Abstract
The O-linked β-N-acetylglucosamine (O-GlcNAc) post-translational modification was first discovered thirty years ago and is highly concentrated in the nuclear pore. In the years since the discovery of this single sugar modification, substantial progress has been made in understanding the biochemistry of O-GlcNAc and its regulation. Nonetheless, O-GlcNAc modification of proteins continues to be overlooked, due in large part to the lack of reliable methods available for its detection. Recently, a new crop of immunological and chemical detection reagents has changed the research landscape. Using these tools, approximately 1000 O-GlcNAc-modified proteins have been identified. While other forms of glycosylation are typically associated with extracellular proteins, O-GlcNAc is abundant on nuclear and cytoplasmic proteins. In particular, phenylalanine-glycine (FG) nucleoporins (NUPs) are heavily O-GlcNAc-modified. Recent experiments are beginning to provide insight into the functional implications of O-GlcNAc modification on certain proteins, but its role in the nuclear pore has remained enigmatic. However, tantalizing new results suggest that O-GlcNAc may play roles in regulating nucleocytoplasmic transport.
Keywords: O-GlcNAc, nucleoporin, FG repeat, stress, hexosamine, nucleocytoplasmic transport, photcrosslinking, diazirine, Huisgen cycloaddition, unstructured proteins
Introduction: the O-GlcNAc post-translational modification
The O-GlcNAc modification consists of a single sugar, N-acetylglucosamine (GlcNAc), found attached to the side chain oxygen of serine or threonine residues on nuclear and cytoplasmic proteins [1]. Like phosphorylation, O-GlcNAc-ylation is reversible. However, in contrast to the large number of kinases and phosphatases that add and remove phosphate, mammalian genomes encode a single intracellular O-GlcNAc transferase (OGT) [2] and a single O-GlcNAc hydrolase (OGA) [3]. OGT produces the O-GlcNAc modification by transferring GlcNAc from UDP-GlcNAc to a protein substrate, while OGA removes O-GlcNAc by hydrolytically cleaving the glycosidic bond [4]. OGT genes have been identified in the genomes of all multicellular organisms, including filamentous fungi, worms, insects, plants, and mammals [5]. O-GlcNAc has not been detected in the yeasts Saccharomyces cerevisiae or Schizosaccharomyces pombe, but some prokaryotes do harbor enzymes that can produce a chemically identical modification [6, 7]. In addition to the intracellular form of O-GlcNAc discussed in this review, extracellular O-GlcNAc has been recently discovered [8]; a second OGT (EOGT) catalyzes its production within the secretory pathway of mammalian cells [9].
The GlcNAc donor, UDP-GlcNAc, is an abundant central metabolite that integrates signals from multiple metabolic pathways – glucose metabolism, amino acid metabolism, fatty acid metabolism, and nucleotide metabolism (Figure 1) [10-12]. Typically, cells take up glucose and produce UDP-GlcNAc through the hexosamine biosynthetic pathway (HBP). Upon entering the HBP, glucose is first phosphorylated to glucose-6-phosphate, followed by isomerization to fructose-6-phosphate. Next, glutamine fructose-6-phosphate amidotransferase (GFAT) converts fructose-6-phosphate to glucosamine-6-phosphate by incorporating a nitrogen atom from the amino acid glutamine. Acetyl-CoA then serves as a donor for acetylation of the amine group to yield GlcNAc-6-phosphate, thereby reporting on the state of pyruvate and fatty acid metabolism. N-acetylglucosamine-phosphate mutase (AGM1) isomerizes GlcNAc-6-phosphate to GlcNAc-1-phosphate. Finally, UDP-N-acetylglucosamine pyrophosphorylase (UAP1) couples GlcNAc-1-phosphate with UTP, producing UDP-GlcNAc. This step incorporates information about the nucleotide and high-energy phosphate pool. Because of the overall integration of metabolic signals, the HBP is often described as a “nutrient-sensing” pathway. The extent of O-GlcNAc-ylation depends on UDP-GlcNAc levels, making this post-translational modification responsive to nutritional cues [13, 14].
Figure 1. O-GlcNAc modification integrates metabolic information.
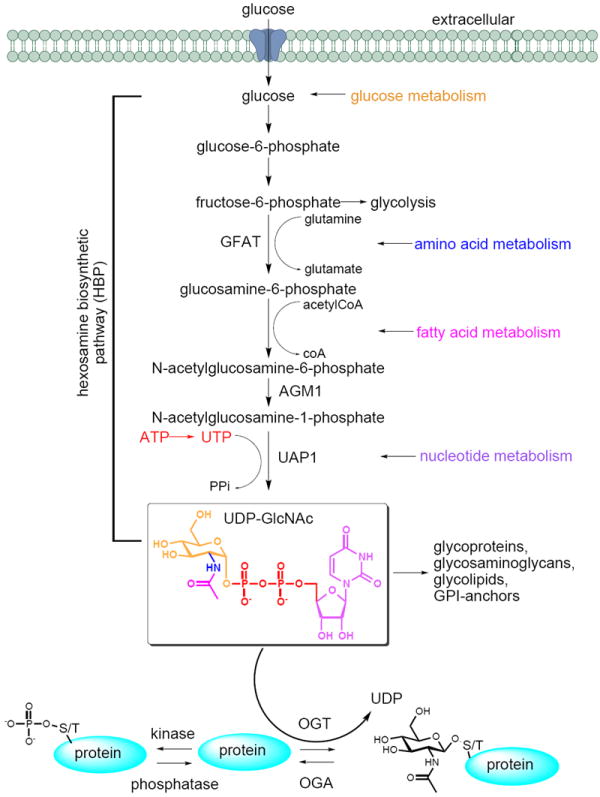
The nucleotide-sugar UDP-GlcNAc is produced through the hexosamine biosynthetic pathway (HBP). This pathway integrates metabolic signals from carbohydrate (glucose), amino acid (glutamine), fatty acid (acetyl-CoA), and nucleotide (UTP) metabolism. Because ATP is required for UTP biosynthesis, the HBP also reports on energy charge. UDP-GlcNAc is required for various forms of glycosylation, including the production of the O-GlcNAc modification. O-GlcNAc is produced by the enzyme OGT and removed from proteins by the enzyme OGA. Sites of O-GlcNAc modification are often competitively phosphorylated by kinases.
O-GlcNAc-ylation is found on a diverse range of nuclear and cytoplasmic proteins [1]. In many cases O-GlcNAc-ylation and phosphorylation compete to occupy the same Ser/Thr sites [15-18]. However, the number of known O-GlcNAc-ylated proteins is relatively small compared with known phosphoproteins. To date, approximately 1300 O-GlcNAc-modified proteins have been identified from all organisms and sites of O-GlcNAc-ylation have been mapped in only a small subset of known O-GlcNAc-ylated proteins. But existing data likely underrepresent the true extent of O-GlcNAc-ylation [19], since reliable methods to detect this modification have only recently become available. There is no consensus sequence that can be used to predict sites of O-GlcNAc modification, although new computation tools are attempting to fill this gap [20].
Within existing lists of O-GlcNAc-ylated proteins, nucleoporins (NUPs) are highly represented (Figure 2). Indeed, NUPs are the most heavily O-GlcNAc-modified proteins and were some of the first O-GlcNAc-modified proteins to be identified [21-24]. Nucleoporins identified as O-GlcNAc modified include NUP62 [21-25], NUP153 [25, 26], NUP214/CAN [25, 26], NUP358 [26], POM121 [27], NUP98 [25], NUP155 [25], p58 [23], p54 [25], p45 [23], NUP93 [25], NUP210 [25], NUP205 [25], NUP160 [25], NUP107 [25], NUP188 [25], NUP88 [25], NUP85 [25], and NUP35 [25].
