Abstract
Protein motions underlie conformational and entropic contributions to enzyme catalysis, however relatively little is known about the ways in which this occurs. Studies of the mitogen-activated protein kinase, ERK2, by hydrogen-exchange mass spectrometry (HX-MS) suggest that activation enhances backbone flexibility at the linker between N- and C-terminal domains, while altering nucleotide binding mode. Here, we address the hypothesis that enhanced backbone flexibility within the hinge region facilitates kinase activation. We show that hinge mutations that enhance flexibility promote changes in the nucleotide binding mode consistent with domain movement, without requiring phosphorylation. They also lead to the activation of monophosphorylated ERK2, a form which is normally inactive. The hinge mutations bypass the need for pTyr but not pThr, suggesting that Tyr phosphorylation controls hinge motions. In agreement, monophosphorylation of pTyr enhances both hinge flexibility and nucleotide binding mode, measured by HX-MS. Our findings demonstrate that regulated protein motions underlie kinase activation. Our working model is that constraints to domain movement in ERK2 are overcome by phosphorylation at pTyr, which increases hinge dynamics to promote the active conformation of the catalytic site.
INTRODUCTION
The activation of MAP kinases is controlled by phosphorylation at Thr-Xxx-Tyr sequences within the activation loop, catalyzed by dual specificity MAP kinase kinases (MKKs). Phosphorylation of both Thr and Tyr residues is required, and negligible activation is seen with phosphorylation of either residue alone, or mutation of either or both residues to acidic amino acids. Solvent viscometric steady state rate measurements have shown that the mechanism of activation by phosphorylation is dominated by rate enhancement of steps involving phosphoryl group transfer (1).
X-ray structures of ERK2 in its inactive, unphosphorylated (0P) and active, dual phosphorylated (2P) forms provide important insights into the structural changes underlying ERK2 activation (2,3). Dual phosphorylation rearranges the activation loop from an inactive conformation which precludes substrate binding, to an active conformation which enables recognition of the Ser/Thr-Pro phosphorylation motif (3). In addition, ion pair interactions between pThr183 in the activation loop and Arg65 and Arg68 in helix αC enable communication between N- and C-terminal domains. Finally, activation loop rearrangement opens a high affinity binding site for a docking motif found in substrates and scaffold proteins (4,5).
Biophysical measurements suggest that ERK2 is also regulated at the level of protein dynamics. Hydrogen exchange mass spectrometry (HX-MS) revealed changes in hydrogen-deuterium exchange (HX) rates within localized regions of the kinase upon activation by phosphorylation (6). In particular, HX increases within residues LMETD109, which form the hinge region between N- and C-terminal domains. Structural differences between 0P- and 2P-ERK2 in this region are not obvious, suggesting that phosphorylation does not affect conformation, but instead alters conformational mobility. In accordance, site directed spin label-electron paramagnetic resonance spectroscopy measurements of ERK2 showed changes in correlation rates at the hinge upon ERK2 phosphorylation, without changes in the local environment (7). Together, these observations suggest that ERK2 activation modulates protein motions at the hinge.
Studies of protein kinases have shown the importance of domain movements for catalytic function. For example, in the catalytic (C) subunit of cAMP-dependent protein kinase (PKA), nucleotide and substrate binding elicits N- and C-terminal domain rotation to form a closed conformation (Fig. 1A) (8,9). By contrast, X-ray structures of both 0P- and 2P-ERK2 show open conformations, raising questions about how the necessary domain movements needed for closure could be achieved. One clue is that 0P- and 2P-ERK2 bind with similar affinities to the nucleotide analog, AMP-PNP, yet differ in the extent to which AMP-PNP binding protects from hydrogen exchange with solvent, measured by HX-MS (Fig. 1B). In particular, 2P-ERK2 shows a greater extent of HX protection by AMP-PNP binding within the Mg2+ positioning loop (DFG motif), located at the interface between N- and C-terminal domains (10). Thus, nucleotide has two binding modes which distinguish the 0P and 2P-kinase activity states.
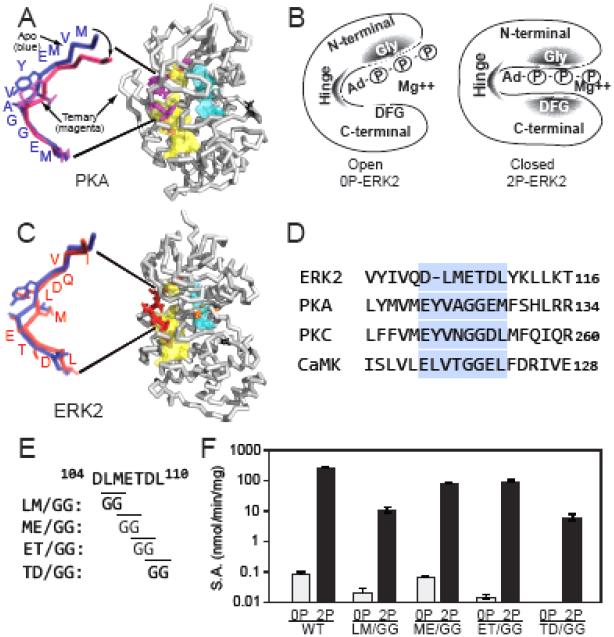
Fig. 1. Mutations modulating hinge flexibility in ERK2.
(A) X-ray structure of PKA, showing the closed domain ternary complex with bound MnATP and PKI inhibitor peptide (1ATP, with ligands removed). The hydrophobic R and C spines are shown in blue and yellow, respectively. The expanded view overlays the hinge regions of PKA in the ternary complex (magenta) with the open domain apo form (1J3H, blue), showing the angle change between closed and open conformations. (B) Summary of previous findings (10). Left: In 0P-ERK2, shaded regions highlight the N-terminal Gly-rich loop and hinge region, where AMP-PNP binding reduces HX, reflecting steric protection from solvent in the nucleotide binding pocket. Right: 2P-ERK2 shows the same degree of HX protection in the Gly loop and hinge region, but HX protection in the DFG motif is 10-fold higher in 2P-ERK2 than 0P-ERK2. These patterns suggest domain movements that distinguish 0P-ERK2 and 2P-ERK2. They are illustrated as open and closed conformations, although other structural models are possible. (C) X-ray structure of ERK2 (1ERK) showing the hinge region (red) and conserved DFG motif (orange). M106 in the hinge interacts with residues in the C spine of ERK2. The expanded view overlays the hinge regions of apo-PKA (blue) and ERK2 (red), both which correspond to open domain conformations. (D) Sequence alignment of hinge regions in ERK2, PKA, protein kinase C (PKC) and Ca2+-calmodulin protein kinase II (CaMKII). PKA, PKC and CaMKII show Gly-Gly residues at the hinge and are active as kinase domains. (E) In ERK2, Gly-Gly mutations were placed at positions along the hinge where HX was observed to increase in response to activation loop phosphorylation and activation. (F) Specific activities (±s.d., n=3) of wild-type and hinge mutants of ERK2 show significant activity of each mutant after dual phosphorylation.
