Abstract
In multicellular organisms, patterns of gene expression are established in response to gradients of signaling molecules. During fly development in early Drosophila embryos, the Bicoid (Bcd) morphogen gradient is established within the first hour after fertilization. Bcd acts as a transcription factor, initiating the expression of a cascade of genes that determine the segmentation pattern of the embryo, which serves as a blueprint for the future adult organism. A robust understanding of the mechanisms that govern this segmentation cascade is still lacking, and a new generation of quantitative measurements of the spatiotemporal concentration dynamics of the individual players in this cascade is necessary for further progress. Here we describe a series of methods that represent the beginning of the use of Bcd as a quantification example. We describe the generation of a transgenic fly line expressing a Bcd-enhanced green fluorescent protein fusion protein. Using two-photon microscopy, we analyze the Bcd concentration dynamics and measure absolute Bcd expression levels in living fly embryos. These experiments have proven to be fruitful, generating new insights into the mechanisms that lead to the establishment and readout of the Bcd gradient. Generalization of these methods to other genes in the Drosophila segmentation cascade is straightforward and should further our understanding of the early patterning processes and the architecture of the underlying genetic network structure.
INTRODUCTION
Early patterning of multicellular organisms results from the interpretation of morphogen gradients by relatively small genetic regulatory networks, containing only a handful of genes that are able to determine the blueprint for the future adult structure of the entire organism. The inputs and outputs of these networks are protein molecules that are synthesized by the cell and act as “transcription factors,” which bind to the DNA to control downstream network elements. Essential for our understanding of the patterning network are a quantitative mapping of the relationships between the inputs and outputs of the system and a rigorous characterization of the noise present in these regulatory elements. Often times, these patterning networks show very high developmental accuracy and therefore very low noise from biological sources, such that all noise from technical sources must be kept at a minimum to allow for precise quantification. Over the past decade, a picture of the noise in genetic control (Elowitz et al. 2002; Ozbudak et al. 2002; Blake et al. 2003; Raser and O’Shea 2004; Pedraza and van Oudenaarden 2005) and of the global network structure that patterns the embryo (Reinitz and Sharp 1995; Fujioka et al. 1999; Jaeger et al. 2004a; Peter and Davidson 2009) has been fairly well established. Therefore, we can use these data to ask questions about the overall function and design of such networks. Such data also describe the capacity of these networks to transmit positional information; i.e., information of individual cells about their spatial location within the organism.
Our current understanding of such networks is derived mainly from genetic manipulations and static images of fixed tissue (Jaeger et al. 2004b). To fully describe the spatiotemporal regulatory interactions that determine patterning, however, a complete dynamic view is needed. Development is an intrinsically dynamic process during which spatial and temporal components are intimately tied together. Characterizing the dynamics of development is important both for gaining insights into complex developmental processes and for testing the possible mechanisms and models for gradient formation (Crick 1970; Bergmann et al. 2007; Coppey et al. 2007; DeLotto et al. 2007; Kicheva et al. 2007; Hecht et al. 2009) and gene regulation (von Dassow et al. 2000; Bialek and Setayeshgar 2005; Tostevin et al. 2007; Manu et al. 2009). Furthermore, for a fully quantitative understanding of the genetic regulation that determines the early patterning processes, we need to make high precision measurements of the relevant protein concentrations in living embryos. Such measurements require high image resolution, high sensitivity, and low variability, which are most easily achieved through higher intensities and slow acquisition modes. However, high energies usually result in photobleaching of the specimen, and slow acquisition times are incompatible with developmental dynamics. Overexposure of the embryo to light energy might interfere with the measured quantity and with the natural course of development. Finally, carefully determining the correct correlation between the number of photons collected and the protein concentration being measured is important. Here, we describe the method by which high precession measurements of morphogen gradients can be made to measure reproducibility between individuals for a single developmental stage: the beginning of nuclear cycle 14. However, the method can be extended to any number of times to obtain a more dynamic understanding of the patterns’ spatiotemporal profile.