Figure 2. Many nuclear pore proteins are modified with O-GlcNAc.
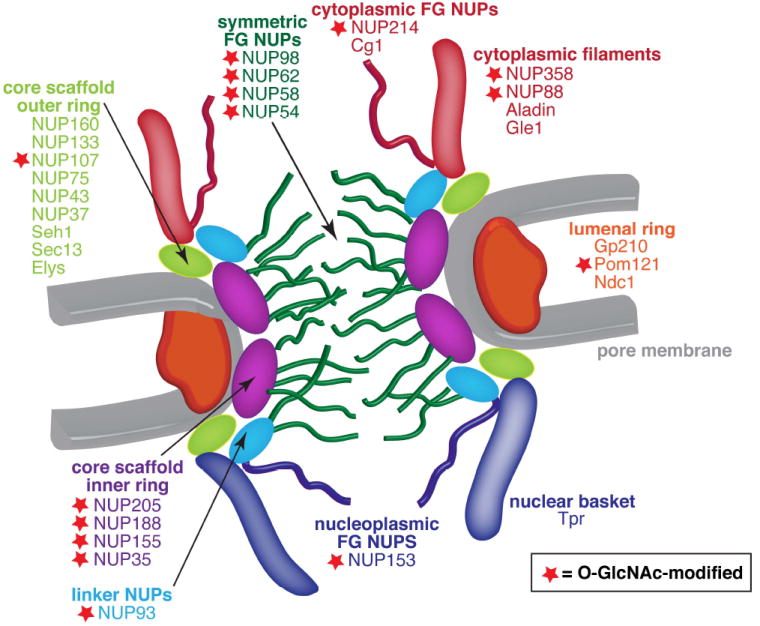
Red stars in this diagram of the nuclear pore mark nuclear pore proteins demonstrated to be O-GlcNAc-ylated. The highest densities of O-GlcNAc are found on unstructured FG repeat regions, which project into the center of the nuclear pore and extend on both the nuclear and cytoplasmic sides of the pore. O-GlcNAc-modified FG repeat regions are also found in NUP358, the major component of cytoplasmic filaments. In addition to modifying NUPs rich in FG repeats, O-GlcNAc has also been detected on inner ring, outer ring, and linker NUPs, and even on the transmembrane NUP POM121.
O-GlcNAc modification of nucleoporins was first reported in 1987, but early experiments gave conflicting views of the role of glycosylation in nuclear pore function. Since that time, the O-GlcNAc field and the field of nucleocytoplasmic transport have developed in parallel. This review reports on the nexus between these fields. First, we outline the availability of new methods to label and isolate O-GlcNAc-modified proteins, as well as the implementation of mass spectrometry-based proteomics approaches to identify O-GlcNAc-modified proteins and sites of modification. By revealing specific sites of O-GlcNAc-ylation and how site occupancy changes in response to stimuli, these methods promise to revolutionize studies of O-GlcNAc function. Turning to the nuclear pore, we describe early experiments to understand the function of nuclear pore glycosylation and detail the ubiquity of this modification among multicellular eukaryotes. We survey conditions that alter O-GlcNAc-ylation, noting the correlation between changes in O-GlcNAc levels and alterations in nucleocytoplasmic transport. Next, we summarize data suggesting that O-GlcNAc modification can affect protein-protein interactions in the nuclear pore and also describe a new photocrosslinking method that can be used to discover O-GlcNAc-mediated interactions. Then, we describe how results from an in vitro transport assay suggest a role for O-GlcNAc in modulating pore selectivity. Finally, we speculate on the potential for O-GlcNAc to affect the behavior of nucleoporins functioning in roles outside of the nuclear pore. Based on technical developments in the both the O-GlcNAc and nucleoporin fields, we expect to see the emergence of new insights into the function of nuclear pore glycosylation in coming years.
Methods for detecting O-GlcNAc
O-GlcNAc-ylation is widespread and found on proteins of diverse function, but in most cases the functional impact of this modification is unknown. Progress in this area has been hindered by the lack of reagents and methods to specifically detect and isolate O-GlcNAc-ylated molecules. However, dramatic improvements have occurred in the past decade and a variety of O-GlcNAc detection and purification reagents are now available (Table 1) [28]. These methods fall into a several classes: (1) enzymatic labeling, in which the O-GlcNAc residue is enzymatically modified, (2) affinity reagents, including lectins and antibodies, that recognize O-GlcNAc, and (3) metabolic labeling, in which a bioorthogonal functional group is metabolically incorporated into the O-GlcNAc modification. Use of these reagents for purification of O-GlcNAc-modified proteins further enables mass spectrometry-based proteomics methods that can be used to discover novel O-GlcNAc-ylated proteins and identify sites of modification.
Table 1.
Methods to detect O-GlcNAc modification of proteins
| Method | Key reagents | Can be used with animal and/or human samples? | Applied in combination with mass spectrometry |
|---|---|---|---|
| Lectin binding | WGA [22, 27, 37] | Yes | [40, 52] |
|
| |||
| O-GlcNAc antibodies | RL2 [24], CTD110.6 [41] | Yes | [43, 119, 120] |
| 9D1E4(10), 18B10.C7(3), and 1F5.D6(14) [43] | |||
|
| |||
| Metabolic labeling | Ac4GlcNAz [44], Ac4GalNAz [45], Ac4GlcNAlk [46] | No | [44-46, 121, 122] |
|
| |||
| Enzymatic tagging | UDP-3H-Gal [29, 39, 123], GalT1 (Y289L) [31], UDP-ketoGal [32, 124], aminooxy-PEG [35], UDP-GalNAz [33, 55, 56] | Yes | [33, 55, 56] |
Enzymatic labeling of O-GlcNAc
The O-GlcNAc modification was first discovered using bovine milk galactosyltransferase GalT1 to enzymatically transfer 3H-galactose from UDP-3H-galactose to O-GlcNAc-modified proteins [29]. This radiolabeling method also enables identification of specific O-GlcNAc-ylated proteins [30]. While the radioisotope provides a useful means for labeling O-GlcNAc-modified proteins, purification of labeled proteins is less straightforward. To address this challenge, GalT1 was engineered to allow transfer of galactose analogs bearing a different chemical handles.
The mutant GalT1(Y289L) [31] can accept either UDP-ketogalactose (UDP-ketoGal) [32] or UDP-N-azidoacetylgalactosamine (UDP-GalNAz) [33] to enzymatically tag O-GlcNAc residues in cell and tissue lysates. O-GlcNAc-ylated proteins modified with ketoGal can be further functionalized with aminooxy reagents. For example, selective biotinylation of O-GlcNAc-modified proteins facilitates proteomics applications [34], while functionalization with polyethylene glycol (PEG) can reveal the stoichiometry of O-GlcNAc-ylation [35]. Similarly, O-GlcNAc-ylated proteins modified with GalNAz can be labeled with alkyne reagents [33] through an azide-alkyne Huisgen cycloaddition reaction (often termed “click chemistry” [36]). Various alkyne reagents are commercially available.
O-GlcNAc binding reagents
Much early work [22, 37-39] in the O-GlcNAc field relied on a carbohydrate-binding lectin, wheat germ agglutinin (WGA), as a detection reagent. WGA binds GlcNAc and has been used to identify O-GlcNAc-modified proteins and to enrich them for proteomics analysis [40]. WGA shows strong binding to some O-GlcNAc-ylated proteins by binding multiple GlcNAc moieties at the same time, but its utility is limited by its low affinity for single GlcNAc residues. Another limitation of WGA arises from its affinity for another sugar, sialic acid, which results in poor specificity.
