Abstract
Objective
To identify new genes and risk factors associated with frontotemporal dementia (FTD). Several genes and loci have been associated with different forms of FTD, but a large number of families with dementia do not harbor mutations in these genes.
Design
Whole-exome sequencing and whole-genome genotyping were performed in all patients. Genetic variants obtained from whole-exome sequencing were integrated with the data obtained from whole-genome genotyping.
Setting
Database of the Behavioral Neurology Outpatient Clinic of the Department of Neurology, Istanbul Faculty of Medicine, Istanbul, Turkey.
Patients
Forty-four Turkish patients with an FTD-like clinical diagnosis were included in the study. Relatives were screened when appropriate.
Main Outcome Measure
Mutations in the triggering receptor expressed on myeloid cells 2 gene (TREM2).
Results
In 3 probands with FTD-like disease, we identified different homozygous mutations in TREM2 that had previously been associated with polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy (PLOSL). None of these 3 patients had a typical clinical presentation of PLOSL: they presented with behavioral change and subsequent cognitive impairment and motor features but without any bone cysts or bone-associated phenotypes. Imaging showed white matter abnormalities as well as frontal atrophy in all 3 patients.
Conclusions
Our results show that TREM2 is responsible for an unexpectedly high number of dementia cases in our cohort, suggesting that this gene should be taken into account when mutations in other dementia genes are excluded. Even for complex syndromes such as dementia, exome sequencing has proven to be a rapid and cost-effective tool to identify genetic mutations, allowing for the association of clinical phenotypes with unexpected molecular underpinnings.
Genetic analysis has identified several genes and loci as underlying frontotemporal dementia (FTD). These include mutations in the granulin precursor gene (GRN, OMIM 138945)1,2 and microtubule-associated protein tau gene (MAPT, OMIM 157140).3 Most recently, a hexanucleotide repeat expansion in the noncoding region of the chromosome 9 open reading frame 72 gene (C9ORF72, OMIM 614260) has been shown to be the genetic cause of disease in a group of patients with FTD.4,5 Nonetheless, a large part of the genetic cause of FTD remains unknown. This is particularly evident in families with ages at onset between 55 and 70 years where mendelian variants in yet to be identified genes are thought to underlie the disease. Because of the low frequency of these variants, they will not be identifiable through genome-wide association studies. Technological advances now allow the analyses of complete exomes and genomes, placing us in a position to study these families. In fact, several recent studies have succeeded in identifying the genetic basis of different disorders by using exome and genome sequencing.6–9 These studies demonstrate that even by using a small number of samples, it is now possible to uncover genetic alterations not only in mendelian diseases but also in multifactorial complex disorders. This has great clinical utility with major implications for disease gene discovery, diagnosis, and, ultimately, therapeutic approaches.
As part of an ongoing study aimed at characterizing, from a genetic perspective, a large sample of Turkish families with dementia, we sequenced the whole exome of several cases. In 3 of these families, we identified homozygous mutations in the triggering receptor expressed on myeloid cells 2 gene (TREM2, OMIM605086). Mutations in this gene have previously been associated with polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy (PLOSL) (also known as Nasu-Hakola disease). This is a rare autosomal recessive disease characterized by early-onset progressive dementia and bone cysts. Herein, we describe 3 Turkish patients with TREM2 homozygous mutations, presenting clinically with an FTD-like syndrome but without any bone cysts or other bone phenotype, suggesting that mutations in this gene may be a more frequent cause of dementia than previously considered and showing that this diagnosis should be considered even in the absence of bone problems.