EXPERIMENTAL PROCEDURES
Microscopy Apparatus
A custom-built, two-photon excitation laser scanning microscope (Denk et al. 1990) is used for all in vivo imaging of Drosophila embryos described here. The microscope is made of both commercial and custom parts, adapted to increase light collection through simultaneous detection of both epi- and transfluorescence (Svoboda et al. 1997; Mainen et al. 1999). Figure 1 shows the objective, stage, and condenser of the experimental apparatus. Samples are excited by light from a mode-locked pulsed Ti: sapphire laser (Mira 900; Coherent, Inc., Santa Clara, CA) (~100-fsec pulses at 80 MHz), whose wavelength is tuned to ~920 nm by a custom-made set of midband filters. The laser power can be varied with an opto-electric light modulator or Pockels cell (model 350-80LA; Conoptics, Inc., Danbury, CT). Coupled scanning mirrors are used to keep the beam stationary with respect to the stage, which is capable of translation in the x, y, and z directions through the use of a modified Sutter MP-285 micromanipulator (Sutter Instrument, Novato, CA). Detection efficiency is increased through the use of both a condenser and a 25×/0.8 Imm Korr DIC (differential interference contrast) objective (Carl Zeiss, Germany), whose signals are then amplified by two separate photomultiplier tubes (Hamamatsu R3896 and C6270; Hamamatsu Corporation, Bridgewater, NJ). The advantage of utilizing this additional transfluorescence lies in the larger numerical aperture (NA) of the oil-immersion condenser (NA = 1.4), which provides an increased collection efficiency resulting in an increased signal-to-noise ratio by a factor of ~2. This configuration also prevents the loss of collection efficiencies due to scattering, as the signal loss in the epi-channel sustained with an increased tissue depth is compensated by the corresponding increase in the transchannel signal, leaving the sum of epi- and transfluorescent signals approximately constant with variations in tissue depths (Mainen et al. 1999). The microscope is controlled by customized ScanImage software (Janelia Farm Research Campus, Ashburn, VA; Pologruto et al. 2003).
FIGURE 1.
A two-photon microscope setup showing the objective, the stage, and a high-NA oil-immersion condenser with trans- and epidetection systems indicated. Insert shows a typical slide of embryos for mounting. PMT, photomultiplier tube; A/D converter, analog-to-digital converter.
Generation of Fly Strain
The Bcd morphogen gradient in the fruit fly Drosophila melanogaster is an ideal system for starting to decipher in vivo transcription factor dynamics. As the primary input into the gene regulatory network, it determines the ultimate anterior patterning of the embryo; a qualitative static picture of its form and function is well established (Driever and Nüsslein-Volhard 1988a,b; 1989); and the major part of its activity occurs >1 h after its expression begins. Thus, sufficient time is left for completion of the maturation processes of the relevant proteins.
To visualize the spatiotemporal dynamics of Bcd, we generated transgenic Drosophila embryos in which endogenous Bcd was replaced with a green fluorescent fusion protein (Bcd–GFP) (Gregor et al. 2007b). To ensure the biological relevance of protein level measurements made with this fly strain, its Bcd expression levels need to resemble endogenous wild-type levels as closely as possible. This precaution is particularly important in high precision measurements of low protein levels and their fluctuations. To generate this fly strain, we used a plasmid carrying a transcript that codes for a recombinant Bcd protein fused to eGFP (Tsien 1998; Mavrakis et al. 2010) at its amino terminus. The fusion construct (Hazelrigg et al. 1998) had a size of 6.5 kb and contained endogenous bcd 5′ and 3′ UTRs (untranslated regions), which are known to mediate anterior localization and translation of bcd mRNA. This construct completely rescues embryos from bcd-mutant mothers. Qualitatively, no developmental defects are detected throughout the entire life cycle. Quantitatively, measured cues that directly follow from the embryo’s biological and physical properties are identical to wild type (e.g., the position of the cephalic furrow and the gradient’s length are constant) (Gregor et al. 2007b). The latter measures ensure that both the protein concentration levels and protein dynamics imaged in the transgenic rescue embryo mimic their natural counterparts in wild-type embryos, justifying the relevance of their subsequent quantification.