Recently, protein dynamics in ERK2 were analyzed by Carr-Purcell-Meiboom-Gill (CPMG) NMR relaxation dispersion experiments, measuring exchange between conformational states in Ile, Val and Leu side chain methyl groups (11). In 0P-ERK2, relaxation dispersion measurements reported fast conformational exchange processes (e.g., A ⇌ B interconversion) in Ile/Leu/Val residues, with little or no evidence for coupling between these residues. However, in 2P-ERK2, residues throughout the kinase core could be fit globally, consistent with a single exchange process with rate constant kex = kAB + kBA = 300 s−1 and pA and pB populations of 20% and 80%, respectively. The residues appeared throughout the N-terminal and C-terminal domains, except for the MAP kinase insert. Thus, changes in dynamics accompany phosphorylation and activation of ERK2, where internal motions become dominated by a slow-exchange process characterized by interconversion between two major conformational states. The fact that the global process involves residues throughout the N-terminal and C-terminal domains suggests that the underlying motion involves domain movements. This is consistent with phosphorylation-induced changes in the interactions between Mg2+-AMP-PNP and the DFG motif, although other structural models are possible. Taken together, the findings imply that in solution, 0P-ERK2 is constrained from hinge motions, which in turn interferes with domain movement. Phosphorylation to form 2P-ERK2 bypasses these constraints, allowing optimized configuration of the catalytic site.
Here, we address the hypothesis that regulation of hinge motions in ERK2 is relevant to the mechanism of kinase activation by phosphorylation. We show that introducing hinge mutations which increase backbone flexibility allows adoption of the active state nucleotide binding mode, without activation loop phosphorylation. Monophosphorylation of ERK2 at Tyr185 is sufficient to increase HX at the hinge, and also reduces HX in the Mg2+ positioning DFG motif upon AMP-PNP binding, suggesting that pTyr phosphorylation enhances hinge flexibility and domain movements needed for active state nucleotide binding. The hinge mutations promote activation of a normally inactive monophosphorylated form of ERK2, where pThr is phosphorylated and Tyr is mutated to Glu. Thus, mutations which enhance hinge flexibility bypass the need for pTyr phosphorylation, arguing that that hinge flexibility is coupled to activation. We propose that Tyr phosphorylation has the distinct role of controlling hinge dynamics in ERK2 in order to promote the switch in nucleotide binding and/or domain movement, events that are crucial for kinase activation.
RESULTS
Hinge sequences in ERK2 and other protein kinases
In previous studies, AMP-PNP binding led to steric protection from HX within the N-terminal domain and hinge regions, to levels that were comparable between 0P-ERK2 and 2P-ERK2 (10). In contrast, greater protection from HX was observed in 2P-ERK2 compared to 0P-ERK2 within the conserved DFG motif, which is located within the C-terminal domain where it forms a coordination site for nucleotide-bound Mg2+. A model to explain the HX patterns proposed domain movements leading to differing interdomain conformations for 0P-ERK2 and 2P-ERK2, respectively (Fig. 1B). Interconversion between open and closed conformations would be consistent with these movements, although other models are possible. We hypothesized that constraints at the hinge region prevents domain movement in 0P-ERK2, whereas activation loop phosphorylation releases these constraints and increases hinge backbone flexibility in 2P-ERK2.
X-ray structures of PKA C-subunit show the apo enzyme in an open conformation, where the N- and C-terminal lobes are too far apart to enable catalysis (8). In the ternary complex with nucleotide and substrate, the structure undergoes domain closure to a closed conformation, involving hinge bending and sideways rotation about the lobes (9, Fig. 1A). Based on NMR and molecular dynamics, this has been attributed to underlying motions of the Gly loop and ATP binding pocket and requires integrity of “hydrophobic spine” architecture conserved throughout protein kinases (12-14). Structural and sequence comparisons show that the hinge residues in ERK2 where altered conformational mobility was observed by HX-MS (METDL110; 6) align with the sequence Gly-Gly within the PKA hinge (Fig. 1C,D). The Gly-Gly sequence is also found in protein kinase C (PKC) and Ca2+-calmodulin-dependent protein kinase II (CaMKII) (Fig. 1D), which like PKA are constitutively active as isolated kinase domains. This suggests that kinases with Gly-Gly hinge sequences may have lowered constraints to domain movement.
In order to examine the effect of hinge flexibility on domain interactions in ERK2, Gly-Gly substitutions were introduced into the LMETD109 sequence, yielding mutants LM/GG, ME/GG, ET/GG, and TD/GG (Fig. 1E), each produced in unphosphorylated and dual phosphorylated forms. Initial rate measurements showed that the hinge mutations had little effect on ERK2 activity when unphosphorylated (Fig. 1F). After dual phosphorylation, each mutant showed lower activity than wild-type kinase, but still allowed significant activation by phosphorylation (Fig. 1F). Thus, the specific activities of the dual phosphorylated forms served as benchmarks for full activation of each hinge mutant.
HX-MS of wild-type and mutant ERK2
HX-MS was used to analyze the four hinge mutants, each in their unphosphorylated forms. Thirty-two peptides were identified in ERK2 after pepsin digestion, covering 91% of residues and 87% of exchangeable amides (Fig. S1A, Table S1). Cleavages in the hinge region varied between the mutants due to the Gly-Gly substitutions (Fig. S1B). In each mutant, the cleavage sites in the activation loop yielded peptides identical to those in wild type (WT) 0P-ERK2, which became protected from cleavage in WT 2P-ERK2, as previously observed (6, 10). The cleavage sites in the rest of the protein sequence were identical among all ERK2 forms.