Today, WGA is less commonly used for O-GlcNAc detection, because antibodies provide higher affinity and better specificity. Two anti-O-GlcNAc antibodies, RL2 and CTD110.6, are commercially available and are commonly used to detect O-GlcNAc modification by immunoblot. RL2 is an IgG antibody that was raised specifically against O-GlcNAc modified components of the nuclear pore [24]. While RL2 binding depends on the presence of O-GlcNAc, it also requires protein determinants. Thus, RL2 preferentially recognizes O-GlcNAc-modified NUPs. It can also detect some other O-GlcNAc modified proteins, although typically less strongly. CTD110.6 is an IgM antibody that was produced using the O-GlcNAc-ylated carboxy-terminal domain (CTD) of RNA polymerase II as the antigen [41]. CTD110.6 has a broader reactivity than RL2, recognizing a variety of O-GlcNAc-modified proteins. A downside to the broadened substrate scope is that CTD110.6 can also bind certain truncated N-linked glycoproteins that are produced under conditions of glucose deprivation [42]. In addition, because CTD110.6 is an IgM antibody, it cannot easily be used in immunoprecipitation experiments.
To overcome the shortcomings associated with existing detection reagents, Teo et al. used synthetic chemistry to prepare an O-GlcNAc antigen that enabled them to obtain a panel of monoclonal anti-O-GlcNAc IgG antibodies [43]. The high affinities of these antibodies allowed the researchers to isolate O-GlcNAc-ylated proteins. These samples were analyzed by mass spectrometry to identify more than 200 mammalian O-GlcNAc modified proteins. One can also imagine using these antibodies in combination with MS/MS to identify both the O-GlcNAc modified proteins and sites of modification. Three of these new O-GlcNAc antibodies (9D1E4(10), 18B10.C7(3), and 1F5.D6(14)) are now commercially available.
Metabolic labeling of O-GlcNAc
As a complement to chemoenzymatic labeling and affinity reagents, Bertozzi and co-workers utilized the hexosamine salvage pathway to metabolically deliver azides to O-GlcNAc-modified proteins [44, 45]. They cultured cells with a cell-permeable azide-bearing monosaccharide, Ac4GlcNAz [44]. After deacetylation by intracellular enzymes, GlcNAz can enter the HBP and be activated to UDP-GlcNAz. GlcNAz is then transferred to serine and threonine residues of substrate proteins by OGT. The resulting azide-modified proteins can be further functionalized with biotin or fluorophores using azide-alkyne cycloaddition chemistry. However, the labeling achieved by this approach is limited by the inefficient production of UDP-GlcNAz. Thus, the group developed a more faithful and robust metabolic reporter for O-GlcNAc using a cell-permeable, azide-modified analog of N-acetylgalactosamine (GalNAc) [45]. In this approach, cells are cultured with Ac4GalNAz, which is activated to UDP-GalNAz and then epimerized to UDP-GlcNAz. Again, GlcNAz is transferred to substrate proteins by OGT. This approach provides a more robust way to metabolically label O-GlcNAc modified proteins with the azide functional group. Using the azide handle for purification, the group was able to perform proteomics analysis, identifying about 200 potential O-GlcNAc-modified proteins. Most recently Pratt and co-workers reported another metabolic labeling strategy, relying on a cell-permeable alkyne analog of GlcNAc (Ac4GlcNAlk) [46]. An important advantage of the alkyne analog is that it is less readily epimerized, providing selective labeling of GlcNAc-containing molecules. O-GlcNAlk-labeled proteins can be further functionalized by cycloaddition reaction with azide-containing compounds, enabling fluorescence microscopy and proteomics applications.
Proteomics methods for O-GlcNAc discovery
Since the discovery of O-GlcNAc, efforts have been underway to employ mass spectrometry to identify O-GlcNAc-modified proteins and map sites of modification on O-GlcNAc-modified peptides. But standard mass spectrometric methods have yielded limited information. Two challenges stand in the way: (1) methods for enriching for O-GlcNAc-modified peptides are inadequate and (2) commonly used mass spectrometry methods are not optimal for O-GlcNAc detection (although typical mass spectrometry analysis has revealed some sites of modification [19, 47, 48]). Efficient enrichment is essential because O-GlcNAc modification is typically substoichiometric and unmodified peptides can suppress the signal of O-GlcNAc peptides. Alternative mass spectrometry methods are required because of the notorious lability of the O-GlcNAc glycosidic bond. In particular, electron transfer dissociation (ETD) fragmentation has been an enabling technology for mapping O-GlcNAc sites [49-53].
While affinity purification using WGA [40, 52] or an anti-O-GlcNAc antibody [43] can be used to obtain material for mass spectrometry analysis, achieving adequate enrichment is difficult. As an alternative, methods that rely on chemical derivatization to enrich O-GlcNAc-modified proteins or peptides have been developed. Early progress came in 2002, when Hart and co-workers used beta-elimination followed by Michael addition with dithiothreitol (BEMAD) to enrich for O-GlcNAc peptides, followed by affinity chromatography and LC-MS/MS to identify sites of O-GlcNAc modification on proteins including NUP155 [54]. More recently a chemical/enzymatic photochemical cleavage (CEPC) method was developed to enrich for O-GlcNAc peptides [55, 56]. This method combines enzymatic tagging, to add the azide functional group to the O-GlcNAc peptides [33], with chemoselective ligation, to add a photocleavable biotin-alkyne tag (PC-biotin-alkyne). The resulting biotinylated O-GlcNAc peptides can be enriched by avidin chromatography; subsequent photocleavage releases the peptides for mass spectrometry analysis. The portion of the tag that remains attached to the O-GlcNAc peptide bears a charged moiety, which facilitates detection of O-GlcNAc-modified peptides. Implementation of this purification method in combination with mass spectrometry analysis that relies on electron transfer dissociation (ETD) fragmentation enabled identification of 274 O-GlcNAc-ylated proteins and 458 O-GlcNAc sites, many of which were previously unknown [56]. The emergence of routine methods to map sites of O-GlcNAc modification promises to revolutionize functional studies of the role of this modification.
Discovery of O-GlcNAc in the nuclear pore
One of the first places that O-GlcNAc was detected was on nuclear pore proteins [21-24]. Indeed, nucleoporins are the most heavily O-GlcNAc-modified proteins; for example, up to ten O-GlcNAc residues have been detected on NUP62 [23, 35]. Many of the O-GlcNAc-ylated NUPs are so-called FG NUPs, which contain the extensive repeats of phenylalanine-glycine (FG); the FG regions are common sites of O-GlcNAc-ylation. In fact, all known metazoan FG NUPs are O-GlcNAc-modified in the FG regions. These same FG regions interact directly with the karyopherins that shuttle through the nuclear pore complex (NPC) and play important roles in nuclear transport [57, 58].
The abundance of O-GlcNAc in the nuclear pore suggests that it plays a functional role, but exactly what that function is has remained unclear. In 1987, Finlay et al. first addressed this topic [37]. Using an in vitro nuclear transport assay, they discovered that the GlcNAc-binding lectin WGA effectively blocks the nuclear transport, while the addition of free competitor GlcNAc restores activity. Furthermore, experiments performed with radiolabeled WGA provided evidence for a direct interaction between WGA and nuclear pore proteins. Akey et al. also observed binding of WGA to NUPs and suggested that interactions between cargo and O-GlcNAc-modified nucleoporins might comprise an early step in nuclear transport [59]. In later work, Finlay et al. observed that nuclear pores depleted of WGA-binding proteins lost the ability to perform nuclear import, but activity could be restored by addition of O-GlcNAc-modified nuclear pore glycoproteins from Xenopus or rat [60]. Miller and Hanover performed a similar depletion experiment, but replaced the WGA-binding proteins with rat nuclear glycoproteins whose terminal GlcNAc residues had been enzymatically galactosylated [61]. However, they observed that galactosylation of O-GlcNAc does not affect the nuclear transport. Taken together, these early studies indicated that the O-GlcNAc-modified nucleoporins played critical roles in nuclear transport, but argued against a role for direct binding of the O-GlcNAc residues. Notably, none of these studies examined mammalian nuclear pores that contained the full complement of nucleoporins, yet lacked O-GlcNAc.
