active CMV infection was probably the result of reactivation of latent virus.
Comparison of the qualitative results of antigenemia and qPCR showed a significant number of positive samples for PCR analysis, which is due to the greater sensitivity of this technique, resulting in a reduced correlation between these tests. However, comparison of the results of viral load values between the samples with positive or negative antigenemia showed a correlation between these values, where patients with positive CMV antigenemia had higher viral loads. This difference was statistically significant and allowed seeking a cutoff value for this viral load with a high positive predictive value, similar to that already known for antigenemia.
Many factors are associated with the risk of active CMV infection, such as immunological status of the patient and donor, conditioning regimens, aGvHD prophylaxis, and underlying disease.12 These conditions can probably explain the different frequencies and severities in the reports of CMV disease in various HSCT centers. Even within a same center, there are patients with more severe CMV disease than others. Therefore, further studies comparing different CMV diagnosis methods should be carried out in groups of patients according to their clinical characteristics. However, it would take a very long time before we achieved adequate sample sizes.
qPCR results from whole blood and plasma samples showed a moderate correlation between the absolute values obtained from viral loads, but in logarithmic scale, there was a difference of 0.19 between the logarithmic average, differences less than 0.7 log10 copies/ml are not considered significant, and the results can be interpreted as equivalent.13,14 Previous studies demonstrated that tests on whole blood samples are more sensitive than those on plasma or leukocyte.15–17 Other authors reported that tests could be carried out on blood fractions (leukocytes or plasma) and whole blood. Nevertheless, detection of CMV DNA on whole blood was found to occur earlier and measure greater amounts of CMV DNA.5,18,19 The extraction of nucleic acids in samples of whole blood is easier; it can be automated and dispense earlier steps, such as the separation of plasma or buffy coat concentration minimizing hands-on time. Thus, we decided to use whole blood to investigate CMV DNAemia and introduced this methodology in our routine laboratory.
Comparison of qPCR and antigenemia results at different days after transplantation showed a significant difference, with poor agreement between the results obtained from days +22 to +63, which is precisely the period of highest risk for CMV reactivation. This occurred mainly because the qPCR was positive earlier than antigenemia and remained positive for a longer period. Currently, a positive antigenemia has a high predictive value for active infection: thus, pre-emptive therapy is recommended. However, regarding qPCR, new tests should be performed on a new sample, and if the second sample remains positive and shows increased viral load, pre-emptive therapy should be recommended.20 The same was shown in studies with animal models.21,22
CMV pp65 antigen detection has been routinely performed in our HSCT patients for many years to guide pre-emptive therapy of CMV reactivation, preventing its progression to disease. However, HSCT is becoming a very complex procedure, there is a greater number of unrelated donors, as well as HLA mismatched donors and recipients of T cell-depleted allograft are being performed. Consequently, many patients have presented early CMV disease, when they present leukopenia and this has been one of the pitfalls of antigen detection test in this center.
All the patients that evolved to death presented severe diarrhea, abnormal liver function, and suspected bacterial infection. Intestinal aGvHD was confirmed in two patients. All patients had positive CMV qPCR a few weeks before their death, and two of them remained CMV-positive with increased viral loads until their death. Antigenemia was positive in one patient in the same period. The test could not be performed in the other patient due to leukopenia. Antigenemia was negative and viral load decreased significantly in the third patient. These patients were treated with broad-spectrum antibiotics, steroids, and ganciclovir, but progressed to multiple organ failure and death. No autopsy was performed. Based on our laboratory findings, we suspect that in two patients, who had increasing viral load, death may be a consequence of CMV indirect effect.
Considering pre-emptive therapy criteria based on pp65 antigenemia no less than one positive cell, a ROC curve analysis indicated a value of 3.03 log10 (1067.5 copies/ml) of DNA CMV as the best predictor of CMV infection progression, indicating the patients who required pre-emptive therapy. The observed high sensitivity allows recommending pre-emptive therapy use in patients with viral load values above the cutoff value in patients at high risk for CMV disease. This result was very similar to the threshold of 3.00 log10 and 3.17 log10 previously obtained.23,24 This study allowed the validation of the molecular assay for monitoring active CMV infection in patients undergoing HSCT. However, it is worth mentioning that the observed values should be prospectively validated using a larger sample size.