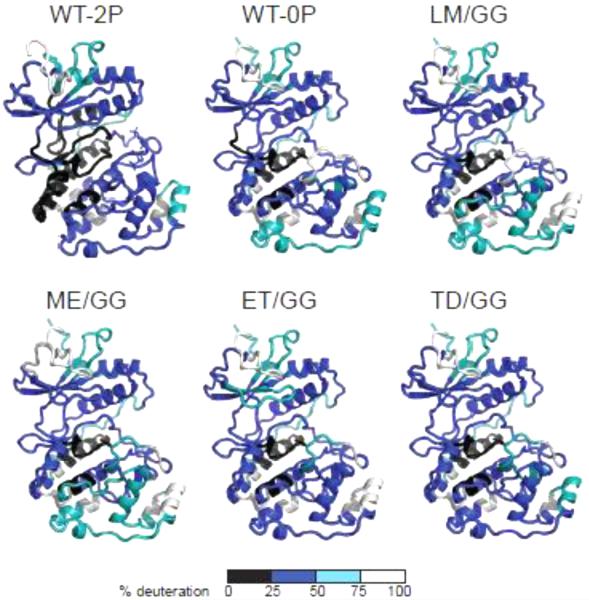
HX patterns among the four hinge mutants were similar to those of wild-type (WT) 0P- and 2P-ERK2 (Fig. 2), and consistent with the extent of solvent accessibility suggested by the WT X-ray structures. Solvent-exposed loop regions showed high rates of exchange, and N- and C-terminal core regions showed slower exchange, indicating that each mutant adopted a solution conformation similar to that of WT enzyme. The Gly-Gly substitutions enhanced HX in the hinge region (Fig. S1B), consistent with increased backbone flexibility expected from the reduction in side chain size.
Fig. 2. ERK2 mutants show comparable solvent accessibility measured by HX-MS.
The extent of deuteration measured after 4 h in D2O, mapped against X-ray structures of 0P- or 2P-ERK2 (both forms apo). Colors distinguish peptides deuterated to 0-25% (black), 26-50% (blue), 51-75% (teal), and 76-100% (white) of the total number of exchangeable amides.
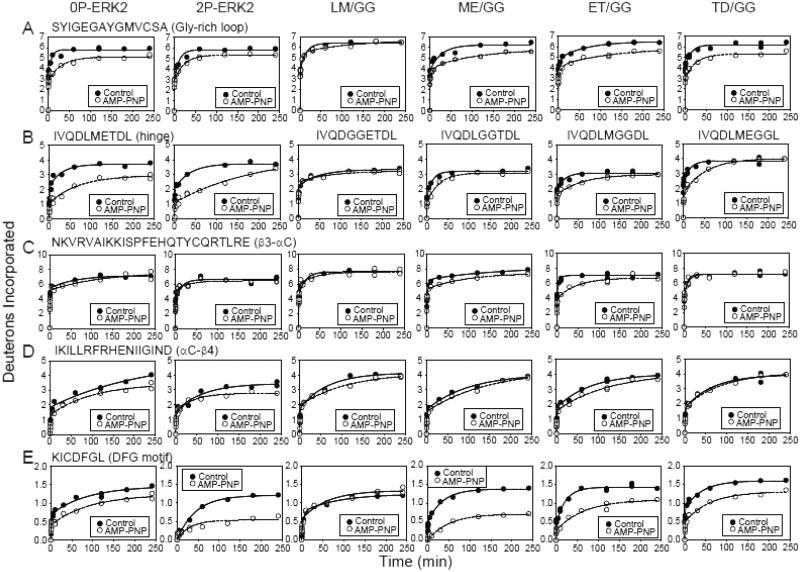
HX-MS was then performed on each ERK2 form, preincubated in the presence vs absence of AMP-PNP (Fig. 3, Table S2). In both 0P-ERK2 and 2P-ERK2, AMP-PNP binding sterically interfered with HX in peptides containing residues known to contact nucleotide, as reported previously (10). For example, HX protection was observed within the Gly-rich loop (SYIGEGAYGMVCSA, Fig. 3A), which forms backbone hydrogen bond interactions with Pβ-Pγ groups on ATP; the hinge region (IVQDLMETDL, Fig. 3B), which hydrogen bonds with nucleotide base and sugar; and strand β3-helix αC-strand β4 (NKVRVAIKKISPFEHQTYCQRTLRE and IKILLRFRHENIIGIND, Figs. 3C,D), which together stabilize nucleotide binding through conserved Lys-Glu ion pair interactions with Pα-Pβ. In contrast, HX protection in the C-terminal DFG motif (KICDFGL, Fig. 3E) increased in 2P-ERK2 over 0P-ERK2, reflecting greater interdomain movement in activated WT-ERK2.
Fig. 3. HX protection in ERK2 upon AMP-PNP binding.
HX time courses comparing WT and mutant ERK2 in the presence (○) or absence (●) of AMP-PNP. All mutants except LM/GG showed HX protection in regions previously found to interact with nucleotide. (A) Peptide SYIGEGAYGMVCSA comprises the Gly-rich loop, where AMP-PNP binding protects against HX by >1 Da. (B) Peptide IVQDLMETDL contains the hinge region where AMP-PNP binding yields lower HX protection in the hinge mutants than WT-ERK2. (C,D) Peptides NKVRVAIKKISPFEHQTYCQRTLRE and IKILLRFRHENIIGIND comprise the β3-αC-β4 region, which coordinates Mg2+-ATP. HX protection occurs within αC-β4 in WT-ERK2 and within β3-αC in mutants ME/GG- and ET/GG-ERK2. (E) Peptide KICDFGL corresponds to the DFG motif in the C-terminal domain. HX protection by nucleotide binding is greater in 2P-ERK2 and in mutants ME/GG- and ET-GG-ERK2 compared to 0P-ERK2. The results suggest that ME/GG, and to a lesser extent ET/GG, promote interdomain interactions in ERK2, even without activation loop phosphorylation.
Differing patterns of HX protection by AMP-PNP were observed in the hinge mutants ME/GG-, ET/GG- and TD/GG-ERK2, each in their unphosphorylated form. All three mutants showed protection of the Gly-rich loop by nucleotide, comparable to that of 0P- and 2P-WT-ERK2 (Fig. 3A). In the hinge, protection from HX by AMP-PNP binding was reduced compared to WT-ERK2 (Fig. 3B), consistent with disrupted nucleotide interactions due to the Gly-Gly mutations. Steric protection was observed within the β3-αC region in mutants ME/GG- and ET/GG-ERK2 (Fig. 3C). Mutant LM/GG-ERK2 showed little protection by AMP-PNP, suggesting overall reduced binding affinity, as corroborated below.
The most striking differences between WT and mutant forms of ERK2 were observed in the Mg2+ positioning DFG motif (Fig. 3E). Here, ME/GG-ERK2 displayed higher protection of HX upon nucleotide binding than 0P-ERK2, to a degree comparable to that of 2P-ERK2. ET/GG-ERK2 also showed higher protection of HX, although to a lesser extent than ME/GG-ERK2, while TD/GG-ERK2 showed comparable protection to 0P-WT-ERK2. The behavior suggested that the ME/GG-ERK2 mutant (and to a lesser extent, ET/GG) adopts a nucleotide binding mode resembling 2P-ERK2, even when its activation loop is unphosphorylated. Thus, hinge residue mutations that would be expected to enhance backbone flexibility led to changes in nucleotide binding mode, as measured by protection of HX. This was consistent with a mechanism involving the control of domain movements by modulating hinge flexibility. The differing behavior between ME/GG- and TD/GG-ERK2 reveals site specificity in the placement of the Gly residues, evidence that regulation of hinge flexibility within the Met-Glu-Thr sequence is most critical for overcoming the constraint to dynamics in 0P-ERK2.
Because the hinge region in protein kinases contacts nucleotide, we measured the affinity of each ERK2 mutant for AMP-PNP using isothermal titration calorimetry. The Kd for AMP-PNP binding to LM/GG-ERK2 exceeded 1 mM (Table 1, Fig. S2), consistent with HX-MS measurements indicating little or no protection by 1 mM nucleotide (Fig. 3C). The Kd for AMP-PNP binding to ME/GG-, ET/GG- and TD/GG-ERK2 ranged between 120-860 μM, comparable to 0P- and 2P-WT-ERK2. Importantly, although ME/GG-ERK2 showed the greatest HX protection of the DFG region by AMP-PNP binding, it showed the weakest binding affinity for AMP-PNP (Fig. S2, Table 1); therefore, the higher HX protection in this mutant (Fig. 3E) was not due to higher binding site occupancy. Instead, the HX protection patterns of ME/GG- and ET/GG-ERK2 were consistent with altered nucleotide binding mode and/or domain movements.
Table 1.
Thermodynamics of AMP-PNP binding to ERK2.
| ERK2 form | Kd ± s.d. (mM) |
ΔG° ± s.d. (kcal/mol) |
ΔH° ± s.d. (kcal/mol) |
ΔS (kcal/mol/deg) |
% Bound |
|---|---|---|---|---|---|
| WT-0P | 0.20 ± 0.01 | −5.12 ± 0.04 | −6.09 ± 0.13 | −0.003 | 83 |
| WT-2P | 0.42 ± 0.06 | −4.68 ± 0.09 | −3.23 ± 0.14 | 0.005 | 71 |
| LM/GG | >1 | N.D. | 0 | ||
| ME/GG | 0.86 ± 0.02 | −4.25 ± 0.02 | −4.84 ± 0.23 | −0.002 | 54 |
| ET/GG | 0.37 ± 0.05 | −4.75 ± 0.08 | −5.08 ± 0.13 | −0.001 | 73 |
| TD/GG | 0.12 ± 0.02 | −5.44 ± 0.09 | −3.44 ± 0.17 | 0.006 | 89 |
Domain movement is mediated by ERK2 monophosphorylation
We asked whether domain movements in ERK2 were associated with monophosphorylation at either pThr or pTyr. Activation loop mutants were constructed by substituting the wild-type TEY sequence with AEY, EEY, TEF, and TEE, and then reacted with active MKK1 to generate the monophosphorylated forms, AEpY, EEpY, pTEF and pTEE. Each monophosphorylated form showed low catalytic activity (Fig. S3A) as expected, given that ERK2 activation normally requires dual phosphorylation at both pThr or pTyr residues.
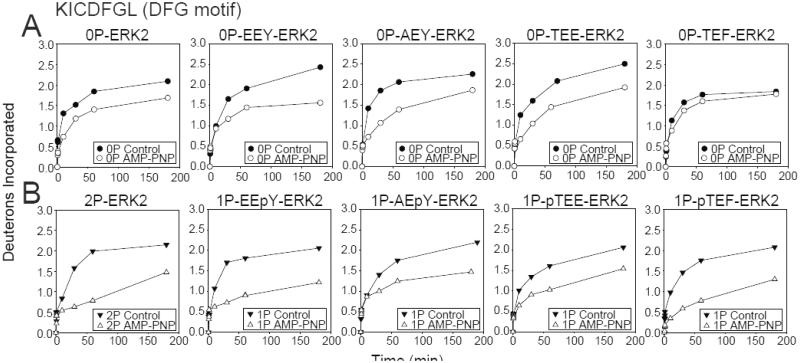
Each form was examined by HX-MS. Unphosphorylated, all mutant forms of ERK2 showed HX protection by AMP-PNP in the DFG motif to a degree comparable to or less than that of 0P-ERK2 (Fig. 4A), suggesting that each adopted an inactive state conformation. In contrast, the HX protection of DFG by nucleotide was higher for monophosphorylated mutant EEpY-ERK2 than 0P-ERK2, and comparable to that of 2P-ERK2 (Fig. 4B), suggesting that pTyr phosphorylation promotes the nucleotide binding mode characteristic of the active state. AEpY-ERK2 and pTEE-ERK2 both showed lower HX protection than EEpY-ERK2 and pTEF-ERK2, respectively. Interestingly, HX protection of pTEF-ERK2 was comparable to that of 2P-ERK2. Therefore, monophosphorylation at either Tyr185 or Thr183 enhanced HX protection of the DFG motif by nucleotide, but the characteristics differed, where Tyr185 phosphorylation enhanced HX protection only when Thr183 was mutated to Glu, whereas Thr183 phosphorylation enhanced HX protection in a manner that was suppressed when Tyr185 was mutated to Glu.
Fig. 4. Monophosphorylation of ERK2 allows domain closure.
Time courses for WT and activation loop mutants, EEY-, AEY-, TEF- and TEE-ERK2, in (A) unphosphorylated and (B) monophosphorylated states. EEpY- and pTEF-ERK2 show significant HX protection by AMP-PNP in the DFG motif to a level comparable with 2P-ERK2, consistent with the active state nucleotide binding mode and domain movement. In contrast, AEpY- and pTEE-ERK2 more closely resemble 0P-ERK2, consistent with the inactive state nucleotide binding mode. Only EEpY-ERK2 enhanced HX within the hinge region, consistent with increased hinge flexibility (see Fig. S4).
pThr and pTyr phosphorylation differentially affected hinge conformational mobility, as measured by HX of hinge peptide METDL110 (carried out in absence of nucleotide; Fig. S4). Only EEpY-ERK2 enhanced HX at the hinge in a manner similar to 2P- ERK2, while pTEF-ERK2 and all other forms reduced HX at the hinge. This suggests that EEpY-ERK2 coordinately increased hinge flexibility as measured by HX at the hinge in the apo form, and also increased domain movement, as measured by HX protection of the DFG motif upon AMP-PNP binding. In contrast, pTEF-ERK2 reduced hinge flexibility in a manner inconsistent with its effect on HX protection of the DFG motif, suggesting that the latter reflects alternate mechanisms, e.g. ion pairing between pThr and helix αC.
Hinge mutations activate ERK2
Finally we asked whether hinge flexibility contributes to the mechanism of kinase activation by phosphorylation. If this were true, the hinge mutations that enhanced flexibility should bypass constraints that restrict ERK2 activation, even without dual phosphorylation. Therefore, ME/GG, ET/GG and TD/GG mutations were combined with pT183- or pY185- monophosphorylated activation loop mutants and assayed for catalytic activity.
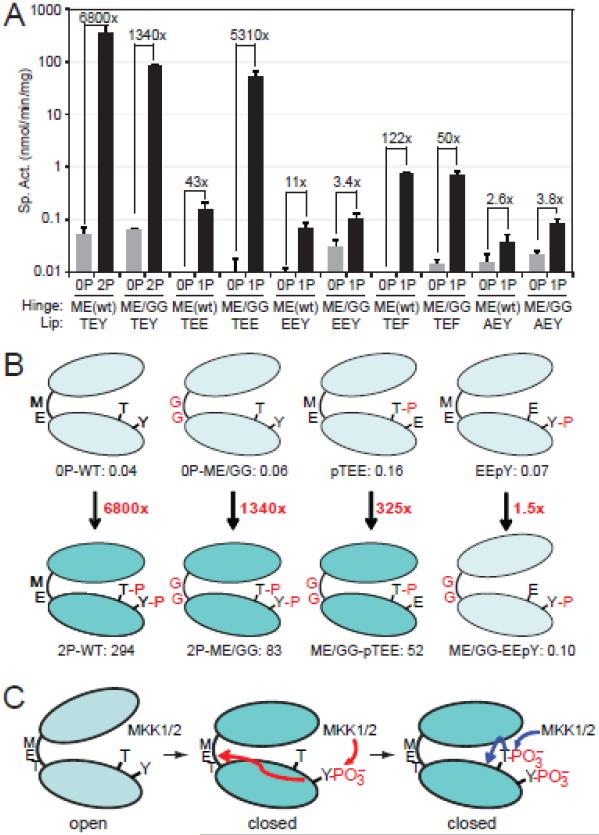
The results are shown in Fig. 5A,B. ME/GG-ERK2 and pTEE-ERK2 showed specific activities (0.06 and 0.16 nmol/min/mg, respectively) comparable to the activity of 0P-WT-ERK2 (0.05 nmol/min/mg). However, the combination of ME/GG + pTEE substantially enhanced ERK2 activity, increasing to 52 nmol/min/mg. This was comparable to the activity of dual phosphorylated (pTEpY) ME/GG-ERK2 (83 nmol/min/mg), our benchmark for full activation. This activity was 5,310-fold higher than the same mutant in its unphosphorylated form, and 325-fold higher than pTEE-ERK2 without the hinge mutation. Thus, synergy between ME/GG and pTEE led to significant activation of monophosphorylated ERK2, to a level comparable to that of the dual phosphorylated form.
Fig. 5. Hinge mutations activate ERK2 monophosphorylated at Thr183.
(A) Specific activities of WT- and ME/GG-ERK2, harboring activation loop mutations TEE, EEY, TEF, and AEY. Mutants were assayed in their unphosphorylated (−) forms or following phosphorylation by active MKK1 (+). Bars show average ± s.d. of triplicate measurements. Significant kinase activation is observed in ME/GG-pTEE-ERK2, compared to monophosphorylated pTEE-ERK2 or unphosphorylated ME/GG-ERK2. Data for other mutant forms are shown in Fig. S3. (B) Summary showing ERK2 activation by ME/GG in combination with pTEE but not EEpY. (C) Model for ERK2 activation by regulated dynamics. Prior to phosphorylation, ERK2 is constrained from domain movement and adopts an inactive solution conformation. Phosphorylation at Tyr185 by MKK1 enhances backbone flexibility in the hinge region, allowing domain movement and adoption of interactions needed to optimize the catalytic site. Phosphorylation at Thr183 stabilizes the activation loop conformation and modulates other events needed for catalytic site configuration.
The requirement for monophosphorylation was specific for pT183, because ME/GG did not significantly enhance the activity of EEpY-ERK2 or AEpY-ERK2 (Fig. 5A,B). Therefore, modulating hinge flexibility bypassed the need for pY185, but not pT183. This suggests that phosphorylation of Tyr185, not Thr183, regulates domain movement by controlling hinge flexibility. ME/GG failed to activate pTEF-ERK2, suggesting that kinase activation still requires a negatively charged residue at position 185. Activation was not observed with hinge mutants LM/GG- or TD/GG-ERK2 or with activation loop mutants TED- or DEY-ERK2, while mutants EEE- and DED-ERK2 completely inactivated the kinase with no further effect of the hinge mutations (Fig. S3B,C). Thus, phosphate occupancy at Thr183 was required for activation. Together, our results indicate that Tyr185 phosphorylation controls domain movement, while Thr183 phosphorylation controls catalytic site reorganization and/or other steps in activation.
DISCUSSION
Our study shows that in ERK2, kinase activation is linked to the regulation of protein dynamics by phosphorylation. By introducing Gly mutations at the hinge between N- and C-terminal domains, ERK2 in its unphosphorylated form bypasses constraints which normally interfere with activation. These mutations enable ERK2 to adopt a conformation that is consistent with the active form, leading to elevated activity upon monophosphorylation at Thr183, which is normally insufficient for activation. It is reasonable to expect that Gly mutations should increase backbone flexibility, due to the enhanced degrees of freedom when bulky hydrophobic side chains are removed. In particular, M106 forms hydrophobic interactions with residues in the proposed “C-spine” of ERK2. These would be disrupted in the ME/GG mutant, thus, the effects of mutation on domain movement might be attributed to changes in conformation as well as conformational mobility. However, conformational differences around M106 are not apparent between the X-ray structures of wild-type 0P and 2P-ERK2. Taken together, our results argue that the mechanism of ERK2 activation by dual phosphorylation involves altered dynamics at the hinge, allowing the enzyme to shift populations from inactive to active conformations (Fig. 5C). One way to envision these motions involves interdomain movements between open and closed conformers, although other models are possible. These are consistent with the global exchange process induced by phosphorylation that we observed by NMR (11).
Importantly, kinase activation occurred when the hinge mutants were combined with monophosphorylated Thr183, but not monophosphorylated Tyr185. Nonequivalence towards activation of the two phosphorylation sites was previously suggested by steady state kinetic measurements, where higher activity was shown in AEpY compared to pTEF (15). In X-ray studies of 2P-ERK2, pT183 makes extensive ion pair and hydrogen bonding contacts with Arg residues located in the N-terminal helix C, as well as the catalytic site, the activation loop, and the C-terminal L16 extension (3), thus forming contacts with the N-terminal domain and influencing the orientation of residues needed for catalysis. In contrast, pY185 rotates from a buried position to a solvent accessible position, where it forms ion pair interactions with Arg residues to form the P+1 recognition site, providing contacts and an extended surface for trans-proline selectivity. Therefore, we initially expected that pT183 would play a greater role in modulating domain movement through its interactions with the N-terminal domain. The fact that the hinge mutations circumvented the need for pY185, but not pT183, reveals a role for Y185 phosphorylation in domain movement that was not obvious from the X-ray structure.
It was also intriguing that activation was observed when combining ME/GG with monophosphorylated activation loop sequence pTEE, but not pTEF or pTED. This implies that combinatorial activation does not rely on pThr alone, but instead requires Glu at residue 185, perhaps to accommodate a specific geometry for ion pair interactions. In X-ray studies, replacing Y185 with Glu destabilized the activation loop, causing extensive disorder within the loop and P+1 site and yielding an inactive enzyme (16). We speculate that the ME/GG hinge mutation reduces this disorder, by stabilizing the activation loop via domain movements. Overall, our findings expand our understanding of how each phosphorylation site in ERK2 controls distinct intramolecular processes involved in kinase activation, by showing that in addition to proximal interactions with substrate, pY185 affects allosteric transitions at the hinge region, located ~20 Å from the sites of phosphorylation.
How these allosteric transitions are controlled is a fascinating question. Sequence alignments (Fig. 1D) show Gly residues at the hinge in several protein kinases. In PKA, the hinge Gly-Gly residues correspond to the pivot point for domain rotation between apo and ternary forms, and structurally align with the Met-Glu-Thr residues in ERK2. Importantly, only ME/GG and ET/GG mutations favored domain movement suggested by the HX data, while TD/GG did not. This suggests that the region in the hinge most important for the constraint to activation involves just a few residues. Substituting Gly into this very localized region in ERK2 might enhance the number of preorganized conformers available for sampling the active conformation, which cannot be mimicked by perturbing flexibility at adjacent positions.
Like ERK2, the importance of hinge motions for kinase activation and allosteric communication between the activation loop and hinge has been suggested for other protein kinases, including FGFR2, ZAP-70, and EGFR (17-19). However, in ERK1, which shares the same hinge sequence with ERK2, both unphosphorylated and phosphorylated forms show HX protection by AMP-PNP characteristic of the nucleotide binding mode corresponding to the active state of ERK2 (20). Therefore, the hinge sequence alone is not sufficient to predict whether linker dynamics limit activation. Instead, motions at side chains that contact the Met-Glu-Thr residues might also be important for controlling conformational mobility at the hinge. Such motions in ERK2 presumably originate from the phosphorylation of Tyr185, and may be propagated over long distances. We speculate that an intramolecular pathway of side chain and backbone connectivities might transmit information from pY183 to the backbone flexibility at the hinge, allowing conformational selection of closed structures. Targeting the hinge region for control of catalytic activity may provide an effective way to develop more selective allosteric inhibitors for kinases such as ERK2.
MATERIALS AND METHODS
Protein expression and purification
Rat ERK2 mutants containing Gly-Gly substitutions at hinge residues L105-M106 (LM/GG), M106-E107 (ME/GG), E107-T108 (ET/GG), and T108-D109 (TD/GG), and mutants where activation loop residues Thr183 and/or Tyr185 were substituted with Glu, Asp, Ala or Phe were constructed in NpT7-5 His6-ratERK2. Proteins were expressed in E. coli-BL21(DE3)pLysS by induction with isopropyl β-D-1-thiogalactopyranoside (IPTG, 0.4 mM), and purified with Ni2+-NTA affinity resin (Qiagen, Valencia, CA) followed by MonoQ FPLC as described (10,21). Active dual phosphorylated WT-ERK2 (2P-ERK2) was expressed using the plasmid NpT7-5 MKK1R4F-His6-ratERK2 (22,23). Other ERK2 phosphoforms were generated by in vitro phosphorylation with active mutant MKK1 (G7B:ΔN4/S218D/M219D/N221D/S222D) as described (24).
