Interactions between aromatic amino acid residues in the pore helix and S5 transmembrane domain control gating of the Ca2+-activated potassium channel KCa3.1.
Abstract
The Ca2+-activated potassium channel KCa3.1 is emerging as a therapeutic target for a large variety of health disorders. One distinguishing feature of KCa3.1 is that the channel open probability at saturating Ca2+ concentrations (Pomax) is low, typically 0.1–0.2 for KCa3.1 wild type. This observation argues for the binding of Ca2+ to the calmodulin (CaM)–KCa3.1 complex, promoting the formation of a preopen closed-state configuration leading to channel opening. We have previously shown that the KCa3.1 active gate is most likely located at the level of the selectivity filter. As Ca2+-dependent gating of KCa3.1 originates from the binding of Ca2+ to CaM in the C terminus, the hypothesis of a gate located at the level of the selectivity filter requires that the conformational change initiated in the C terminus be transmitted to the S5 and S6 transmembrane helices, with a resulting effect on the channel pore helix directly connected to the selectivity filter. A study was thus undertaken to determine to what extent the interactions between the channel pore helix with the S5 and S6 transmembrane segments contribute to KCa3.1 gating. Molecular dynamics simulations first revealed that the largest contact area between the pore helix and the S5 plus S6 transmembrane helices involves residue F248 at the C-terminal end of the pore helix. Unitary current recordings next confirmed that modulating aromatic–aromatic interactions between F248 and W216 of the S5 transmembrane helical segment and/or perturbing the interactions between F248 and residues in S6 surrounding the glycine hinge G274 cause important changes in Pomax. This work thus provides the first evidence for a key contribution of the pore helix in setting Pomax by stabilizing the channel closed configuration through aromatic–aromatic interactions involving F248 of the pore helix. We propose that the interface pore helix/S5 constitutes a promising site for designing KCa3.1 potentiators.
INTRODUCTION
The voltage-insensitive calcium-activated potassium channel of intermediate conductance, KCa3.1, has been documented to play a prominent role in a large variety of physiological processes including immune reactions involving memory B and T cells (Wulff et al., 2004), transepithelial ion transport in Cl−-secreting epithelial cells (Hayashi et al., 2012), control of vascular tone (Félétou and Vanhoutte, 2007), and proliferation/migration in various cell types (Cruse et al., 2006; Bradding and Wulff, 2009; Gao et al., 2010). It follows that KCa3.1 is now recognized as a promising therapeutic target to treat life-threatening health disorders (Singh et al., 2001; Szkotak et al., 2004; Murthy et al., 2005; Roth et al., 2011). In line with this proposal are several reports demonstrating that a pharmacological activation of KCa3.1 and KCa2.3 channels constitutes a potential unique endothelium-specific antihypertensive therapy (Sheng et al., 2009; Damkjaer et al., 2012). Of interest is also the observation that KCa3.1 activation could improve cAMP-induced Cl− secretion in tissues coming from cystic fibrosis patients with partial CFTR function, thus identifying KCa3.1 activation as a complementary strategy to correct the basic ion transport defect in cystic fibrosis (Roth et al., 2011).
KCa3.1 channels are tetrameric membrane proteins with each subunit organized in six transmembrane segments, S1–S6, with a pore motif between segments 5 (S5) and 6 (S6). The channel assembly and trafficking are regulated by the constitutively bound calmodulin (CaM) molecule, which also confers Ca2+ sensitivity (Joiner et al., 2001). The crystal structure of the KCa3.1 channel is not known. On the basis of homology modeling data and substituted cysteine accessibility method (SCAM) experiments, we proposed that residues V275, T278, A279, V282, and A286 were lining the channel pore with residues C276, C277, L280, and L281, oriented opposite to the pore lumen (Simoes et al., 2002; Klein et al., 2007). Additional experiments subsequently demonstrated that the bundle-crossing region located at the level of residues V282–A286 could not form a tight seal impermeable to K+ ions for the closed KCa3.1 channel. This conclusion came from experiments in which we showed that the accessibility of the thiol-modifying agent Ag+ to cysteines engineered in the channel cavity was independent of the channel-conducting state (Garneau et al., 2009). In agreement with these results, Ba2+-based experiments on the Ca2+-activated KCa2.2 channel demonstrated that the channel gate was most likely located deep in the channel central cavity (Bruening-Wright et al., 2007). In addition, data were presented indicating that negative gating of the KCa2.1 and KCa2.3 channels by NS8593 occurs through interactions with gating structures at a position close to the selectivity filter, deep in the pore inner vestibule (Jenkins et al., 2011). It was also suggested that the selectivity region of KCa2.x channels represents a binding site for potentiators such as GW542573X and CM-TPMF, with the residue equivalent to L215 in the KCa3.1 S5 transmembrane helix playing a determinant role (Hougaard et al., 2009, 2012). Therefore, these observations tend to support a model where the selectivity filter region constitutes a key determinant to the action of KCa3.1 regulators while demonstrating that residues distinct from the CaM-binding domain in the C terminus play a pivotal role in determining apparent Ca2+ affinity and gating properties. However, channel gating is a dynamic process that involves concerted conformational changes in many parts of the channel protein. As Ca2+-dependent gating of KCa3.1 originates from the binding of Ca2+ to CaM in the C terminus, the hypothesis of a gate located at the level of the selectivity filter requires that the conformational change initiated in the C terminus be transmitted to the S6 transmembrane helix and to the channel pore helix directly connected to the selectivity filter. The exact molecular mechanism underlying KCa3.1 opening in response to Ca2+ binding to the CaM–KCa3.1 complex in the C terminus remains to be elucidated. Structural information pertinent to channel gating was, however, obtained through the crystallization of CaM bound to the rat KCa2.2–CaM-binding domain in the presence of Ca2+ (Schumacher et al., 2001). On the basis of this structure, it was proposed that a large-scale conformational rearrangement is taking place in the presence of Ca2+, where the N-lobe of CaM binds to a C-terminal segment of an adjacent channel monomer resulting in a dimerization of contiguous subunits. This rearrangement would in turn lead to a rotation/translation of the associated S6 transmembrane domains and to the opening of the ion-conducting pore (Schumacher et al., 2001, 2004; Wissmann et al., 2002; Maylie et al., 2004).
As the CaM-binding domain of KCa3.1 is directly connected to the S6 transmembrane helix, activation of the channel gate at the level of the selectivity filter could depend upon the coupling between each of the channel pore helices and the associated S6 transmembrane segment. SCAM experiments have already provided evidence for widespread conformational changes in the pore helix of the cyclic nucleotide–gated channel during gating (Liu and Siegelbaum, 2000). These observations were interpreted as evidence for a gate localized at the selectivity filter of the channel (Liu and Siegelbaum, 2000). Mutations that perturb the pore helix of the Kir6.2 channel were similarly reported to affect the fast gating kinetics of the channel (Proks et al., 2001). The importance to gating of the interactions between the selectivity filter and the pore helix in channels was also confirmed in a study on KirBac3.1, which showed that changes at the level of the cytoplasmic domain were relayed to the selectivity filter (Gupta et al., 2010). Similarly, changes in the conductivity of the KirBac selectivity filter were found to be correlated with remote conformational changes of the cytoplasmic domains, in agreement with the selectivity filter playing a role in gating by directly responding to global conformational changes of the channel (Clarke et al., 2010). Finally, work on the KcsA channel has led to the conclusion that C-type inactivation could arise from mechanical deformations propagating through a network of steric contacts between the channel inner helical bundle (equivalent to T278 KCa3.1) and the C-terminal end of the pore helix (equivalent to L249–T250 KCa3.1) (Cordero-Morales et al., 2007). Collectively, these observations suggest that residues located deep in the KCa3.1 channel pore, close to the selectivity filter, could not only contribute to the CaM–KCa3.1 complex to gating but also represent target sites for a pharmacological control of KCa3.1 activity.
How the selectivity filter region in KCa3.1 is involved in gating remains unknown. However, one of the distinguishing features of the KCa3.1 Ca2+ dependence is that Pomax, the channel open probability at saturating Ca2+ concentrations (>20 µM), remains low, typically 0.1–0.2 for the wild-type channel, in contrast to a Pomax of 0.8 reported for the KCa2.2 channel in the high activity mode (Hirschberg et al., 1998). The observation of a low Pomax at saturating Ca2+ conditions argues for the binding of Ca2+ to the CaM–KCa3.1 complex, promoting the formation of a closed-state configuration from which the channel transits to an open configuration. Such behavior has been documented in numerous ligand-gated channels where the agonist does not alter the open state but brings the channel to a preopen configuration. It is likely that the setting of Pomax depends on the energetics governing the interactions taking place at the selectivity filter level.
In this study, we tested the hypothesis that interactions of the KCa3.1 pore helix with the S5 and S6 transmembrane segments contribute in setting Pomax. More specifically, we intended to identify which residues of the pore helix are involved in functionally coupling the pore helix to the S5 and S6 transmembrane segments during gating. Our results demonstrate that important changes in Pomax can be obtained by modulating aromatic–aromatic interactions between F248 of the pore helix and W216 of the S5 transmembrane helical segment, and/or by perturbing the interactions between F248 and the S6 transmembrane segment at the level of the G274 hinge residue.
MATERIALS AND METHODS
Computer-based homology modeling
A first class of 3-D structures for the KCa3.1 pore region (F190–R287) was generated through homology modeling (Šali and Blundell, 1993) using as template the crystal structure of the Kv1.2 channel (Protein Data Bank accession no. 2A79). The choice of the Protein Data Bank 2A79 structure was essentially based on the analysis provided by the I-TASSER server, which identified Protein Data Bank 2A79 as the template with the best TM score of 0.869, knowing that a TM score >0.5 corresponds approximately to two structures of similar topology (Roy et al., 2010). The model includes part of the S4–S5 cytosolic linker plus the segment extending from the N terminus of the S5 transmembrane helix to the C terminus of the S6 transmembrane helix. A second class of models was obtained from the SAM-T08 server, which identified the Protein Data Bank accession number 3BEH (bacterial cyclic nucleotide–regulated channel) structure as the top-ranking template (Karplus, 2009). Both templates were thus used to generate models of the KCa3.1 channel in the open (Protein Data Bank 2A79) and closed (Protein Data Bank 3BEH) configuration.
Molecular dynamics (MD) simulations
MD simulations were performed using the CHARMM-CGENFF force field with the channel pore region incorporated into a DPPC lipid bilayer in contact at top and bottom with an identical explicit water medium. Channel incorporation was accomplished according to the procedure implemented in the Charmm-Gui package (Jo et al., 2008). Overall, the system contains 55,732 atoms including 161 DPPC lipid molecules, 9,576 TIP3P model water molecules, plus 19 K+ and 33 Cl− ions to ensure electroneutrality at near physiological concentration. Cut-on and cutoff parameters needed to define nonbonded interactions were set to 10 and 12 Å, respectively, and SHAKE constraints were used to fix lengths of bonds involving hydrogen atoms. The system was equilibrated according to the six-cycle scheme implemented in the Charmm-Gui output for a total equilibration period of 500 ps. Trajectories were generated for 32–64 ns using a time step of 2 fs, and electrostatic and van der Waals interaction energies were computed from trajectories sampled at 0.2 ns. MD simulations were performed for a system at constant pressure (1 atm) and constant temperature (300°K). Solvent accessibility surface areas (SASAs) were averaged over a 64-ns trajectory using a standard probe of 1.6 Å.
Channel expression and molecular biology
Wild-type and mutant KCa3.1 channels were expressed in Xenopus laevis oocytes by injection of their RNAs transcribed in vitro from pT7TS vector, which contains 5′ and 3′ untranslated regions of Xenopus β-globin mRNA. Approximately 1–10 ng KCa3.1 RNA was injected into each oocyte. Recordings were performed 1–7 d after injection. All the mutants were generated using the Q5 Site-Directed Mutagenesis kit (New England Biolabs, Inc.) and verified by sequencing at the Université de Montréal genomics platform. Before patch clamping, the defolliculated oocyte was briefly incubated in a hyperosmotic solution containing (mM) 250 KCl, 1 MgSO4, 1 EGTA, 50 sucrose, and 10 HEPES, buffered at pH 7.4 with KOH and the vitelline membrane peeled off using fine forceps. The oocyte was then transferred to a superfusion chamber for patch-clamp measurements.
Solutions
The bath and patch pipette solutions contained (mM) 200 K2SO4, 1.8 MgCl2, 0.025 CaCl2, and 25 HEPES, buffered at pH 7.4 with KOH (referred to 200 mM K2SO4). Sulfate salts were used to minimize the contamination from endogenous Ca2+-dependent chloride channels while enabling the chelation of contaminant divalent cations such as Ba2+. Calcium-free solutions were prepared by adding 1 mM EGTA to 200 mM K2SO4 solutions without CaCl2. Bath solution changes were performed as described previously using a rapid solution changer system (RSC-160; Biological) (Banderali et al., 2004). The solution exchange time was <10 ms. bis(2-mercaptoethyl)sulfone (BMS; EMD Millipore) was directly dissolved in the 200-mM K2SO4 solution at a concentration of 8 mM.
