The ability of membrane proteins to sense and respond to changes in membrane voltage is critical for a vast array of biological processes, from generating and propagating the nerve impulse to excitation–secretion coupling. Hodgkin and Huxley (1952) were the first to characterize voltage-activated potassium (Kv) and sodium (Nav) currents, but the common architecture of voltage-activated cation channels only became apparent after the genes encoding these proteins were identified (Noda et al., 1986; Tanabe et al., 1987; Timpe et al., 1988). We now appreciate that the voltage sensitivity of these channels can be ascribed to modular voltage-sensing domains comprised of S1–S4 helices (Lu et al., 2001, 2002; Jiang et al., 2003; Long et al., 2007; Bezanilla, 2008; Swartz, 2008). Bioinformatic searches subsequently identified S1–S4 voltage-sensing domains in other protein families, including voltage-sensitive phosphatases (VSPs; Kumánovics et al., 2002; Murata et al., 2005), where the S1–S4 domain regulates an intracellular enzyme, and voltage-activated proton channels (Hv1; Ramsey et al., 2006; Sasaki et al., 2006), in which the S1–S4 domain forms a stand-alone pore. Although the sequences of S1–S4 domains vary considerably, the mechanisms of these domains appear to be so highly conserved that Kv channels from humans and hyperthermophilic archebacteria are both sensitive to voltage sensor toxins from tarantula venom (Swartz, 2008). S1–S4 domains also adopt similar structures in the few Kv and Nav channels that have been successfully crystalized (Jiang et al., 2003; Long et al., 2007; Payandeh et al., 2011), suggesting that there is a common blueprint, or design principle, for constructing a voltage sensor. In this issue of The Journal of General Physiology, Palovcak et al. computationally analyze the thousands of examples of S1–S4 domains present in all three kingdoms of life to identify the key design features common to all S1–S4 domains.
Palovcak et al. (2014) begin by identifying S1–S4 domain sequences in the National Center for Biotechnology Information database using a hidden Markov model (HMM; Eddy, 2004) trained on an initial set, or seed, of well known, phylogenetically diverse sequences. This training seed allows the HMM to estimate the probability distribution of amino acids at each position, and because the HMM accounts for differences in evolutionary pressure, it is typically better able to detect distantly related sequences than a simple BLAST (Basic Local Alignment Search Tool) search. Additionally, an HMM can automatically provide the most probable alignment, and its likelihood, for each residue in the sequence, which is especially helpful when the alignment appears ambiguous. Using this approach, the authors’ HMM identified >6,600 sequences from all known branches of the family of S1–S4 domains and, after grouping similar sequences, were left with 3,821 effectively independent sequences, a colossal number compared with those obtained through previous multiple sequence alignment (MSA) analyses of voltage-activated ion channels.
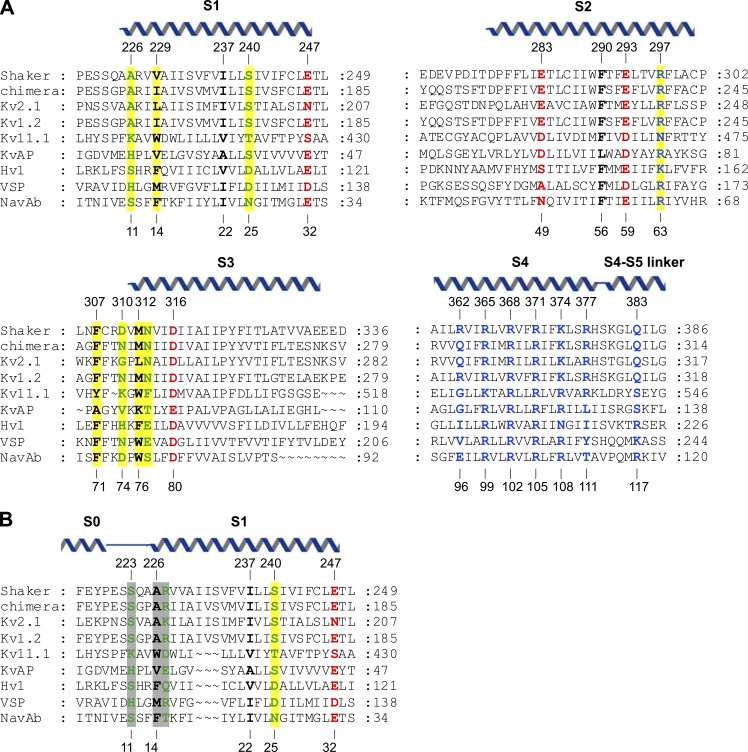