Linearity of Antibody Staining
Previously, gene expression levels in Drosophila embryos have been quantified using fluorescent antibody staining of the gene product (Kosman et al. 1998; Houchmandzadeh et al. 2002; Jaeger et al. 2004b). Such quantification relies on the assumption that the actual protein concentrations detected by the antibodies, and the fluorescence intensity, are linearly related to the embryo’s natural protein concentration levels. The Bcd–GFP fusion construct introduced into the bcd-mutant background flies allows for a direct test of this linearity of antibody staining as a method of quantifying relative protein concentrations. This test is achieved by staining Bcd–GFP embryos with an antibody against GFP (or Bcd) and measuring the autofluorescence of GFP and the intensity of the antibody staining in fixed tissue. The principal difficulty in this process is to avoid damaging the Bcd–GFP protein during the staining protocol. Therefore, to avoid the severe attenuation of GFP autofluorescence that occurs with the usual methanol treatment during the fixation process, embryos were fixed in paraformaldehyde and subsequently hand-peeled to remove the vitelline membrane. Next, the embryos were stained with an anti-GFP antibody, allowing simultaneous imaging of GFP autofluorescence and antibody staining in the same embryo. Figure 2 shows a comparison of these two probes at the surface and at the midsagittal plane of a single embryo. In both cases, the fluorescence intensity is linearly related to the protein concentration. This proportionality shows that antibody staining can be reliably used to measure relative Bcd concentrations in Drosophila embryos. These antibodies will be important in quantitatively studying the subsequent gene network involved in the embryogenesis of Drosophila, particularly the gap genes and the pair-rule genes for which fluorescent fusion proteins have not yet been developed for in vivo imaging.
FIGURE 2.
Linearity of antibody staining. (A–D) A single embryo was fixed in formaldehyde during nuclear cycle 14 (6.7% paraformaldehyde in 1× phosphate-buffered saline for 45 min) and stained with rabbit anti-GFP primary antibody (Millipore [previously Chemicon], Billerica, MA) following published protocols (Wieschaus and Nüsslein-Volhard 1986). The secondary antibody was conjugated with infrared Alexa-647 (Molecular Probes/Invitrogen Corporation, Carlsbad, CA), maximally reducing spectral overlap with the green GFP autofluorescence. The embryo was imaged both at the surface (A, C) and at the midsagittal plane (B, D) using confocal microscopy (Leica SP5, 20× oilimmersion objective plan apochromat, NA = 0.7; Leica Microsystems, Germany). GFP-autofluorescence (A, B) and anti-GFP staining (C, D) were recorded in consecutive runs. Scale bars, 100 µm. Typical dimensions of Drosophila eggs are 500 × 1800 µm. (E) Extracted raw fluorescence intensity profiles from (A) and (C) projected on the embryo’s anteroposterior axis. Each point corresponds to a single nucleus (for details, see Gregor et al. 2007b). (F) Raw fluorescence intensity profiles from (B) and (D) projected on the embryo’s anteroposterior axis, extracted by sliding a circular averaging area along the edge of the embryo (for details, see Houchmandzadeh et al. 2002). (G) Scatter plot of GFP-autofluorescence intensities versus fluorescence antibody staining intensities extracted from (A) and (C) for all nuclei. Each point corresponds to a single nucleus; the curves are normalized by nuclei of maximal and minimal intensities. (H) Scatter plot of fluorescence intensities extracted from (B) and (D). The blue line corresponds to the dorsal profile and the red line corresponds to the ventral profile.
Live Imaging of Drosophila Embryos
Genetically modified flies are kept in various containers, the bottoms of which are removable and contain a yeasted agar medium for oviposition. Typically, the flies are allowed to lay eggs for~1 h after changing the oviposition plate before the embryos are harvested. However, this time can be increased to ensure a larger collection of embryos on each plate. Replacing the oviposition plates of various cups in staggered time intervals, each lasting ~15 min, is often useful to control the maximal developmental progress of each plate of embryos. It will be beneficial during the actual imaging session, as it helps to vary the time at which the embryos enter nuclear cycle 14.
Harvested embryos are treated with pure bleach (8% hypochloride solution) for 15 sec to remove the outer chorion membrane. After the embryos are rinsed, they are sorted using a stereomicroscope to select for various characteristics, such as size or developmental stage. If the time allotted for oviposition is >2 h, this sorting becomes important to ensure that the embryos have not already matured past the desired developmental stage. Once sorted, all embryos are oriented identically on an agar substrate. Their anteroposterior (head–tail) axes are aligned with the y-scan of the light beam, and their dorsoventral (back–front) axes are aligned with the x-scan. The embryos are then mounted by carefully pushing a prepared glass slide that is coated with transparent glue onto the agar. Finally, embryos are immersed in either halocarbon oil or water, depending on the microscope objective used.
Although planar localization of excitation remains one of the key benefits of in vivo two-photon microscopy, excitation within this focal plane is nonuniform owing to the decrease in scanning laser intensity with an increased distance from the focal plane’s center. To correct for this effect, a uniformly fluorescent slide is imaged to produce a flat-field correction for the later acquisitions.