Patch-clamp recordings
Multiple- and single-channel inside-out recordings were performed using an amplifier (Axopatch 200A; Molecular Devices). Patch pipettes were pulled from borosilicate capillaries using a pipette puller (model PP-83; Narishige), which was used uncoated. The resistance of the patch electrodes ranged from 2 to 5 MΩ. Data acquisition was performed using a Digidata 1320A acquisition system (Molecular Devices) at a sampling rate of 1.0 kHz unless specified otherwise. Single-channel analysis was carried out using the QUB package (Qin et al., 1996, 1997). Dwell-time measurements were performed on data that were idealized according to the segmental-k means method based on a hidden Markov model–type analysis.
Unnatural amino acids incorporation
Incorporation of unnatural amino acids was performed according to the procedure described by Wang et al. (2006). In brief, we used a pU6-pMpa plasmid (provided by P.G. Schultz, Scripps Research Institute, La Jolla, CA), which encodes two genes: Bacillus stearothermophilus Tyr-tRNA (with anti-codon mutated to CUA, complement to the TAG stop codon) and Escherichia coli Tyrosyl-tRNA-synthetase, which was mutated in Y37V/D182S/F183M to make the plasmid available to specifically recognize the unnatural amino acid p-methoxy-l-phenylalanine (pMpa). The plasmid was coexpressed with the KCa3.1 channel mutated at position F248 into an amber stop codon (TAG) in Xenopus oocytes, in which 5 mM pMpa (Sigma-Aldrich) had been injected.
Statistical analysis
Statistical significance was analyzed using unpaired Student’s t test. P < 0.05 was considered statistically significant. Data are expressed as mean ± SD.
Online supplemental material
Fig. S1 shows a Western blot of full-length expression of a KCa3.1 channel tagged with 6-His at position 132 in the external linker S1–S2, where residue F248 has been replaced by the unnatural amino acid pMpa. Fig. S2 presents a Western blot obtained for the F248W–W216F and W216F mutants, confirming channel expression. Fig. S3 presents the open and closed dwell-time distributions computed for the wild-type, F248A, and F248W KCa3.1 channels. The online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.201311097/DC1.
RESULTS
Structural properties of the pore helix region
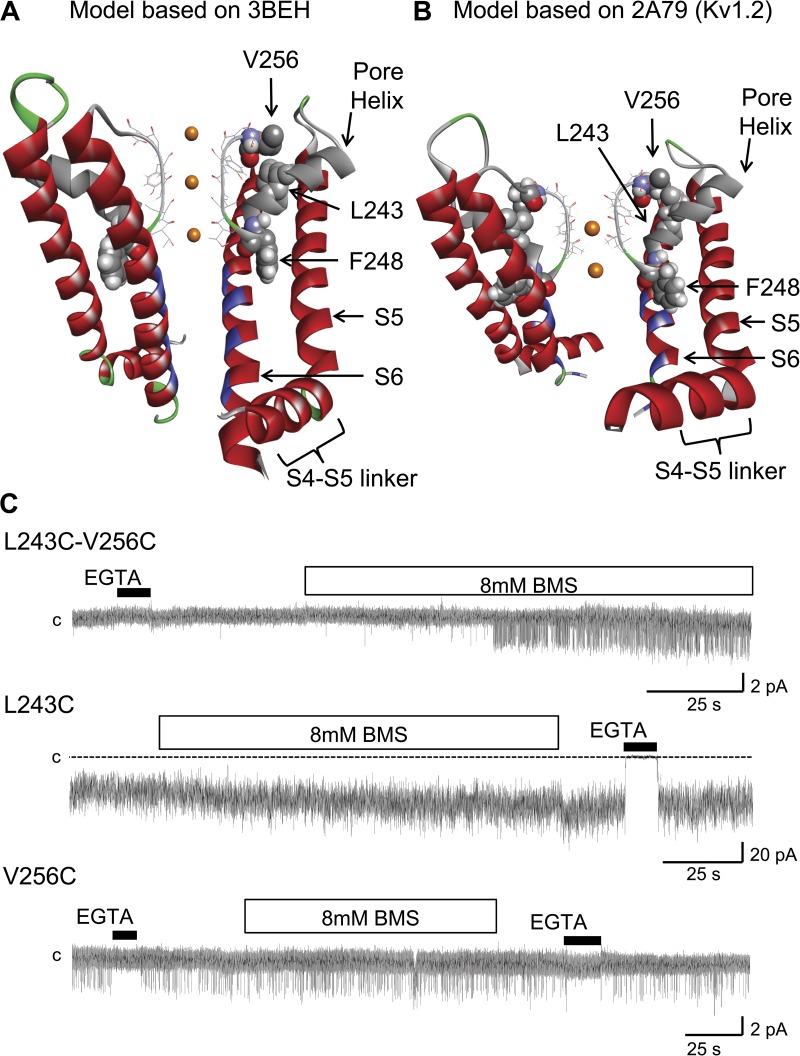
The proposed structures for the 3BEH-based (bacterial cyclic nucleotide–regulated channel) and 2A79-based (Kv1.2 channel) KCa3.1 channel pore region are presented in Fig. 1 (A and B). The selectivity filter is seen to be extending from T250 to T260 and is connected to the pore helix comprising amino acids G235 to L249. In both models, the residues V275, T278, V282, and A286 (blue) are predicted to line the channel pore, in agreement with our previous SCAM studies (Klein et al., 2007). An analysis of the distance between the Cα of corresponding residues along S6 on opposite subunits indicates that the narrowest part of the conduction pathway should be located at the level of V282, with a van der Waals pore diameter of ∼4.5 Å for the 3BEH-based model compared with >12 Å for the model derived from the 2A79 template. It follows that the structure obtained from the 3BEH template is likely to be more representative of the KCa3.1 channel in the closed conformation relative to the 2A79-based model, which better accounts for the KCa3.1 open configuration. Both models predict that residues I244 and F248 of the pore helix should be projecting between the S5 and S6 transmembrane helices, whereas L243 should be oriented toward the selectivity filter in proximity of V256 at the C-terminal end of the selectivity filter region. The proposed configuration for the pore helix was investigated in experiments where the spatial proximity of the L243 and V256 residues was tested through disulfide bond formation between Cys engineered at L243 and V256, respectively. The inside-out patch-clamp recordings presented in Fig. 1 C confirmed in this regard that channel activity of the L243C–V256C double mutant was sensitive to the addition of the small reducing agent BMS to the bathing solution. These observations confirmed that disulfide bond formation between L243C and V256C tends to stabilize the channel closed state configuration by constricting the selectivity filter. This effect was specific to the double mutation L243C–V256C system, as demonstrated in the multi-channel (L243C) and single-channel (V256C) recordings presented in Fig. 1 C where channel activity appeared insensitive to BMS application for single mutation channels.
Figure 1.
Ribbon representation of the KCa3.1 pore region, including the S4–S5 cytosolic linker, the S5 and S6 transmembrane helices, the pore helix (gray), and the selectivity filter regions (stick representation). (A) Model structure generated by MODELLER9.11 using as template the crystal structure solved for a bacterial cyclic nucleotide–regulated channel MlotiK1 (Protein Data Bank accession no. 3BEH). Also shown is a space-filling representation of residues L243, F248, and V256, with the L243 side chain projecting opposite to the S5 and S6 transmembrane segments toward V256 of the selectivity filter. In contrast, F248 is predicted to be oriented with its side chain making contact with both the S5 and S6 helices. The residues documented to be facing the channel pore are colored in blue (Klein et al., 2009). (B) Model structure generated by MODELLER9.11 using as template the crystal structure solved for the mammalian Kv1.2 channel (Protein Data Bank accession no. 2A79). As in A, residue F248 is seen as projecting between the transmembrane helices S5 and S6. Only two subunits are presented for clarity. (C) Inside-out patch recordings supporting the formation of a disulfide bond between Cys engineered at 256 and 243. The reducing agent BMS caused channel activation when applied onto the L243C–V256C double mutation channel but failed to affect channel activity when applied to the V256C (single-channel) and L243C (multi-channel) single mutation channels. These observations support the orientation of the pore helix relative to the selectivity filter proposed in A and B. Channels were expressed in Xenopus oocytes, and current records were obtained at −60 mV in symmetrical 200-mM K2SO4 conditions at an internal Ca2+ concentration of 25 µM. The label “c” refers to the zero current level. Illustration by Discovery Studio Visualizer (Accelrys).
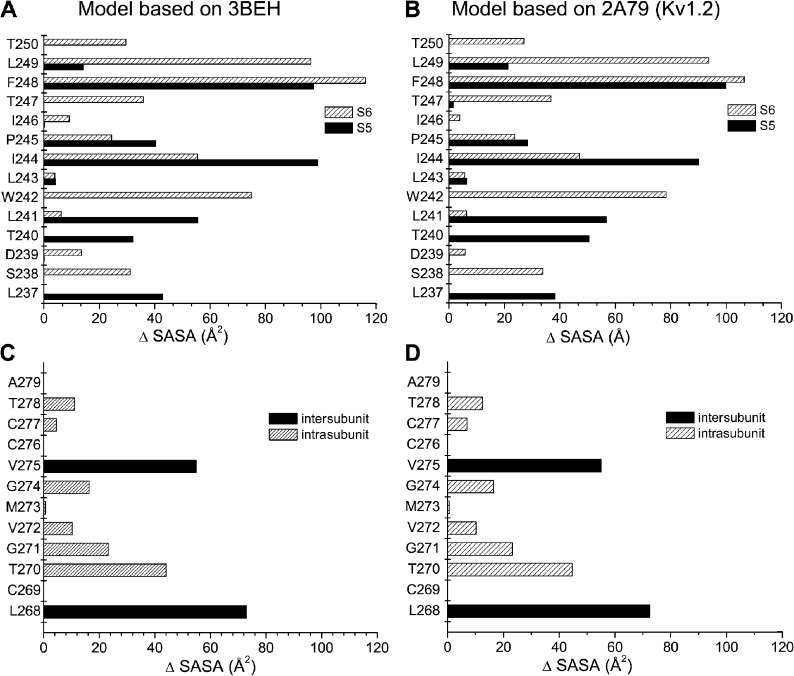
A computer analysis was next undertaken to determine which residues of the KCa3.1 pore helix form the interface with the S5 and S6 transmembrane segments. Our approach consisted of computing for each residue of the pore helix the difference in SASA (ΔSASA) with and without the S5 and S6 helices. It is expected that KCa3.1 residues not located at the interface formed by the pore helix with the S5 and S6 transmembrane segments will have SASA values unaffected by the presence of the S5 or S6 helix (ΔSASA = 0). In contrast, residues of the pore helix contributing to the interface with the S5 and S6 helices should be characterized by a reduction of their SASA values when calculations include either the S5 or S6 transmembrane segment. Fig. 2 (A and B) summarizes the predicted contribution of each pore helix residue to the interface, with the S5 and S6 transmembrane helices averaged over a 64-ns MD simulation. As seen, the largest contact areas with both the S5 and S6 helices involve residue F248 at the C-terminal end of the pore helix regardless of the template used. In contrast, residues T240, L241, and I244 of the pore helix appeared to contribute more to the S5 than S6 transmembrane segment interface, with ΔSASA values remaining overall smaller relative to F248. Notably, calculations for residues L249 and W242 revealed contact areas almost entirely limited to the S6 transmembrane helix, an effect coming from W242 and L249 contributing to intersubunit contacts. As expected, ΔSASA values for L243 and I246 predicted to be oriented facing the selectivity filter were not affected by the presence of the S5 and S6 helices, indicating small if any interface contact surfaces. Collectively, this analysis showed that the largest contribution to the pore helix interface with both the S5 and S6 transmembrane segments is coming from residue F248.
Figure 2.
Characterization of the interface formed by the pore helix with the S5 and S6 transmembrane segments based on the difference in SASA (ΔSASA) for the KCa3.1 model derived from the 3BEH (A and C) or 2A79 (B and D) templates. ΔSASA values were computed from MD 64-ns trajectories using the Charmm37 force field. The contribution of individual pore helix residue to the interface formed by the pore helix and S5 transmembrane segment or pore helix and S6 transmembrane helix (A and B) was estimated as described in the Results. The results in A and B show that the average contact area with either the S5 or S6 transmembrane segment is maximal for residue F248 independently of the template used. In contrast, residues T240, L241, and I244 contributed more to the interface between the pore helix and the S5 than the S6 segment, whereas residues W242 and L249 appeared to form an interface with S6 exclusively. Notably, residues L243 and I246 failed to contribute to the interface formed by the pore helix with the S5 and S6 transmembrane segments, in accordance with these residues being oriented toward the channel selectivity filter. A similar procedure applied to the interface formed by the pore helix and the S6 transmembrane segment (C and D) revealed important contributions coming from T278, C277, V275, G274, V272, G271, T270, and L268, with the V275 and L268 contribution coming essentially from intersubunit interactions. It is concluded that the interface between the pore helix and the S5 plus S6 segments of each subunit essentially involves F248 at the C-terminal end of the pore helix.