Distinct cutoff values were previously reported depending on whether they were HSCT or solid organ transplant patients. In HSCT recipients, cutoff values of 300 copies/ml,25 1000 copies/ml,20 9960 copies/ml,7 and 418.4 copies/104,10 have been reported in other studies. These studies included patients with various diseases and who underwent different type of transplant (related and unrelated donors). In transplant of solid organ, higher values have been reported, such as 200 000 genomes/ml,6 3000 copies/ml,20 and 24 000 copies/ml.7 Nevertheless, all included patients underwent different organ transplant, such as kidney, lung, and liver, which are associated to distinct immunosuppression profile. As shown, these parameters are extremely variable among different centers and patients’ clinical profiles, then comparisons have little validity. Moreover, further studies with a much larger sample size and the availability of international standards or external control panels are needed to perform valid comparisons between different transplant settings.5,14
The significant variability in CMV load results among currently used molecular methods limits the interlaboratory comparison of these results. Seeking to override this limitation, the World Health Organization Expert Committee on Biological Standardization has recently established the 1st International Standard for CMV DNA testing. The results will be reported in international units (IU)/ml that can be compared across transplants centers. The availability of these controls will allow transplants centers to assess the performance of the methodologies used, and the results obtained will enable to define common guidelines for CMV pre-emptive therapy.26,27
The methodology of real-time PCR showed an excellent performance for this population and must be performed routinely to monitor these patients. Nonetheless, besides the clinical benefit of replacement of pp65 CMV antigen detection by qPCR, it should be also considered its financial impact on the setting of HSCT in low- and middle-income countries, such as Brazil. As all RT-PCR commercial kits need to be imported, the costs of these tests are too high for implementation in public health hospitals, where the most procedures transplants are performed in the country. Thus, currently, we suggest routine qPCR in patients at high risk for CMV disease, especially those with persistent leukopenia after transplantation and in underweight children, which often makes it impossible to collect a larger amount of blood to perform CMV antigenemia. In leukopenic patients, the definition of a DNA threshold to viral load with high sensitivity and specificity could allow combining the two analyses in the patients’ monitoring, performing qPCR in the leukopenia period, and switching to pp65 antigen detection when the patient has a while blood cell count sufficient to permit this test. However, further controlled studies that include a larger number of patients are needed to determine the clinical impact of this proposal on the monitoring of CMV infection.
Human CMV infection represents an important complication in HSCT patients, and methods to identify patients at high risk of CMV infection have been a challenge in the care of these individuals.6,28,29 To identify these patients, knowing the impact of this pathogen in the setting and implementing methodologies for early CMV detection is of utmost importance to improve prognosis of HSCT patients.
Author Contributions
G. Breda and B. Almeida contributed equally to this work; S. M. Raboni and S. M. Almeida designed the research; S. Carstensen, M. B. Nogueira, and L. R. Vidal contributed with analytic tools; S. M. Raboni analyzed the data and wrote the paper.
Partial results of these data were presented at the 2011 Bone Marrow Transplant Society Congress in Rio de Janeiro, Brazil.
Conflict of Interest
The authors declare no potential conflict of interest.