One benefit of simultaneously comparing a large number of evolutionarily diverse sequences is that the ambiguity of individual alignments can be directly quantified. For S1–S4 domains, the alignment of S2 and S3 is relatively clear because of the highly conserved acidic residues; however, published binary sequence alignments have differed considerably for S1 and S4 (Lacroix and Bezanilla, 2012; Mishina et al., 2012; Kulleperuma et al., 2013). Aligning S4, the helix that moves in response to changes in membrane voltage, has been particularly difficult because this helix contains a repeating triad of Arg and two hydrophobic residues (e.g., ArgXXArgXXArgXXArgXX) that can vary in length between three and six Arg residues (and can even contain gaps). Thus, whenever two S4 helices have different numbers of Arg residues, there will be several different registers (each shifted by three residues) that equally optimize the number of aligned Arg residues (Kulleperuma et al., 2013). However, the authors demonstrate that because an HMM weights each position by its variability, one can define a most-probable alignment for S4 and compare it with other alignments in terms of the likelihood of finding each specific residue at a particular position (called a posterior probability; Wolfsheimer et al., 2012). In Fig. 1 A, we show the output MSA from Palovcak et al. (2014) with representatives from several branches of the family, including the Shaker (Timpe et al., 1988) and Kv2.1 channels, for which extensive functional data exist (Bezanilla, 2008; Swartz, 2008); the Kv1.2/2.1 paddle chimera, the only eukaryotic voltage-activated channel for which an x-ray structure has been solved (Long et al., 2007); and NavAb, a Nav channel from Arcobacter buzleri for which an x-ray structure has been solved (Payandeh et al., 2011) and that the authors use as a reference. This output MSA clearly provides a valuable starting point for comparisons between any two S1–S4 domains, in particular those where x-ray structures are not available for a closely related S1–S4 domain.
Figure 1.
Sequence alignment of S1–S4 voltage-sensing domains. (A) Sequence alignment from the output MSA of Palovcak et al. (2014), including Shaker Kv (UniProt accession no. P08510), Kv1.2/2.1 paddle chimera (Protein Data Bank accession no. 2R9R_B), rat Kv2.1 (UniProt accession no. P15387), rat Kv1.2 (UniProt accession no. P63142), human Kv11.1 (UniProt accession no. Q12809), KvAP (UniProt accession no. Q9YDF8), human Hv1 (UniProt accession no. Q96D96), Ciona VSP (UniProt accession no. Q4W8A1), and NavAb (UniProt accession no. A8EVM5). Conserved positions are colored as follows: polar, green; hydrophobic, black; acidic, red; and basic, blue. Positions highlighted in yellow have not been extensively studied previously but are identified as highly conserved by Palovcak et al. (2014). Helices are positioned according to the structure of the Kv1.2/2.1 paddle chimera (2R9R; Long et al., 2007). (B) Alternative alignment of S1 that takes into consideration the longer S1 helix in Kv channels (Long et al., 2007) compared with Nav channels (Payandeh et al., 2011). In this alignment, positions 11 and 14 in NavAb are equally or more highly conserved than in A, and position 15 becomes highly conserved.