In a single imaging session, ~100 embryos are mounted on a slide and viewed using ScanImage (see Fig. 1, insert). By memorizing embryo positions in software, a cycle loop permits all embryos to be imaged in quick succession to monitor their developmental progress. Although these images are low resolution (256 × 256-pixel frames; 2 msec/line; 0.5 sec total acquisition time per embryo), they are adequate to show the size and density of the visible nuclei, allowing for the developmental stage of the embryo to be monitored to determine the embryos’ nuclear cycles. The completion of nuclear envelope degradation at the end of nuclear cycle 13 serves as a developmental marker that is used to ensure that each embryo is imaged at the same stage of development. Typically, images are acquired during early nuclear cycle 14, or 18 min after the above marker is reached.
On reaching the desired developmental stage, the imaging configuration is changed to a higher resolution with 8 msec/line, and three 512 × 512-pixel frames are taken and Kalman-averaged for each acquisition, resulting in a total acquisition time of 36.9 sec per embryo. Beginning with the anterior end, the embryo is imaged in three sections, with each successive image shifted 200 µm along the anteroposterior axis of the embryo. These three images are stitched together during the data analysis to recover an image of the entire embryo in this zoomed configuration.
The laser power at the sample is adjusted to ~5–40 mW(10 mW here corresponds to 5 × 1010 W/ m2 for a point-spread-function width of 0.5 µm) with the Pockels cell. To assess the amount of photobleaching that occurs at a given laser power, 10 high-resolution frame scans of the previously described specifications are made in quick succession before the imaging session, and the averaged nuclear concentrations at each point on the AP axis are compared over subsequent exposures, allowing for the quantification of the loss of intensity because of photobleaching at each point along this AP axis. The laser power for a given imaging session is eventually chosen such that this effect is minimized. Customized batch image-processing routines can be developed using ImageJ open source software (http://rsbweb.nih.gov/ij/).
Calculating Absolute Bicoid Concentration
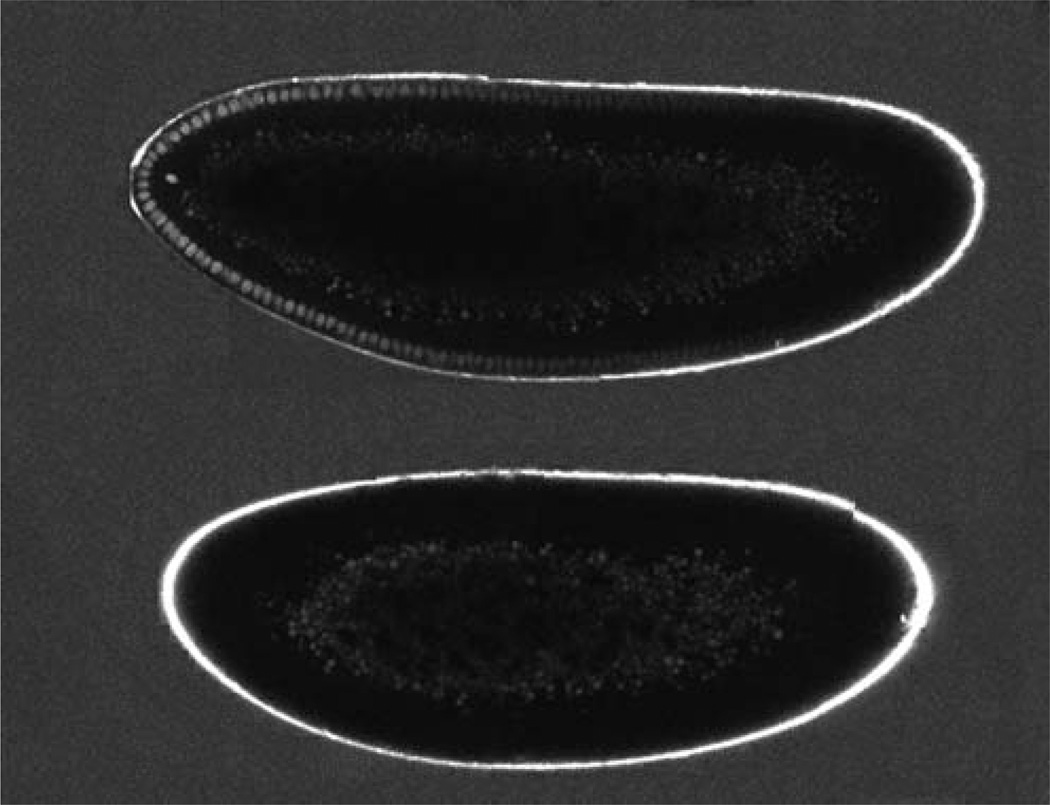
Both wild-type and Bcd-GFP-expressing embryos are immersed in a water solution that contains a known quantity of purified GFP molecules. They are then imaged (Fig. 3). An automated custom algorithm in MATLAB (The MathWorks, Natick, MA) identifies each nucleus within the embryo and calculates its average fluorescence intensity as follows. A mask of the embryo is obtained through a threshold that is determined by