References
- 1.Boeckh M, Leisenring W, Riddell SR, Bowden RA, Huang ML, Myerson D, et al. Late cytomegalovirus disease and mortality in recipients of allogeneic hematopoietic stem cell transplants: importance of viral load and T-cell immunity. Blood. 2003;101:407–14 [DOI] [PubMed] [Google Scholar]
- 2.Freeman RB., Jr The ‘indirect’ effects of cytomegalovirus infection. Am J Transplant. 2009;9:2453–8 [DOI] [PubMed] [Google Scholar]
- 3.Gentile G, Picardi A, Capobianchi A, Spagnoli A, Cudillo L, Dentamaro T, et al. A prospective study comparing quantitative cytomegalovirus (CMV) polymerase chain reaction in plasma and pp65 antigenemia assay in monitoring patients after allogeneic stem cell transplantation. BMC Infect Dis. 2006;6:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varani S, Landini MP. Cytomegalovirus-induced immunopathology and its clinical consequences. Herpesviridae. 2011;2(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerna G, Lilleri D, Furione M, Baldanti F. Management of human cytomegalovirus infection in transplantation: validation of virologic cut-offs for preemptive therapy and immunological cut-offs for protection. New Microbiol. 2011;34:229–54 [PubMed] [Google Scholar]
- 6.Allice T, Cerutti F, Pittaluga F, Varetto S, Franchello A, Salizzoni M, et al. Evaluation of a novel real-time PCR system for cytomegalovirus DNA quantitation on whole blood and correlation with pp65-antigen test in guiding pre-emptive antiviral treatment. J Virol Methods. 2008;148(1–2):9–16 [DOI] [PubMed] [Google Scholar]
- 7.Cariani E, Pollara CP, Valloncini B, Perandin F, Bonfanti C, Manca N. Relationship between pp65 antigenemia levels and real-time quantitative DNA PCR for Human Cytomegalovirus (HCMV) management in immunocompromised patients. BMC Infect Dis. 2007;7:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan J, Ma WH, Yang MF, Xue H, Gao HN, Li LJ. Real-time fluorescent quantitative PCR assay for measuring cytomegalovirus DNA load in patients after haematopoietic stem cell transplantation. Chin Med J (Engl). 2006;119:871–4 [PubMed] [Google Scholar]
- 9.Kim DJ, Kim SJ, Park J, Choi GS, Lee S, Kwon CD, et al. Real time PCR assay compared with antigenemia assay for detecting cytomegalovirus infection in kidney transplant recipients. Transplant Proc. 2007;39:1458–60 [DOI] [PubMed] [Google Scholar]
- 10.Peres RM, Costa CR, Andrade PD, Bonon SH, Albuquerque DM, de Oliveira C, et al. Surveillance of active human cytomegalovirus infection in hematopoietic stem cell transplantation (HLA sibling identical donor): search for optimal cutoff value by real-time PCR. BMC Infect Dis. 2010;10:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ljungman P, Griffiths P, Payas C. Definitions of cytomegalovirus infection and disease in transplant recipients. Clin Infect Dis. 2002;34:1094–7 [DOI] [PubMed] [Google Scholar]
- 12.George B, Pati N, Gilroy N, Ratnamohan M, Huang G, Kerridge I, et al. Pre-transplant cytomegalovirus (CMV) serostatus remains the most important determinant of CMV reactivation after allogeneic hematopoietic stem cell transplantation in the era of surveillance and preemptive therapy. Transpl Infect Dis. 2010;12:322–9 [DOI] [PubMed] [Google Scholar]
- 13.Kotton CN, Kumar D, Caliendo AM, Asberg A, Chou S, Snydman DR, et al. ; Transplantation Society International CMV Consensus Group. International consensus guidelines on the management of cytomegalovirus in solid organ transplantation. Transplantation. 2010;89:779–95 [DOI] [PubMed] [Google Scholar]
- 14.Lilleri D, Lazzarotto T, Ghisetti V, Ravanini P, Capobianchi MR, Baldanti F, et al. ; SIV-AMCLI Transplant Surveillance. Multicenter quality control study for human cytomegalovirus DNAemia quantification. New Microbiol. 2009;32:245–53 [PubMed] [Google Scholar]