Intuitively, the most striking feature of any MSA is the degree of conservation at particular positions. Indeed, previous studies have identified a host of critical residues within S1–S4 domains, including the highly conserved periodic motif of basic residues within the S4 helix (Fig. 1 A, blue residues), acidic residues in S1–S3 that serve as stabilizing countercharges for the S4 Arg residues (Fig. 1 A, red residues), and highly conserved bulky hydrophobic residues near the middle of the S1–S4 domain that S4 Arg residues move past as the domain changes conformation between resting and activated states (Fig. 1 A, black residues; Noda et al., 1986; Tanabe et al., 1987; Timpe et al., 1988; Papazian et al., 1995; Aggarwal and MacKinnon, 1996; Seoh et al., 1996; Jiang et al., 2003; Long et al., 2007; Bezanilla, 2008; Swartz, 2008; Tao et al., 2010; Lacroix and Bezanilla, 2011). In Palovcak et al. (2014), the authors take an unbiased approach by calculating the Kullback-Leibler divergence (DKL) for each position in their large MSA. Essentially, DKL compares the distribution of amino acids at each position with those typically found in that environment (e.g., a lipid-facing inner or outer membrane interface), thereby identifying sites that are evolutionarily constrained. For example, position 25 in the S1 of NavAb has a high DKL because it is commonly occupied by polar residues (Asn and Ser), which are uncommon in the middle of the membrane. In Hv channels, this position (D112) plays a crucial role in proton conduction (Musset et al., 2011). In the end, this analysis identifies 21 positions within S1–S4 domains that have high DKL scores and thus have been subjected to particularly strong evolutionary pressure. Reassuringly, 13 out of 21 of these positions have been previously shown to play critical roles in the hydrophobic core of the domain, the acidic residue clusters that stabilize S4, or the periodic ArgXX motif within S4 (Fig. 1 A). Most of the eight new positions identified with this approach are located in the intracellular half of S1–S3 (e.g., residues 11, 14, 63, 71, 74, 76, and 77 in NavAb) and are typically occupied by polar, aromatic, and positively charged residues. Although the precise mechanistic significance of these positions remains unclear, the authors’ analysis strongly suggests that further investigation is warranted.
Palovcak et al. (2014) then use their gargantuan MSA to search for key interactions within S1–S4 domains by identifying positions undergoing coevolution. Conceptually, the idea behind this analysis is straightforward. If two positions, A and B, make an essential interaction in any conformation, variations in the amino acid at position A (e.g., acidic to polar) should be correlated with variations at position B (e.g., basic to polar). It is important to note that there is not a simple link between pairs of residues that exhibit direct structural interactions and those that coevolve. First, two positions can have a very strong structural interaction but show no signs of coevolution if one or both positions are invariant. For example, there is strong experimental evidence that Arg residues in the S4 helix of Kv channels interact strongly with the charge-transfer center (F56 in NavAb and F290 in Shaker; Tao et al., 2010; Lacroix and Bezanilla, 2011), but the Arg residues and F290 vary so infrequently that they show no sign of coevolution. Conversely, two positions that play critical roles in stabilizing the same state could undergo coevolution even if there is no direct structural interaction between them. However, coevolution is strongly suggestive of a structural interaction between two positions, and to detect such sites, the authors performed a direct-coupling analysis (DCA), which quantifies the degree to which variations of the amino acid at one position are correlated with a second.