Glycosylation of the nuclear pore is ubiquitous in multicellular organisms
To directly address the functional role of O-GlcNAc in nuclear transport, it would be advantageous to create cell lines that lack this modification. However, obtaining viable mammalian cells lacking O-GlcNAc remains a challenge. The Marth group attempted to delete OGT in mice using a genetic approach, but no viable OGT-null offspring were obtained [62, 63]. Deletion of OGT is also lethal in cell culture: cells undergo approximately two rounds of division and then enter senescence. However, an inducible OGT-knockout in fibroblast culture has been reported [64], allowing study of O-GlcNAc-deficient cells during a limited time window. The inability to generate OGT-null animals or cells suggests that O-GlcNAc plays an essential role in mammalian cell function. But, surprisingly, Hanover and co-workers were able to delete OGT from C. elegans with no obvious developmental defect [65], offering an exciting opportunity to assess the functional role of O-GlcNAc-ylation. Analysis of OGT-deficient nematodes revealed that nuclear pore complexes lacking O-GlcNAc still mediate nuclear transport of transcription factors. OGT-null worms do exhibit other phenotypes, including impaired stress tolerance, decreased lipid storage, increase in carbohydrate stores, and suppressed dauer formation.
Glycosylation of the nuclear pore also occurs in plants [66]. In Arabidopsis, as well as other plants, two OGT genes are present: SPINDLY (SPY) and SECRET AGENT (SEC). SEC/SPY double mutant embryos don’t survive to seed development, but the individual mutants are viable [67-70]. Analysis of mutant plants reveals roles for SPY in gibberellin (GA) responses [68-70], cytokine signaling [71], and circadian rhythms [72]. SEC-null plants have more subtle phenotypes that include slower leaf production and shorter shoot at flowering [67]. Intriguingly, Taoka et al. found that the O-GlcNAc modification is critical to the interaction between NCAPs (non-cell-autonomous proteins) and Nt-NCAPP1 (Nicotiana tabacum non-cell-autonomous pathway protein 1) [73]. Specifically, O-GlcNAc-ylation of Ser-66 on NCAP Cm-PP16-1 is required for effective interaction with Nt-NCAPP1. Mutation of this residue blocks their interaction and affects plasmodesmatal trafficking, a cell-to-cell trafficking process in plants that exhibits multiple similarities to transport through the nuclear pore [74].
O-GlcNAc levels are regulated by stress and by cell cycle
As described above, yeast lack O-GlcNAc, yet have a functional NPC, suggesting that O-GlcNAc is not essential for nuclear transport per se. Nonetheless, nuclear pore O-GlcNAc might play roles in the transport of specific factors or in transport that occurs under specific conditions. For example, while canonical nuclear import is inhibited by stress, some nuclear import does occur under stressful conditions and may be crucial to the stress response. Indeed, a recent report showed that a novel transport protein Hikeshi is essential for resolving thermal stress, by facilitating accumulation of a heat shock protein (HSP) in the nucleus [75]. Hikeshi interacts directly with FG-repeat NUPs and is responsible for selective nuclear import of heat shock protein 70 (HSP70). These results raise the possibility that stress-induced changes in O-GlcNAc modification of FG NUPs could facilitate certain nuclear transport pathways, while interfering with others. Consistent with this idea, diverse stressful stimuli have been reported to affect O-GlcNAc levels; these include hypoxia [76-78], cell cycle arrest [79, 80], oxidative or reductive conditions [38, 81, 82], UV irradiation [38, 83], ethanolic stress [38, 77], osmotic stress [38, 84], ER stress [38, 85], trauma hemorrhage [84, 86-89], ischemia reperfusion injury [76-78, 89-95], and heat shock [38, 96] (Figure 3). At the same time, modulation of O-GlcNAc levels affects stress tolerance. Overall, decreasing O-GlcNAc levels results in cells that are less tolerant [38, 77, 78], whereas increasing the levels results in cells that are more tolerant [38, 76, 90, 97].
Figure 3. O-GlcNAc levels are regulated by a variety of stimuli.
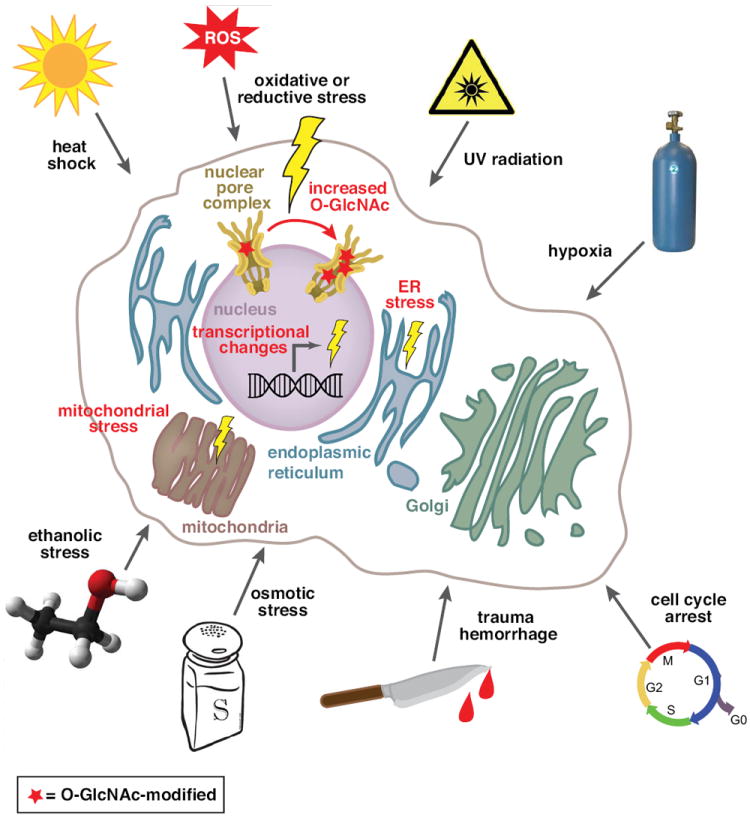
A variety of stressful stimuli result in increased O-GlcNAc levels, including increased O-GlcNAc-ylation of NUPs. Stress is also associated with changes in nucleocytoplasmic transport, induced transcription of stress-induced genes, and alterations in mitochondrial and endoplasmic reticulum function. Mechanistic roles for O-GlcNAc in mediating stress-induced changes in cell function have been proposed, but remain largely unexplored.
O-GlcNAc regulation is also essential for cell cycle progression. Galactosylation of O-GlcNAc-modified proteins results in severe defects in M- to S-phase transition [98]. Perturbing O-GlcNAc levels by altering expression of OGT or OGA also causes cell cycle defects [79]. Similarly, pharmacological alteration of O-GlcNAc levels affects the ability of cells re-enter the cell cycle and progress to the next stage [79]. Thus, cell cycle progression depends on O-GlcNAc modification of proteins; however, more work is required to uncover the identities of the key O-GlcNAc-regulated proteins and determine how O-GlcNAc affects their function. One possibility is that changes in O-GlcNAc-ylation of NUPs play a regulatory role in the alterations in NPC structure and function that occur throughout the cell cycle [99, 100].