METHODS
HUMAN SUBJECTS
We reviewed the database of the Behavioral Neurology Outpatient Clinic of the Department of Neurology, Istanbul Faculty of Medicine, Istanbul, Turkey to identify all patients with a primary clinical syndrome within the FTD spectrum10 and an available blood sample. Forty-four patients were identified who had presented initially with either a change in personality and behavioral symptoms (consistent with either probable or possible behavioral variant FTD following the Frontal Temporal Dementia Consortium criteria11) or with a progressive aphasia (consistent with either semantic dementia or progressive nonfluent aphasia), although a number of patients subsequently developed atypical features including seizures. A diagnosis of FTD was made on the basis of clinical and neuropsychological assessment and independent of any neuroimaging. Seventy-six samples from healthy controls (mainly caregivers and spouses accompanying patients to the clinic) were also collected. Written consent for participation was obtained in accordance with institutional review board standards. Most of the families had Turkish ethnic background (n = 39) and suspected (n = 23) or known (n = 4) consanguinity. The general features of the patients' cohort studied are presented in eTable 1. Genomic DNA from these samples was prepared from venous blood samples by standard procedures. Samples from the human genome resequencing panel of the Centre d'Etude du Polymorphisme Humain–Human Genome Diversity Cell Line Panel (n = 275) were used to assess the frequency of genetic variation in exon 2 of TREM2.12
EXOME SEQUENCING
Sequences corresponding to all annotated human exons were enriched by hybridization using the SeqCap EZ Exome Library version 1.0 (Roche NimbleGen), as per manufacturers' protocols. The DNA samples were sequenced in 1 flow cell lane each, on paired-end 50–base pair HiSeq 2000 runs (Illumina Inc), yielding an average of about 6 billion high-quality bases per sample. Image analysis and base calling were performed using the Illumina pipeline (version 1.7.1) with default parameters. Sequence reads were mapped to the reference genome (human genome 18) using the Burrows-Wheeler aligner.13 SAMtools was used to generate BAM files.14 Single-nucleotide polymorphisms (SNPs) and indels were called using the Genome Analysis Toolkit.15 Visual inspection of variants was performed, when necessary, using the Integrative Genomics Viewer.16
SANGER SEQUENCING
To confirm the mutations found by exome sequencing in the probands, test family members, and screen control individuals, ExonPrimer (http://ihg.gsf.de/ihg/ExonPrimer.html) was used to generate primers for amplification of TREM2 exon 2. This exon was polymerase chain reaction amplified using Roche FastStart PCR Master Mix polymerase (Roche Diagnostics Corp) and the polymerase chain reaction products were sequenced using the same forward and reverse primers with Applied Biosystems BigDye terminator version 3.1 sequencing chemistry and run on an ABI3730XL genetic analyzer as per the manufacturer's instructions (Applied Biosystems). The sequences were analyzed using Sequencher software, version 4.2 (Gene Codes).
ILLUMINA SNP BEADCHIP ANALYSIS
To exclude the presence of large structural variants and to perform homozygosity mapping, the available DNA samples from the 3 families in whom mutations in TREM2 were found were run on HumanOmniExpress BeadChips as per the manufacturer's instructions (Illumina Inc). Data were visualized and analyzed using the GenomeStudio Data Analysis Software (Illumina Inc).
RESULTS
GENETIC ANALYSIS
No pathogenic mutations in the known FTD genes were found in this cohort; however, in 1 sample, a novel variant in MAPT was identified (data not shown).
By applying a series of filters, we were able to identify a previously described homozygous nonsense mutation (p.Q33X) in TREM2 in case 1 (eTable 2). This led us to screen the whole cohort for variants inTREM2. Two more homozygous changes were identified. For these 3 cases, whole-exome sequencing resulted in the identification of an average of 1225 novel (not present in dbSNP version 132 or the 1000 Genomes project and including variants located in exon/intron boundaries ±4 base pairs) single-nucleotide variants, of which an average of 70 were homozygous (eTable 2). SIFT was used as a first-pass filter to predict how the amino acid substitutions found would affect protein function, taking into account sequence homology and the physical properties of amino acids, predicting 17 homozygous changes to be damaging in case 2 (including TREM2 p.T66M) and 30 homozygous changes in case 3 (including TREM2 p.Y38C). From the analysis of the results corresponding to stop-loss and stop-gain variants in the 3 probands (Table), the stop-loss mutation p.X152R in PRB1, found in probands 1 and 3, could be considered as a possible cause of disease. However, this variant is now known to be a polymorphism (rs111877208) with a high minor allele frequency in the 1000 Genomes project (minor allele frequency A = 0.273).
Table.