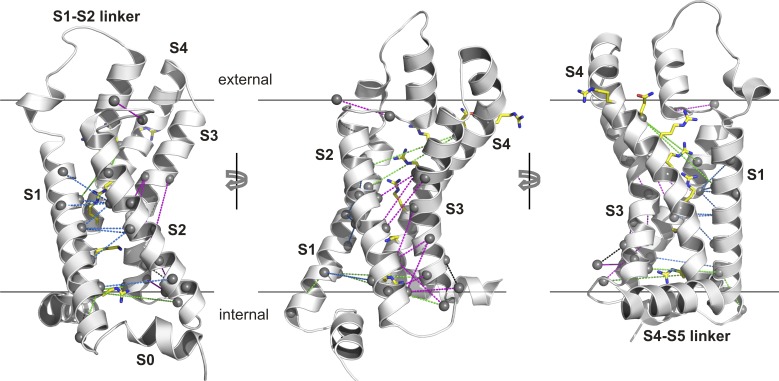
Using this approach, the authors uncovered 24 pairs of residues within the S1–S4 domains that are strongly coupled and mapped these pairs onto the activated-state structure of NavAb (Payandeh et al., 2011). For readers more familiar with the structure of the Kv1.2/2.1 paddle chimera (Long et al., 2007), we provide the equivalent map onto that structure (Fig. 2). Of the top 24 pairs of residues identified, 20 are positioned to interact directly in the NavAb structure, and 19 are so positioned in the paddle chimera structure. Most of these coupled pairs of residues are positioned between S1 and S2 (Fig. 2, dashed blue lines) or between S2 and S3 (Fig. 2, dashed magenta lines), and only two pairs are between S4 and the other three helices. In effect, this DCA supports the idea that S1–S3 forms a relatively stationary scaffold against which the S4 helix moves as it changes conformation between resting and activated states. Inspection of the contact maps for the structures of NavAb and the Kv1.2/2.1 paddle chimera reveals a large number of interactions between the S1–S3 helices, supporting this idea. Those structures also show numerous contacts between S4 and the S1–S3 helices, many of which are not seen in the DCA. One explanation for this apparent discrepancy might be that some of the interactions between S4 and the other three helices are invariant between S1–S4 domains in the MSA and thus cannot be seen in the DCA. Stabilizing interactions between Arg residues in S4 and acidic residues in S1–S3 are likely candidates for the types of interactions that DCA might miss. It is also possible that some interactions between S4 and S1–S3 differ between subfamilies of S1–S4 domains and that the DCA cannot identify these because it was performed on a large and diverse MSA of all known S1–S4 domains. If subfamily-specific interactions between S4 and S1–S3 do exist, they are unlikely to be essential because the paddle motif, a helix-turn-helix motif composed of S3b and S4 helices, can be transplanted between many different types of proteins that contain S1–S4 domains, including Kv channels, Nav channels, Hv1 channels, and VSPs, without disrupting voltage-sensing functions (Alabi et al., 2007; Bosmans et al., 2008). Moreover, it has also recently been reported that coexpression of constructs encoding the N terminus through S3 of the Shaker Kv channel with those encoding S4 through the C terminus gives rise to functional voltage sensors (Priest et al., 2013).
Figure 2.
Mapping of evolutionarily coupled residues onto the x-ray structure of the Kv1.2/Kv2.1 paddle chimera. The Cα atoms of all coupled residues are identified with small spheres, and S4 Arg residues are shown as sticks. 19 coupled pairs that are positioned to interact directly are connected with dashed blue lines between S1 and S2 and with dashed magenta lines between S2 and S3. Five coupled pairs whose positions in the structure are not compatible with direct structural interactions are connected with dashed green lines. The 24 coupled pairs are those listed in Table S3 in Palovcak et al. (2014).
At present, the available x-ray structures of Kv and Nav channels have provided a detailed picture of the activated state of the S1–S4 domains in these channels, but we currently lack structures of these proteins in the resting states that are populated at negative membrane voltages where the channels are closed. Although most of the pairs of coevolving residues identified in the DCA coupling results could plausibly interact directly in the activated state structures, several are too far apart, raising the possibility that they interact in the resting state. Two of these coevolving pairs involve the first Arg position of the S4 helix (E96 in NavAb and R362 in Shaker); in the first pair, this position couples with N25 in NavAb (S240 in Shaker) within the S1 helix, and in the second it couples with N49 in NavAb (E283 in Shaker) within the S2 helix. Cd2+ bridging experiments in the Shaker Kv channel have shown that R262C can bridge with either I241C in S1 or I287C in S2, and in both cases the bridges form in the resting state (Campos et al., 2007). These bridging residues would be nearby those identified in the coupling analysis; in the case of the S4 bridge with S1, the coupling analysis and Cd2+ bridge differ by one residue within S1, and in the case of the S4 bridge with S2, the two approaches differ by one turn of the S2 helix. In addition, Palovcak et al. (2014) also point out that these two coevolving pairs are compatible with several computational models (Vargas et al., 2012) for the resting states of Kv and Nav channels.