- 15.Garrigue I, Boucher S, Couzi L, Caumont A, Dromer C, Neau-Cransac M, et al. Whole blood real-time quantitative PCR for cytomegalovirus infection follow-up in transplant recipients. J Clin Virol. 2006;36:72–5 [DOI] [PubMed] [Google Scholar]
- 16.Smith TF, Paya CV. The clinical use of various blood compartments for cytomegalovirus (CMV) DNA quantitation in transplant recipients with CMV disease. Transplantation. 2002;73:968–73 [DOI] [PubMed] [Google Scholar]
- 17.von Müller L, Hinz J, Bommer M, Hampl W, Kluwick S, Wiedmann M, et al. CMV monitoring using blood cells and plasma: a comparison of apples with oranges? Bone Marrow Transplant. 2007;39:353–7 [DOI] [PubMed] [Google Scholar]
- 18.Caliendo AM, Ingersoll J, Fox-Canale AM, Pargman S, Bythwood T, Hayden MK, et al. Evaluation of real-time PCR laboratory developed test using analyte-specific reagents for cytomegalovirus quantification. J Clin Microbiol. 2007;45:1723–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalpoe JS, Kroes AC, de Jong MD, Schinkel J, de Brouwer CS, Beersma MF, et al. Validation of clinical application of cytomegalovirus plasma DNA load measurement and definition of treatment criteria by analysis of correlation to antigen detection. J Clin Microbiol. 2004;42:1498–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchetti S, Santangelo R, Manzara S, D’onghia S, Fadda G, Cattani P. Comparison of real-time PCR and pp65 antigen assays for monitoring the development of Cytomegalovirus disease in recipients of solid organ and bone marrow transplants. New Microbiol. 2011;34(2):157–64 [PubMed] [Google Scholar]
- 21.Regoes RR, Staprans SI, Feinberg MB, Bonhoeffer S. Contribution of peaks of virus load to simian immunodeficiency virus pathogenesis. J Virol. 2002;76:2573–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Regoes RR, Bowen EF, Cope AV, Gor D, Hassan-Walker AF, Prentice HG, et al. Modelling cytomegalovirus replication patterns in the human host: factors important for pathogenesis. Proc Biol Sci. 2006;273(1596):1961–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boeckh M, Ljungman P. How we treat cytomegalovirus in hematopoietic cell transplant recipients. Blood. 2009;113:5711–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halfon P, Berger P, Khiri H, Martineau A, Pénaranda G, Merlin M, et al. Algorithm based on CMV kinetics DNA viral load for preemptive therapy initiation after hematopoietic cell transplantation. J Med Virol. 2011;83:490–5 [DOI] [PubMed] [Google Scholar]
- 25.Kanda Y, Yamashita T, Mori T, Ito T, Tajika K, Mori S, et al. A randomized controlled trial of plasma real-time PCR and antigenemia assay for monitoring CMV infection after unrelated BMT. Bone Marrow Transplant. 2010;45:1325–32 [DOI] [PubMed] [Google Scholar]
- 26.Cardenoso L, Pinsky BA, Lautenschlager I, Aslam S, Cobb B, Vilchez R, et al. CMV antigenemia and quantitative viral load assessments in hematopoietic stem cell transplant recipients. J Clin Virol. 2013;56:108–12 [DOI] [PubMed] [Google Scholar]
- 27.Hirsche HH, Lautenschlager I, Pinsky BA, Cardenoso L, Aslam S, Cobb B, et al. An international multicenter performance analysis of cytomegalovirus load tests. Clin Infect Dis. 2013;56:367–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Razonable RR, Brown RA, Wilson J, Groettum C, Kremers W, Espy M, et al. The clinical use of various blood compartments for cytomegalovirus (CMV) DNA quantitation in transplant recipients with CMV disease. Transplantation. 2002;73(6):968–73 [DOI] [PubMed] [Google Scholar]
- 29.Razonable RR, Paya CV, Smith TF. Role of the laboratory in diagnosis and management of cytomegalovirus infection in hematopoietic stem cell and solid organ transplant recipients. J Clin Microbiol. 2002;40:746–52 [DOI] [PMC free article] [PubMed] [Google Scholar]