Homozygous Stop Changes (Including Stop Loss and Stop Gain) in the 3 Cases Where Homozygous TREM2 Mutations Were Found
| Chr | Gene | Change Function | Change | |
|---|---|---|---|---|
| Proband 1 | 6 | TREM2 | Stop gain | NM_018955:c.C97T:p.033X |
| 7 | TSGA13 | Stop gain | NM_052933:c.C183G:p.Y61X | |
| 12 | PEB1 | Stop loss | NM_005039:c.T454C:p.X152R | |
| Proband 2 | 17 | CCDC144NI | Stop gain | NM_001004306:c.C533A:p.S178X |
| 19 | IL28A | Stop gain | NM_172133:c.C562T:p.R188X | |
| Proband 3 | 12 | PRB1 | Stop loss | NM_005039:c.T454C:p.X152R |
| 14 | PLEKHH1 | Stop gain | NM_020715:c.C4042T:p.R1348X | |
| 14 | YLPM1 | Stop gain | NM_019539:c.G4950T:p.E1654X | |
| 19 | HAS1 | Stop gain | NM_001523:c.C42A:p.C14X |
Abbreviation: Chr, chromosome.
Samples from unaffected family members were available for genetic testing for the 3 families (Figure 1). This screening revealed no other homozygous mutations in TREM2 (eFigure 1). Similarly, no other family members shared regions of homozygosity encompassing TREM2 on chromosome 6 (eFigure 2).
Figure 1.
Pedigrees of the 3 families where triggering receptor expressed on myeloid cells 2 gene (TREM2) mutations were found. Arrowheads indicate probands. Black filled symbols represent affected subjects with dementia. White symbols represent unaffected family members and white symbols with a black dot represent individuals where dementia has been reported but not clinically confirmed. The genotypes (N indicates normal and M, mutation found) are represented for those individuals for whom DNA samples were available.
To exclude that the variants found were common polymorphisms, we sequenced exon 2 of the TREM2 gene in samples from 351 healthy control individuals from different regions of the world (including 76 Turkish samples). Four changes were found in 12 samples (eTable 3). From these, the p.T96K/M variant was found in the homozygous state in 1 sample and in heterozygosity, in 6 samples. This seems to be a highly variant locus because 2 SNPs (p.T96K and p.T96R [rs2234253], with an established minor allele frequency of 0.04 in dbSNP version 135) have been previously reported, and herein, we describe the novel change p.T96M found in 3 samples. The homozygous mutation found in family 2 (p.T66M) was found in heterozygosity in a Pakistani sample. These data confirm that the homozygous variants herein associated with disease (p.Q33X, p.T66M, and p.Y38C) are not common in healthy populations from different regions of the world. In fact, to our knowledge, no homozygous mutations have been described in the literature in healthy individuals, suggesting a complete penetrance for this type of mutation.
CLINICAL DETAILS OF THE FAMILIES
Family 1
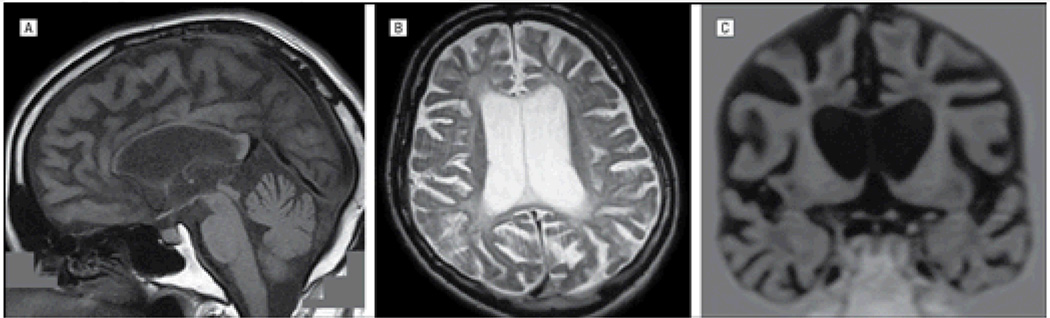
The proband originates from Trabzon, a city on the Black Sea coast of Anatolia in Turkey. He was said to have had a brief period of hypoxia during birth and a few febrile seizures over the course of his first 3 years of life, but following this, he had normal development and no other medical problems during his childhood. At the age of 20 years, he developed a change in his personality with aggressive behavior. Behavioral problems of aggression, impulsivity, and perseverative behavior progressively worsened over the following years and increasing apathy was also noted. During this same period, he also developed generalized tonic-clonic seizures and, in his late 20s, cognitive impairment. Ten years into his illness, at the age of 30 years, he was seen in the Neurology Department at Istanbul University. He was dysarthric and a neurological examination revealed an ophthalmoplegia with bradykinesia and brisk deep tendon reflexes throughout. Cognitive testing showed marked executive dysfunction and memory impairment. Routine serum and cerebrospinal fluid testing as well as electromyogram were normal. A skeletal survey did not show any radiography abnormalities. Magnetic resonance imaging (MRI) of the brain showed focal atrophy of the frontal and temporal lobes, as well as the corpus callosum. There were diffuse confluent white matter abnormalities, especially adjacent to both the anterior and posterior horns of the ventricles, which were disproportionally enlarged (Figure 2). Technetium Tc 99m single-photon emission computed tomography showed hypoperfusion bilaterally in the frontal and temporal lobes, predominantly on the right, as well as the basal ganglia. Ultrastructural examination of a cutaneous biopsy specimen, performed by electron microscopy, showed granular osmophilic deposits in eccrine sweat gland epithelial cells as seen in aging cells but also frequently found in Kufs disease. He is a dizygotic twin and has 2 other brothers, all healthy. His parents were cousins, his father died at age 58 years of tuberculosis, and all family members were from the same village. Although there was no family history of dementia, an aunt and uncle both had been diagnosed with a psychotic disorder and 1 nephew had epilepsy (Figure 1).
Figure 2.
Magnetic resonance images from the proband of family 1. Sagittal T1-weighted (A), axial T2-weighted (B), and coronal T1-weighted (C) images show marked thinning of the corpus callosum; diffuse confluent white matter abnormalities in the periventricular white matter, especially adjacent to the anterior and posterior horns of the lateral ventricle; cortical atrophy; and disproportionally enlarged ventricles.
Family 2
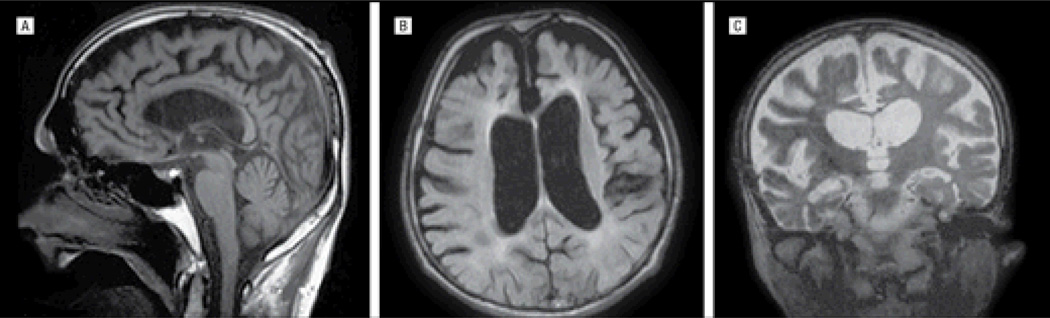
The proband originates from Agri, a city in the Eastern Anatolian region of Turkey. He developed a change in personality in his late 30s with aggressive and perseverative behavior. At the age of 40 years, he developed impairment in cognition, particularly affecting memory. His behavioral and cognitive impairments both progressed over the next few years and he was first assessed at the age of 45 years in the Neurology Department at Istanbul University. Neurological examination at the time revealed bradykinesia, apraxia, and postural instability. Mini-Mental State Examination score was 22 of 30. Results of routine serum and cerebrospinal fluid testing were normal. An MRI scan of the brain revealed atrophy particularly affecting the frontal lobes with marked ventricular enlargement and thinning of the corpus callosum. Confluent periventricular white matter abnormalities were also seen, especially adjacent to the lateral ventricle (Figure 3). The patient continued to deteriorate over the next few years, with progressive behavioral and cognitive impairment (Mini-Mental State Examination score was 13 of 30 at the age of 48 years) and progressive cerebral atrophy on MRI affecting predominantly the frontal lobes. A skeletal survey at this time did not show any radiography abnormalities. He was the second of 5 siblings from consanguineous parents. One of his brothers, his paternal uncle, and his maternal grandfather also had memory impairment, although all had an age at onset older than 60 years (Figure 1).
Figure 3.