The predictive power of the authors’ analysis of sequence conservation and coevolution will be clearer after the functional impact of their newly identified conserved residues and coevolving pairs has been investigated experimentally and x-ray structures of S1–S4 voltage-sensing domains in resting states have been solved. However, it is reassuring that established structural features of S1–S4 domains, such as the S4 Arg residues, acidic countercharges, and hydrophobic core, appear naturally. The lack of coevolving pairs between S4 and S1–S3 is also largely consistent with our current understanding. The present DCA does not detect several previously defined structural interactions between elements within S1–S4 domains. For instance, this analysis did not detect interactions between S4 Arg residues and residues in either the charge-transfer center or the acidic residue clusters, likely because the participating residues are too highly conserved. The present DCA also failed to detect interactions between the outer portions of S4 with S3b, where the S3b helix has been shown to interact with S4 in the activated state and serve as a hydrophobic stabilizer of the S4 helix (Xu et al., 2013). In this instance, the interactions may be too nonspecific (i.e., hydrophobic interactions) to be detected using DCA. Indeed, these examples nicely illustrate the types of structural features MSA analyses can detect and those that must be found by other methods.
In Palovcak et al. (2014), the authors have lumped together all known S1–S4 domains, which is reasonable when the goal is to find universal common features. Although S1–S4 domains share common structural features and mechanisms, it is likely there will be important differences between subfamilies. For example, comparison of the structures of the Kv1.2/2.1 paddle chimera and NavAb reveals that the S1 helix is one helical turn longer in Kv channels compared with Nav channels. As a result, an evolutionarily conserved pairing between residues 9 and 14 in NavAb are positioned to interact locally in that x-ray structure, but on opposite sides of the S1 helix in the structure of the paddle chimera (Fig. 2, left). If we introduce this difference into the authors’ MSA for S1 (Fig. 1 B), this pair of residues can interact locally (not depicted), the conservation of two positions identified by Palovcak et al. (2014) improves, and a neighboring position also becomes highly conserved (Fig. 1 B, gray shading). It would be valuable to undertake comparable analyses specifically comparing different subfamilies of S1–S4 domains to correlate sequence differences with functional specialization. For example, a conserved Asp in S1 of Hv1 is required for proton selectivity (Musset et al., 2011), yet there must be additional critical adaptations because that position is conserved in VSPs that do not conduct protons. It would also be interesting to compare S1–S4 domains from voltage-activated channels with those found in CNG and transient receptor potential (TRP) channels, two types of tetrameric cation channels that lack strong voltage sensitivity. At least in TRPV1 channels, the S1–S4 domain adopts a similar fold to that discussed here, and it does not appear to change conformation as the channel opens and closes in response to activating ligands (Cao et al., 2013). One might predict that there would be a larger number of coevolving residues within the S1–S4 domains of TRP channels and that more of these would occur between the S4 helix and S1–S3. Finally, could such an analysis shed light on how conformational changes in S1–S4 domains couple to and control the conformation of the pore domain in voltage-activated cation channels? Interactions between the S4–S5 linker and the C-terminal end of S6 are known to be crucial for coupling voltage-sensing and pore domains (Lu et al., 2001, 2002), and the essential differences between channels that are activated by membrane depolarization compared with those activated by hyperpolarization may reside in this region (Kwan et al., 2012). However, the sequences in these regions vary considerably, and the underlying mechanisms of coupling voltage-sensing and pore domains remain to be uncovered.
Acknowledgments
We thank J. Kalia, D. Krepkiy, and members of the Swartz laboratory for helpful discussions.
This work was supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke, National Institutes of Health, to K.J. Swartz.
The authors declare no competing financial interests.