Does O-GlcNAc regulate protein-protein interactions in the nuclear pore?
Stress and cell cycle progression are stimuli that alter O-GlcNAc at a global level; these changes in O-GlcNAc levels can be observed specifically on FG NUPs. Further, the same stimuli that alter the degree of O-GlcNAc modification of nucleoporins can also result in changes in nuclear transport. For example, oxidative stress increases O-GlcNAc modification of multiple FG NUPs [38, 81] and also inhibits both classical nuclear import and CRM1-mediated nuclear export [81]. Oxidant treatment also modulates binding interactions between CRM1 and Ran, between CRM1 and NUPs, and among FG NUPs. These data are consistent with a model in which O-GlcNAc modification in response to oxidative stress affects NUP stability and/or interactions with other proteins [101]. Thus, glycosylation-induced alterations in FG NUP behavior could provide a mechanism for stress-induced changes in nuclear transport.
A report published earlier this year examined the role of O-GlcNAc in NUP-NUP interactions. Mizuguchi-Hata et al. decreased O-GlcNAc levels by silencing OGT and observed enhanced association between NUP62 and NUP88 relative to control cells [102]. Since both NUP62 and NUP88 are known to be O-GlcNAc-ylated, this result suggests that O-GlcNAc modification of one or both of the proteins enhances NUP62-NUP88 association and could represent the discovery of an O-GlcNAc-mediated binding interaction.
Photocrosslinking offers an opportunity to discover O-GlcNAc-mediated interactions
Although one possible role for O-GlcNAc modification is to mediate novel protein-protein interactions, so far there is little direct evidence for O-GlcNAc-dependent complex formation. To facilitate the discovery of O-GlcNAc-dependent interactions, we reported a method to photochemically capture molecules that are proximal to O-GlcNAc modification sites [103]. This method relies on metabolic incorporation of the diazirine photocrosslinking group onto O-GlcNAc residues (Figure 4). Unlike traditional crosslinking methods (such as formaldehyde), metabolic incorporation of a photoactivatable crosslinker enables specific crosslinking of interactions proximal to the crosslinker, in this case an O-GlcNAc residue [104].
Figure 4. Photochemical crosslinking to discover O-GlcNAc-mediated interactions.
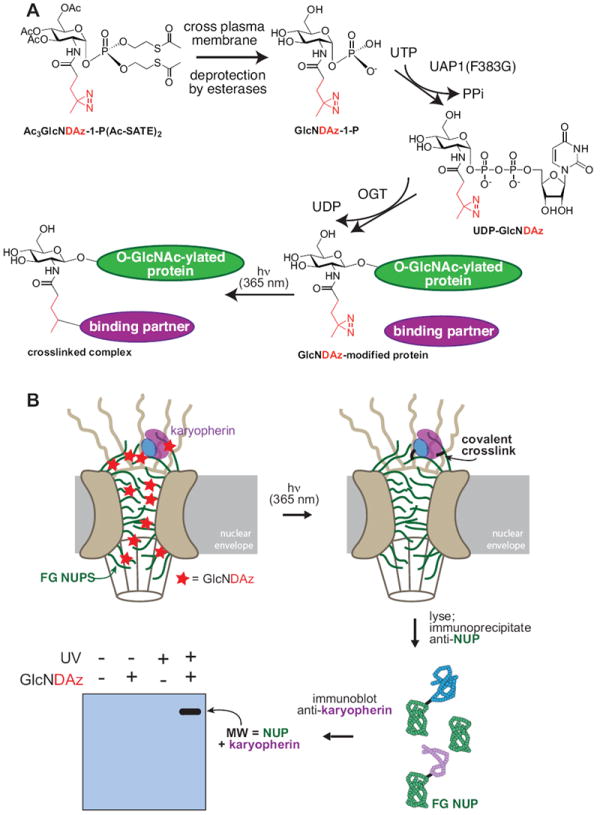
(A) Cells can be induced to produce a diazirine-modified form of O-GlcNAc by stably expressing a mutant form of the UDP-GlcNAc pyrophosphorylase (UAP1(F383G)) and adding a cell-permeable, diazirine-modified analog of GlcNAc-1-P (Ac3GlcNDAz-1-P(AcSATE)2) to the culture medium. The diazirine functional group is activated by UV irradiation, resulting in covalent crosslinks between O-GlcNAc-modified proteins and neighboring molecules. (B) Crosslinked complexes were isolated using an antibody specific for FG NUPs. Subsequent LC-MS/MS analysis of the crosslinked complexes revealed evidence for direct interactions between O-GlcNAc-modified FG NUPs and karyopherins, specifically that TNPO1 interacts with both NUP153 and NUP358.
In-cell production of diazirine-modified O-GlcNAc (O-GlcNDAz) is achieved by an approach that relies on chemical synthesis and enzyme engineering (Figure 4A) [103]. A cell-permeable, diazirine-modified analog of GlcNAc-1-phosphate (Ac3GlcNDAz-1-P(Ac-SATE)2) is prepared synthetically. Ac3GlcNDAz-1-P(Ac-SATE)2 enters cells, where it is deprotected to GlcNDAz-1-phosphate. A mutant form of the UDP-GlcNAc pyrophosphorylase (UAP1) is required to activate GlcNDAz-1-phosphate to UDP-GlcNDAz. OGT then transfers GlcNDAz from UDP-GlcNDAz to substrate proteins, forming the O-GlcNDAz modification. Intact, O-GlcNDAz-containing cells are subjected to UV irradiation, which activates the diazirine crosslinker, resulting in covalent bonds between O-GlcNAc-modified proteins and nearby molecules. After lysis, the covalently crosslinked complexes can be characterized by immunoprecipitation and immunoblot analysis, or by mass spectrometry. Our initial report of this technology showed that it could be used to covalently capture interactions between FG NUPs and karyopherins (Figure 4B). Specifically, we observed evidence for O-GlcNAc-mediated crosslinking between transportin-1 (TNPO1; karyopherin β2) and both NUP358 (RanBP2) and NUP153. These interactions can be mapped onto the emerging models of the NPC [58], revealing that TNPO1 interacts directly with FG repeats on both the nuclear and cytoplasmic faces of the pore. Our crosslinking results suggest that O-GlcNAc residues are intimately associated with essential recognition events in nuclear transport. Further work is required to assess whether and how O-GlcNAc modification affects the affinity of these interactions.
Impact of O-GlcNAc on FG NUP function
Nucleocytoplasmic transport depends on direct interactions between FG NUPs and karyopherins The O-GlcNAc-modified regions of FG NUPs project into the center of the nuclear pore and form “bristling extensions” that protrude from both faces of the NPC [105]. The FG regions are natively disordered, forming a meshwork that facilitates karyopherin transport [58, 106]. Several models have been proposed to explain the mechanism by which the FG repeat regions regulate selective transport – allowing passage of appropriate cargo while excluding other similarly-sized macromolecules – but the molecular details remain poorly defined and actively debated [58]. In one experimental approach to investigate the mechanism of selective transport, the Görlich group devised an in vitro model system that is amenable to detailed manipulation at the molecular level. Using purified FG regions of NUPs, these researchers produced elastic hydrogels that recapitulate transport properties of intact NPCs [107]. FG NUP hydrogels are transparent, enabling visualization of the transit of fluorescently labeled transport molecules through the hydrogel. O-GlcNAc modification of a hydrogel made from Xenopus NUP98 made the gel more dynamic and facilitated entry of large nuclear transport complexes, while efficiently excluding “inert” proteins [108]. Overall, the O-GlcNAc-modified NUP98 hydrogel was more selective than any non-glycosylated NUP hydrogel. Examination of hydrogel structure by solid-state NMR spectroscopy showed that non-glycosylated gels are rigid, while an O-GlcNAc-modified hydrogel lacks amyloid-like β-structures. Thus, O-GlcNAc modification makes FG NUP hydrogels more elastic and more dynamic. This change in structure translates into altered permeability properties that allow selective transit through the hydrogel. Further work is required to determine whether O-GlcNAc exerts similar effects on the activity of FG NUPs functioning in a native NPC.