Because the CaM–KCa3.1 complex is directly connected to the S6 transmembrane helix, a similar procedure was applied to identify residues of the S6 segment contributing to the interface with the pore helix. The calculations presented in Fig. 2 (C and D) show that residues T278, C277, V275, G274, V272, G271, T270, and L268 in S6 are susceptible to interact with the channel pore helix in both models. Interactions involving V275 and T268 appeared unique, however, as they represent intersubunit contacts exclusively. Of interest, we noted that residue V275 can interface with L249 at the C-terminal end of the pore helix on an adjacent subunit. A close proximity between V275 and an adjacent pore helix would be in agreement with the observed strong channel inhibition by MTS reagents of the V275C mutant channel (Garneau et al., 2009), and with the contribution of V275 to the TRAM-34–blocking action (Wulff et al., 2000).
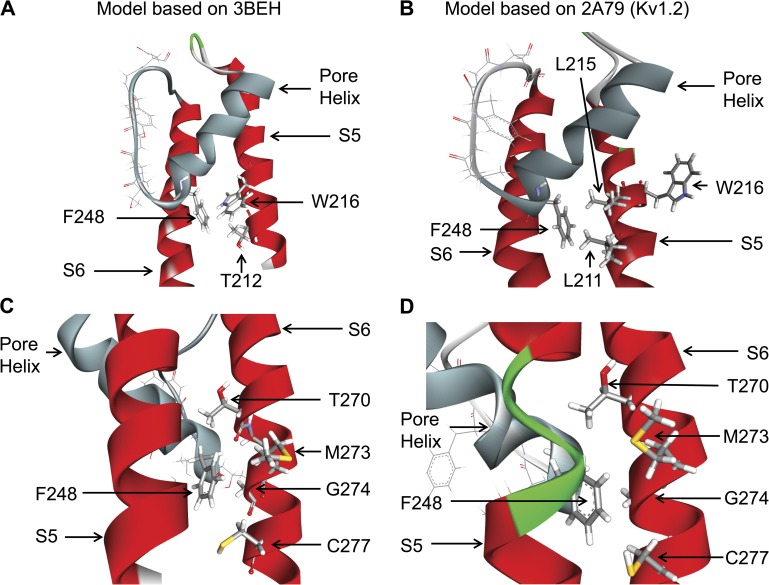
Detailed representations of the interaction network between F248 and residues in S5 and S6 derived either from the 3BEH (bacterial nucleotide–activated potassium channel; A and C) or 2A79 (Kv1.2; B and D) templates are presented in Fig. 3. In both models, the side chains of T270, G274, and C277 of the S6 transmembrane helix are seen to be oriented toward the pore helix at the level of F248 (Fig. 3, C and D). However, the interaction pattern of the pore helix–S5 residues was found to be model dependent, with T212 and W216 facing F248 in the 3BEH-based model (Fig. 3 A), in contrast to the structure derived from the Kv1.2 (2A79) template where F248 essentially makes contact with L211 of S5 (Fig. 3 B).
Figure 3.
Detailed representations of the KCa3.1 channel pore helix region (gray) derived either from the 3BEH (bacterial nucleotide–activated potassium channel) or 2A79 (Kv1.2) templates. A and B illustrate that the interface between F248 and the S5 transmembrane helix is template dependent, with F248 predicted to be interacting with W216 and T212 of S5 for the 3BEH-derived model but with L211 for the model obtained using the Kv1.2 structure (2A79) as template. C and D show in contrast a structural organization of the interface between F248 and the S6 transmembrane segment in which F248 is in both cases within proximity of the Gly hinge at 274 on S6 added to potential contacts with C277, M273, and T270. Illustrations by Discovery Studio Visualizer (Accelrys).
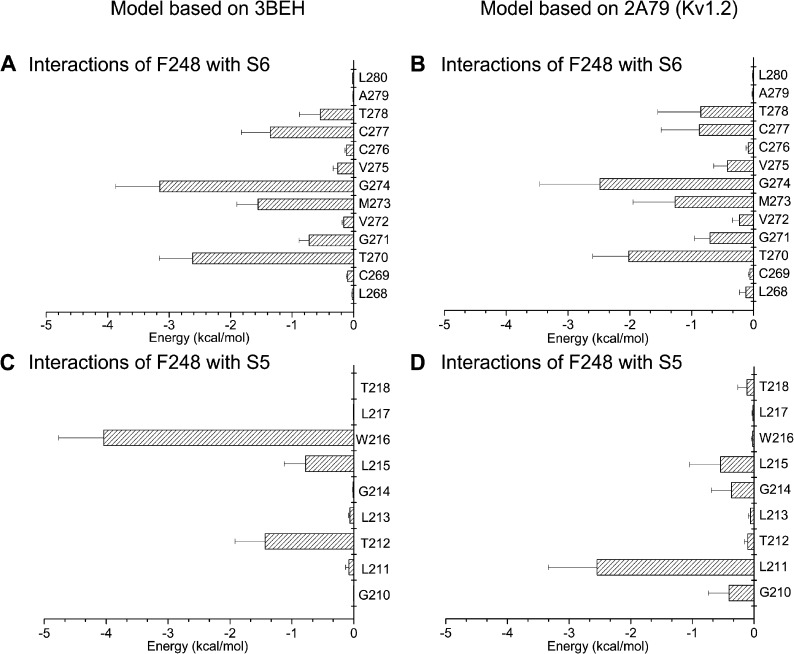
A detailed analysis of the nonbonded van der Waals plus electrostatic energy between F248 of the pore helix and residues extending from L268 to L280 along the S6 transmembrane segment is presented in Fig. 4 (A and B). The most important interactions were seen between F248 and the glycine hinge G274, arguing for F248 being within close proximity of the G274 backbone. Strong interactions were similarly estimated between F248 and T270, G271, M273, C277, and T278. These predictions appeared template independent and could be obtained for the 3BEH- and 2A79-based models. Interaction energy between residue F248 and residues of the S5 transmembrane helix is presented in Fig. 4 (C and D). The resulting interaction pattern appeared in this case model dependent, with F248 favorably interacting with W216, T212, and L215 in the 3BEH-derived structure, and with L211 for the Kv1.2-based open configuration.
Figure 4.
Electrostatic and van der Waals interaction energy between F248 and residues L268 to L280 of S6 (A and B) or residues G210 to T218 of S5 (C and D). Energy calculations were based on 64-ns MD trajectories obtained for the KCa3.1 model derived either from the 3BEH (nucleotide-activated) or 2A79 (Kv1.2) templates. The bar graphs in A and B indicate that F248 of the pore helix strongly interacts with the Gly hinge at 274 for both models and to a lesser extent with T270, M273, C277, and T278. The interaction energy pattern remained overall template independent. In contrast, strong interaction energies were estimated between F248 and W216 for the KCa3.1 model derived from the 3BEH template, whereas the maximum interaction energy for the KCa3.1 2A79-derived model involves residues F248 and L211. This analysis thus indicates that the interaction energy pattern between F248 and residues in S5 varies according to the template used.
Which residues of the pore helix affect the channel maximum open probability?
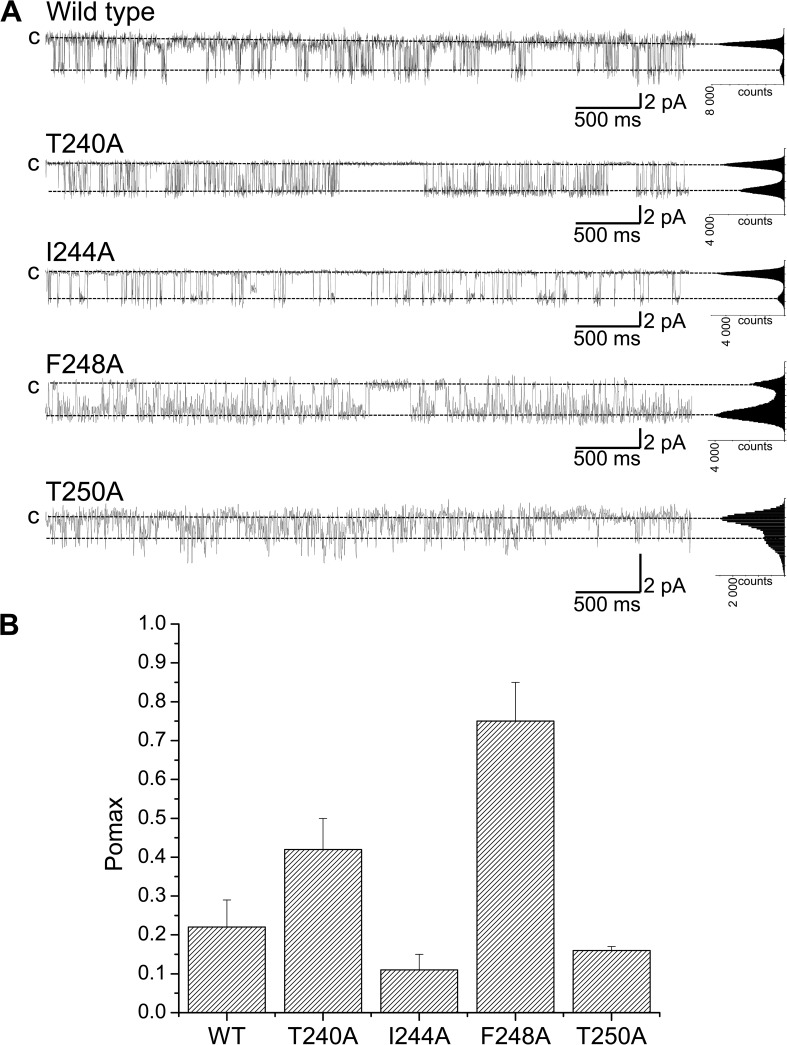
The previous analysis has led to the identification of residues predicted to line the interface formed by the pore helix with the S5 and S6 transmembrane segments. An alanine scan was thus performed to determine how important these residues are in setting Pomax. Typical single-channel recordings are presented in Fig. 5 A. We note that the mutation F248A resulted in a Pomax of 0.75 ± 0.10 (n = 3) compared with 0.22 ± 0.07 (n = 8) for the wild-type channel (Fig. 5 B, WT). The mutation T240A led to a less drastic increase in Pomax with a value of 0.42 ± 0.08 (n = 4) compared with I244A, which resulted in a Pomax decrease to 0.10 ± 0.04 (n = 4). The mutation T250A had a minor impact on the channel Pomax but resulted in an important decrease in the channel unitary conductance, from 30 pS for KCa3.1 wild type to 9 pS for the T250A mutant (not depicted). Collectively, these results show that the mutation F248A is unique, as it caused a 3.2-fold increase in Pomax.
Figure 5.
Alanine scan of the KCa3.1 pore helix region. (A) Inside-out current recordings obtained in symmetrical 200-mM K2SO4 conditions at a pipette potential of 100 mV (T240A, I244A, and F248A) or 150 mV (T250A) from KCa3.1 mutants expressed in Xenopus oocytes. The label “c” refers to zero current level. The internal Ca2+ concentration was 25 µM throughout. The mutation F248A caused a highly significant increase of the channel open probability at saturating internal Ca2+ concentration (Pomax) relative to wild type (P < 0.0005) compared with T240A (P < 0.01). The substitution I244A caused a nonsignificant variation in Pomax relative to wild type. Because of its strategic position at the N-terminal end of the selectivity filter, the mutation of T250 to Ala resulted in an important decrease in channel conductance (9 pS compared with 30 pS), with Pomax being not significantly different from wild type. These results are summarized in B with Pomax values of 0.42 ± 0.08 (n = 4), 0.10 ± 0.04, 0.75 ± 0.01 (n = 3), and 0.16 ± 0.01 (n = 2) for the T240A, I244A, F248A, and T250A channels, respectively. Our results point toward the interactions involving F248 with the S5 and S6 helices as being determinant in setting Pomax. Also shown are all-point histograms computed from the entire recording.