Sharona E. Gordon served as editor.
References
- Aggarwal S.K., MacKinnon R. 1996. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 16:1169–1177 10.1016/S0896-6273(00)80143-9 [DOI] [PubMed] [Google Scholar]
- Alabi A.A., Bahamonde M.I., Jung H.J., Kim J.I., Swartz K.J. 2007. Portability of paddle motif function and pharmacology in voltage sensors. Nature. 450:370–375 10.1038/nature06266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla F. 2008. How membrane proteins sense voltage. Nat. Rev. Mol. Cell Biol. 9:323–332 10.1038/nrm2376 [DOI] [PubMed] [Google Scholar]
- Bosmans F., Martin-Eauclaire M.F., Swartz K.J. 2008. Deconstructing voltage sensor function and pharmacology in sodium channels. Nature. 456:202–208 10.1038/nature07473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos F.V., Chanda B., Roux B., Bezanilla F. 2007. Two atomic constraints unambiguously position the S4 segment relative to S1 and S2 segments in the closed state of Shaker K channel. Proc. Natl. Acad. Sci. USA. 104:7904–7909 10.1073/pnas.0702638104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao E., Liao M., Cheng Y., Julius D. 2013. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 504:113–118 10.1038/nature12823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. 2004. What is a hidden Markov model? Nat. Biotechnol. 22:1315–1316 10.1038/nbt1004-1315 [DOI] [PubMed] [Google Scholar]
- Hodgkin A.L., Huxley A.F. 1952. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 117:500–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Lee A., Chen J., Ruta V., Cadene M., Chait B.T., MacKinnon R. 2003. X-ray structure of a voltage-dependent K+ channel. Nature. 423:33–41 10.1038/nature01580 [DOI] [PubMed] [Google Scholar]
- Kulleperuma K., Smith S.M., Morgan D., Musset B., Holyoake J., Chakrabarti N., Cherny V.V., DeCoursey T.E., Pomès R. 2013. Construction and validation of a homology model of the human voltage-gated proton channel hHV1. J. Gen. Physiol. 141:445–465 10.1085/jgp.201210856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumánovics A., Levin G., Blount P. 2002. Family ties of gated pores: evolution of the sensor module. FASEB J. 16:1623–1629 10.1096/fj.02-0238hyp [DOI] [PubMed] [Google Scholar]
- Kwan D.C., Prole D.L., Yellen G. 2012. Structural changes during HCN channel gating defined by high affinity metal bridges. J. Gen. Physiol. 140:279–291 10.1085/jgp.201210838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix J.J., Bezanilla F. 2011. Control of a final gating charge transition by a hydrophobic residue in the S2 segment of a K+ channel voltage sensor. Proc. Natl. Acad. Sci. USA. 108:6444–6449 10.1073/pnas.1103397108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix J.J., Bezanilla F. 2012. Tuning the voltage-sensor motion with a single residue. Biophys. J. 103:L23–L25 10.1016/j.bpj.2012.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S.B., Tao X., Campbell E.B., MacKinnon R. 2007. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 450:376–382 10.1038/nature06265 [DOI] [PubMed] [Google Scholar]
- Lu Z., Klem A.M., Ramu Y. 2001. Ion conduction pore is conserved among potassium channels. Nature. 413:809–813 10.1038/35101535 [DOI] [PubMed] [Google Scholar]
- Lu Z., Klem A.M., Ramu Y. 2002. Coupling between voltage sensors and activation gate in voltage-gated K+ channels. J. Gen. Physiol. 120:663–676 10.1085/jgp.20028696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina Y., Mutoh H., Knöpfel T. 2012. Transfer of Kv3.1 voltage sensor features to the isolated Ci-VSP voltage-sensing domain. Biophys. J. 103:669–676 10.1016/j.bpj.2012.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y., Iwasaki H., Sasaki M., Inaba K., Okamura Y. 2005. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 435:1239–1243 10.1038/nature03650 [DOI] [PubMed] [Google Scholar]