Beyond the nuclear pore: does O-GlcNAc affect transcriptional roles of nucleoporins?
Nucleoporins have roles outside of nuclear transport [109]. An intimate connection between nuclear transport and transcription exists [110]. In 1985, Günter Blobel formulated the “gene gating” hypothesis, which proposes that the three-dimensional structure of the genome is organized to facilitate direct interactions between NPCs and transcribable chromatin [111]. Such interactions were hypothesized to facilitate expedient cytoplasmic export of new transcripts. More recently, genome-wide chromatin immunoprecipitation (ChIP) studies have shown that NUPs are preferentially associated with active chromatin [112]. In addition to facilitating mRNA export, evidence suggests that FG-repeat NUPs play an active role in regulating transcriptional programs [113]. In 2010, two groups reported that certain FG-repeat NUPs (including NUP98 and NUP62) are associated with Drosophila chromatin and directly influence transcriptional outcomes [114, 115]. These mobile NUPs are dynamically recruited to active chromatin [116] by as-yet-unknown mechanisms and regulate genes involved in development and the cell cycle. Intriguingly, oncogenic fusion proteins that contain FG NUP regions can also functional as transcriptional regulators [117]. However, no studies have investigated whether the O-GlcNAc modification plays a role in either NUP recruitment to chromatin or transcriptional activation. NUPs share a key similarity with many transcription factors: modification with O-GlcNAc. Indeed, the O-GlcNAc-binding lectin WGA was historically used to purify transcription factors [118]. Insight into any role that O-GlcNAc may play in the transcriptional functions of NUPs awaits investigation.
Summary and Outlook
O-GlcNAc modification of the nuclear pore was first observed more than 25 years ago but its function remains a mystery. Slowly but surely, the technical barriers that have limited study of O-GlcNAc-ylation of nucleoporins are being surmounted. Now, new chemical biology and mass spectrometry methods offer ways to identify specific sites of O-GlcNAc modification. Once these sites are mapped, it will be possible to embark on mechanistic studies to test the functional roles of individual O-GlcNAc sites. An in vitro model of nuclear transport that relies on nucleoporin hydrogels provides a readily manipulable framework to investigate functional impacts of O-GlcNAc-ylation. Indeed, hydrogel analysis indicates that O-GlcNAc modification can significantly modulate permeability to karyopherin-cargo complexes, but it is not yet clear if the same effects will be observed in NPCs functioning in a cell. Several suggestive pieces of data indicate that O-GlcNAc may promote certain protein-protein interactions and a new, O-GlcNAc-specific photocrosslinking method provides a strategy to identify such glycan-dependent binding events. While OGT silencing in mammalian cells remains a challenge, the availability of OGT-null C. elegans offers an important model system for studying the role of O-GlcNAc in a living, multi-cellular organism. Experiments performed in this model system indicate that nucleocytoplasmic transport can occur in the absence of O-GlcNAc, but leave open the possibility that O-GlcNAc might modulate transport under certain conditions. Indeed, the extent of O-GlcNAc modification is highly responsive to various stressors, stimuli that also alter nucleocytoplasmic transport. In addition, emerging data indicate that nucleoporins moonlight, playing roles in processes including transcription and mitosis. Even if O-GlcNAc-ylation of nucleoporins is dispensable for nucleocytoplasmic transport, this modification might still play important roles in the secondary functions of nucleoporins. We predict that future studies will integrate diverse technological developments to define the roles of O-GlcNAc in modulating the multiple functions of nucleoporins.
Acknowledgments
We thank Yuh Min Chook, Beatriz Fontoura, Akiko Fujita, Andrea Rodriguez, and Amberlyn Wands for comments on the manuscript and gratefully acknowledge support from the NIH (R01GM090271) and the Welch Foundation (I-1686).
References
- 1.Hart GW, Housley MP, Slawson C. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 2.Haltiwanger RS, Holt GD, Hart GW. Enzymatic addition of O-GlcNAc to nuclear and cytoplasmic proteins. Identification of a uridine diphospho-N-acetylglucosamine:peptide beta-N-acetylglucosaminyltransferase. J Biol Chem. 1990;265:2563–2568. [PubMed] [Google Scholar]
- 3.Dong DL, Hart GW. Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. J Biol Chem. 1994;269:19321–19330. [PubMed] [Google Scholar]
- 4.Vocadlo DJ. O-GlcNAc processing enzymes: catalytic mechanisms, substrate specificity, and enzyme regulation. Curr Opin Chem Biol. 2012;16:488–497. doi: 10.1016/j.cbpa.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 5.Olszewski NE, West CM, Sassi SO, Hartweck LM. O-GlcNAc protein modification in plants: Evolution and function. Biochim Biophys Acta. 2010;1800:49–56. doi: 10.1016/j.bbagen.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen A, Kamp HD, Gründling A, Higgins DE. A bifunctional O-GlcNAc transferase governs flagellar motility through anti-repression. Genes Develop. 2006;20:3283–3295. doi: 10.1101/gad.1492606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selzer Jr, Hofmann F, Rex G, Wilm M, Mann M, Just I, Aktories K. Clostridium novyi alpha-toxin-catalyzed incorporation of GlcNAc into Rho subfamily proteins. J Biol Chem. 1996;271:25173–25177. doi: 10.1074/jbc.271.41.25173. [DOI] [PubMed] [Google Scholar]
- 8.Matsuura A, Ito M, Sakaidani Y, Kondo T, Murakami K, Furukawa K, Nadano D, Matsuda T, Okajima T. O-linked N-acetylglucosamine is present on the extracellular domain of Notch receptors. J Biol Chem. 2008:1–10. doi: 10.1074/jbc.M806202200. [DOI] [PubMed] [Google Scholar]
- 9.Sakaidani Y, Nomura T, Matsuura A, Ito M, Suzuki E, Murakami K, Nadano D, Matsuda T, Furukawa K, Okajima T. O-linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell-matrix interactions. Nature Comm. 2011;2:583–589. doi: 10.1038/ncomms1591. [DOI] [PubMed] [Google Scholar]
- 10.Wells L, Vosseller K, Hart GW. A role for N-acetylglucosamine as a nutrient sensor and mediator of insulin resistance. Cell Mol Life Sci. 2003;60:222–228. doi: 10.1007/s000180300017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Love DC, Hanover JA. The hexosamine signaling pathway: Deciphering the “O-GlcNAc Code”. Sci STKE. 2005;2005:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 12.Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol. 2012;13:270–276. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- 13.Hanover JA, Krause MW, Love DC. Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat Rev Mol Cell Biol. 2012;13:312–321. doi: 10.1038/nrm3334. [DOI] [PubMed] [Google Scholar]
- 14.Bond MR, Hanover JA. O-GlcNAc cycling: A link between metabolism and chronic disease. Ann Rev Nutrition. 2013;33:205–229. doi: 10.1146/annurev-nutr-071812-161240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou T-Y, Hart GW, Dang CV. c-Myc Is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J Biol Chem. 1995;270:18961–18965. doi: 10.1074/jbc.270.32.18961. [DOI] [PubMed] [Google Scholar]