Magnetic resonance images from the proband of family 2. Sagittal T2-weighted (A), coronal T1-weighted (B), and axial fluid-attenuated inversion recovery–weighted (C) images show thinning of the corpus callosum with cortical atrophy and periventricular white matter abnormalities.
Family 3
The proband originates from Erzincan, a city in the Eastern Anatolian region of Turkey. He was well until the age of 33 years when he had a generalized tonic-clonic seizure. He had no further seizures over the next few years, but at the age of 36 years, he developed a change in personality. He became more socially withdrawn and was at times aggressive. He had progressive behavioral problems over the following years with apathy predominating and subsequently developing mild memory impairment. Neurological examination at this time revealed bradykinesia and mild postural instability. At the age of 41 years, he had a further generalized tonic-clonic seizure and, when assessed at this time, was felt to have an akinetic-rigid syndrome with cognitive impairment and apathy. He was first assessed at the age of 43 years in the Neurology Department at Istanbul University by which time he had had a total of 8 generalized tonic-clonic seizures during the preceding 8 years. More recently, he had developed visual hallucinations and urinary incontinence. Neurological examination revealed a marked akinetic-rigid syndrome. He was severely cognitively impaired with a Mini-Mental State Examination score of 8 of 30. Results of routine serum and cerebrospinal fluid testing were normal as was electromyogram. An MRI scan of the brain showed cortical atrophy predominantly affecting the frontal lobes with some white matter abnormalities, particularly periventricular (Figure 4). No calcification of the basal ganglia was seen on a computed tomographic scan. The patient died 2 years later at the age of 45 years. He was the third of 4 siblings from consanguineous parents. There had been no family history of cognitive or behavioral impairment (Figure 1).
Figure 4.
Magnetic resonance images from the proband of family 3. Sagittal T1-weighted (A), axial fluid-attenuated inversion recovery–weighted (B), and coronal T2-weighted (C) images reveal significant thinning of the corpus callosum; diffuse confluent white matter abnormalities in the periventricular white matter, especially adjacent to the anterior and posterior horns of the lateral ventricle; global cortical atrophy; and enlarged ventricles.
COMMENT
By exome sequencing patients with dementia originating from Turkey, we were able to identify homozygous mutations in TREM2 in 3 probands from unrelated families. One of the mutations found (p.Q33X) has been previously described17,18 and is predicted to result in the generation of a truncated protein lacking the transmembrane and cytoplasmic domains, resulting in a complete loss of function of the protein. The 2 other homozygous mutations found (p.Y38C and p.T66M) are predicted to alter the protein function. In particular, p.Y38C alters the flanking consensus sequence of one of the cysteines potentially involved in generating the interchain disulfide bridge of the Ig-SF V-type fold (Figure 5). TREM2 is a single-pass type I membrane protein that forms a receptor-signaling complex with the TYRO protein tyrosine kinase-binding protein (TYROBP) and triggers activation of the immune responses in macrophages and dendritic cells.20 TREM2 is expressed on immature dendritic cells, osteoclasts, and microglia.21 It may be involved in chronic inflammation by triggering the production of constitutive rather than inflammatory chemokines and cytokines.22
Figure 5.
The triggering receptor expressed on myeloid cells 2 gene (TREM2) structure spans 41126246 to 41130922 base pairs (bp) on chromosome (chr) 6p21.1. A, Schematic of TREM2 (drawn using FancyGene19), the mutation nomenclature given with respect to RefSeq identifier NM_018965.2. Exons are drawn to scale and represented with gray boxes with untranslated regions (UTR) in red. Mutation locations are approximate: mutations identified in this study are represented below and previously described mutations are represented on top. B, The conservation of mutations identified in the present study. GenBank accession numbers: NP_061838.1, Homo sapiens; XP_001174113.1, Pan troglodytes;XP_851231.1, Canis lupus; NP_001073048.1, Bos taurus; NP_112544.1, Mus musculus; XP_217335.3,Rattus norvegicus; and NP_001032921.1, Gallus gallus.