visual inspection. The original image is filtered so that each pixel within the mask is replaced by the mean intensity of a nucleus-sized disk of the corresponding pixels in the original image. From this averaged image, a ring of pixels (centered on the nuclei and roughly two nuclear diameters wide) is created by eroding the mask. The average intensity of each pixel segment perpendicular to the ring is determined to account for the fact that not all nuclei are located the same distance from the edge of the embryo. The algorithm, using the average values, finds the local maxima around the ring, with aminimum spacing determined by eye, to provide a rough idea of the location of the nuclei. The center of the nucleus is determined as the location of the point of maximum intensity in a square of nuclear size in the averaged image centered on each peak from the ring. The intensity value for each nucleus is the average intensity of a nuclei-sized disk in the original image centered on this point. The algorithm identifies the anteroposterior axis of the embryo and outputs a plot of the calculated intensity of each nucleus versus its position in a fractional egg length along the anteroposterior axis. The ratio of the average intensity of a nucleus-sized region in the GFP solution to the concentration of the GFP solution is used to convert the intensity values from arbitrary units into nuclear concentrations of Bcd–GFP in the embryo. To correct for background, the average intensity values of nucleus-sized regions within the wild-type embryos are calculated throughout the embryo and compared across three embryos (green points with error bars in Fig. 4A). This value is then subtracted from the calculated nuclear intensities to determine the absolute concentration of Bcd–GFP molecules in individual nuclei (Gregor et al. 2007a).
FIGURE 3.
Absolute concentration measurements. An embryo expressing a Bcd–GFP fusion protein (top) and a wild-type embryo (bottom) immersed in a solution of 34 ± 3 nM GFP. Both embryo images were taken during the same imaging session; each embryo was imaged in three pieces, which were reassembled by software. The two resulting embryo images were joined for display. (Note that the difference in embryo size reflects naturally varying egg sizes in the wild-type population.)
FIGURE 4.
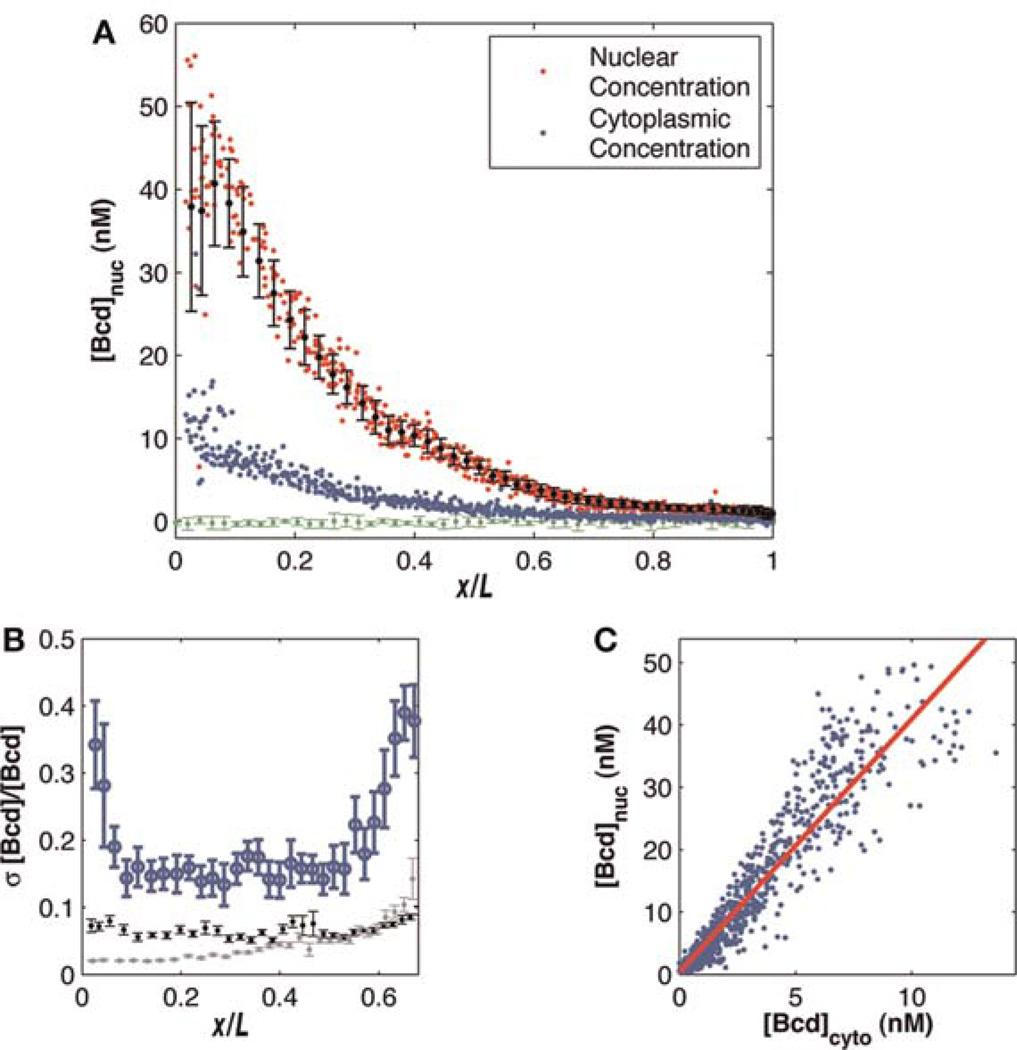