- 16.Cheng X, Hart GW. Alternative O-glycosylation/O-phosphorylation of serine-16 in murine estrogen receptor β: Post-translational regulation of turnover and transactivation activity. J Biol Chem. 2001;276:10570–10575. doi: 10.1074/jbc.M010411200. [DOI] [PubMed] [Google Scholar]
- 17.Kelly WG, Dahmus ME, Hart GW. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J Biol Chem. 1993;268:10416–10424. [PubMed] [Google Scholar]
- 18.Du XL, Edelstein D, Dimmeler S, Ju Q, Sui C, Brownlee M. Hyperglycemia inhibits endothelial nitric oxide synthase activity by posttranslational modification at the Akt site. J Clin Invest. 2001;108:1341–1348. doi: 10.1172/JCI11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahne H, Gholami AM, Kuster B. Discovery of O-GlcNAc-modified proteins in published large-scale proteome data. Mol Cell Prot. 2012;11:843–850. doi: 10.1074/mcp.M112.019463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Torii M, Liu H, Hart G, Hu Z-Z. dbOGAP - An integrated bioinformatics resource for protein O-GlcNAcylation. BMC Bioinform. 2011;12:91. doi: 10.1186/1471-2105-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis LI, Blobel G. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc Natl Acad Sci U S A. 1987;84:7552–7556. doi: 10.1073/pnas.84.21.7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanover JA, Cohen CK, Willingham MC, Park MK. O-linked N-acetylglucosamine is attached to proteins of the nuclear pore. Evidence for cytoplasmic and nucleoplasmic glycoproteins. J Biol Chem. 1987;262:9887–9894. [PubMed] [Google Scholar]
- 23.Holt GD, Snow CM, Senior A, Haltiwanger RS, Gerace L, Hart GW. Nuclear pore complex glycoproteins contain cytoplasmically disposed O-linked N-acetylglucosamine. J Cell Biol. 1987;104:1157–1164. doi: 10.1083/jcb.104.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snow CM, Senior A, Gerace L. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J Cell Biol. 1987;104:1143–1156. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD, Shabanowitz J, Hunt DF, Hart GW. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci Signal. 2010;3:ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Favreau C, Worman HJ, Wozniak RW, Frappier T, Courvalin J-C. Cell cycle-dependent phosphorylation of nucleoporins and nuclear pore membrane protein Gp210. Biochemistry. 1996;35:8035–8044. doi: 10.1021/bi9600660. [DOI] [PubMed] [Google Scholar]
- 27.Hallberg E, Wozniak R, Blobel G. An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. J Cell Biol. 1993;122:513–521. doi: 10.1083/jcb.122.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cecioni S, Vocadlo DJ. Tools for probing and perturbing O-GlcNAc in cells and in vivo. Curr Opin Chem Biol. 2013;17:719–728. doi: 10.1016/j.cbpa.2013.06.030. [DOI] [PubMed] [Google Scholar]
- 29.Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 1984;259:3308–3317. [PubMed] [Google Scholar]
- 30.Holt GD, Haltiwanger RS, Torres CR, Hart GW. Erythrocytes contain cytoplasmic glycoproteins. O-linked GlcNAc on Band 4.1. J Biol Chem. 1987;262:14847–14850. [PubMed] [Google Scholar]
- 31.Ramakrishnan B, Qasba PK. Structure-based Design of β1,4-Galactosyltransferase I (β4Gal-T1) with equally efficient N-acetylgalactosaminyltransferase activity: Point mutation broadens βGal-T1 donor specificity. J Biol Chem. 2002;277:20833–20839. doi: 10.1074/jbc.M111183200. [DOI] [PubMed] [Google Scholar]
- 32.Khidekel N, Arndt S, Lamarre-Vincent N, Lippert A, Poulin-Kerstien KG, Ramakrishnan B, Qasba PK, Hsieh-Wilson LC. A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications. J Amer Chem Soc. 2003;125:16162–16163. doi: 10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- 33.Clark PM, Dweck JF, Mason DE, Hart CR, Buck SB, Peters EC, Agnew BJ, Hsieh-Wilson LC. Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J Amer Chem Soc. 2008;130:11576–11577. doi: 10.1021/ja8030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc Natl Acad Sci U S A. 2004;101:13132–13137. doi: 10.1073/pnas.0403471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rexach JE, Rogers CJ, Yu S-H, Tao J, Sun YE, Hsieh-Wilson LC. Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat Chem Biol. 2010;6:645–651. doi: 10.1038/nchembio.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thirumurugan P, Matosiuk D, Jozwiak K. Click chemistry for drug development and diverse chemical-biology applications. Chem Rev. 2013;113:4905–4979. doi: 10.1021/cr200409f. [DOI] [PubMed] [Google Scholar]
- 37.Finlay DR, Newmeyer DD, Price TM, Forbes DJ. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J Cell Biol. 1987;104:189–200. doi: 10.1083/jcb.104.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zachara NE, O’Donnell N, Cheung WD, Mercer JJ, Marth JD, Hart GW. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. J Biol Chem. 2004;279:30133–30142. doi: 10.1074/jbc.M403773200. [DOI] [PubMed] [Google Scholar]
- 39.Schindler M, Hogan M, Miller R, DeGaetano D. A nuclear specific glycoprotein representative of a unique pattern of glycosylation. J Biol Chem. 1987;262:1254–1260. [PubMed] [Google Scholar]
- 40.Vosseller K, Trinidad JC, Chalkley RJ, Specht CG, Thalhammer A, Lynn AJ, Snedecor JO, Guan S, Medzihradszky KF, Maltby DA, Schoepfer R, Burlingame AL. O-Linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity fhromatography and mass spectrometry. Mol Cell Prot. 2006;5:923–934. doi: 10.1074/mcp.T500040-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Comer FI, Vosseller K, Wells L, Accavitti MA, Hart GW. Characterization of a mouse monoclonal antibody specific for O-linked N-acetylglucosamine. Anal Biochem. 2001;293:169–177. doi: 10.1006/abio.2001.5132. [DOI] [PubMed] [Google Scholar]
- 42.Isono T. O-GlcNAc-specific antibody CTD110.6 cross-reacts with N-GlcNAc2-modified proteins induced under glucose deprivation. PLoS ONE. 2011;6:e18959. doi: 10.1371/journal.pone.0018959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teo CF, Ingale S, Wolfert MA, Elsayed GA, Nöt LG, Chatham JC, Wells L, Boons G-J. Glycopeptide-specific monoclonal antibodies suggest new roles for O-GlcNAc. Nat Chem Biol. 2010;6:338–343. doi: 10.1038/nchembio.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc Natl Acad Sci U S A. 2003;100:9116–9121. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyce M, Carrico IS, Ganguli AS, Yu S-H, Hangauer MJ, Hubbard SC, Kohler JJ, Bertozzi CR. Metabolic cross-talk allows labeling of O-linked β-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc Natl Acad Sci U S A. 2011;108:3141–3146. doi: 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaro BW, Yang Y-Y, Hang HC, Pratt MR. Chemical reporters for fluorescent detection and identification of O-GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4-1. Proc Natl Acad Sci U S A. 2011;108:8146–8151. doi: 10.1073/pnas.1102458108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reason AJ, Blench IP, Haltiwanger RS, Hart GW, Morris HR, Panico M, Dell A. High-sensitivity FAB-MS strategies for O-GlcNAc characterization. Glycobiology. 1991;1:585–594. doi: 10.1093/glycob/1.6.585. [DOI] [PubMed] [Google Scholar]
- 48.Greis KD, Hayes BK, Comer FI, Kirk M, Barnes S, Lowary TL, Hart GW. Selective detection and site-analysis of O-GlcNAc-modified glycopeptides by β-elimination and tandem electrospray mass spectrometry. Anal Biochem. 1996;234:38–49. doi: 10.1006/abio.1996.0047. [DOI] [PubMed] [Google Scholar]