Interestingly, mutations in both these genes have been associated with PLOSL or Nasu-Hakola disease. This disorder is associated with pathological fractures due to polycystic osseous lesions (often in the 20s) followed by a change in personality and progressive behavioral symptoms (consistent with a behavioral variant FTD syndrome) in the following decade. Subsequent progressive cognitive impairment (predominantly affecting memory, although aphasia and other cognitive domains may be involved particularly as the disease progresses) and motor symptoms occur with death usually before the age of 50 years.23 Generalized tonic-clonic seizures and/or myoclonus may also be seen in some patients. All 3 patients presented with a change in personality and behavioral symptoms and subsequently developed cognitive impairment. Motor features were seen in all 3 patients, most commonly features of an akinetic-rigid syndrome, which is not uncommon in patients with FTD, particularly as the disease progresses. Clinical diagnosis in all 3 mutation carriers was consistent with probable behavioral FTD following the Frontal Temporal Dementia Consortium criteria.11 However, 2 of the patients developed seizures, which would be atypical for FTD, and MRI findings were not only limited to frontotemporal lobar atrophy but also showed evidence of extensive white matter abnormalities and thinning of the corpus callosum, unusual imaging features for FTD. Importantly, none of the patients had any clinical or radiological evidence of bone involvement.
Computed tomographic brain imaging in PLOSL usually shows calcification of the basal ganglia (although this was not seen in the 1 patient to have computed tomographic imaging from family 3).18 Consistent with this, MRI may show decreased signal intensity in the basal ganglia on T2 imaging. T2 imaging also commonly shows diffusely and relatively symmetrically increased signal intensity in the white matter, although some cases have been reported with more limited changes. Brain imaging also may show cerebral (predominantly frontal and temporal lobe), and to a lesser extent cerebellar, atrophy23,24 as well as corpus callosum thinning.18 The imaging of the 3 patients was consistent with these previous findings with all patients showing white matter changes (although the extent was variable between the patients) and all having frontal and/or temporal atrophy as well as thinning of the corpus callosum. White matter abnormalities on standard MRI sequences are uncommon in patients with FTD caused by tau, TAR DNA-binding protein 43, or fused in sarcoma pathology and such findings in a patient with a clinical FTD syndrome would suggest further investigation for other pathological causes.
Individuals with a single mutated TREM2 allele have been shown to have subclinical changes, with 1 study demonstrating deficits in visuospatial memory associated with mild hypoperfusion in the basal ganglia in 2 heterozygotes.25 Heterozygous carriers in our families had no abnormalities on clinical or imaging examination.
Given the low prevalence of PLOSL in the population, TREM2 and TYROBP are very rarely considered in the genetic analysis of patients with dementia unless bone cysts are found during the clinical evaluation. Mutations in TREM2 leading to pure early-onset dementia without bone cysts have only been previously reported in a single Lebanese family where a deletion in the 5′ consensus donor splice site in intron 1 was identified.26 Similar to our families, the dementia in this family resembled behavioral FTD clinically, although with some atypical features and the presence of white matter changes on MRI. This report points to a role of TREM2 in cases of dementia without the presence of bone cysts. Our results reinforce this and suggest that TREM2 and possibly TYROBP should be considered when mutations in the other dementia genes are not found, particularly in patients with a behavioral FTD phenotype and white matter abnormalities seen on MRI.
In summary, herein we demonstrate that exome sequencing is a valid, rapid, and cost-effective tool to identify disease-causing mutations in complex syndromes like dementia. Using exome sequencing, we were able to identify the genetic cause of disease in 3 patients who otherwise would have never been screened for TREM2 because of their nontypical clinical presentation.
Supplementary Material
Acknowledgments
Funding/Support: This work was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services, project Z01 AG000950-06, Alzheimer's Research UK, an Istanbul Neuropsychiatric Hospital (NPIstanbul) research grant, the Motor Neurone Disease Association, and an anonymous donor.
Additional Contributions: We thank all the patients and their families. Nihan Unaltuna-Erginel, PhD, helped with DNA management and Ali Bayram, BSc, provided technical help.
Footnotes
Author Contributions: Drs Guerreiro and Lohmann contributed equally to this work. Drs Hardy, Guerreiro, and Lohmann had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: Hanagasi, Emre, Singleton, and Hardy. Acquisition of data: Guerreiro, Lohmann, Gurunlian, Dursun, Bilgic, Hanagasi, Gurvit, and Singleton. Analysis and interpretation of data: Guerreiro, Brás, Gibbs, Rohrer, and Hardy. Drafting of the manuscript: Lohmann, Dursun, and Hardy. Critical revision of the manuscript for important intellectual content: Guerreiro, Lohmann, Brás, Gibbs, Rohrer, Gurunlian, Bilgic, Hanagasi, Gurvit, Emre, Singleton, and Hardy. Obtaining funding: Emre, Singleton, and Hardy. Administrative, technical, and material support: Guerreiro, Lohmann, Brás, Gibbs, Gurunlian, Dursun, Bilgic, and Hanagasi. Study supervision: Emre, Singleton, and Hardy.