Proteins used in HX-MS experiments were dialyzed into 10 mM potassium phosphate pH 7.5, 50 mM KCl, 1 mM dithiothreitol (DTT), and stored in aliquots at -80°C. LC-MS showed that >95% of 2P-ERK2 was dual phosphorylated at Thr183 and Tyr185, between 92-100% of different 1P-ERK2 forms were monophosphorylated, and >95% of 0P-ERK2 was completely unphosphorylated. For kinase activity measurements, ERK2 proteins were purified by Ni2+-NTA, and dialyzed into 10 mM HEPES, pH 7.5, 0.1 M NaCl, 1 mM DTT.
Hydrogen-exchange mass spectrometry
Proteins were analyzed by HX-MS following pepsin proteolysis as described (10,25). ERK2 hinge mutants (5 μg) were incubated in 85% (v/v) D2O and HX data collection performed using a QStar Pulsar QqTOF mass spectrometer interfaced with an Agilent Cap1100 HPLC (POROS R1 20, 500 μm i.d. × 10 cm) (10,25). ERK2 activation loop mutants (3 μg) were incubated in 85% (v/v) D2O and HX data collection performed using a Waters Synapt G2 mass spectrometer interfaced with a Waters Acquity UPLC (POROS R1 20, 500 μm i.d. × 10 cm). Peptides were identified by LC-MS/MS performed on the QStar generating peaklists using Analyst QS 2.0 software, and on an LTQ Orbitrap instrument generating peaklists using readw 4.3.1 software (25). Peptides were identified using Mascot v.1.9 with “no enzyme” specified and Mowse ≥ 30, and all assignments were validated by manual analysis (Table S1). Mass tolerances were 2.5 Da (parent) and 1.2 Da (fragment) for QStar datasets, and 1.2 Da (parent) and 0.8 Da (fragment) for LTQ-Orbitrap datasets. Time-zero measurements of background in-exchange during proteolysis were made by acidifying the reaction before D2O. Three replicate runs were performed after 1 min deuteration for each ERK2 form, in order to estimate the standard error of peptide deuteration (0.12 Da).
Data analysis and weighted average mass (WAM) calculations were performed as described (25), correcting deuteration measurements for background in-exchange and back-exchange (Table S2). Time courses were fit by nonlinear least squares (NLSQ) to the equation Y = N − Ae −k1t − Be −k2t − Ce −k3t, where Y equals the number of deuterons exchanged at time t; A, B, and C are the number of backbone amides exchanging with rate constants k1, k2, and k3; and N equals the maximal deuteration over the experimental period (N = A + B + C). Subtracting N from the total number of exchangeable backbone amides yields NE, the number of amides which are non-exchanging during the experimental period (26-28). Curve fitting was performed using SigmaPlot 9.0 (Systat Software, Inc.).
Protein kinase assays
Initial rates were measured by phosphorylation of bovine myelin basic protein (MBP, Sigma, St. Louis, MO). In experiments comparing phosphorylated to unphosphorylated proteins, ERK2 (12 μg) was preincubated with or without active mutant MKK1 (1 μg) (MKK1-ΔN4/S218D/M219D/N221D/S222D, ref. 24) for 2 h at 30°C in 20 mM HEPES pH 7.4, 20 mM MgCl2, 4 mM ATP, 4 mM DTT (70 μL final). Reactions were initiated by adding 25 mM Hepes, 25 mM MgCl2, 10 mM DTT, 1 mM ATP, and 2.5 μCi [γ-32P] ATP to a mixture of 1-100 ng ERK2 and 2 μg MBP (25 μL final), and incubating for 15-60 min at 30°C corresponding to initial rate conditions. Proteins were separated by SDS-PAGE, and activities quantified by phosphoimager analysis of MBP phosphorylation (Typhoon 9400, GE Healthcare, Piscataway, NJ).
Isothermal titration calorimetry
ERK2 proteins (100 μM) were titrated with AMP-PNP (1.37 μL, 5 mM, 28 injections) at 30 °C in 50 mM Tris pH 7.5, 150 mM NaCl, 5 mM MgSO4, 2% (v/v) glycerol, 0.1% (w/v) EDTA using an ITC200 calorimeter (MicroCal, Piscataway, NJ). Blank runs without ERK2 were subtracted from experimental runs. Thermograms were integrated and Kd and thermodynamic parameters were estimated using Origin 7.0 software.
Supplementary Material
▶ Regulated protein motions contribute to the activation of the MAP kinase, ERK2.
▶ Enhanced backbone flexibility within the hinge region accompanies ERK2 activation.
▶ Mutations which enhance backbone flexibility within the hinge region mimic domain movements observed upon activation lip phosphorylation.
▶ Mutations which enhance backbone flexibility also facilitate kinase activation without dual phosphorylation.
ACKNOWLEDGEMENTS
We are indebted to Thomas Lee and Adam Ring for many valuable discussions. This work was supported by NIH grant R01GM074134 (NGA).