Nuclear and cytoplasmic Bcd gradient measurements. (A) Nuclear and cytoplasmic Bcd–GFP profiles of 12 embryos. Each red dot represents the average concentration in a single nucleus at the midsagittal plane of the embryo (on average 80 nuclei per embryo). Each blue dot represents the average concentration in a region outside each nucleus as described in the text. All nuclei from all embryos are binned in 50 bins over which the mean and standard deviation were computed (black points with error bars). The green curve corresponds to wild-type (no GFP) intensity levels. The vertical axis shows raw Bcd–GFP concentration in nanomolars (nM). (B) For each bin in (A), standard deviation divided by the mean is plotted as a function of relative egg length (blue). Error bars are computed by bootstrapping more than seven embryos. Gray and black lines show estimated contributions to measurement noise from imaging noise and focal plane adjustment noise, respectively (as described in the text). (C) Scatter plot for data in (A) shows a linear relationship between nuclear and cytoplasmic Bcd–GFP concentrations. Each dot represents a single nucleus; the red line is a linear fit to all points (R2 = 0.89) with a slope of 4.12 ± 0.2 (mean ± standard deviation over five independent data sets).
Measuring Reproducibility Across Embryos
Imaging multiple live embryos using the methods described here makes it possible to measure the reproducibility of the Bcd gradient across these embryos. Initially, nuclear Bcd gradients are extracted from each embryo using the same algorithm that was used to determine absolute Bcd concentrations. These gradients are plotted together on a single graph (red dots in Fig. 4A). Variability on this graph is determined by partitioning the anteroposterior axis into 50 bins and calculating the mean and standard deviation of the intensities in each bin (black points and error bars). The mean fluorescence background of a given image is calculated by determining the average of all intensities of the entire data set from 90% to 95% egg length, and this background is subtracted from the data. Intensities are converted to absolute concentrations as described above. This allows us to quantify the reproducibility r, defined as r = σ/(μcorr), where σ is the standard deviation and μcorr is the mean intensity with the background subtracted at each location on the anteroposterior axis. In this case, the mean and standard deviation are taken over intensities from all embryos located within the given bin. The reproducibility is plotted along the anteroposterior axis with error bars determined by bootstrapping (Fig. 4B) (Gregor et al. 2007a). To measure cytoplasmic Bcd concentrations, a custom algorithm takes nucleus-sized disks centered at the algorithmically determined nuclei and extends each of their edges a set number of pixels (determined by visual inspection) normal to the embryo mask to create a large region around each nucleus. The cytoplasmic intensity (blue points in Fig. 4A) is determined by the average intensity in this region, excluding the nucleus and a small buffer around the nucleus.