Aromatic residues control Pomax
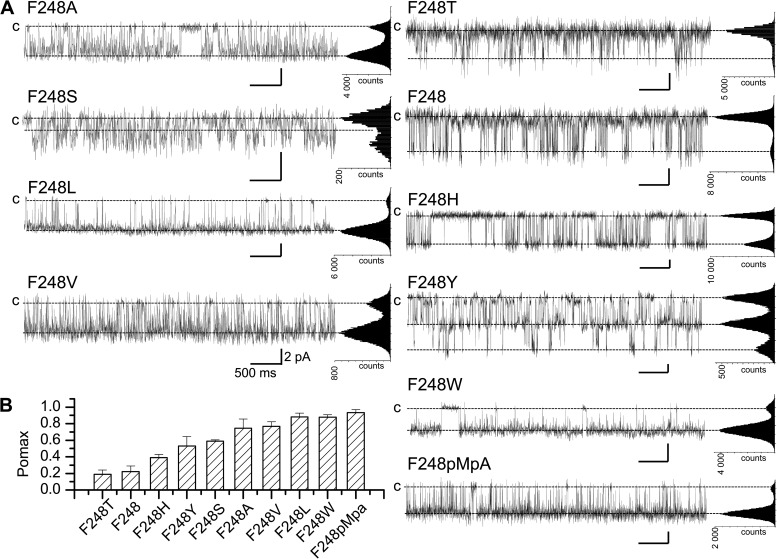
To determine if hydrophobicity and/or volume of the residue at position 248 control Pomax, experiments were performed in which F248 was substituted by residues differing in size and/or hydrophobicity. Typical unitary current traces are presented in Fig. 6. These recordings demonstrate that among all the mutants tested, F248T only yielded Pomax values (0.19 ± 0.05; n = 3) not superior to wild type (F248, 0.22 ± 0.07; n = 8). Clearly, there was no direct correlation between Pomax and the volume of the substituting residue, as replacing F248 by a smaller (F248A) or larger amino acid (F248W) resulted in both cases in an increased Pomax relative to wild type. In addition, we noted that although Val (V) and Thr (T) are of roughly the same shape and volume, there was a significant difference in channel activity, with Pomax of 0.77 ± 0.05 (n = 3) and 0.19 ± 0.05 (n = 3) for the F248V and F248T mutants, respectively. We concluded that the substitution of a methyl (V) by a hydroxyl group (T) modified the interaction energy pattern involving residue 248 as to cause a decrease in Pomax value. The results of this analysis are summarized in Fig. 6 B, which shows that the overall Pomax ranking reads T ≈ F < H < Y < S < A < V < L ≈ W.
Figure 6.
Examples of inside-out current recordings (A) and calculated Pomax (B) for KCa3.1 channels after the substitution of F248 by residues differing in volume size and/or hydrophobicity. Recordings were performed in symmetrical 200-mM K2SO4 conditions in saturating internal Ca2+ (25 µM) at a pipette potential of 100 mV. Low Pomax values compared with wild type were obtained with the F248T mutant only with a mean value of 0.19 ± 0.05 (n = 3). The substitution of F248 by either a smaller (F248A) or larger (F248W) residue led systematically to a Pomax increase. Also shown is an example of inside-out current trace obtained in conditions where F248 was replaced by the unnatural amino acid pMpa, a tyrosine analogue obtained by substituting the hydroxyl moiety of Tyr by an O-CH3 group. This substitution led to a drastic increase in Pomax with a mean value of 0.94 ± 0.03 (n = 4), compared with 0.22 ± 0.07 (n = 8) for wild type. (B) Bar graph summarizing the effect of substituting F248 by nonpolar residues differing in volume size and/or hydrophobicity. The resulting Pomax ranking reads: T ≈ F < H < Y < S < A < V < L ≈ W < pMpa, confirming the absence of correlation between Pomax and the volume size and/or hydrophobicity (A < S < T < H < V < Y < L < F < W) (Wimley et al., 1996) of the substituting residue. Also shown are all-point histograms computed from the entire recording. Bars, 2 pA and 0.5 s.
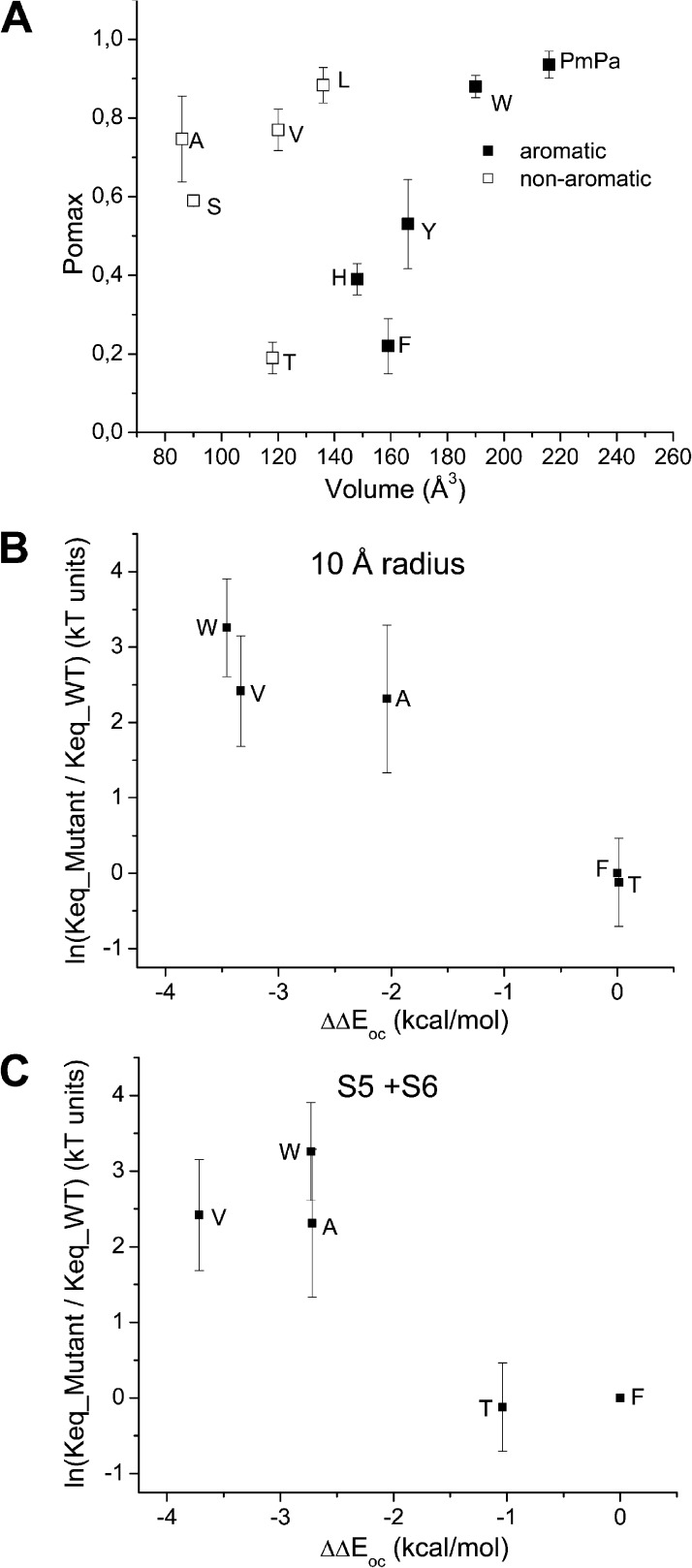
A scatter plot of Pomax as a function of residue 248 volume size is presented in Fig. 7 A. This analysis reveals two distinct sets of data. Whereas there was a modest but significant increase in Pomax as a function of volume size for Ala, Val, and Leu, drastic changes were observed with the aromatic (Phe, Tyr, His, and Trp) residues. Clearly, increasing the size of the aromatic side chain engineered at position 248 resulted in higher Pomax values. Importantly, aromatic residues such as Phe and Tyr showed significant lower Pomax values compared with nonaromatic amino acids of similar sizes such as Val or Leu, suggesting specific effects related to the presence of aromatic rings in these cases. To confirm that the size of aromatic residues at 248 is a key determinant to Pomax, experiments were undertaken in which we used the unnatural amino acid pMpa, a tyrosine analogue obtained by substituting the hydroxyl moiety of Tyr by a O-CH3 group. The resulting aromatic amino acid has an estimated volume of 227 Å3 compared with 192 Å3 for Tyr and 203 Å3 for Trp. Single-channel recordings confirmed that the substitution of F248pMpa caused an important increase in Pomax, with a mean value of 0.94 ± 0.03 (n = 4), in agreement with the proposal of a strong correlation between Pomax and the volume of the aromatic residue engineered at 248 (see Fig. 7 A).
Figure 7.
(A) Effect on Pomax of substituting F248 by either nonaromatic or aromatic residues. Scatter plot of Pomax as a function of the residue volume. This analysis reveals two distinct sets of data. Whereas there was a modest but significant increase in Pomax as a function of volume size for Ala (A), Val (V), and Leu (L), drastic changes were observed with the aromatic (Phe [F], Tyr [Y], His [H], Trp [W], and pMpa) residues. Importantly, aromatic residues such as Phe (F) and Tyr (Y) showed significant lower Pomax values compared with nonaromatic amino acids of similar sizes, such as Val (V) or Leu (L), suggesting specific effects related to the presence of an aromatic residue at 248. (B and C) Ln(Keq_mutant/Keq_wild_type), where Keq = Pomax/(1 − Pomax) plotted as a function of the energy perturbation ΔΔEoc coming from mutating residue 248 for the F248T, F248V, F248A, and F248W mutants. ΔΔEoc was calculated as ΔEoc_mutant − ΔEoc_wild_type, where ΔEoc is the difference in nonbonded energy for residue 248 between the open and closed state. Nonbonded energies were computed from >32–64-ns MD trajectories by averaging the van der Waals plus electrostatic interactions between the residue at 248 and the surrounding atoms within a 10-Å radius (excluding lipids and water) (B), or between the residue at 248 and the S5 and S6 helices (C). The resulting analysis is in agreement with the Pomax ranking F248T ≈ F248 < F248A ≈ F248V < F248W observed experimentally, while establishing an energy equivalence (ΔΔEoc ≈ 0) between a Thr (T) and a Phe (F) at 248. The latter result argues for π–π interactions involving F248 and the S5 transmembrane segment in the closed state as being determinant in setting Pomax for the KCa3.1 wild-type channel. These simulations also predict an increase in Pomax relative to wild type with the F248W mutant despite the possibility of π–π interactions with S5, an effect attributable to the mutation F248W stabilizing to a greater extent the channel open than closed configurations. In contrast, the Pomax increase seen with the F248A and F248V mutants appeared to be related to a stronger destabilization for the channel closed than open state.
S5 transmembrane helix control of Pomax via F248
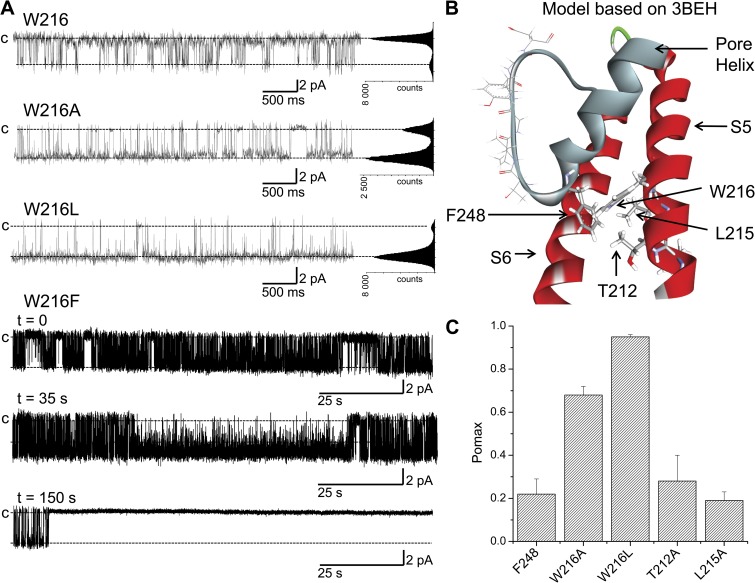
The observation of a lower Pomax value for aromatic compared with nonaromatic residues engineered at 248 could be indicative of specific interactions involving aromatic–aromatic interactions potentially contributing to the energy barrier controlling channel opening. Aromatic amino acids are known to play a prominent role in determining protein structure through π–π or hydrogen–π interactions. For polar hydrogen–π interactions, the donors are hydrogen atoms connected to electronegative atoms (for instance, R-OH, R-NH2), and the acceptors may be represented by various aromatic molecules and conjugate π groups (Du et al., 2013). More importantly, we note that the 3BEH-based model of KCa3.1 illustrated in Fig. 3 A shows possible face-edge interactions between F248 and W216 of the S5 transmembrane helix. As Tyr, Trp, His, and Phe constitute polar hydrogen acceptors, which can potentially interact with a polar hydrogen donor such as Trp (NH) (Du et al., 2013), the effect on Pomax seen with the substitutions F248Y/H/W could be indicative of polar hydrogen–π interactions and/or π–π interactions between the residue 248 in the pore helix and W216 in S5. Such interactions would be absent with the F248A/V/L mutations, leading to higher Pomax values as seen experimentally. As Trp can also behave as a hydrogen acceptor in polar hydrogen–π interactions, a strong interaction between F248T (hydrogen donor) and W216 could explain the unexpected observation of F248T leading to a smaller Pomax compared with F248A despite a larger side-chain volume. However, this interaction would be size dependent, as the Pomax for the F248S mutant is similar to the Pomax of F248A. It follows that substituting W216 (hydrogen donor) by nonaromatic residues such as Ala or Leu should impair aromatic–aromatic interactions involving F248 and reproduce the effects observed when F248 was substituted by a nonaromatic amino acid. Examples of single-channel recordings obtained with the W216A and W216L mutants are illustrated in Fig. 8 A. Clearly, replacing W216 by a nonaromatic residue caused a drastic increase in Pomax, with values of 0.68 ± 0.04 (n = 3) and 0.95 ± 0.01 (n = 3) for W216A and W216L, respectively (see Fig. 8 C). The effect observed with W216A cannot be attributed to a decrease of the side-chain volume, as the substitution W216L led to a higher Pomax value relative to W216A. Collectively, these results support a model in which polar hydrogen–π and/or π–π interactions involving residue 248 of the pore helix and W216 on S5 (Fig. 8 B) contribute to stabilize the gate closed configuration, while confirming the prominent role of the selectivity filter region to the KCa3.1 gating process. In line with this proposal is the observation that substituting W216 by the smaller Phe aromatic residue (W216F) resulted either in the absence of detectable channel activity (50% of the 25 oocytes tested) despite Western blot evidence for channel expression (see Fig. S2), or in single-channel records where the channel gate switched within <3 min in 50% of the recordings considered, from a high Pomax state (0.7 ± 0.2; n = 3) to a permanent closed configuration (Fig. 8 A). Such behavior was not seen when W216 was mutated to a nonaromatic residue such as Ala or Leu (Fig. 8 A). These observations are compatible with aromatic–aromatic interactions involving F248 and W216F significantly increasing the likelihood of forming a stable closed state. Notably, experiments in which the Phe at 248 and Trp at 216 were interchanged (F248W–W216F) failed to yield detectable channel activity despite biochemical evidence of channel expression (see Fig. S2).