Financial Disclosure: None reported.
REFERENCES
- 1.Baker M, Mackenzie IR, Pickering-Brown SM, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442(7105):916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 2.Cruts M, Gijselinck I, van der Zee J, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442(7105):920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 3.Hutton M, Lendon CL, Rizzu P, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393(6686):702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 4.Renton AE, Majounie E, Waite A, et al. ITALSGEN Consortium. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42(1):30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lupski JR, Reid JG, Gonzaga-Jauregui C, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362(13):1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zimprich A, Benet-Pagès A, Struhal W, et al. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am J Hum Genet. 2011;89(1):168–175. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vilariño-Güell C, Wider C, Ross OA, et al. VPS35 mutations in Parkinson disease. Am J Hum Genet. 2011;89(1):162–167. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neary D, Snowden JS, Gustafson L, et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51(6):1546–1554. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- 11.Rascovsky K, Hodges JR, Knopman D, et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134(Pt 9):2456–2477. doi: 10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cann HM, de Toma C, Cazes L, et al. A human genome diversity cell line panel. Science. 2002;296(5566):261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Handsaker B, Wysoker A, et al. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative Genomics Viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soragna D, Papi L, Ratti MT, Sestini R, Tupler R, Montalbetti L. An Italian family affected by Nasu-Hakola disease with a novel genetic mutation in the TREM2 gene. J Neurol Neurosurg Psychiatry. 2003;74(6):825–826. doi: 10.1136/jnnp.74.6.825-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klünemann HH, Ridha BH, Magy L, et al. The genetic causes of basal ganglia calcification, dementia, and bone cysts: DAP12 and TREM2. Neurology. 2005;64(9):1502–1507. doi: 10.1212/01.WNL.0000160304.00003.CA. [DOI] [PubMed] [Google Scholar]
- 19.Rambaldi D, Ciccarelli FD. FancyGene: dynamic visualization of gene structures and protein domain architectures on genomic loci. Bioinformatics. 2009;25(17):2281–2282. doi: 10.1093/bioinformatics/btp381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paloneva J, Manninen T, Christman G, et al. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet. 2002;71(3):656–662. doi: 10.1086/342259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colonna M. TREMs in the immune system and beyond. Nat Rev Immunol. 2003;3(6):445–453. doi: 10.1038/nri1106. [DOI] [PubMed] [Google Scholar]
- 22.Bouchon A, Dietrich J, Colonna M. Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol. 2000;164(10):4991–4995. doi: 10.4049/jimmunol.164.10.4991. [DOI] [PubMed] [Google Scholar]
- 23.Paloneva J, Autti T, Raininko R, et al. CNS manifestations of Nasu-Hakola disease: a frontal dementia with bone cysts. Neurology. 2001;56(11):1552–1558. doi: 10.1212/wnl.56.11.1552. [DOI] [PubMed] [Google Scholar]
- 24.Bianchin MM, Capella HM, Chaves DL, et al. Nasu-Hakola disease (polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy—PLOSL): a dementia associated with bone cystic lesions. from clinical to genetic and molecular aspects. Cell Mol Neurobiol. 2004;24(1):1–24. doi: 10.1023/B:CEMN.0000012721.08168.ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montalbetti L, Ratti MT, Greco B, Aprile C, Moglia A, Soragna D. Neuropsychological tests and functional nuclear neuroimaging provide evidence of subclinical impairment in Nasu-Hakola disease heterozygotes. Funct Neurol. 2005;20(2):71–75. [PubMed] [Google Scholar]
- 26.Chouery E, Delague V, Bergougnoux A, Koussa S, Serre JL, Mégarbané A. Mutations in TREM2 lead to pure early-onset dementia without bone cysts. Hum Mutat. 2008;29(9):E194–E204. doi: 10.1002/humu.20836. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.