Quantification of Errors
Four main sources of measurement noise have been determined: (1) imaging noise due to the microscope, (2) nuclear identification noise caused by incorrectly centering the averaging region on the center of each nucleus, (3) focal plane adjustment noise from slight differences between the imaged plane and the actual center plane of the embryo, and (4) rotational asymmetry around the anteroposterior axis. Imaging noise is quantified by taking five consecutive images of a small section of an embryo and by calculating the reproducibility with the mean and standard deviation taken over intensities from the five images of the same embryo. Error is introduced by the photobleaching of GFP caused by repeated excitation. Photobleaching effects can be controlled by imaging an embryo repeatedly and by analyzing the significance of nuclear intensity decay with each successive image acquisition. All data presented here were obtained at a laser power where the photobleaching effect was negligible (~1%–2% during the imaging process). Nuclear identification noise is obtained by artificially displacing the nuclear centers found algorithmically. For nine such centers forming a 3 × 3-pixel area around the algorithmically determined nuclear center, a new nuclear intensity is calculated using the same averaging disk. For each nucleus, reproducibility is computed with the mean and standard deviation taken at the nine locations. Focal plane adjustment noise is calculated by taking nine images, each 0.3 µm apart, with the chosen focal plane as the center image and calculating the reproducibility with the mean and standard deviation taken over intensities from the nine images of different focal planes. An error caused by rotational asymmetry is estimated by comparing dorsal and ventral gradients in individual embryos to determine an upper bound on the error. The gray and black lines in Figure 4B represent the estimated contributions to noise from imaging noise and focal plane adjustment noise, respectively.
OUTLOOK
The methods described here allow for imaging and quantification of a particular GFP fusion protein in living fly embryos. Over the next decade, this approach is likely to be extended to zygotic transcription factors and other concentration measurements of the proteome, hopefully leading to a complete dynamic description of the early fly patterning cascade. Extending this approach to related species, such as houseflies or wasps, should be fairly straightforward, provided that the transgenic lines can be generated to incorporate a fusion protein containing a GFP derivative and that this fusion protein is expressed at wild-type levels in a knockout background. It would be particularly exciting to visualize multiple GFP derivatives of different colors in the same embryo, all tagging different transcription factors of the same small regulatory network. Cross-correlation analyses of data generated from such embryos would give us direct access to the network and help to elucidate its design principles.
ACKNOWLEDGMENTS
We are grateful to our collaborators for aiding in the collection and analysis of some of the data presented here, and for comments on the article: William Bialek, Shawn Little, Feng Liu, Alistair McGregor, David Tank, Stephan Thiberge, and Eric Wieschaus. Our work has been supported by Princeton University, National Institutes of Health (NIH) grant R01 GM077599, and Howard Hughes Medical Institute (HHMI).
REFERENCES
- Bergmann S, Sandler O, Sberro H, Shnider S, Schejter E, Shilo BZ, Barkai N. Pre-steady-state decoding of the Bicoid morphogen gradient. PLoS Biol. 2007;5:e46. doi: 10.1371/journal.pbio.0050046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek W, Setayeshgar S. Physical limits to biochemical signaling. Proc Natl Acad Sci. 2005;102:10040–10045. doi: 10.1073/pnas.0504321102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake WJ, Kaern M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- Coppey M, Berezhkovskii AM, Kim Y, Boettiger AN, Shvartsman SY. Modeling the bicoid gradient: Diffusion and reversible nuclear trapping of a stable protein. Dev Biol. 2007;312:623–630. doi: 10.1016/j.ydbio.2007.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. Diffusion in embryogenesis. Nature. 1970;225:420–422. doi: 10.1038/225420a0. [DOI] [PubMed] [Google Scholar]
- DeLotto R, DeLotto Y, Steward R, Lippincott-Schwartz J. Nucleocytoplasmic shuttling mediates the dynamic maintenance of nuclear dorsal levels during Drosophila embryogenesis. Development. 2007;134:4233–4241. doi: 10.1242/dev.010934. [DOI] [PubMed] [Google Scholar]
- Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluo-rescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Driever W, Nüsslein-Volhard C. A gradient of bicoid protein in Drosophila embryos. Cell. 1988a;54:83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- Driever W, Nüsslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988b;54:95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- Driever W, Nüsslein-Volhard C. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature. 1989;337:138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Fujioka M, Emi-Sarker Y, Yusibova GL, Goto T, Jaynes JB. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development. 1999;126:2527–2538. doi: 10.1242/dev.126.11.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, Tank DW, Wieschaus EF, Bialek W. Probing the limits to positional information. Cell. 2007a;130:153–164. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, Wieschaus EF, McGregor AP, Bialek W, Tank DW. Stability and nuclear dynamics of the bicoid morphogen gradient. Cell. 2007b;130:141–152. doi: 10.1016/j.cell.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelrigg T, Liu N, Hong Y, Wang S. GFP expression in Drosophila tissues: Time requirements for formation of a fluorescent product. Dev Biol. 1998;199:245–249. doi: 10.1006/dbio.1998.8922. [DOI] [PubMed] [Google Scholar]
- Hecht I, Rappel WJ, Levine H. Determining the scale of the Bicoid morphogen gradient. Proc Natl Acad Sci. 2009;106:1710–1715. doi: 10.1073/pnas.0807655106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchmandzadeh B, Wieschaus E, Leibler S. Establishment of developmental precision and proportions in the early Drosophila embryo. Nature. 2002;415:798–802. doi: 10.1038/415798a. [DOI] [PubMed] [Google Scholar]
- Jaeger J, Blagov M, Kosman D, Kozlov KN, Manu, Myasnikova E, Surkova S, Vanario-Alonso CE, Samsonova M, Sharp DH, et al. Dynamical analysis of regulatory interactions in the gap gene system of Drosophila melanogaster. Genetics. 2004a;167:1721–1737. doi: 10.1534/genetics.104.027334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J, Surkova S, Blagov M, Janssens H, Kosman D, Kozlov KN, Manu, Myasnikova E, Surkova S, Vanario-Alonso, et al. Dynamic control of positional information in the early Drosophila embryo. Nature. 2004b;430:368–371. doi: 10.1038/nature02678. [DOI] [PubMed] [Google Scholar]
- Kicheva A, Pantazis P, Bollenbach T, Kalaidzidis Y, Bittig T, Jülicher F, González-Gaitán M. Kinetics of morphogen gradient formation. Science. 2007;315:521–525. doi: 10.1126/science.1135774. [DOI] [PubMed] [Google Scholar]
- Kosman D, Small S, Reinitz J. Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Dev Genes Evol. 1998;208:290–294. doi: 10.1007/s004270050184. [DOI] [PubMed] [Google Scholar]
- Mainen ZF, Maletic-Savatic M, Shi SH, Hayashi Y, Malinow R, Svoboda K. Two-photon imaging in living brain slices. Methods. 1999;18:181, 231–239. doi: 10.1006/meth.1999.0776. [DOI] [PubMed] [Google Scholar]
- Manu, Surkova S, Spirov AV, Gursky VV, Janssens H, Kim A-R, Radulescu O, Vanario-Alonso CE, Sharp DH, Samsonova M, et al. Canalization of gene expression in the Drosophila blastoderm by gap gene cross regulation. PLoS Biol. 2009;7:e1000049. doi: 10.1371/journal.pbio.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis M, Pourquie O, Lecuit T. Lighting up developmental mechanisms: How fluorescence imaging heralded a new era. Development. 2010;137:373–387. doi: 10.1242/dev.031690. [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- Pedraza JM, van Oudenaarden A. Noise propagation in gene networks. Science. 2005;307:1965–1969. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. Genomic control of patterning. Int J Dev Biol. 2009;53:707–716. doi: 10.1387/ijdb.072495ip. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pologruto TA, Sabatini BL, Svoboda K. ScanImage: Flexible software for operating laser scanning microscopes. Biomed Eng Online. 2003;2:13. doi: 10.1186/1475-925X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O’Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinitz J, Sharp DH. Mechanism of eve stripe formation. Mech Dev. 1995;49:133–158. doi: 10.1016/0925-4773(94)00310-j. [DOI] [PubMed] [Google Scholar]
- Svoboda K, Denk W, Kleinfeld D, Tank DW. In vivo dendritic calcium dynamics in neocortical pyramidal neurons. Nature. 1997;385:161–165. doi: 10.1038/385161a0. [DOI] [PubMed] [Google Scholar]
- Tostevin F, ten Wolde PR, Howard M. Fundamental limits to position determination by concentration gradients. PLoS Comput Biol. 2007;3:e78. doi: 10.1371/journal.pcbi.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien R-Y. The green fluorescent protein. Ann Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- von Dassow G, Meir E, Munro EM, Odell GM. The segment polarity network is a robust developmental module. Nature. 2000;406:188–192. doi: 10.1038/35018085. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Nüsslein-Volhard C. Looking at embryos. In: Roberts D, editor. Drosophila: A practical approach. Oxford: IRL Press; 1986. pp. 199–228. [Google Scholar]