Figure 8.
Evidence indicating potential aromatic–aromatic interactions involving residue 248 of the pore helix and W216 of S5. (A) Inside-out current recordings demonstrating that the absence of aromatic–aromatic interactions resulting from the substitution of W216 by the nonaromatic Ala or Leu residue caused a strong increase in channel activity, with Pomax values of 0.68 ± 0.04 (n = 3) and 0.95 ± 0.01 (n = 3), respectively, compared with 0.22 ± 0.07 (n = 8) for the wild-type channel (W216). However, such behavior was not observed with the L215A mutant nor the T212A (not depicted). Substituting W216 by the smaller Phe aromatic residue (W216F) resulted in a single-channel fluctuation pattern characterized by a period of high activity (Pomax = 0.70 ± 0.2; n = 3), ending within <3 min in 50% of the recordings (n = 10) by a transition to stable closed state. Such behavior is compatible with strong aromatic–aromatic F248–W216F interactions maintaining the channel in the closed configuration. These results are summarized in the bar graph presented in C. These observations agree with a model whereby aromatic–aromatic interactions between F248 of the pore helix and W216 of S5 tend to stabilize the channel closed configuration, as suggested by the 3BEH-based model illustrated in B. Recordings were performed in symmetrical 200-mM K2SO4 conditions in saturating internal Ca2+ (25 µM) at a pipette potential of 100 mV. All-point histograms were computed from the entire recording. Illustration by Discovery Studio Visualizer (Accelrys).
Other residues in S5 could be interacting with F248
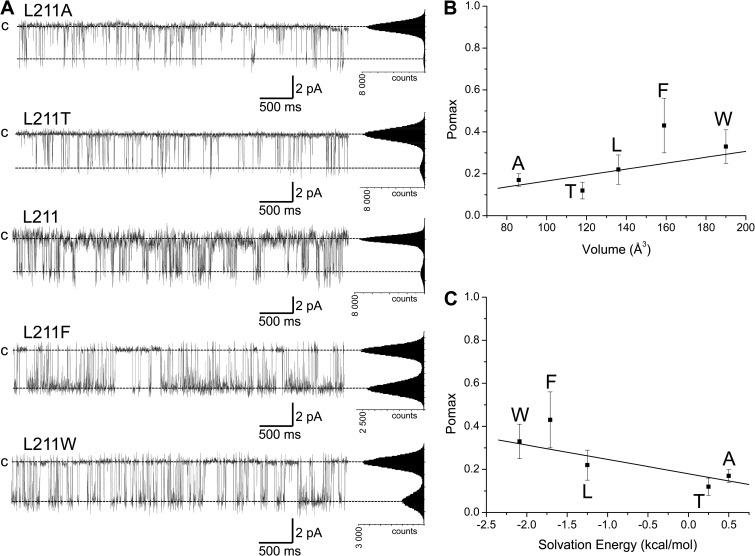
The structural model and the interaction energy diagram illustrated in Figs. 8 B and 4 C, respectively, point toward T212 and L215 in S5 as potential residues susceptible to be within the interaction range of F248. The contribution of these two residues in setting Pomax was investigated in single-channel experiments using the T212A and L215A mutant channels. In contrast to the mutation W216A, the substitution T212A or L215A failed to significantly affect channel activity, with Pomax values of 0.28 ± 0.15 (n = 3) and 0.19 ± 0.04 (n = 3), respectively (Fig. 8 C). These observations do not support a model whereby OH–π interactions involving T212 and F248 would participate in the control of Pomax. In contrast to the model derived from the 3BEH template, the model based on the Kv1.2 (2A79) structure predicts strong interaction energy between F248 and L211, and to a lesser extent with L215. The importance of these interactions in setting Pomax was investigated in inside-out patch-clamp experiments where L211 was substituted by residues of variable volume size and hydrophobicity. The results of these experiments are summarized in Fig. 9. On the basis of the model derived from the Kv1.2 template, one would expect a potential stabilization of the channel open state by favoring π–π or OH–π interactions between F248 and Phe (F), Trp (W), or Thr (T) residues engineered at 211. Our results indicate a small increase in Pomax with the L211F mutant relative to wild type, whereas the mutations L211T and L211W appeared ineffective in modifying Pomax. At best, we found a correlation between the volume size of the residue engineered at 211 and Pomax, with a Pearson correlation coefficient of 0.75 ± 0.15 (Fig. 9 B). A stronger correlation was obtained between Pomax and the solvation energy of the residue engineered at 211 with a Pearson coefficient of 0.84 ± 0.07 (Wimley et al., 1996) (Fig. 9 C). Collectively, these observations suggest that the interactions between F248 and the S5 transmembrane helix are essentially governed by W216 for the channel in the closed configuration (3BEH-based model), with no significant contribution from L211, T212, and L215.
Figure 9.
Effects of mutating residue L211 in S5 on KCa3.1 Pomax. (A) Inside-out current recordings demonstrating that increasing the volume size of the substituting residue at position 211 in S5 has no significant impact on Pomax relative to wild type. A significant increase was, however, observed with the L211F, suggesting possible aromatic–aromatic interactions between L211F and F248. The results of this analysis are presented in the scatter plot illustrated in B, which shows a Pearson correlation coefficient of 0.75 ± 0.15 between the volume size of the residue engineered at 211 and Pomax. (C) A better correlation was obtained between Pomax and the solvation energy of the residue at 211 with a Pearson correlation coefficient of 0.84 ± 0.07. This analysis confirms that L211 can come in close proximity of F248 during gating, as revealed by the Pomax increase seen with the L211F mutant. Recordings were performed in symmetrical 200-mM K2SO4 conditions in saturating internal Ca2+ (25 µM) at a pipette potential of 100 mV. All-point histograms were computed from the entire recording.
F248 interactions with the S6 transmembrane helix
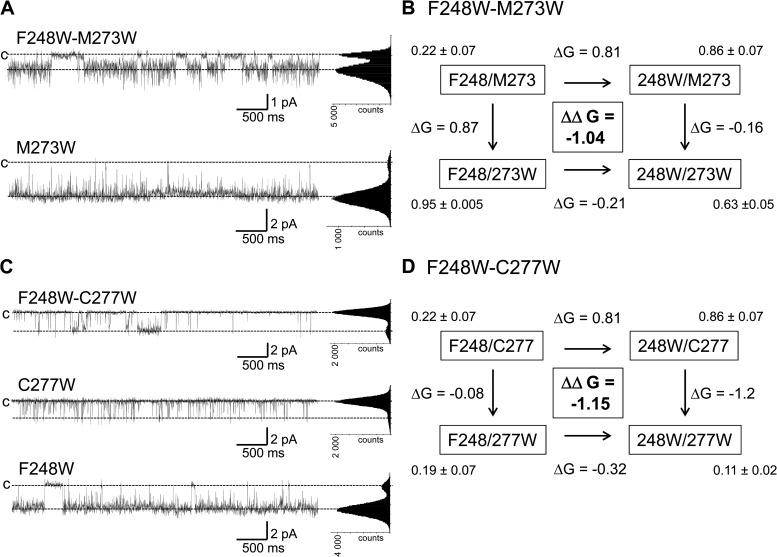
The results presented in Fig. 4 indicate that F248 can interact preferentially with the Gly hinge at 274 and with residues T270, M273, C277, and T278 of the S6 transmembrane helix. MD simulations further suggest that the strong interactions with G274 arise in part from van der Waals contacts indicating a very close proximity between the G274 backbone and the F248 side chain. As the CaM–KCa3.1 complex is directly connected to the S6 transmembrane segment, a double mutant cycle analysis was performed to determine how these interactions could contribute to Pomax. In this analysis, the coupling energy ΔΔG was computed as (ΔGmutation1-mutation2 + ΔGWT-WT) − (ΔGmutation1-WT + ΔGmutation2-WT), with ΔG given by kT ln(Pomax), with k the Boltzmann constant and T the temperature (Horovitz, 1996; Wall-Lacelle et al., 2011). Examples of cycle analyses are shown in Fig. 10. We note first the absence of coupling with either F248 or F248W for the C277A, M273A, and T270A mutants with ΔΔG of 0.02 ± 0.28 kcal/mol, −0.27 ± 0.30 kcal/mol, and −0.01 ± 0.10 kcal/mol for the F248W–C277A, F248W–M273A, and F248W–T270A double mutant cycle systems, respectively (not depicted). In addition, the observation that the mutations T270A and T270S were ineffective in modifying Pomax when added to the F248 or F248W template (not depicted) was seen as evidence for T270 not being involved in OH–π-type interactions with F248 or F248W. Strong ΔΔG of −1.04 ± 0.26 kcal/mol and −1.15 ± 0.56 kcal/mol was obtained, however, with the F248W–M273W and F248W–C277W double mutants (Fig. 10). No current could be detected with the T270W mutant (not depicted). Of interest, the mutation F248W failed to cause an increase in Pomax when added to the C277W template, an effect not seen with C277A (not depicted). These observations suggest potential aromatic–aromatic interactions between F248W and C277W, as seen with W216 (see Fig. 11). We also note that the mutation M273W (Fig. 10 A) led to a Pomax of 0.95 ± 0.005 (n = 3) with F248 and to 0.63 ± 0.05 (n = 4) when F248 was substituted by a Trp (F248W–M273W). With a Pomax of 0.86 ± 0.07 (n = 5) for F248W–M273, 0.80 ± 0.10 (n = 3) for F248W–M273A (not depicted), and 0.63 ± 0.05 (n = 4) for F248W–M273W (Fig. 10 B), there is no apparent correlation between the volume size of residue 273 and Pomax in conditions where F248 has been mutated into a Trp. Such behavior was not found when mutations of M273 were performed on the wild-type KCa3.1 template (F248) with a Pomax of 0.22 ± 0.02 (n = 3) and 0.95 ± 0.005 (n = 3) for the F248–M273A (not depicted) and F248–M273W (Fig. 10 B) channels, respectively. Clearly, the presence at 248 of a Phe with a smaller side-chain volume compared with Trp caused the channel Pomax to become more sensitive to the volume of the residue substituting for M273. These results confirm the close proximity between F248 and M273, as predicted from the homology-based representations of the KCa3.1 open and closed configurations (see Fig. 11).
Figure 10.
Mutant cycle analyses demonstrating functional coupling between F248W of the pore helix and C277W or M273W of the S6 transmembrane segment. Energies are expressed in kilocalorie/mole. (A) Example of inside-out current recordings illustrating the effect of the M273W mutation on the change in Pomax resulting from the substitution at 248 of a Phe by a Trp. These observations are summarized in the cycle diagram illustrated in B, which shows that the mutation M273W resulted in a drastic Pomax increase (0.95 ± 0.005; n = 3), an effect that was partly impaired with a Trp at 248 (Pomax of 0.63 ± 0.05; n = 4). (C) Examples of inside-out current recordings illustrating the coupling between Trp at 248 and 277, respectively. The cycle diagram in D indicates in this regard that the mutation C277W totally prevented the Pomax increase normally resulting from the substitution F248W, while being ineffective in modifying Pomax with F248. These observations would be compatible with Trp–Trp interactions between F248W at the pore helix and C277W on S6 stabilizing the closed state (Fig. 11). On the basis of this analysis, it is concluded that both C277 and M273 are in proximity of F248. Recordings were performed in symmetrical 200-mM K2SO4 conditions in saturating internal Ca2+ (25 µM) at a pipette potential of 100 mV. All-point histograms were computed from the entire recording.
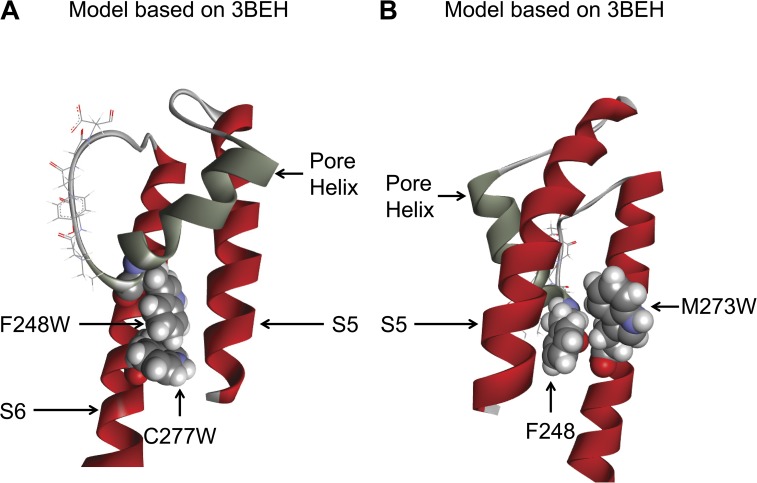
Figure 11.
Structure representations of single and double mutants supporting close proximity between C277 and M273 of S6 and F248 of the pore helix. Representation of the channel in the closed state (3BEH template). (A) The relative orientation of the Thr (W) for the F248W–C277W mutant suggests possible π–π interactions that would stabilize the pore helix in the closed configuration, thus accounting for the mutation C277W preventing the Pomax increase coming from the F248W mutation. (B) Also illustrated is the tight packing coming from the mutation M273W, which confirms the proximity of F248 and M273, as demonstrated through the double mutant cycle analysis in Fig. 10. Illustrations by Discovery Studio Visualizer (Accelrys).
DISCUSSION
The results presented here demonstrate that aromatic–aromatic interactions between F248 of the pore helix and W216 of the S5 transmembrane segment play a crucial role in setting the channel maximum open probability, while supporting the proposal of an active gate located at the level of the selectivity filter. It is also shown that F248 is within the interaction range of C277 and M273 adjacent to the Gly hinge of the S6 transmembrane segment and could thus contribute to the functional link between the CaM–KCa3.1 complex and the pore helix. These results suggest that the channel closed configuration is stabilized by aromatic–aromatic interactions between F248 at the C-terminal end of the pore helix and the S5 transmembrane segment, and support a model where the interface between the pore helix and the S5 transmembrane segment constitutes a potential target site for the design of KCa3.1 potentiators.
Modeling of the KCa3.1 pore region
Our homology modeling analysis has led to the identification of two templates susceptible to account for the KCa3.1 pore region: the mammalian Shaker Kv1.2 channel (2A79) (Long et al., 2005a) and the bacterial cyclic nucleotide–regulated MlotiK1 channel (3BEH) (Clayton et al., 2008). The resulting structures likely represent two distinct configurations of the KCa3.1 channel with the 2A79- and 3BEH-based structures referring to the channel open and closed states, respectively. SASA calculations have also revealed that the largest contact areas with both the S5 and S6 helices involve residue F248 at the C-terminal end of the pore helix regardless of the template used. The results of these analyses strongly suggest that F248 participates in conveying the structural changes induced by the binding of Ca2+ to the CaM–KCa3.1 complex to the pore helix. This proposal is in line with data coming from mass spectroscopy measurements where it was demonstrated that residue M94 of KirBac3.1 (equivalent to F248 in KCa3.1) displays among the largest changes in solvent accessibility when changing from a closed to an open configuration (Gupta et al., 2010). These observations were taken as evidence for a channel gate located close to the selectivity filter (Gupta et al., 2010), in accordance with the present results demonstrating a key contribution of F248 in setting the channel Pomax. Changes in solvent accessibility during KCa3.1 gating are also expected on the basis of our SASA calculations, with the surface contribution of I244 and F248 to the interface formed by the pore helix with the S5 and S6 transmembrane segments varying from 114 and 173 Å2 for the closed 3BEH-based configuration to 102 and 162 Å2 for the open-like configuration (unpublished data). In contrast, the surface contribution of L249 did not change appreciably between the two channel model structures, with respective values of 99.4 and 99.2 Å2 (unpublished data). Thus, these calculations support an increase in solvent accessibility at I244 and F248 when changing from a closed to an open state, an effect not seen at L249, which is more exposed to the channel central cavity.
Aromatic interactions between the pore helix and the S5 transmembrane segment
Whereas the 3BEH- and 2A79-based models led to similar interaction energy profiles between F248 and S6 residues, important differences were seen in the interaction energy profiles between F248 and residues of the S5 transmembrane helix. Of interest is the observation that F248 is mostly interacting with W216 and T212 of S5 for the 3BEH-based configuration, in contrast to the 2A79-based structure where F248 is predicted to interact for the most part with L211 and L215. More importantly, our results support a model whereby interactions between aromatic residues at 248 and W216 control Pomax. Substituting either F248 or W216 by nonaromatic amino acids systematically led to an increase in Pomax, arguing for aromatic–aromatic interactions stabilizing the channel closed configuration. Aromatic rings have been documented to interact preferably in a stacked offset mode or T-shape mode in which the H atoms on one ring points toward the π–electron cloud of the second ring (Samanta et al., 1999). Our results would be in accordance with π–π and/or polar hydrogen–π-type interactions accounting for Phe, His, and Tyr at position 248 in the pore helix showing significant lower Pomax values compared with nonaromatic amino acids of similar sizes, such as Val or Leu. These aromatic–aromatic interactions are likely to stabilize the selectivity filter closed configuration leading to low Pomax values. A decrease in Pomax relative to wild type was also observed with the F248T mutant. On the basis of the side-chain volume alone, the F248T mutation should have led to a Pomax comparable to F248V, which is clearly not the case (see Fig. 6). Additional forces might contribute in this case to the interactions with Trp; in particular, a hydroxyl group close to the π face of W216 could be responsible for polar hydrogen–π interactions. It is also apparent from the data presented in Figs. 7 A and 8 that increasing the volume size of the residues at 248 resulted in higher Pomax values, an effect that was particularly strong when coupled to interactions involving aromatic rings.
To get further insights on how nonbonded interactions involving F248 control Pomax, 32–64-ns MD simulations were performed for the wild-type (F248), F248T, F248A, F248V, and F248W mutant channels in the closed (3BEH template) and open (2A79 template) configurations. The resulting trajectories were used to compute the energy perturbation ΔΔEoc coming from mutating residue 248, where ΔΔEoc = ΔEoc_mutant-ΔEoc_wild_type, with ΔEoc as the difference in nonbonded energy for residue 248 between the channel open and closed states. ΔEoc values were obtained by averaging the nonbonded interaction energy (van der Waals and electrostatic) between residue 248 and the surrounding atoms within a 10-Å radius (excluding lipid and water) (Fig. 7 B) or between residue 248 and the S5 plus S6 transmembrane helices exclusively (Fig. 7 C). It follows that the ratio Keq_mutant/Keq_wild type, where Keq = Pomax/(1 − Pomax) (Fig. 7 A) should vary according to exp(−ΔΔEoc/kT), as long as mutation at 248 does not affect to a significant degree the nonbonded interactions between residues other than 248. Furthermore, ΔΔEoc values refer to energy changes for a single subunit and do not include a possible contribution coming from a combination of subunits. Therefore, negative ΔΔEoc values reflect an increase in Pomax relative to wild type as the ratio Keq_mutant/Keq_wild type becomes >1 under these conditions. The results of these calculations are illustrated in Fig. 7 (B and C), where the logarithm of the ratio Keq_mutant/Keq_wild type is plotted as a function of ΔΔEoc. Despite limitations coming from the use of homology-derived structures describing the selectivity filter region only, our trajectory analysis demonstrates a clear correlation between ΔΔEoc and the logarithm of the ratio Keq_mutant/Keq_wild_type, in agreement with our experimental observation of a Pomax ranking corresponding to F248 ≈ F248T << F248A ≈ F248V < F248W. This correlation holds for ΔΔEoc computed for residue 248 interacting with all the atoms within a 10-Å radius and for 248 interacting with the S5 and S6 helices exclusively. Furthermore, these calculations indicated that in contrast to Ala (A) and Val (V), the mutation F248T caused an equal stabilization of the channel closed (Eclosed_F248T − Eclosed_F248 = −7.91 kcal/mol, with Eclosed_mutant = van der Waals + electrostatic interaction energy between residue 248 and atoms in a 10-Å radius for the closed configuration) and open state (Eopen_F248T − Eopen_F248 = −7.96 kcal/mol), in accordance with ΔΔEoc ≈ 0 (Fig. 7 B) in this case and a Pomax (F248T) ≈ Pomax (wild type) as observed experimentally (Fig. 7 A). The absence of a significant energy difference ΔΔEoc with F248T and not with the F248A and F248V mutants supports of a model whereby aromatic–aromatic interactions involving F248 are determinant in setting Pomax for the KCa3.1 wild-type channel. We note, however, that the mutation F248S has led to a Pomax value comparable to F248A despite the possibility of OH–π interaction as for F248T. MD simulations confirmed in this regard that the F248S mutant behaves as F248T and should be characterized by a low Pomax, as observed with F248T. However, the mutation F248S has appeared unique among the mutant channels tested, including F248T, as it not only affected the channel Pomax but also caused a major decrease (threefold) of the channel unitary conductance (10 pS), arguing for a direct effect on the selectivity filter. Similarly, the F248C mutant where the O-H group of Ser is now replaced by S-H failed to yield detectable channel activity (not depicted), suggesting again a robust impact on the selectivity filter region. We speculate that these additional interactions and how they can be related to the channel Pomax cannot be properly taken into account by our current models, thus limiting the predictive value of MD simulations for the F248S mutant. The results in Fig. 7 (B and C) equally show that the energy difference ΔΔEoc for the F248W channel is within the energy range computed for the nonaromatic Ala (A) or Val (V) residues despite the possibility in this case of π–π interactions as previously discussed. An analysis of the associated MD trajectories revealed that the mutation F248W better stabilized the channel open (Eopen_F248W − Eopen_F248 = −7.5 kcal/mol) than the closed state (Eclosed_F248W − Eclosed_F248 = −4.0 kcal/mol), resulting in ΔΔEoc = −3.4 kcal/mol for an energy balance favoring an increase in Pomax relative to wild type as observed experimentally. This effect could be linked to the mutation F248W leading to an increase in stabilizing van der Waals interactions with the S5 and S6 helices in the open state, a mechanism that would account for the results presented in Fig. 7 A, where Pomax is seen to increase as a function of the residue volume for aromatic amino acids. A stabilization of the open state would also be compatible with the conclusions coming from a dwell-time analysis that confirmed that the mutation F248W results in a drastic right displacement of the open-time distribution curve from a mean value of 7 ms for wild type to 285 ms for F248W (Fig. S3). Changes in the closed-time distributions were equally observed characterized by the appearance of a dominant short-time distribution centered at 3 ms in support of a main destabilization of the channel closed configuration (Fig. S3). The Pomax augmentation observed with the F248A mutant seems, however, to arise from a different mechanism involving a greater destabilization of the channel closed state (Eclosed_F248A − Eclosed_F248 = 2.0 kcal/mol) relative to the open state (Eopen_F248A − Eopen_F248 = −0.01 kcal/mol), an effect caused in part by the absence in this case of robust stabilizing electrostatic interactions with the S5 and S6 transmembrane helices in the closed configuration. A dwell-time analysis showed in this regard that the mutation F248A caused a left displacement of the closed-time distribution curve with the main distribution centered at 30 ms for wild type shifted to 6 ms for F248A (Fig. S3). These changes were coupled to a right shift of the open-time distribution from 7 ms for wild type to 50 ms for the F248A mutant (Fig. S3). However, the effects observed on the open-time distribution remains substantially less important than the changes seen with the F248W mutant. This conclusion can be extended to the F248V mutant. Collectively, the correlation illustrated in Fig. 7 (B and C) between ln(Keq_mutant/Keq_wild_type) and ΔΔEoc tends to support a model whereby the pore helix is a major determinant in setting Pomax through nonbonded interactions involving F248 in the closed state. Of interest, a gating mechanism that takes into account possible interactions between the channel pore helix and the S5 transmembrane segment is in line with the observations reported for the hERG1 potassium channel where the interaction (hydrogen bonding) between H562 of S5, equivalent to W216 in KCa3.1, and the amino acids T218 (I244 pore helix in KCa3.1) and S221 (T247 pore helix in KCa3.1) of the pore helix is functionally important for deactivation and V1/2 of activation (Lees-Miller et al., 2009). It was also proposed that hydrophobic interaction between L273 (L215 residue in S5-KCa 3.1) and V310 (F248 in KCa3.1) in the KCNQ1 potassium channel stabilizes the open state of the selectivity filter, so that disruption of these interactions by mutating V310 to the small residues Ala or Gly could destabilize the pore open state, causing enhanced rate and extent of inactivation (Seebohm et al., 2005). However, such behavior contrasts with the current proposal of F248 interacting with the S5 transmembrane helix leading to a stabilization of the channel closed state.
Additional evidence in favor of wild-type Pomax being governed by F248 interacting with S5 in the closed state came from measuring the Pomax sensitivity to L211 mutations, as interaction energy calculations and the structural model presented in Fig. 3 predicted a strong coupling energy between F248 and L211 for the 2A79-based KCa3.1 structure. Although a positive correlation was observed between the side-chain volume of the residue at position 211 in S5 and Pomax with a Pearson correlation coefficient of 0.75 ± 0.15, our mutation study failed to show a strong perturbing effect of L211 mutation on Pomax. However, a significant increase in Pomax relative to wild type was observed with L211F, suggesting that L211F can come in proximity of F248 and stabilizes the channel open configuration through aromatic–aromatic interactions. Clearly, the wild-type KCa3.1 Pomax is governed more by the interactions between F248 and W216 for the channel closed configuration than by the interactions between F248 and L211 for the channel in the open configuration. These interactions constitute potential targets for a pharmacological control of KCa3.1 in accordance with the observation that mutating in KCa2.x channels the residue equivalent to L215 in S5 affects the action of the KCa2.x activators GW542573X and CM-TPMF (Hougaard et al., 2012). However, an F248-based model cannot totally account for the lower Pomax reported for KCa3.1 compared with members of the KCa2.x channel family, as both KCa2.x channels and KCa3.1 have a Phe and a Trp at positions equivalent to 248 and 216 in KCa3. A lower Pomax could still arise from differences in the overall interaction between the pore helix and the S5 segment if one considers that the amino acid composition of the pore helix and S5 transmembrane segment for KCa2.x and KCa3.1 differs by 46 and 70%, respectively, with four of the seven pore helix residues at the interface with S5 not conserved between the KCa2.x and KCa3.1 channels. In addition, we showed in a previous work that Pomax could be increased by mutating residue S367 in the KCa3.1 CaM-binding domain to the equivalent Thr residue in KCa2.x channels (Morales et al., 2013). The difference in Pomax seen between KCa3.1 and KCa2.x channels likely reflects a global difference in the channel open configuration, despite an identical CaM-based mechanism at the origin of channel activation.
Coupling F248 to the S6 transmembrane helix
Although the proposed analysis highlights the importance of F248 interacting with the S5 transmembrane helix in the closed state to Pomax, MD-based calculations of nonbonded energy between F248 and residues in S6 showed strong attractive interactions with the Gly hinge residue at 274 and to a lesser extent with C277, M273, and T270 on S6 in both model structures. The strong interactions with G274 essentially reflect the tight packing between the F248 side chain and the G274 backbone, suggesting a possible allosteric coupling between the conformational changes induced by Ca2+ at the level of the CaM–KCa3.1 complex and the gating events at the selectivity filter. Significant coupling energy values were also obtained in double mutant cycle experiments using the F248W–M273W and F248W–C277W systems. The extent of the coupling depended on the size of the residue at 273 and 277, as no significant ΔΔG values were obtained with the F248W–M273A and F248W–C277A double mutants. In fact, Pomax values for the C277W and C277A (not depicted) mutants did not appear statistically different from wild type, thus ruling out perturbing interactions of the C277W or C277A (see Fig. 10) with F248. We noted, however, that the mutation C277W could reverse the Pomax increase resulting from the substitution of F248W, suggesting that aromatic–aromatic interactions could in this case stabilize the pore helix closed configuration (Fig. 11). MD simulations demonstrated in this regard that the mutation C277W on the KCa3.1–F248W template caused a greater stabilization of the closed (Eclosed_F248W_C277W-Eclosed_F248W = −2.3 kcal/mol) than open (Eopen_F248W_C277W-Eopen_F248W = −1.3 kcal/mol) configuration for a ΔΔEoc relative to F248W of +1.0 kcal/mol. These simulations are thus in agreement with the observed decrease in Pomax coming from the mutation C277W on the KCa3.1–F248W template, an effect caused for the most part by an increase in electrostatic interactions between F248W and C277W (Fig. 11). However, the conclusions of our MD simulations need to be interpreted taking into account that our models do not include the transmembrane segments S1–S4 and thus provide a partial description only of the residues in the vicinity of C277. Although our observations confirmed that C277W is within the interaction range of F248W, we cannot conclude that there is an important contribution of C277 interacting with F248 in setting Pomax for KCa3.1 wild type. Our results also show an important increase in channel activity caused by the M273W substitution with a Pomax of 0.95 ± 0.005 (n = 3). These observations are in line with results obtained for the Kir3.4 (S143T) channel, where mutating to either a Val or Iso the residue L174 equivalent to M273 in KCa3.1 resulted in constitutively active channels (Rosenhouse-Dantsker and Logothetis, 2006). On the basis of these observations, it was argued that the position above the central Gly could play the role of the gating hinge and control gating.
Toward a global gating mechanisms for KCa3.1
Our results added new elements to the complexity of the KCa3.1 gating mechanism by highlighting the important role played by the interactions between the S5 transmembrane segment and the channel pore helix in setting Pomax. In contrast to voltage-gated channels where gating originates from the movement of the S4 transmembrane helix, KCa3.1 gating is initiated by the formation of a CaM–KCa3.1 complex in the C terminus directly connected to the S6 transmembrane helix. The formation of the CaM–KCa3.1 complex most likely involves the formation of the dimeric structure between two adjacent KCa3.1 subunits and potentially the formation of a dimer of dimers, as discussed in a previous work (Morales et al., 2013). Several studies on Kv channels have demonstrated that interaction between the S6 transmembrane segments and the S4–S5 linker at the cytoplasmic side of the membrane is essential for electromechanical coupling (Lu et al., 2002; Ferrer et al., 2006; Prole and Yellen, 2006; Choveau et al., 2011; Labro et al., 2011). This interaction would be crucial to couple the voltage-sensor movement in S4 with the activation gate at the C-terminal end of the S6 transmembrane segment (Long et al., 2005b; Labro et al., 2011). Given that the Ca2+-dependent CaM–KCa3.1 complex is connected to the S6 and not S4 transmembrane segment, one would not expect a significant contribution of the S4–S5 linker region to the KCa3.1 gating process. However, the closed- and open-state models of KCa3.1 illustrated in Fig. 1 (A and B) indicate that the S4–S5 linker encircles the channel pore at the level of the bundle-crossing region, so that outward movements of the S6 transmembrane segment can be transmitted to the S4–S5 linker. A displacement of the S4–S5 linker could in turn produce a rotation and translation of the S5 transmembrane helix and affect the interactions between S5 and the channel pore helix. Our results clearly show that these interactions are crucial to the energy balance between the channel open and closed configurations. A possible gating process for KCa3.1 would thus consist of the formation of a CaM–KCa3.1 complex in the C terminus leading to a displacement of the S6 transmembrane helices that would induce a movement of the S4–S5 linkers, resulting in a rotation/displacement of the S5 transmembrane segments and to a modification of the interactions between S5 and the pore helix. This process would work in parallel with a direct coupling of S6 with the pore helix and the selectivity filter region as documented in this work. This mechanism could explain how the conformational change induced by Ca2+ binding to CaM can be transmitted to the selectivity filter region, insuring gating. Validation of this model would, however, require an in-depth evaluation of the role played by the S4–S5 linker in KCa3.1 gating.
Conclusion
This work demonstrates for the first time that aromatic–aromatic interactions between the pore helix and the S5 transmembrane segment in KCa3.1 control the channel Pomax. Thus, interfering with these interactions represents a promising approach to modulate KCa3.1 channel activity. Therefore, the interface formed by the pore helix and the S5 transmembrane segment represents a valuable site that can be used for the design of KCa3.1 potentiators.
Supplementary Material
Acknowledgments
We are grateful to Calcul Québec for the computer facilities. We would like to acknowledge the work of Ms. Julie Verner for expert oocyte preparation.
This work was supported by grants from the Canadian Institutes of Health Research (to R. Sauvé) and from Cystic Fibrosis Canada (to R. Sauvé and E. Brochiero). R. Sauvé and L. Parent are members of the FRQS-funded GEPROM group.
The authors declare no competing financial interests.
Lawrence G. Palmer served as editor.
Footnotes
Abbreviations used in this paper:
- BMS
- bis(2-mercaptoethyl)sulfone
- CaM
- calmodulin
- MD
- molecular dynamics
- pMpa
- p-methoxy-l-phenylalanine
- SASA
- solvent accessibility surface area
- SCAM
- substituted cysteine accessibility method
References
- Banderali U., Klein H., Garneau L., Simoes M., Parent L., Sauvé R. 2004. New insights on the voltage dependence of the KCa3.1 channel block by internal TBA. J. Gen. Physiol. 124:333–348 10.1085/jgp.200409145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradding P., Wulff H. 2009. The K+ channels K(Ca)3.1 and K(v)1.3 as novel targets for asthma therapy. Br. J. Pharmacol. 157:1330–1339 10.1111/j.1476-5381.2009.00362.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruening-Wright A., Lee W.S., Adelman J.P., Maylie J. 2007. Evidence for a deep pore activation gate in small conductance Ca2+-activated K+ channels. J. Gen. Physiol. 130:601–610 10.1085/jgp.200709828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choveau F.S., Rodriguez N., Abderemane Ali F., Labro A.J., Rose T., Dahimène S., Boudin H., Le Hénaff C., Escande D., Snyders D.J., et al. 2011. KCNQ1 channels voltage dependence through a voltage-dependent binding of the S4-S5 linker to the pore domain. J. Biol. Chem. 286:707–716 10.1074/jbc.M110.146324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke O.B., Caputo A.T., Hill A.P., Vandenberg J.I., Smith B.J., Gulbis J.M. 2010. Domain reorientation and rotation of an intracellular assembly regulate conduction in Kir potassium channels. Cell. 141:1018–1029 10.1016/j.cell.2010.05.003 [DOI] [PubMed] [Google Scholar]
- Clayton G.M., Altieri S., Heginbotham L., Unger V.M., Morais-Cabral J.H. 2008. Structure of the transmembrane regions of a bacterial cyclic nucleotide-regulated channel. Proc. Natl. Acad. Sci. USA. 105:1511–1515 10.1073/pnas.0711533105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero-Morales J.F., Jogini V., Lewis A., Vásquez V., Cortes D.M., Roux B., Perozo E. 2007. Molecular driving forces determining potassium channel slow inactivation. Nat. Struct. Mol. Biol. 14:1062–1069 10.1038/nsmb1309 [DOI] [PubMed] [Google Scholar]
- Cruse G., Duffy S.M., Brightling C.E., Bradding P. 2006. Functional KCa3.1 K+ channels are required for human lung mast cell migration. Thorax. 61:880–885 10.1136/thx.2006.060319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damkjaer M., Nielsen G., Bodendiek S., Staehr M., Gramsbergen J.B., de Wit C., Jensen B.L., Simonsen U., Bie P., Wulff H., Köhler R. 2012. Pharmacological activation of KCa3.1/KCa2.3 channels produces endothelial hyperpolarization and lowers blood pressure in conscious dogs. Br. J. Pharmacol. 165:223–234 10.1111/j.1476-5381.2011.01546.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q.S., Wang Q.Y., Du L.Q., Chen D., Huang R.B. 2013. Theoretical study on the polar hydrogen-pi (Hp-pi) interactions between protein side chains. Chem. Cent. J. 7:92 10.1186/1752-153X-7-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félétou M., Vanhoutte P.M. 2007. Endothelium-dependent hyperpolarizations: past beliefs and present facts. Ann. Med. 39:495–516 10.1080/07853890701491000 [DOI] [PubMed] [Google Scholar]
- Ferrer T., Rupp J., Piper D.R., Tristani-Firouzi M. 2006. The S4-S5 linker directly couples voltage sensor movement to the activation gate in the human ether-a’-go-go-related gene (hERG) K+ channel. J. Biol. Chem. 281:12858–12864 10.1074/jbc.M513518200 [DOI] [PubMed] [Google Scholar]
- Gao Y.D., Hanley P.J., Rinné S., Zuzarte M., Daut J. 2010. Calcium-activated K(+) channel (K(Ca)3.1) activity during Ca(2+) store depletion and store-operated Ca(2+) entry in human macrophages. Cell Calcium. 48:19–27 10.1016/j.ceca.2010.06.002 [DOI] [PubMed] [Google Scholar]
- Garneau L., Klein H., Banderali U., Longpré-Lauzon A., Parent L., Sauvé R. 2009. Hydrophobic interactions as key determinants to the KCa3.1 channel closed configuration. An analysis of KCa3.1 mutants constitutively active in zero Ca2+. J. Biol. Chem. 284:389–403 10.1074/jbc.M805700200 [DOI] [PubMed] [Google Scholar]
- Gupta S., Bavro V.N., D’Mello R., Tucker S.J., Vénien-Bryan C., Chance M.R. 2010. Conformational changes during the gating of a potassium channel revealed by structural mass spectrometry. Structure. 18:839–846 10.1016/j.str.2010.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Wang J., Hede S.E., Novak I. 2012. An intermediate-conductance Ca2+-activated K+ channel is important for secretion in pancreatic duct cells. Am. J. Physiol. Cell Physiol. 303:C151–C159 10.1152/ajpcell.00089.2012 [DOI] [PubMed] [Google Scholar]
- Hirschberg B., Maylie J., Adelman J.P., Marrion N.V. 1998. Gating of recombinant small-conductance Ca-activated K+ channels by calcium. J. Gen. Physiol. 111:565–581 10.1085/jgp.111.4.565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz A. 1996. Double-mutant cycles: a powerful tool for analyzing protein structure and function. Fold. Des. 1:R121–R126 10.1016/S1359-0278(96)00056-9 [DOI] [PubMed] [Google Scholar]
- Hougaard C., Jensen M.L., Dale T.J., Miller D.D., Davies D.J., Eriksen B.L., Strøbaek D., Trezise D.J., Christophersen P. 2009. Selective activation of the SK1 subtype of human small-conductance Ca2+-activated K+ channels by 4-(2-methoxyphenylcarbamoyloxymethyl)-piperidine-1-carboxylic acid tert-butyl ester (GW542573X) is dependent on serine 293 in the S5 segment. Mol. Pharmacol. 76:569–578 10.1124/mol.109.056663 [DOI] [PubMed] [Google Scholar]
- Hougaard C., Hammami S., Eriksen B.L., Sørensen U.S., Jensen M.L., Strøbæk D., Christophersen P. 2012. Evidence for a common pharmacological interaction site on K(Ca)2 channels providing both selective activation and selective inhibition of the human K(Ca)2.1 subtype. Mol. Pharmacol. 81:210–219 10.1124/mol.111.074252 [DOI] [PubMed] [Google Scholar]
- Jenkins D.P., Strøbæk D., Hougaard C., Jensen M.L., Hummel R., Sørensen U.S., Christophersen P., Wulff H. 2011. Negative gating modulation by (R)-N-(benzimidazol-2-yl)-1,2,3,4-tetrahydro-1-naphthylamine (NS8593) depends on residues in the inner pore vestibule: pharmacological evidence of deep-pore gating of K(Ca)2 channels. Mol. Pharmacol. 79:899–909 10.1124/mol.110.069807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S., Kim T., Iyer V.G., Im W. 2008. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29:1859–1865 10.1002/jcc.20945 [DOI] [PubMed] [Google Scholar]
- Joiner W.J., Khanna R., Schlichter L.C., Kaczmarek L.K. 2001. Calmodulin regulates assembly and trafficking of SK4/IK1 Ca2+-activated K+ channels. J. Biol. Chem. 276:37980–37985 [DOI] [PubMed] [Google Scholar]
- Karplus K. 2009. SAM-T08, HMM-based protein structure prediction. Nucleic Acids Res. 37:W492–W497 10.1093/nar/gkp403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein H., Garneau L., Banderali U., Simoes M., Parent L., Sauvé R. 2007. Structural determinants of the closed KCa3.1 channel pore in relation to channel gating: Results from a substituted cysteine accessibility analysis. J. Gen. Physiol. 129:299–315 10.1085/jgp.200609726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein H., Garneau L., Trinh N.T., Privé A., Dionne F., Goupil E., Thuringer D., Parent L., Brochiero E., Sauvé R. 2009. Inhibition of the KCa3.1 channels by AMP-activated protein kinase in human airway epithelial cells. Am. J. Physiol. Cell Physiol. 296:C285–C295 10.1152/ajpcell.00418.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labro A.J., Boulet I.R., Choveau F.S., Mayeur E., Bruyns T., Loussouarn G., Raes A.L., Snyders D.J. 2011. The S4-S5 linker of KCNQ1 channels forms a structural scaffold with the S6 segment controlling gate closure. J. Biol. Chem. 286:717–725 10.1074/jbc.M110.146977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Miller J.P., Subbotina J.O., Guo J., Yarov-Yarovoy V., Noskov S.Y., Duff H.J. 2009. Interactions of H562 in the S5 helix with T618 and S621 in the pore helix are important determinants of hERG1 potassium channel structure and function. Biophys. J. 96:3600–3610 10.1016/j.bpj.2009.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Siegelbaum S.A. 2000. Change of pore helix conformational state upon opening of cyclic nucleotide-gated channels. Neuron. 28:899–909 10.1016/S0896-6273(00)00162-8 [DOI] [PubMed] [Google Scholar]
- Long S.B., Campbell E.B., Mackinnon R. 2005a. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–903 10.1126/science.1116269 [DOI] [PubMed] [Google Scholar]
- Long S.B., Campbell E.B., Mackinnon R. 2005b. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 309:903–908 10.1126/science.1116270 [DOI] [PubMed] [Google Scholar]
- Lu Z., Klem A.M., Ramu Y. 2002. Coupling between voltage sensors and activation gate in voltage-gated K+ channels. J. Gen. Physiol. 120:663–676 10.1085/jgp.20028696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maylie J., Bond C.T., Herson P.S., Lee W.S., Adelman J.P. 2004. Small conductance Ca2+-activated K+ channels and calmodulin. J. Physiol. 554:255–261 10.1113/jphysiol.2003.049072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales P., Garneau L., Klein H., Lavoie M.F., Parent L., Sauvé R. 2013. Contribution of the KCa3.1 channel–calmodulin interactions to the regulation of the KCa3.1 gating process. J. Gen. Physiol. 142:37–60 10.1085/jgp.201210933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy M., Pedemonte N., MacVinish L., Galietta L., Cuthbert A. 2005. 4-Chlorobenzo[F]isoquinoline (CBIQ), a novel activator of CFTR and DeltaF508 CFTR. Eur. J. Pharmacol. 516:118–124 10.1016/j.ejphar.2005.04.037 [DOI] [PubMed] [Google Scholar]
- Proks P., Capener C.E., Jones P., Ashcroft F.M. 2001. Mutations within the P-loop of Kir6.2 modulate the intraburst kinetics of the ATP-sensitive potassium channel. J. Gen. Physiol. 118:341–353 10.1085/jgp.118.4.341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prole D.L., Yellen G. 2006. Reversal of HCN channel voltage dependence via bridging of the S4-S5 linker and Post-S6. J. Gen. Physiol. 128:273–282 10.1085/jgp.200609590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F., Auerbach A., Sachs F. 1996. Estimating single-channel kinetic parameters from idealized patch-clamp data containing missed events. Biophys. J. 70:264–280 10.1016/S0006-3495(96)79568-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F., Auerbach A., Sachs F. 1997. Maximum likelihood estimation of aggregated Markov processes. Proc. Biol. Sci. 264:375–383 10.1098/rspb.1997.0054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenhouse-Dantsker A., Logothetis D.E. 2006. New roles for a key glycine and its neighboring residue in potassium channel gating. Biophys. J. 91:2860–2873 10.1529/biophysj.105.080242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth E.K., Hirtz S., Duerr J., Wenning D., Eichler I., Seydewitz H.H., Amaral M.D., Mall M.A. 2011. The K+ channel opener 1-EBIO potentiates residual function of mutant CFTR in rectal biopsies from cystic fibrosis patients. PLoS ONE. 6:e24445 10.1371/journal.pone.0024445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Kucukural A., Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šali A., Blundell T.L. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- Samanta U., Pal D., Chakrabarti P. 1999. Packing of aromatic rings against tryptophan residues in proteins. Acta Crystallogr. D Biol. Crystallogr. 55:1421–1427 10.1107/S090744499900726X [DOI] [PubMed] [Google Scholar]
- Schumacher M.A., Rivard A.F., Bächinger H.P., Adelman J.P. 2001. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature. 410:1120–1124 10.1038/35074145 [DOI] [PubMed] [Google Scholar]
- Schumacher M.A., Crum M., Miller M.C. 2004. Crystal structures of apocalmodulin and an apocalmodulin/SK potassium channel gating domain complex. Structure. 12:849–860 10.1016/j.str.2004.03.017 [DOI] [PubMed] [Google Scholar]
- Seebohm G., Westenskow P., Lang F., Sanguinetti M.C. 2005. Mutation of colocalized residues of the pore helix and transmembrane segments S5 and S6 disrupt deactivation and modify inactivation of KCNQ1 K+ channels. J. Physiol. 563:359–368 10.1113/jphysiol.2004.080887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J.Z., Ella S., Davis M.J., Hill M.A., Braun A.P. 2009. Openers of SKCa and IKCa channels enhance agonist-evoked endothelial nitric oxide synthesis and arteriolar vasodilation. FASEB J. 23:1138–1145 10.1096/fj.08-120451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes M., Garneau L., Klein H., Banderali U., Hobeila F., Roux B., Parent L., Sauvé R. 2002. Cysteine mutagenesis and computer modeling of the S6 region of an intermediate conductance IKCa channel. J. Gen. Physiol. 120:99–116 10.1085/jgp.20028586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Syme C.A., Singh A.K., Devor D.C., Bridges R.J. 2001. Benzimidazolone activators of chloride secretion: potential therapeutics for cystic fibrosis and chronic obstructive pulmonary disease. J. Pharmacol. Exp. Ther. 296:600–611 [PubMed] [Google Scholar]
- Szkotak A.J., Murthy M., MacVinish L.J., Duszyk M., Cuthbert A.W. 2004. 4-Chloro-benzo[F]isoquinoline (CBIQ) activates CFTR chloride channels and KCNN4 potassium channels in Calu-3 human airway epithelial cells. Br. J. Pharmacol. 142:531–542 10.1038/sj.bjp.0705846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall-Lacelle S., Hossain M.I., Sauvé R., Blunck R., Parent L. 2011. Double mutant cycle analysis identified a critical leucine residue in the IIS4S5 linker for the activation of the Ca(V)2.3 calcium channel. J. Biol. Chem. 286:27197–27205 10.1074/jbc.M111.237412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Xie J., Schultz P.G. 2006. A genetically encoded fluorescent amino acid. J. Am. Chem. Soc. 128:8738–8739 10.1021/ja062666k [DOI] [PubMed] [Google Scholar]
- Wimley W.C., Creamer T.P., White S.H. 1996. Solvation energies of amino acid side chains and backbone in a family of host-guest pentapeptides. Biochemistry. 35:5109–5124 10.1021/bi9600153 [DOI] [PubMed] [Google Scholar]
- Wissmann R., Bildl W., Neumann H., Rivard A.F., Klöcker N., Weitz D., Schulte U., Adelman J.P., Bentrop D., Fakler B. 2002. A helical region in the C terminus of small-conductance Ca2+-activated K+ channels controls assembly with apo-calmodulin. J. Biol. Chem. 277:4558–4564 10.1074/jbc.M109240200 [DOI] [PubMed] [Google Scholar]
- Wulff H., Miller M.J., Hansel W., Grissmer S., Cahalan M.D., Chandy K.G. 2000. Design of a potent and selective inhibitor of the intermediate-conductance Ca2+-activated K+ channel, IKCa1: a potential immunosuppressant. Proc. Natl. Acad. Sci. USA. 97:8151–8156 10.1073/pnas.97.14.8151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff H., Knaus H.G., Pennington M., Chandy K.G. 2004. K+ channel expression during B cell differentiation: implications for immunomodulation and autoimmunity. J. Immunol. 173:776–786 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.