- Musset B., Smith S.M., Rajan S., Morgan D., Cherny V.V., Decoursey T.E. 2011. Aspartate112 is the selectivity filter of the human voltage-gated proton channel. Nature. 480:273–277 10.1038/nature10557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda M., Ikeda T., Suzuki H., Takeshima H., Takahashi T., Kuno M., Numa S. 1986. Expression of functional sodium channels from cloned cDNA. Nature. 322:826–828 10.1038/322826a0 [DOI] [PubMed] [Google Scholar]
- Palovcak E., Delemotte L., Klein M.L., Carnevale V. 2014. Evolutionary imprint of activation: The design principles of VSDs. J. Gen. Phys. 143:145–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazian D.M., Shao X.M., Seoh S.A., Mock A.F., Huang Y., Wainstock D.H. 1995. Electrostatic interactions of S4 voltage sensor in Shaker K+ channel. Neuron. 14:1293–1301 10.1016/0896-6273(95)90276-7 [DOI] [PubMed] [Google Scholar]
- Payandeh J., Scheuer T., Zheng N., Catterall W.A. 2011. The crystal structure of a voltage-gated sodium channel. Nature. 475:353–358 10.1038/nature10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest M.F., Lacroix J.J., Villalba-Galea C.A., Bezanilla F. 2013. S3-s4 linker length modulates the relaxed state of a voltage-gated potassium channel. Biophys. J. 105:2312–2322 10.1016/j.bpj.2013.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey I.S., Moran M.M., Chong J.A., Clapham D.E. 2006. A voltage-gated proton-selective channel lacking the pore domain. Nature. 440:1213–1216 10.1038/nature04700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M., Takagi M., Okamura Y. 2006. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 312:589–592 10.1126/science.1122352 [DOI] [PubMed] [Google Scholar]
- Seoh S.A., Sigg D., Papazian D.M., Bezanilla F. 1996. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 16:1159–1167 10.1016/S0896-6273(00)80142-7 [DOI] [PubMed] [Google Scholar]
- Swartz K.J. 2008. Sensing voltage across lipid membranes. Nature. 456:891–897 10.1038/nature07620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe T., Takeshima H., Mikami A., Flockerzi V., Takahashi H., Kangawa K., Kojima M., Matsuo H., Hirose T., Numa S. 1987. Primary structure of the receptor for calcium channel blockers from skeletal muscle. Nature. 328:313–318 10.1038/328313a0 [DOI] [PubMed] [Google Scholar]
- Tao X., Lee A., Limapichat W., Dougherty D.A., MacKinnon R. 2010. A gating charge transfer center in voltage sensors. Science. 328:67–73 10.1126/science.1185954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpe L.C., Schwarz T.L., Tempel B.L., Papazian D.M., Jan Y.N., Jan L.Y. 1988. Expression of functional potassium channels from Shaker cDNA in Xenopus oocytes. Nature. 331:143–145 10.1038/331143a0 [DOI] [PubMed] [Google Scholar]
- Vargas E., Yarov-Yarovoy V., Khalili-Araghi F., Catterall W.A., Klein M.L., Tarek M., Lindahl E., Schulten K., Perozo E., Bezanilla F., Roux B. 2012. An emerging consensus on voltage-dependent gating from computational modeling and molecular dynamics simulations. J. Gen. Physiol. 140:587–594 10.1085/jgp.201210873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfsheimer S., Hartmann A., Rabus R., Nuel G. 2012. Computing posterior probabilities for score-based alignments using ppALIGN. Stat. Appl. Genet. Mol. Biol. 11:Article 1. [DOI] [PubMed] [Google Scholar]
- Xu Y., Ramu Y., Shin H.G., Yamakaze J., Lu Z. 2013. Energetic role of the paddle motif in voltage gating of Shaker K+ channels. Nat. Struct. Mol. Biol. 20:574–581 10.1038/nsmb.2535 [DOI] [PMC free article] [PubMed] [Google Scholar]