ABBREVIATIONS
- HX-MS
hydrogen exchange mass spectrometry
- ERK2
extracellular-regulated protein kinase 2
- 0P-ERK2
unphosphorylated ERK2
- 2P-ERK2
dual phosphorylated ERK2
- PKA
cAMP-dependent protein kinase
- PKC
protein kinase C
- MBP
myelin basic protein
- UPLC
ultra-high pressure liquid chromatography
- ITC
isothermal titration calorimetry
- WAM
weighted average mass
- QqTOF
quadrupole time-of-flight
- DTT
dithiothreitol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Prowse NC, Lew J. Mechanism of ERK2 activation by dual phosphorylation. J. Biol. Chem. 2001;276:99–103. doi: 10.1074/jbc.M008137200. [DOI] [PubMed] [Google Scholar]
- 2.Zhang F, Strand A, Robbins D, Cobb MH, Goldsmith EJ. Atomic structure of the MAP kinase ERK2 at 2.3 Å resolution. Nature. 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- 3.Canagarajah BJ, Khokhlatchev A, Cobb MH, Goldsmith EJ. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90:859–869. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- 4.Lee T, Hoofnagle AN, Kabuyama Y, Stroud J, Min X, Goldsmith EJ, Chen L, Resing KA, Ahn NG. Docking motif interactions in MAP kinases revealed by hydrogen exchange mass spectrometry. Molecular Cell. 2004;14:43–55. doi: 10.1016/s1097-2765(04)00161-3. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs D, Glossip D, Xing H, Muslin AJ, Kornfeld K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 1999;13:163–75. [PMC free article] [PubMed] [Google Scholar]
- 6.Hoofnagle AN, Resing KA, Goldsmith EJ, Ahn NG. Changes in protein conformational mobility upon activation of extracellular regulated protein kinase-2 as detected by hydrogen exchange. Proc. Natl. Acad. Sci. USA. 2001;98:956–961. doi: 10.1073/pnas.98.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoofnagle AH, Stoner JW, Lee T, Eaton SS, Ahn NG. Phosphorylation-dependent changes in structure and dynamics in ERK2 detected by SDSL and EPR. Biophys. J. 2004;86:395–403. doi: 10.1016/S0006-3495(04)74115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akamine P, Madhusudan, Wu J, Xuong NH, Ten Eyck LF, Taylor SS. Dynamic features of cAMP-dependent protein kinase revealed by apoenzyme crystal structure. J. Mol. Biol. 2003;327:159–171. doi: 10.1016/s0022-2836(02)01446-8. [DOI] [PubMed] [Google Scholar]
- 9.Zheng J, Trafny EA, Knighton DR, Xuong NH, Taylor SS, Ten Eyck LF, Sowadski JM. 2.2 Å refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr. Sect. D. 1993;49:362–365. doi: 10.1107/S0907444993000423. [DOI] [PubMed] [Google Scholar]
- 10.Lee T, Hoofnagle AN, Resing KA, Ahn NG. Hydrogen exchange solvent protection by an ATP analogue reveals conformational changes in ERK2 upon activation. J. Mol. Biol. 2005;353:600–612. doi: 10.1016/j.jmb.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 11.Xiao Y, Lee T, Latham MP, Warner LR, Tanimoto A, Pardi A, Ahn NG. Phosphorylation releases constraints to domain motion in ERK2. Proc. Natl. Acad. Sci. USA. 2014 doi: 10.1073/pnas.1318899111. In press. doi: 10.1073/pnas.1318899111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masterson LR, Cheng C, Yu T, Tonelli M, Kornev A, Taylor SS, Veglia G. Dynamics connect substrate recognition to catalysis in protein kinase A. Nat. Chem. Biol. 2010;6:821–828. doi: 10.1038/nchembio.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu B, Wong CF, McCammon JA. Release of ADP from the catalytic subunit of protein kinase A: A molecular dynamics simulation study. Protein Sci. 2005;14:159–168. doi: 10.1110/ps.04894605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robbins DJ, Zhen E, Owaki H, Vanderbilt CA, Ebert D, Geppert TD, Cobb MH. Regulation and properties of extracellular signal regulated-protein kinases 1 & 2 in vitro. J Biol. Chem. 1993;268:5097–5106. [PubMed] [Google Scholar]
- 16.Zhang J, Zhang F, Ebert D, Cobb MH, Goldsmith EJ. Activity of the MAP kinase ERK2 is controlled by a flexible surface loop. Structure. 1995;3:299–307. doi: 10.1016/s0969-2126(01)00160-5. [DOI] [PubMed] [Google Scholar]
- 17.Chen H, Ma J, Wi W, Eliseenkova AV, Xu C, Neubert TA, Miller WT, Mohammadi M. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol. Cell. 2007;27(5):717–730. doi: 10.1016/j.molcel.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deindl S, Kadlecek TA, Cao X, Kuriyan J, Weiss A. Structural basis for the inhibition of tyrosine kinase activity of ZAP-70. Cell. 2007;129:735–746. doi: 10.1016/j.cell.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 19.Shan Y, Arkhipov A, Kim ET, Pan AC, Shaw DE. Transitions to catalytically inactive conformations in EGFR kinase. Proc Natl Acad Sci U S A. 2013;110:7270–7275. doi: 10.1073/pnas.1220843110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ring AY, Sours KM, Lee T, Ahn NG. Distinct patterns of activation-dependent changes in conformational mobility between ERK1 and ERK2. Internat. J. Mass Spec. 2010;302:101–109. doi: 10.1016/j.ijms.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emrick MA, Lee T, Starkey P, Mumby MC, Resing KA, Ahn NG. The gatekeeper residue controls autoactivation of ERK2 via a pathway of intramolecular connectivity. Proc. Natl. Acad. Sci. USA. 2006;103(48):18101–18106. doi: 10.1073/pnas.0608849103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilsbacher JL, Cobb MH. Bacterial expression of activated mitogen-activated protein kinases. Methods Enzymol. 2001;332:387–400. doi: 10.1016/s0076-6879(01)32217-6. [DOI] [PubMed] [Google Scholar]
- 23.Goldsmith EJ, Cobb MH, Chang CI. Structure of MAPKs. Methods Mol. Biol. 2004;250:127–144. doi: 10.1385/1-59259-671-1:127. [DOI] [PubMed] [Google Scholar]
- 24.Mansour SJ, Candia JM, Matsuuda J, Manning M, Ahn NG. Interdependent domains controlling the enzymatic activity of mitogen-activated protein kinase kinase 1. Biochemistry. 1996;35:15529–15536. doi: 10.1021/bi961854s. [DOI] [PubMed] [Google Scholar]
- 25.Sours KM, Kwok SC, Rachidi T, Lee T, Ring A, Hoofnagle AN, Resing KA, Ahn NG. Hydrogen-exchange mass spectrometry reveals activation-induced changes in the conformational mobility of p38alpha MAP kinase. J. Mol. Biol. 2008;379:1075–1093. doi: 10.1016/j.jmb.2008.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoofnagle AH, Resing KA, Ahn NG. Protein analysis by hydrogen exchange mass spectrometry. Annu. Rev. Biophys. Biomol. Struct. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 27.Hoofnagle AN, Resing KA, Ahn NG. Practical methods for deuterium exchange/mass spectrometry. Methods Mol. Biol. 2004b;250:283–298. doi: 10.1385/1-59259-671-1:283. [DOI] [PubMed] [Google Scholar]
- 28.Lee T, Hoofnagle AN, Resing KA, Ahn NG. Protein hydrogen exchange measured by electrospray ionization mass spectrometry. In: Celis JE, editor. Cell Biology: A Laboratory Handbook. Third Edition Vol. 4. Elsevier Science; 2006. pp. 443–449. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.