- 49.North SJ, Hitchen PG, Haslam SM, Dell A. Mass spectrometry in the analysis of N-linked and O-linked glycans. Curr Opin Struct Biol. 2009;19:498–506. doi: 10.1016/j.sbi.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Myers SA, Daou S, Affar EB, Burlingame A. Electron transfer dissociation (ETD): The mass spectrometric breakthrough essential for O-GlcNAc protein site assignments—a study of the O-GlcNAcylated protein Host Cell Factor C1. Proteomics. 2013;13:982–991. doi: 10.1002/pmic.201200332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Housley MP, Udeshi ND, Rodgers JT, Shabanowitz J, Puigserver P, Hunt DF, Hart GW. A PGC-1α-O-GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose. J Biol Chem. 2009;284:5148–5157. doi: 10.1074/jbc.M808890200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chalkley RJ, Thalhammer A, Schoepfer R, Burlingame AL. Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc Natl Acad Sci U S A. 2009;106:8894–8899. doi: 10.1073/pnas.0900288106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao P, Viner R, Teo CF, Boons G-J, Horn D, Wells L. Combining high-energy C-trap dissociation and electron transfer dissociation for protein O-GlcNAc modification site assignment. J Prot Res. 2011;10:4088–4104. doi: 10.1021/pr2002726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, Hart GW. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol Cell Proteom. 2002;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- 55.Wang Z, Udeshi ND, O’Malley M, Shabanowitz J, Hunt DF, Hart GW. Enrichment and site mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation mass spectrometry. Mol Cell Proteom. 2010;9:153–160. doi: 10.1074/mcp.M900268-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alfaro J, Gong C, Monroe M, Aldrich J, Clauss T, Purvine S, Wang Z, Camp D, Shabanowitz J, Stanley P, Hart G, Hunt D, Yang F, Smith R. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc Natl Acad Sci U S A. 2012;109:7280–7285. doi: 10.1073/pnas.1200425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Radu A, Moore MS, Blobel Gn. The peptide repeat domain of nucleoporin NUP98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 58.Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harbor Perspectives in Biology. 2010 doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akey CW, Goldfarb DS. Protein import through the nuclear pore complex is a multistep process. J Cell Biol. 1989;109:971–982. doi: 10.1083/jcb.109.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Finlay DR, Forbes DJ. Reconstitution of biochemically altered nuclear pores: Transport can be eliminated and restored. Cell. 1990;60:17–29. doi: 10.1016/0092-8674(90)90712-n. [DOI] [PubMed] [Google Scholar]
- 61.Miller MW, Hanover JA. Functional nuclear pores reconstituted with beta 1-4 galactose-modified O-linked N-acetylglucosamine glycoproteins. J Biol Chem. 1994;269:9289–9297. [PubMed] [Google Scholar]
- 62.Shafi R, Lyer SPN, Ellies LG, O’Donnell N, Marek KW, Chui D, Hart GW, Marth JD. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O’Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol Cell Biol. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kazemi Z, Chang H, Haserodt S, McKen C, Zachara NE. O-linked β-N-acetylglucosamine (O-GlcNAc) regulates stress-induced heat shock protein expression in a GSK-β-dependent manner. J Biol Chem. 2010;285:39096–39107. doi: 10.1074/jbc.M110.131102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hanover JA, Forsythe ME, Hennessey PT, Brodigan TM, Love DC, Ashwell G, Krause M. A Caenorhabditis elegans model of insulin resistance: Altered macronutrient storage and dauer formation in an OGT-1 knockout. Proc Natl Acad Sci U S A. 2005;102:11266–11271. doi: 10.1073/pnas.0408771102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heesepeck A, Cole RN, Borkhsenious ON, Hart GW, Raikhel NV. Plant nuclear pore complex proteins are modified by novel oligosaccharides with terminal N-acetylglucosamine. Plant Cell. 1995;7:1459–1471. doi: 10.1105/tpc.7.9.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hartweck LM, Scott CL, Olszewski NE. Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics. 2002;161:1279–1291. doi: 10.1093/genetics/161.3.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jacobsen SE, Olszewski NE. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell. 1993;5:887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilson RN, Somerville CR. Phenotypic suppression of the gibberellin-insensitive mutant (gai) of Arabidopsis. Plant Physiol. 1995;108:495–502. doi: 10.1104/pp.108.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Silverstone AL, Tseng T-S, Swain SM, Dill A, Jeong SY, Olszewski NE, Sun T-P. Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol. 2007;143:987–1000. doi: 10.1104/pp.106.091025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng T-S, Kieffer M, Eshed Y, Olszewski N, Weiss D. The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell. 2012;24:96–108. doi: 10.1105/tpc.111.093518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tseng T-S, Salomé PA, McClung CR, Olszewski NE. SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell. 2004;16:1550–1563. doi: 10.1105/tpc.019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taoka K-I, Ham B-K, Xoconostle-Cázares B, Rojas MR, Lucas WJ. Reciprocal phosphorylation and glycosylation recognition motifs control NCAPP1 interaction with pumpkin phloem proteins and their cell-to-cell movement. Plant Cell. 2007;19:1866–1884. doi: 10.1105/tpc.107.052522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee J-Y, Yoo B-C, Lucas WJ. Parallels between nuclear-pore and plasmodesmal trafficking of information molecules. Planta. 2000;210:177–187. doi: 10.1007/PL00008124. [DOI] [PubMed] [Google Scholar]
- 75.Kose S, Furuta M, Imamoto N. Hikeshi, a nuclear import carrier for Hsp70s, protects cells from heat shock-induced nuclear damage. Cell. 2012;149:578–589. doi: 10.1016/j.cell.2012.02.058. [DOI] [PubMed] [Google Scholar]
- 76.Ngoh GA, Watson LJ, Facundo HT, Jones SP. Augmented O-GlcNAc signaling attenuates oxidative stress and calcium overload in cardiomyocytes. Amino Acids. 2011;40:895–911. doi: 10.1007/s00726-010-0728-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ngoh GA, Facundo HT, Hamid T, Dillmann W, Zachara NE, Jones SP. Unique hexosaminidase reduces metabolic survival signal and sensitizes cardiac myocytes to hypoxia/reoxygenation injury. Circulation Res. 2009;104:41–U149. doi: 10.1161/CIRCRESAHA.108.189431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ngoh GA, Watson LJ, Jones SP. Loss of O-GlcNAc transferase activity sensitizes cardiac myocytes to post-hypoxic death. FASEB J. 2008;22 [Google Scholar]
- 79.Slawson C, Zachara NE, Vosseller K, Cheung WD, Lane MD, Hart GW. Perturbations in O-linked β-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J Biol Chem. 2005;280:32944–32956. doi: 10.1074/jbc.M503396200. [DOI] [PubMed] [Google Scholar]
- 80.Miller MW, Caracciolo MR, Berlin WK, Hanover JA. Phosphorylation and glycosylation of nucleoporins. Arch Biochem Biophys. 1999;367:51–60. doi: 10.1006/abbi.1999.1237. [DOI] [PubMed] [Google Scholar]
- 81.Crampton N, Kodiha M, Shrivastava S, