Approaches from information theory and probabilistic modeling show that voltage-sensing domain sequences conform to a small set of rules.
Abstract
Voltage-sensor domains (VSDs) are modular biomolecular machines that transduce electrical signals in cells through a highly conserved activation mechanism. Here, we investigate sequence–function relationships in VSDs with approaches from information theory and probabilistic modeling. Specifically, we collect over 6,600 unique VSD sequences from diverse, long-diverged phylogenetic lineages and relate the statistical properties of this ensemble to functional constraints imposed by evolution. The VSD is a helical bundle with helices labeled S1–S4. Surrounding conserved VSD residues such as the countercharges and the S2 phenylalanine, we discover sparse networks of coevolving residues. Additional networks are found lining the VSD lumen, tuning the local hydrophilicity. Notably, state-dependent contacts and the absence of coevolution between S4 and the rest of the bundle are imprints of the activation mechanism on the VSD sequence ensemble. These design principles rationalize existing experimental results and generate testable hypotheses.
INTRODUCTION
First discovered in voltage-gated cation channels and later identified in voltage-sensitive phosphatases and proton channels, the voltage-sensor domain (VSD) is a biological molecular device that responds to changes in transmembrane (TM) electrical potential (Yu and Catterall, 2004; Murata et al., 2005; Ramsey et al., 2006). Moreover, VSDs are demonstrably modular and widely distributed in both prokaryotic and eukaryotic cells, indicating an early origin and consequently vast evolutionary exploration of sequence space (Yu and Catterall, 2004; Arrigoni et al., 2013). Because of their ubiquity in cellular electrical signaling, mutations in VSDs give rise to various heritable diseases (Catterall, 2010).
The VSD consists of a four-helix bundle (denoted as S1–S4) embedded in a membrane (Cuello et al., 2004; Schmidt et al., 2006; Sands and Sansom, 2007). The S4 helix contains a highly conserved motif of three to eight positively charged residues, referred to as “gating charges,” which occur at three-residue intervals and occupy a common helical face (Liman et al., 1991; Aggarwal and MacKinnon, 1996; Seoh et al., 1996). These positive charges are stabilized in the TM position through salt-bridging interactions with the negative “countercharge” residues of S1–S3 (Tiwari-Woodruff et al., 2000; Long et al., 2005; Payandeh et al., 2011). These charges exist in an aqueous-like environment as water molecules protrude into the VSD lumen (Freites et al., 2006; Tombola et al., 2007; Krepkiy et al., 2009). A cluster of bulky hydrophobic residues partitions the lumen into two disconnected intracellular and extracellular regions (Tao et al., 2010; Lacroix and Bezanilla, 2011; Cheng et al., 2013).
In response to changes in membrane potential, S4 translates so as to sequentially transfer the gating charges across the hydrophobic region (Campos et al., 2007; Vargas et al., 2012). Because of the partial solvation of the lumen, the TM electric field is focused on this hydrophobic region (Treptow and Tarek, 2006; Jogini and Roux, 2007). A rather small displacement of S4 is therefore sufficient to achieve a complete transfer of positive charge across the field (Ahern and Horn, 2005). This conformational change in S4 allows the VSD to act as a transducer of electrical signals.
Major mechanistic features of VSDs appear well identified (Börjesson and Elinder, 2008; Catterall, 2010). Even so, the protein sequences encoding VSDs in various families can be very different (Guda et al., 2007). Here, we aim to show how the functional requirements of voltage sensing have shaped the sequence distribution of VSDs during evolution. After addressing the specific problem of aligning the register of S4 helices in a statistically rigorous way, we construct a large multiple sequence alignment (MSA) of VSD sequences and search for patterns and regularities that reflect the evolutionary “design principles” underlying voltage-sensor function. Success in this endeavor implies that from the design principles we will be able to recapitulate experimental findings and, importantly, formulate novel, testable hypotheses concerning the structural and functional properties shared among VSDs.
Site-specific residue frequencies are arguably the most intuitive feature that can be extracted from MSAs. Because the analysis may contain thousands of sequences from long-diverged organisms, a frequency distribution of amino acids can be extracted for each column/position of the MSA. The peculiar nature of this distribution is, in general, informative about the evolutionary pressure acting on that particular position (Dokholyan et al., 2002). For instance, a distribution peaked around tyrosine, phenylalanine, and tryptophan suggests a functional requirement for aromaticity at that position. Here, we use this sort of single-site analysis to detect “evolutionarily important” functional sites.
Much information can also be extracted from joint frequency distributions involving pairs of positions. In the native folded state, each residue of the polypeptide chain engages in residue–residue interactions. The specific mutations allowed at a given position are then highly dependent on the chemical identity of neighboring residues. As a result, frequency distributions of amino acids at different positions are expected to be statistically dependent on each other. Detection of these correlations can potentially unveil the physical interactions defining the native structure. However, residue–residue correlations per se are not able to convey this information; in a highly connected network, such as the contacting residues in a folded protein, an extensive set of pairwise interactions can give rise to long-range indirect statistical correlations (Giraud et al., 1999). As a result, pairs of correlated positions are not necessarily in contact. Reconstructing the network of direct interactions entails the logical process of inference: within a stochastic framework (a probabilistic model), it is assumed that residues interact in pairs (are directly coupled) in such a way as to best reproduce the distributions of sequences observed in the MSA (Weigt et al., 2009). Analysis of these direct couplings (direct-coupling analysis [DCA]) is able to highlight functionally crucial residues as those involved in enzymatic active sites and regions of conformational change (Weigt et al., 2009; Morcos et al., 2011; Hopf et al., 2012). It is important to note that these methodologies rely on a large, confidently aligned MSA.
Anticipating our findings, we show how analysis of site-specific frequencies and direct evolutionary couplings (ECs) unveils the major sequence determinants of voltage sensing. The requirement for a translation of S4 through the electric field results in strong conservation of the gating charges, the countercharges, and the S2 phenylalanine. With DCA, we identify unexpected networks of strongly evolutionarily coupled residues surrounding these conserved positions. We also identify sites that have been under significant evolutionary pressure and have not yet been tested for functional relevance. We suggest these sites as potential targets for new mutagenesis experiments. Additionally, we show that the helical interfaces between S1–S4 and S3–S4 do not feature strong ECs, suggesting that no specific residue–residue contacts were conserved along evolution apart from the canonical salt bridges. This lack of expected coevolution is shown to characterize the entire paddle motif, rationalizing chimeragenesis and deletion analysis experiments highlighting the modular, mobile nature of this region. Lastly, observation of specific evolutionarily coupled residue pairs involving positions 96, namely 25–96 and 49–96 (NavAb numbering), provides independent support for conformational models of the VSD along the activation pathway and suggests that positions 25, 49, and 96 have important roles in tuning the activation properties of the VSD.
MATERIALS AND METHODS
Hidden Markov model (HMM) training: “Seed” MSA
Profile HMMs are among the most important approaches to analyze ensembles of sequences. Conceptually, HMMs characterize sequences as stochastic processes: discrete time random walks across a set of states with defined state and transition probabilities (Eddy, 1998). In practice, HMMs are initialized with a protein sequence of interest and iteratively refined by scanning many millions of known protein sequences. Homologous sequences are then collected in an MSA, an array of statistically significant protein sequence alignments. From an abstract perspective, an MSA can be thought of as the physical record of evolution’s exploratory search for structurally and functionally analogous proteins. Subsequent sequence analysis is highly dependent on the quality and quantity of alignments in the MSA, and for this reason, we give special care to its construction.
To train an HMM to identify and align VSD sequences from a large sequence database, we construct a small, confidently aligned “seed” MSA. In the seed MSA, we included homologues representing phylogenetically diverse VSD-containing families (Yu and Catterall, 2004). Initial multiple alignments were generated using ClustalW2 and manually refined such that the alignment was consistent with structural alignments (in the cases where high resolution structures were available) and TM-helix topology predictions (Zhang and Skolnick, 2005; Larkin et al., 2007; Viklund and Elofsson, 2008). See Table S1 for full seed MSA.
VSDs contain several conserved positive gating charges, which come at three-residue intervals on the S4 helix. However, not all VSDs contain the same number of gating charges, and this may lead to uncertainty in the alignment of S4, as three-residue shifts in the alignment register will still match positive charges (Wood et al., 2012; Kulleperuma et al., 2013). We were interested in quantifying how uncertain such alignments actually were. Position-wise reliability of a sequence alignment can be calculated as the posterior probability of each symbol in the alignment being emitted by a pair HMM based on the Blosum62 substitution matrix (Wolfsheimer et al., 2012). As an example, for the pairwise alignment of human Hv1 S4 to that of human Kv1.2 S4, we calculated the positional posterior probability of (a) an alignment generated by the Needleman–Wunsch algorithm, and (b) alignments “sampled” from a pair HMM with Hv1 S4 in alternate registers using ppAlign (Needleman and Wunsch, 1970; Wolfsheimer et al., 2012). The posterior probability over the S4 helix in (a) is much higher than any of the alignments in (b), suggesting that, given the widely used Blosum62 substitution matrix, the alignment of S4 is unambiguous.
An HMM was trained based on the seed MSA using HMMER3.0 (Eddy, 1998). Scanning this HMM against the NCBI protein sequence database, we collected and aligned 6,652 unique VSD sequences containing all four TM helices (Eddy, 1998). Only alignments with an E-value of <0.01 were included. Sequence logos were generated with Weblogo3 (Crooks et al., 2004).
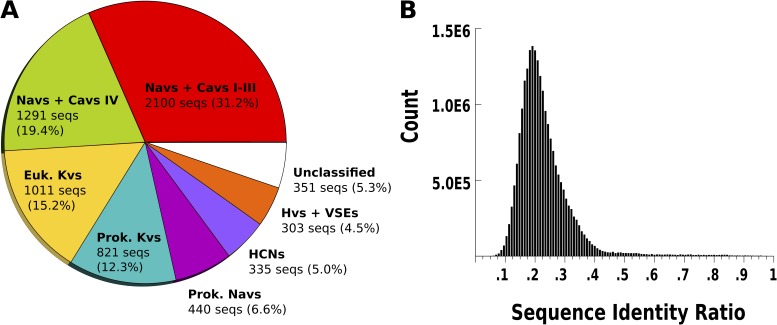
To describe the constitution of the VSD MSA, hierarchical clustering was performed to partition the MSA into protein families. Specifically, we clustered with the neighbor-joining algorithm in the phylogeny inference package (PHYLIP; version 3.695). Neighbor-joining is a fast tree–generating method appropriate for datasets of the size considered here. By identifying sequences in this tree with well-curated annotations from the NCBI protein database, branches can be assigned to known VSD-containing protein families (Yu and Catterall, 2004). This tree is shown in Fig. S1. The seven largest branches corresponded to the first three VSDs in voltage-gated calcium and sodium channel pseudotetramers (Navs + Cavs I-III), the fourth VSD in voltage-gated calcium and sodium channel pseudotetramers (Navs + Cavs IV), eukaryotic voltage-gated potassium channels (Euk. Kvs), prokaryotic voltage-gated potassium channels (Prok. Kvs), hyperpolarization-activated cyclic nucleotide–gated channels (HCNs), and voltage-gated proton channels/voltage-sensitive enzymes (Hvs + VSEs). An additional set of VSD sequences did not cluster with VSD sequences with well-curated annotations, and we labeled them “unclassified.” A piechart of the VSD MSA illustrating these families can be found in Fig. 2 A.
Figure 2.
The sequence sample is correctly stratified and captures the VSD sequence variability. (A) Breakdown of the MSA into protein families. Navs + Cavs I-III, the first three VSDs in the pseudotetrameric voltage-gated sodium and calcium channels; Navs + Cavs IV, the fourth VSD in the pseudotetrameric voltage-gated sodium and calcium channels; Euk. Kvs, eukaryotic voltage-gated potassium channels; Prok. Kvs, prokaryotic voltage-gated potassium channels; Prok. Navs, prokaryotic voltage-gated sodium channels; HCNs, hyperpolarization-activated cyclic nucleotide–gated channels; Hvs + VSEs, voltage-gated proton channels and voltage-sensitive enzymes. (B) Histogram of pairwise sequence identities for the MSA.
To quantify the diversity of sequences in the VSD MSA, we calculated the histogram of pairwise sequence identities for the aligned regions of all sequences (Fig. 2 B). This histogram shows an approximately unimodal distribution peaked around 20% sequence identity. Additionally, we calculated histograms of pairwise sequence identities for MSAs of sequences in each of the seven protein families described above (see Fig. S1). Several histograms showed polymodal distributions, suggestive of VSD subtypes within the identified protein families.
Kullback–Leibler divergence
With such a large quantity of homologous protein sequences in the VSD MSA, site-specific frequency distributions can be determined quantitatively. We can think of the difference between such a distribution and a reference as a measure of the “evolutionary pressure” exerted on that site. The Kullback–Leibler divergence, DKL(i), measures the information that differentiates an observed empirical distribution of amino acids, Pi, from a suitable reference distribution of amino acids, Qi:
Single-site amino acid frequency distributions, Pi, were calculated from the MSA with python scripts. Biases caused by phylogenetic relationships and incomplete sampling were addressed by reweighing sequences in MSA; aligned sequences with high sequence identity (>0.9) were weighted together as a single sequence as described by Morcos et al. (2011). By these criteria, the effective number of sequences in the MSA was calculated to be 3,821.
Reference amino acid distributions, Qi, were calculated for three topologically distinct regions of TM proteins: the inner membrane–water interface, the outer membrane–water interface, and the lipid-facing TM region. The Orientations of Proteins in Membrane (OPM) database consists of membrane protein structures with predicted outer and inner membrane interfaces. From the OPM, 533 polytopic α-helical TM protein structures were downloaded (Lomize et al., 2006). A python script was used to measure the amino acid frequencies within an 11-Å window of each predicted interface (representing the inner and outer membrane–water interfaces) and also between these windows (representing the TM region). These reference distributions and the assignment of topological region to individual positions in the NavAb VSD are available in Tables S1 and S4.
DCA
To accurately reconstruct the network of evolutionarily important interactions from statistical correlations in the MSA, we infer the probabilistic model that makes the least possible number of assumptions about the underlying structure of the data. This criterion is satisfied by the model structure that maximizes the Shannon entropy, namely a Potts model. Additionally, determination of the interactions (the parameter-learning step) must rely on a computationally tractable algorithm. Several solutions to this second problem have been proposed recently (Morcos et al., 2011; Ekeberg et al., 2013). These modeling approaches have been implemented in a family of algorithms known as DCA. DCA has demonstrated unprecedented success in dissecting the correlations present in MSAs into scored pairwise interactions. The set of all possible scored pairs generates an EC score matrix. This matrix appears highly similar to contact maps derived from the crystal structures of members of the protein family and contains sufficient information to allow ab initio structure prediction, identification of interfaces in homo-oligomers, and prediction of conformational changes (Morcos et al., 2011; Hopf et al., 2012).
Using a large MSA as input, DCA infers the parameters of a probabilistic model that reproduces single- and two-site marginals of empirical distribution of sequences contained in the MSA. The least constrained probabilistic model, known as a Potts model, has the following functional form:
P(a1,…,aN) is the probability of observing the sequence {a1,…,aN }, where ai takes values in the alphabet containing the 20 amino acids plus the gap (“-”) symbol. Local fields, hi(a), give rise to the propensity for an amino acid, ai, to be observed at sequence position i, whereas the coupling constants, ei,j(ai, aj), similarly encode the joint propensity of observing amino acids ai and aj at positions i and j, respectively. For a given i,j pair, the set of coupling constants is arranged in a matrix; the Frobenius norm of this matrix is defined as the EC score for the pair i,j. The full set of EC scores composes the EC score matrix. EC score matrices have been shown to be significantly correlated with contact maps. Although a rational interpretation of this correlation would posit that residues engaged in energetically favorable interactions are likely to coevolve, it has yet to be shown to which extent a statistical coupling can be identified with an energetic one. Thus, a strict correspondence between data from double mutant cycle analysis and DCA is not expected a priori. We obtained the EC scores of the VSD using Matlab scripts for pseudolikelihood maximization DCA described in Ekeberg et al. (2013). Molecular visualizations were created with visual molecular dynamics (Humphrey et al., 1996).
Statistical inference to determine direct couplings has been performed using a model structure (Potts model) that assumes a large set of parameters; even considering only 115 positions from the VSD sequences, there are ∼2.5 million parameters corresponding to pairwise coupling constants and local fields. The optimal values of these parameters are found, in general, by maximizing the likelihood of the data, here approximated by a computationally tractable proxy, the “pseudolikelihood” (Ekeberg et al., 2013). The quality of the results is therefore strongly dependent on the size of the dataset and on a correctly stratified sampling. To test whether our MSA of 6,652 unique VSD sequences satisfies these requirements, we calculated the EC score matrices for two disjoint subpopulations in our sample. Specifically, we extracted MSAs from the original dataset corresponding to two distinct phylogenetic lineages, the pseudotetrameric Nav/Cav family and the Kv family. These subpopulations were identified based on the partitioning provided by the hierarchical clustering described above, resulting into 3,391 and 1,832 sequences for Nav/Cav and Kv VSDs, respectively. EC score matrices for both of these families are presented in the supplemental material for comparison with the EC score matrix of the full dataset (Figs. S3, A and B, and 5 C).
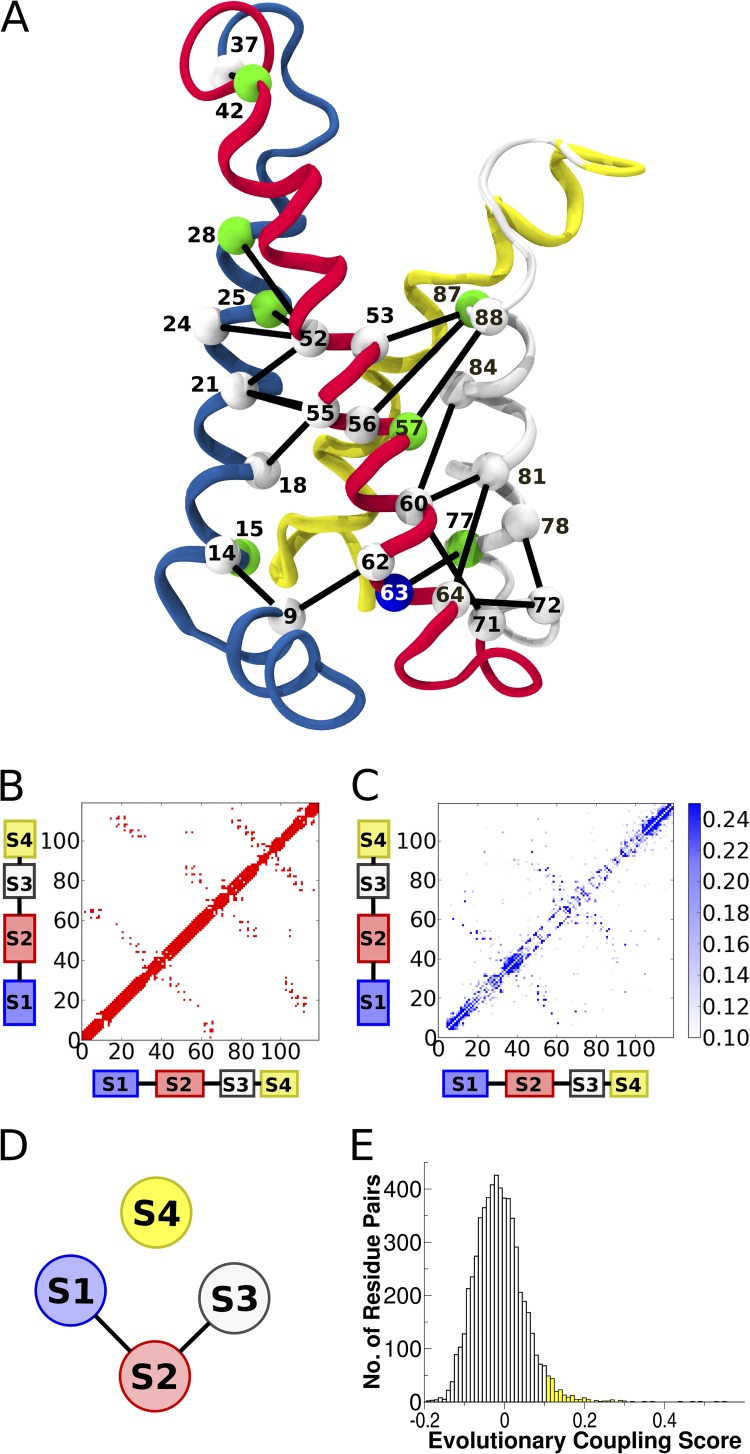
Figure 5.
DCA of the VSD. (A) Cartoon representation of NavAb VSD. Top 20 evolutionarily coupled contacting nonlocal residue pairs (more than five residues apart in the primary structure). C-α atoms are labeled and colored as in Fig. 3. Several top-scoring evolutionarily coupled pairs were excluded, as described in Results. Helices are colored as follows: blue, S1; red, S2; white, S3; yellow, S4. (B) Contact map for the NavAb VSD. A contact is defined between two residues if the respective C-β atoms (C-α in the case of glycine) are within 8 Å. (C) EC map for the VSD MSA. A few couplings were much higher than the 0.25 maximum shown on the colorbar. (D) Schematic showing EC topology of VSD. (E) Histogram of the EC scores for nonlocal residue pairs. All pairs shown in C fall in the yellow region.
Similarity between these EC score matrices suggests that results obtained from the full dataset are not significantly affected by sampling biases. Furthermore, and somewhat more importantly, this similarity indicates that the major features of the sequence ensemble are preserved on passing from Nav/Cav VSDs to Kv VSD ones.
Online supplemental material
The supplemental material contains the seed MSA used to infer the HMM (Table S1). Fig. S1 (A–G) shows pairwise sequence identity histograms for each of the seven protein families identified in our large MSA by hierarchical clustering. Fig. S1 H shows the dendrogram produced by hierarchical clustering, with assigned branches highlighted and labeled. Fig. S2 (A–D) shows the contact maps of four experimentally determined VSD structures and a structural superposition with the NavAb VSD. Fig. S3 (A and B) shows EC score matrices for partitioned MSAs containing either Nav and Cav or Kv VSD sequences. Tables S2 and S3 provide Shaker numbering for positions discussed in this work. Table S4 contains the membrane protein reference distribution. To help the readers map our results on each specific VSD, the MSA is available for download in the supplemental material, as well as a file with the complete set of parameters of the HMM used in this work The file format of the latter (.hmm) is compatible with the HMMER webserver (http://hmmer.janelia.org/search/hmmsearch) and can be used to generate an MSA using a different (possibly the most up to date) sequence database. The online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.201311103/DC1.
RESULTS
The S4 dilemma
To generate the large MSA on which our subsequent analysis is based, we first train a profile HMM to recognize and align VSDs based on a small “seed” alignment (Table S1). This alignment contains representative VSDs from phylogenetically diverse protein families such as the voltage-gated potassium, sodium, and calcium channels, the voltage-gated proton channels, and the voltage-sensitive phosphatases (Yu and Catterall, 2004). Using structural alignment and TM helix topology predictions, we manually refine automatic MSAs for all of the representative sequences included and generate a confident seed alignment, available in the supplemental material and described more fully in Materials and methods. However, recently published homology models of the human voltage-gated proton channel (hHv1) suggest that alignment of the S4 helix among VSDs may be ambiguous (Wood et al., 2012; Kulleperuma et al., 2013). Because hHv1 contains three gating charges and other VSDs contain four to six, alignments that only consider the matching of gating charges produce several different possible registers.
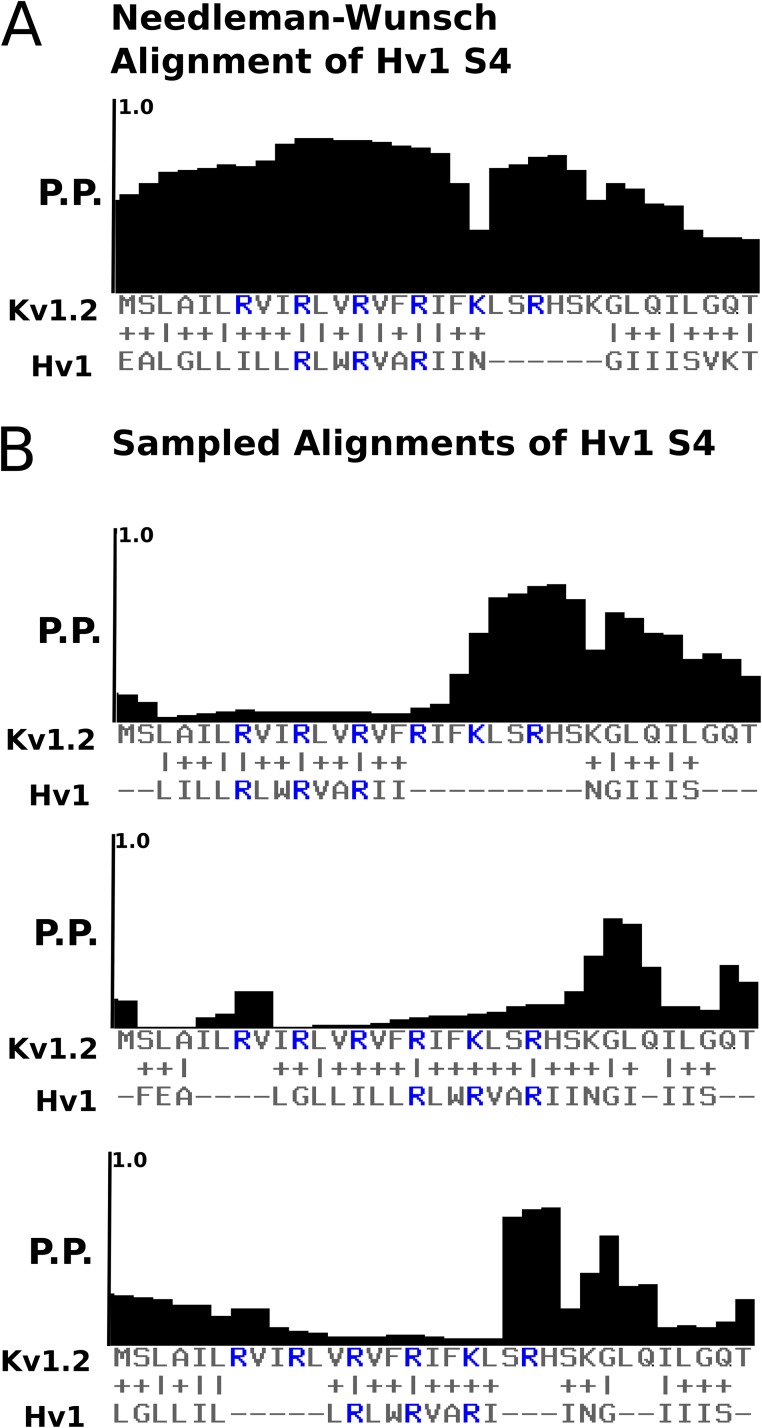
Without structural alignments for all of the VSD representatives included in the seed MSA, we are forced to rely on score-based sequence alignment of S4. A statistically well-founded metric to judge the reliability of score-based alignments is the positional posterior probability (Wolfsheimer et al., 2012). The posterior probability for a certain position in a pairwise alignment is calculated by marginalizing a posterior distribution of scored possible alignments. Simply, the posterior probability that two particular positions in a pair of sequences should be aligned is a measure of how probable it is to see those particular residues aligned if the full distribution of possible alignments is considered. The profile of posterior probabilities for a given pairwise alignment therefore shows where the alignment is probable and where it is uncertain. This is exactly what we would like to see for alternate alignments of S4.
In Fig. 1 A we show the optimal Needleman–Wunsch pairwise alignment of hHv1 to human Kv1.2, that is, the pairwise alignment with the best possible score. Incidentally, the register of this alignment is consistent with that of Kulleperuma et al. (2013). Over the region of the gating charges, the posterior probability is high. Taking advantage of the probabilistic pair HMMs underlying the posterior probability calculation, alternate alignments were sampled with hHv1 in different registers. Fig. 1 B shows that in three alternate registers of S4, the posterior probability of the region covering the gating charges is decimated. This indicates that given the fundamental assumptions of score-based pairwise sequence alignment, the proper register of hHv1 S4 is not ambiguous. This method was used to check the reliability of the S4 register for other pairs of sequences in the seed MSA.
Figure 1.
The register of S4 is not ambiguous. (A) Positional posterior probability of the Needleman–Wunsch optimal pairwise alignment of Kv1.2 and human Hv1. (B) Positional posterior probabilities of alignments sampled from the pair HMMs underlying the ppAlign algorithm. Each alignment represents a different possible register for the S4 helix of Hv1 with respect to Kv1.2.
Collecting and characterizing a large MSA of VSD sequences
With the technical issue of aligning S4 addressed, we train an HMM based on the seed MSA and collect a larger MSA of ∼6,600 VSD sequences as described in Materials and methods. We note that by using an HMM trained on a small but diverse seed MSA, we are able to automatically construct a much larger MSA than those reported previously for the VSD (Guda et al., 2007; Lee et al., 2009; Kulleperuma et al., 2013).
By building an HMM with a phylogenetically diverse seed MSA, we expected to collect a dataset of VSDs as complete as possible. It is important to confirm that the MSA collected does not exclude or over-represent any of the known VSD-containing protein families, and that sequences in each family are sufficiently diverse to capture the VSD sequence variability.
The 6,652 sequences of the VSD MSA were partitioned into clusters representing protein families as described in Materials and methods. In brief, the neighbor-joining hierarchical clustering method was used to construct a dendrogram (PHYLIP). Protein family clusters were labeled by identifying sequences with well-curated annotations in each major branch (see Fig. S1 for tree with family assignments). To assess the sequence variability of the dataset, pairwise sequence identities were calculated for all pairs of sequences in the full MSA and for all pairs within each identified protein family (see Fig. S1 for protein family sequence identity histograms).
As observed in Fig. 2 A, the hierarchical clustering naturally breaks down the MSA into previously described protein families including voltage-gated calcium and sodium channels, eukaryotic and prokaryotic voltage-gated potassium channels, prokaryotic sodium channels, hyperpolarization activated cyclic nucleotide–gated channels, voltage-gated proton channels, and voltage-sensitive enzymes. Interestingly, the first three VSDs of the pseudotetrameric voltage-gated sodium and calcium channels clustered together, and the fourth clustered separately. This may be expected from the findings of Lacroix et al. (2013), in which the functionally distinct kinetics of the first three “fast” domains of Nav1.4 compared with the fourth “slow” domain were shown to arise from specific sequence differences in their respective VSDs.
Fig. 2 B shows the distribution of pairwise sequence identity, where we observe a single large peak around 20% identity. If the dataset consisted of large clusters of mostly identical sequences, we would expect to see additional large peaks at high identity values. Our MSA therefore conforms to our requirements for extensive phylogenetic coverage and sufficient intra-family variability.
We also note the multimodal character of the pairwise sequence identity distributions for individual protein families (see Fig. S1). Multimodal histograms of sequence similarity are a clear indication of substructure in the data. Although our aim here is to confirm sequence diversity in our dataset and not to characterize the VSD sequence composition in every potential subfamily, the presence of modes in the distribution suggests the existence of distinct VSD subtypes within the identified protein families and may warrant further attention.
Single-site analysis of the VSD
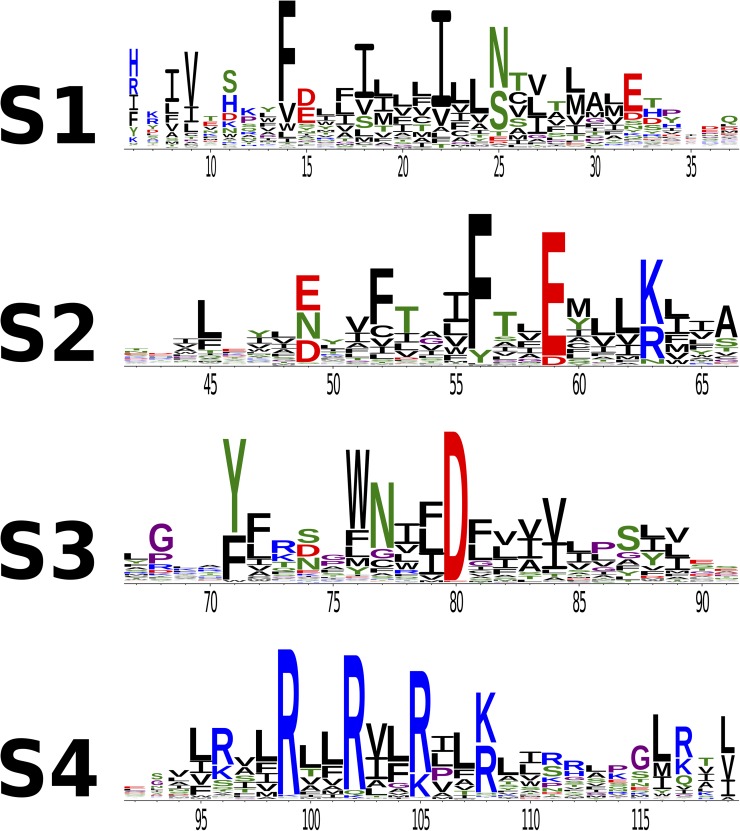
The large VSD MSA is represented as a sequence logo in Fig. 3 (Crooks et al., 2004), with positions numbered according to the Arcobacter butzleri voltage-gated sodium channel (NavAb) sequence (Payandeh et al., 2011).
Figure 3.
Sequence logos of the VSD. Black, CAVILMWF; green, TSNQY; blue, RKH; red, DE; purple, GP. Residue numbering corresponds to homologous positions in NavAb.
The height of the columns in Fig. 3 suggests that conserved residues on the VSD occur with periodicity. This trend was noticed in an early study done with a smaller MSA of VSD sequences (Guda et al., 2007). To quantify this observation, we calculate the Kullback–Leibler divergence (DKL) for each position in the MSA, as described in Materials and methods. In general, DKL values quantify the additional amount of information (in nats) differentiating the empirical distribution of amino acids at a certain position from a suitable reference distribution (Halabi et al., 2009). For instance, residues located in the TM region are expected to be mostly hydrophobic; a TM position with a distribution significantly enriched in hydrophilic residues arguably suggests evolutionary pressure and results in a large value of DKL. Therefore, we use DKL as an empirical site-specific measure of evolutionary pressure.
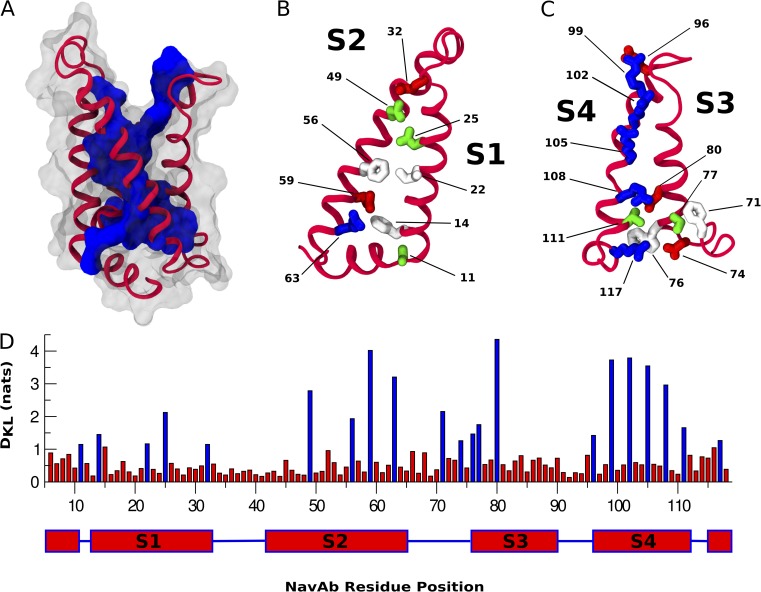
In Fig. 4 A, we map high DKL residues above a threshold (1.1 nats) onto the NavAb VSD and notice that the internal core of the bundle has the highest DKL. The outer surface of the VSD has much lower DKL, consistent with the idea that these positions make nonspecific interactions with membrane lipid tails and are thus subject to less strict evolutionary constraints (Cuello et al., 2004; Schmidt et al., 2006). Interestingly, the top of S3 (termed “S3b,” consisting of positions 86–91) does not exhibit this periodicity (Fig. 4 D), suggesting a lack of evolutionary pressure on this region. Fig. 4 D shows the relative variation of DKL in the VSD.
Figure 4.
Single-site analysis of the VSD. (A) Cartoon representation of the NavAb VSD. Residues with a DKL score greater than the 1.1-nat threshold are represented as a contiguous blue surface. Residues with a DKL score below the threshold are transparent. (B and C) Helices S1–S4 of the NavAb VSD. Residues with a DKL score greater than the 1.1-nat threshold are represented as licorice and colored as in Fig. 1. (D) DKL score by NavAb residue position with residue positions above the 1.1-DKL threshold represented as blue bars.
Unsurprisingly, the S4 gating charges (Fig. 4 C, positions 99, 102, 105, and 108) and the S1–S3 countercharges (Fig. 4, B and C, positions 32, 59, and 80) exhibit high DKL with respect to the rest of the VSD. This is consistent with their conspicuous placement as charged particles in the TM region and with their conserved mechanistic importance among VSDs (Aggarwal and MacKinnon, 1996; Seoh et al., 1996; Tiwari-Woodruff et al., 2000). Extensive mutagenesis studies have identified an S1 isoleucine (Fig. 4 B, position 22) and an S2 phenylalanine (Fig. 4 B, position 33) as the primary constituents of a conserved hydrophobic region present in VSDs (Tao et al., 2010; Lacroix and Bezanilla, 2011, 2012). Mutation of residues in this region modulates both the kinetics and voltage sensitivity of S4 translation. Both of these positions have high DKL (Fig. 4 D). Interestingly, voltage-gated proton channels are not significantly represented in the dataset; the position identified as the selectivity filter in hHv1 (Fig. 4 B, position 25 and hHv1 D112) shows high DKL (43), suggesting a functional significance for this position, even in VSDs that do not conduct protons.
Given that DKL measures evolutionary pressure and that identified functional residues exhibit high DKL, it may be valuable to investigate other high DKL positions throughout the VSD. To the best of our knowledge, residues at the lower membrane interface (Fig. 4, B and C, positions 11, 14, 63, 71, 74, 76, and 77) have not been experimentally characterized. From Fig. 3 we see that these positions take polar, aromatic, and positively charged amino acids. These amino acids are commonly observed at the membrane interface, specifically at the inner leaflet, as they are involved in anchoring the protein to membrane headgroups (von Heijne and Gavel, 1988; Yau et al., 1998). However, in VSDs their relative abundance cannot be explained only by their localization. Indeed, our formulation of DKL explicitly accounts for the propensity for observing these residues at the lower membrane interface. That the presence of these amino acids has been maintained throughout evolution is suggestive of distinct, conserved functional roles.
DCA of the VSD
We performed DCA on the VSD and ranked pairs according to their EC scores (see Materials and methods for the definition of EC scores). Simply, the EC score for a pair of residues is a heuristic measure of the total coupling as inferred from the probabilistic modeling. It is important to note that the distribution of EC scores does not show an obvious threshold of significance. Fig. 5 E shows a histogram of EC scores for non-neighbor pairs (more than five residues apart in sequence). The distribution appears to be approximately Gaussian with a fat right tail. Because we have no prior expectation about the distribution of EC scores, we set an arbitrary threshold of 0.10 (approximately corresponding to a standard deviation from the mean), as depicted by the yellow region in Fig. 5 E. Fig. 5 C only considers those couplings with EC scores >0.10. Considering only positions separated by at least five residues in the primary structure, we find that 20 of the top 23 pairs are in direct physical contact in the crystal structure of the activated NavAb VSD (Fig. 5 A). This gives a true-positive rate of 87% contact prediction for this first small subset of pairs. This is consistent with reported true-positive rates for contact prediction with pseudolikelihood maximization DCA (Ekeberg et al., 2013). These 20 contacting pairs define the interfaces between S1–S2 and S2–S3. Curiously, only two pairs in the top 23 involve S4 coupled to the S1–S3 bundle, and these pairs are not in physical contact in the NavAb VSD structure (Fig. 3, positions 25–96 and 49–96). For comparison with Fig. 5 C, contact maps of four other experimentally determined VSD structures are available in Fig. S2, accompanied by structural superpositions with the NavAb VSD.
To further investigate the spatial distribution of pairs with large EC scores on the VSD structure, we compare the EC score matrix to the contact map of the NavAb VSD (Fig. 5, B and C). The trend is similar to that observed in the top pairs: the contacting interfaces of S1–S2 and S2–S3 are well-defined in Fig. 5 C, whereas the contacting interfaces of S1–S4 and S3–S4 observed in the contact map are completely absent in the EC score matrix (shown schematically in Fig. 5 D). EC score matrices calculated from partitioned MSAs of only Cav/Nav or Kv VSDs show similar features (Fig. S3, A and B). In the next section we consider how design principles of VSD function may have constrained evolution to generate these observations.
DISCUSSION
In this study, we have constructed a large, diverse MSA of VSD sequences and identified patterns with two different but complementary sequence analysis approaches. By calculating the DKL of each position in the alignment, we identified a set of evolutionarily important, core-facing VSD positions (Fig. 3, B and C). Some of these positions are well-known determinants of voltage sensitivity, whereas others remain unexplored (Seoh et al., 1996; Tao et al., 2010; Lacroix and Bezanilla, 2012). We then used DCA to distinguish pairs of evolutionarily coupled residues. The distribution of strongly coupled pairs is inhomogeneous throughout the VSD, suggesting localized regions where specific evolutionary tuning has occurred. Especially strong EC is observed at the S1–S2 and S2–S3 interfaces, whereas the contacting S1–S4 and S3–S4 hydrophobic interfaces show no signal of coevolution. Considering the results, we now attempt to characterize the design principles followed by evolution to shape the observed sequence ensemble of VSDs.
Coevolving, state-dependent residue–residue contacts
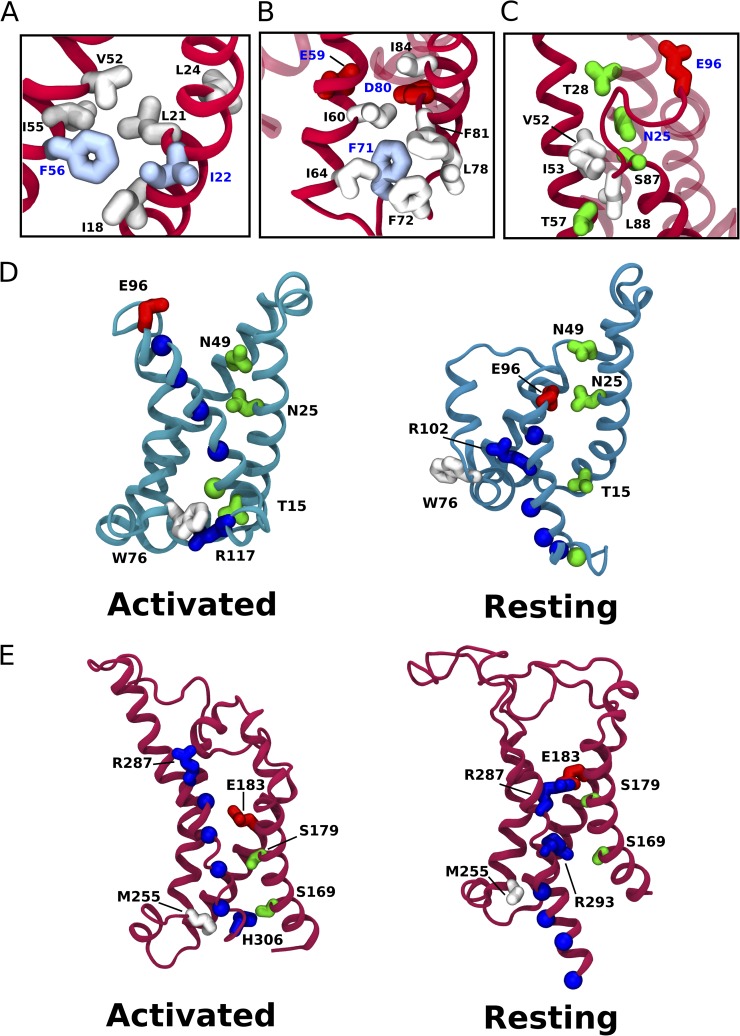
We find that ECs are informative of the conformational transition of S4 upon VSD activation. Specifically, we identified two evolutionarily coupled residue pairs, N49-E96 and N25-E96, which are not in contact in the activated NavAb crystal structure. Several independent lines of evidence show a physical interaction between N49-E96 positions in the resting state of several VSD homologues (DeCaen et al., 2009; Wu et al., 2010). Recent models of NavAb and Kv1.2 deactivation show both pairs in contact in a resting state (Fig. 6, D, N49-E96 and N25-E96, and E, R287-E183) (Bjelkmar et al., 2009; Amaral et al., 2012).
Figure 6.
Coevolving residue–residue contacts. (A) Close-up of the coevolving “hydrophobic nest” that surrounds conserved hydrophobic residues F56 and I22. (B) Close-up of the coevolving “cage” that surrounds F71. (C) Coevolving network of polar and apolar residues at the extracellular mouth of the VSD. (D) Computationally modeled activated and resting states of the NavAb VSD, as described in Amaral et al. (2012). Residues of interest are labeled. (E) Computationally modeled activated and resting (C3) states of the Kv1.2 VSD, as described in Bjelkmar et al. (2009).
Mutations in position 49 have been shown to have dramatic effects on the activation threshold in several VSDs (Papazian et al., 1995; Planells-Cases et al., 1995; Gamal El-Din et al., 2013). Pless et al. (2011) found that of the three countercharges in the Shaker K+ channel (Fig. 3, positions 49, 59, and 80; see Tables S2 and S3 for correspondence to Shaker numbering), alteration of the charge at position 49 had the greatest effect on the activation threshold. This effect can be understood as a modulation of the interaction of position 49 with the gating charges, selectively stabilizing specific states along the activation pathway (DeCaen et al., 2009; Delemotte et al., 2011; Amaral et al., 2012; Henrion et al., 2012; Jensen et al., 2012). We anticipated a coevolution signal between position 49 and all gating charge positions that exhibit some variability (namely, positions 96, 105, and 108). Surprisingly, we found coevolution only between 49 and 96, and this signal is very strong.
Because position 49 and 96 have been shown to interact in resting states of the VSD and mutation of 49 has been shown to stabilize the resting state, we hypothesize that the coevolution between 49 and 96 is the signature of an evolutionary tuning of the resting-state stability.
Position 25 is proximal to 49 in the NavAb VSD (Fig. 6 C), exhibits a qualitatively similar distribution of polar and acidic amino acids (Fig. 3), and also coevolves with 96 (EC score of 0.196). Although position 25 has been shown to determine proton selectivity in voltage-gated proton channels, we are not aware of any studies that probe the effects of mutating 25 on VSD activation threshold (Musset et al., 2011). In analogy to position 49, we hypothesize that it may serve a similar functional role in tuning the relative stability of VSD activation states.
A lack of expected S4 coevolution
Previous reports describing DCA have made strong parallels between the EC score matrix and the contact map (Weigt et al., 2009; Morcos et al., 2011; Hopf et al., 2012; Ekeberg et al., 2013). We were therefore surprised that the S1–S4 and S3–S4 contacting interfaces were absent in the EC score matrix. However, chimeragenesis and deletion analysis experiments suggest that the S3b–S4 paddle is modular (Alabi et al., 2007; Bosmans et al., 2008; Mishina et al., 2012; Kalia and Swartz, 2013; Xu et al., 2013). This modularity implies that as long as conserved features of S4 are maintained among chimeras (such as the gating charges and the hydrophobic faces of the helix), specific coevolution between contacting residue–residue pairs is not required for basic voltage-sensor function. This would be consistent with a free-energy landscape of activation dominated by coupled interactions between gating charges and countercharges, which do exhibit functionally relevant coevolution in at least one case (discussed in detail in the previous section). Although a family-specific tuning of the complementarity between the S1–S4 and S3–S4 interfaces cannot be excluded, the lack of specific coevolution between these interfaces suggests that this is not a necessary feature of VSDs.
Hydrophilic tuning of the VSD lumen
Because the gating charges have to slide into and out of the VSD, the extracellular mouth of the bundle must be solvated. We hypothesize that the chemical character of this “water crevice” depends on a subtle balance between hydrophilic and hydrophobic residues. This is highlighted by the fact that mutations in this region dramatically change the gating kinetics (Lacroix et al., 2013). Compensatory mutations could help maintain a specific hydrophilic character. Coevolution between several pairs of residues supports this hypothesis (Fig. 6 C).
On the opposite, intracellular side of the VSD, R63 and S77 are also observed to be evolutionarily coupled. A stabilizing pair of hydrophilic residues at the intracellular mouth may facilitate the solvation of this side of the VSD. Additionally, W76 and T15 coevolve but are not in physical proximity in the activated NavAb structure, lying on opposite sides of position 117 in the activated states of NavAb and Kv1.2 (Fig. 6, D and E, “Activated” states, and Table S3). In modeled resting states of both NavAb and Kv1.2, another positive charge sits between these two positions (Fig. 6, D and E, “Resting” states). The DKL of 76 is very high, and 15 is right beneath the chosen DKL threshold. We propose that coevolution between these residues tunes the energetics of the S4 transition.
A hydrophobic nest
Although the importance of the conserved gating and countercharges was recognized early in studies of voltage-gated ion channels, the role of the hydrophobic region was recognized much more recently (Tao et al., 2010; Lacroix and Bezanilla, 2011). The experimentally characterized constituents of the hydrophobic region include an S1 isoleucine and an S2 phenylalanine (Fig. 3, I22 and F56). Mechanistically, this region hinders the diffusion of ions and water through the VSD and acts as an energetic barrier for the movement of S4 (Schwaiger et al., 2013). An evolutionarily coupled network of residues surrounds I22 and F56 (Fig. 6 A). We propose that these residues form a “hydrophobic nest” that constrains the geometry in this region. Shape complementarity of residues in the hydrophobic nest is maintained by compensatory mutations, which are in turn detected as ECs by DCA.
A bulky cage
A similar feature is observed at the base of the S1 and S2 helices. Single-site residue frequency analysis highlights F71 as a potentially functionally important residue. This hypothesis is supported by the presence of a highly connected network of couplings surrounding F71 (Fig. 6 B). Residues in this cluster are generally bulky hydrophobic residues. We speculate that F71 and its network of coevolving bulky neighbors define a “cage” that stabilizes the bundle in the region containing the acidic residues E59 and D80. These countercharges are crucial for the activation mechanism and are involved in highly energetic electrostatic interactions; the requirement for their correct positioning resulted in a fine-tuning of the helical interface that separates them.
Concluding remarks
The VSD is a modular molecular machine with a mechanism of activation conserved over billions of years (Yu and Catterall, 2004). The distribution of extant VSD sequences results from an evolutionary exploration of sequence space constrained by the functional requirements of voltage sensing. Sequence analysis of this distribution reveals the nature of these constraints. Here, we applied an information–theoretical approach to describe site-specific frequency distributions of residues. Then, we built a probabilistic model accounting for both site-specific residue propensity and pairwise residue–residue statistical couplings. In both cases, we find that evolutionary constraints are exerted on specific sets of residues or pairs of residues, suggesting a sparse network of residue–residue interactions in which evolution attends to only certain nodes and edges. Previous studies have taken similar sequence analysis approaches and have proven successful in identifying experimentally validated features of membrane proteins (Süel et al., 2003; Lee et al., 2009; Pless et al., 2013). Here, we take a broader perspective and provide a comprehensive picture of the design principles of the VSD: (a) state-dependent coevolving contacts between 49–96 (and possibly 25–96) tune the relative stability of the resting state; (b) absence of specific, tuned interactions along the hydrophobic interfaces of S4 and neighboring S1 and S3 helices enables S4 translation; (c) networks of hydrophilic and hydrophobic residues lining the lumen of the VSD tune the access of penetrating water and gating charges; and (d) conserved residue positions, such as the countercharges I22 or F56, are surrounded by coevolving networks of hydrophobic residues stabilizing their orientation.
Finally, the design principles outlined in this study are encoded in the probabilistic model underlying DCA, the Potts model. This model is capable of sampling sequence space, emitting VSD-like sequences that by construction respect the pairwise correlations observed in real VSD sequences. This raises the exciting possibility of testing the validity of the design principles through the evolutionarily informed de novo design of VSDs.
Supplementary Material
Acknowledgments
The authors thank Cristian Micheletti and Sandipan Chowdhury for critical discussions and review of the manuscript.
This work was supported by the National Institutes of Health, National Institute for General Medical Sciences (grant P01 GM55876 to M.L. Klein). L. Delemotte receives funding from European Union Seventh Framework Program “Voltsens” (grant PIOF-GA-2012-329534).
The authors declare no competing financial interests.
Kenton J. Swartz served as editor.
Footnotes
Abbreviations used in this paper:
- DCA
- direct-coupling analysis
- EC
- evolutionary coupling
- HMM
- hidden Markov model
- MSA
- multiple sequence alignment
- TM
- transmembrane
- VSD
- voltage-sensor domain
References
- Aggarwal S.K., MacKinnon R. 1996. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 16:1169–1177 10.1016/S0896-6273(00)80143-9 [DOI] [PubMed] [Google Scholar]
- Ahern C.A., Horn R. 2005. Focused electric field across the voltage sensor of potassium channels. Neuron. 48:25–29 10.1016/j.neuron.2005.08.020 [DOI] [PubMed] [Google Scholar]
- Alabi A.A., Bahamonde M.I., Jung H.J., Kim J.I., Swartz K.J. 2007. Portability of paddle motif function and pharmacology in voltage sensors. Nature. 450:370–375 10.1038/nature06266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral C., Carnevale V., Klein M.L., Treptow W. 2012. Exploring conformational states of the bacterial voltage-gated sodium channel NavAb via molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 109:21336–21341 10.1073/pnas.1218087109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigoni C., Schroeder I., Romani G., Van Etten J.L., Thiel G., Moroni A. 2013. The voltage-sensing domain of a phosphatase gates the pore of a potassium channel. J. Gen. Physiol. 141:389–395 10.1085/jgp.201210940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelkmar P., Niemelä P.S., Vattulainen I., Lindahl E. 2009. Conformational changes and slow dynamics through microsecond polarized atomistic molecular simulation of an integral Kv1.2 ion channel. PLOS Comput. Biol. 5:e1000289 10.1371/journal.pcbi.1000289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börjesson S.I., Elinder F. 2008. Structure, function, and modification of the voltage sensor in voltage-gated ion channels. Cell Biochem. Biophys. 52:149–174 10.1007/s12013-008-9032-5 [DOI] [PubMed] [Google Scholar]
- Bosmans F., Martin-Eauclaire M.-F., Swartz K.J. 2008. Deconstructing voltage sensor function and pharmacology in sodium channels. Nature. 456:202–208 10.1038/nature07473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos F.V., Chanda B., Roux B., Bezanilla F. 2007. Two atomic constraints unambiguously position the S4 segment relative to S1 and S2 segments in the closed state of Shaker K channel. Proc. Natl. Acad. Sci. USA. 104:7904–7909 10.1073/pnas.0702638104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall W.A. 2010. Ion channel voltage sensors: structure, function, and pathophysiology. Neuron. 67:915–928 10.1016/j.neuron.2010.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.M., Hull C.M., Niven C.M., Qi J., Allard C.R., Claydon T.W. 2013. Functional interactions of voltage sensor charges with an S2 hydrophobic plug in hERG channels. J. Gen. Physiol. 142:289–303 10.1085/jgp.201310992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuello L.G., Cortes D.M., Perozo E. 2004. Molecular architecture of the KvAP voltage-dependent K+ channel in a lipid bilayer. Science. 306:491–495 10.1126/science.1101373 [DOI] [PubMed] [Google Scholar]
- DeCaen P.G., Yarov-Yarovoy V., Sharp E.M., Scheuer T., Catterall W.A. 2009. Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proc. Natl. Acad. Sci. USA. 106:22498–22503 10.1073/pnas.0912307106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delemotte L., Tarek M., Klein M.L., Amaral C., Treptow W. 2011. Intermediate states of the Kv1.2 voltage sensor from atomistic molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 108:6109–6114 10.1073/pnas.1102724108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokholyan N.V., Mirny L.A., Shakhnovich E.I. 2002. Understanding conserved amino acids in proteins. Physica A. 314:600–606 10.1016/S0378-4371(02)01079-8 [DOI] [Google Scholar]
- Eddy S.R. 1998. Profile hidden Markov models. Bioinformatics. 14:755–763 10.1093/bioinformatics/14.9.755 [DOI] [PubMed] [Google Scholar]
- Ekeberg M., Lövkvist C., Lan Y., Weigt M., Aurell E. 2013. Improved contact prediction in proteins: using pseudolikelihoods to infer Potts models. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 87:012707 10.1103/PhysRevE.87.012707 [DOI] [PubMed] [Google Scholar]
- Freites J.A., Tobias D.J., White S.H. 2006. A voltage-sensor water pore. Biophys. J. 91:L90–L92 10.1529/biophysj.106.096065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamal El-Din T.M., Martinez G.Q., Payandeh J., Scheuer T., Catterall W.A. 2013. A gating charge interaction required for late slow inactivation of the bacterial sodium channel NavAb. J. Gen. Physiol. 142:181–190 10.1085/jgp.201311012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud B.G., Heumann J.M., Lapedes A.S. 1999. Superadditive correlation. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 59:4983–4991 [DOI] [PubMed] [Google Scholar]
- Guda P., Bourne P.E., Guda C. 2007. Conserved motifs in voltage-sensing and pore-forming modules of voltage-gated ion channel proteins. Biochem. Biophys. Res. Commun. 352:292–298 10.1016/j.bbrc.2006.10.190 [DOI] [PubMed] [Google Scholar]
- Halabi N., Rivoire O., Leibler S., Ranganathan R. 2009. Protein sectors: evolutionary units of three-dimensional structure. Cell. 138:774–786 10.1016/j.cell.2009.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrion U., Renhorn J., Börjesson S.I., Nelson E.M., Schwaiger C.S., Bjelkmar P., Wallner B., Lindahl E., Elinder F. 2012. Tracking a complete voltage-sensor cycle with metal-ion bridges. Proc. Natl. Acad. Sci. USA. 109:8552–8557 10.1073/pnas.1116938109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf T.A., Colwell L.J., Sheridan R., Rost B., Sander C., Marks D.S. 2012. Three-dimensional structures of membrane proteins from genomic sequencing. Cell. 149:1607–1621 10.1016/j.cell.2012.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- Jensen M.Ø., Jogini V., Borhani D.W., Leffler A.E., Dror R.O., Shaw D.E. 2012. Mechanism of voltage gating in potassium channels. Science. 336:229–233 10.1126/science.1216533 [DOI] [PubMed] [Google Scholar]
- Jogini V., Roux B. 2007. Dynamics of the Kv1.2 voltage-gated K+ channel in a membrane environment. Biophys. J. 93:3070–3082 10.1529/biophysj.107.112540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia J., Swartz K.J. 2013. Exploring structure-function relationships between TRP and Kv channels. Sci. Rep. 3:1523 10.1038/srep01523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krepkiy D., Mihailescu M., Freites J.A., Schow E.V., Worcester D.L., Gawrisch K., Tobias D.J., White S.H., Swartz K.J. 2009. Structure and hydration of membranes embedded with voltage-sensing domains. Nature. 462:473–479 10.1038/nature08542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulleperuma K., Smith S.M.E., Morgan D., Musset B., Holyoake J., Chakrabarti N., Cherny V.V., DeCoursey T.E., Pomès R. 2013. Construction and validation of a homology model of the human voltage-gated proton channel hHV1. J. Gen. Physiol. 141:445–465 10.1085/jgp.201210856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix J.J., Bezanilla F. 2011. Control of a final gating charge transition by a hydrophobic residue in the S2 segment of a K+ channel voltage sensor. Proc. Natl. Acad. Sci. USA. 108:6444–6449 10.1073/pnas.1103397108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix J.J., Bezanilla F. 2012. Tuning the voltage-sensor motion with a single residue. Biophys. J. 103:L23–L25 10.1016/j.bpj.2012.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix J.J., Campos F.V., Frezza L., Bezanilla F. 2013. Molecular bases for the asynchronous activation of sodium and potassium channels required for nerve impulse generation. Neuron. 79:651–657 10.1016/j.neuron.2013.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lee S.-Y., Banerjee A., MacKinnon R. 2009. Two separate interfaces between the voltage sensor and pore are required for the function of voltage-dependent K+ channels. PLoS Biol. 7:e47 10.1371/journal.pbio.1000047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman E.R., Hess P., Weaver F., Koren G. 1991. Voltage-sensing residues in the S4 region of a mammalian K+ channel. Nature. 353:752–756 10.1038/353752a0 [DOI] [PubMed] [Google Scholar]
- Lomize M.A., Lomize A.L., Pogozheva I.D., Mosberg H.I. 2006. OPM: orientations of proteins in membranes database. Bioinformatics. 22:623–625 10.1093/bioinformatics/btk023 [DOI] [PubMed] [Google Scholar]
- Long S.B., Campbell E.B., Mackinnon R. 2005. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 309:903–908 10.1126/science.1116270 [DOI] [PubMed] [Google Scholar]
- Mishina Y., Mutoh H., Knöpfel T. 2012. Transfer of Kv3.1 voltage sensor features to the isolated Ci-VSP voltage-sensing domain. Biophys. J. 103:669–676 10.1016/j.bpj.2012.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcos F., Pagnani A., Lunt B., Bertolino A., Marks D.S., Sander C., Zecchina R., Onuchic J.N., Hwa T., Weigt M. 2011. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl. Acad. Sci. USA. 108:E1293–E1301 10.1073/pnas.1111471108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y., Iwasaki H., Sasaki M., Inaba K., Okamura Y. 2005. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 435:1239–1243 10.1038/nature03650 [DOI] [PubMed] [Google Scholar]
- Musset B., Smith S.M.E., Rajan S., Morgan D., Cherny V.V., Decoursey T.E. 2011. Aspartate 112 is the selectivity filter of the human voltage-gated proton channel. Nature. 480:273–277 10.1038/nature10557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman S.B., Wunsch C.D. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48:443–453 10.1016/0022-2836(70)90057-4 [DOI] [PubMed] [Google Scholar]
- Papazian D.M., Shao X.M., Seoh S.-A., Mock A.F., Huang Y., Wainstock D.H. 1995. Electrostatic interactions of S4 voltage sensor in Shaker K+ channel. Neuron. 14:1293–1301 10.1016/0896-6273(95)90276-7 [DOI] [PubMed] [Google Scholar]
- Payandeh J., Scheuer T., Zheng N., Catterall W.A. 2011. The crystal structure of a voltage-gated sodium channel. Nature. 475:353–358 10.1038/nature10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planells-Cases R., Ferrer-Montiel A.V., Patten C.D., Montal M. 1995. Mutation of conserved negatively charged residues in the S2 and S3 transmembrane segments of a mammalian K+ channel selectively modulates channel gating. Proc. Natl. Acad. Sci. USA. 92:9422–9426 10.1073/pnas.92.20.9422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless S.A., Galpin J.D., Niciforovic A.P., Ahern C.A. 2011. Contributions of counter-charge in a potassium channel voltage-sensor domain. Nat. Chem. Biol. 7:617–623 10.1038/nchembio.622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless S.A., Niciforovic A.P., Galpin J.D., Nunez J.-J., Kurata H.T., Ahern C.A. 2013. A novel mechanism for fine-tuning open-state stability in a voltage-gated potassium channel. Nat Commun. 4:1784 10.1038/ncomms2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey I.S., Moran M.M., Chong J.A., Clapham D.E. 2006. A voltage-gated proton-selective channel lacking the pore domain. Nature. 440:1213–1216 10.1038/nature04700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands Z.A., Sansom M.S.P. 2007. How does a voltage sensor interact with a lipid bilayer? Simulations of a potassium channel domain. Structure. 15:235–244 10.1016/j.str.2007.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D., Jiang Q.-X., MacKinnon R. 2006. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature. 444:775–779 10.1038/nature05416 [DOI] [PubMed] [Google Scholar]
- Schwaiger C.S., Liin S.I., Elinder F., Lindahl E. 2013. The conserved phenylalanine in the K+ channel voltage-sensor domain creates a barrier with unidirectional effects. Biophys. J. 104:75–84 10.1016/j.bpj.2012.11.3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoh S.-A., Sigg D., Papazian D.M., Bezanilla F. 1996. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 16:1159–1167 10.1016/S0896-6273(00)80142-7 [DOI] [PubMed] [Google Scholar]
- Süel G.M., Lockless S.W., Wall M.A., Ranganathan R. 2003. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol. 10:59–69 10.1038/nsb881 [DOI] [PubMed] [Google Scholar]
- Tao X., Lee A., Limapichat W., Dougherty D.A., MacKinnon R. 2010. A gating charge transfer center in voltage sensors. Science. 328:67–73 10.1126/science.1185954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari-Woodruff S.K., Lin M.A., Schulteis C.T., Papazian D.M. 2000. Voltage-dependent structural interactions in the Shaker K+ channel. J. Gen. Physiol. 115:123–138 10.1085/jgp.115.2.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombola F., Pathak M.M., Gorostiza P., Isacoff E.Y. 2007. The twisted ion-permeation pathway of a resting voltage-sensing domain. Nature. 445:546–549 10.1038/nature05396 [DOI] [PubMed] [Google Scholar]
- Treptow W., Tarek M. 2006. Environment of the gating charges in the Kv1.2 Shaker potassium channel. Biophys. J. 90:L64–L66 10.1529/biophysj.106.080754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas E., Yarov-Yarovoy V., Khalili-Araghi F., Catterall W.A., Klein M.L., Tarek M., Lindahl E., Schulten K., Perozo E., Bezanilla F., Roux B. 2012. An emerging consensus on voltage-dependent gating from computational modeling and molecular dynamics simulations. J. Gen. Physiol. 140:587–594 10.1085/jgp.201210873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viklund H., Elofsson A. 2008. OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics. 24:1662–1668 10.1093/bioinformatics/btn221 [DOI] [PubMed] [Google Scholar]
- von Heijne G., Gavel Y. 1988. Topogenic signals in integral membrane proteins. Eur. J. Biochem. 174:671–678 10.1111/j.1432-1033.1988.tb14150.x [DOI] [PubMed] [Google Scholar]
- Weigt M., White R.A., Szurmant H., Hoch J.A., Hwa T. 2009. Identification of direct residue contacts in protein–protein interaction by message passing. Proc. Natl. Acad. Sci. USA. 106:67–72 10.1073/pnas.0805923106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfsheimer S., Hartmann A., Rabus R., Nuel G. 2012. Computing posterior probabilities for score-based alignments using ppALIGN. Stat. Appl. Genet. Mol. Biol. 11:1 10.1515/1544-6115.1702 [DOI] [PubMed] [Google Scholar]
- Wood M.L., Schow E.V., Freites J.A., White S.H., Tombola F., Tobias D.J. 2012. Water wires in atomistic models of the Hv1 proton channel. Biochim. Biophys. Acta. 1818:286–293 10.1016/j.bbamem.2011.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Delaloye K., Zaydman M.A., Nekouzadeh A., Rudy Y., Cui J. 2010. State-dependent electrostatic interactions of S4 arginines with E1 in S2 during Kv7.1 activation. J. Gen. Physiol. 135:595–606 10.1085/jgp.201010408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Ramu Y., Shin H.-G., Yamakaze J., Lu Z. 2013. Energetic role of the paddle motif in voltage gating of Shaker K+ channels. Nat. Struct. Mol. Biol. 20:574–581 10.1038/nsmb.2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau W.M., Wimley W.C., Gawrisch K., White S.H. 1998. The preference of tryptophan for membrane interfaces. Biochemistry. 37:14713–14718 10.1021/bi980809c [DOI] [PubMed] [Google Scholar]
- Yu F.H., Catterall W.A. 2004. The VGL-chanome: a protein superfamily specialized for electrical signaling and ionic homeostasis. Sci. STKE. 2004:re15 10.1126/stke.2532004re15 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Skolnick J. 2005. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33:2302–2309 10.1093/nar/gki524 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.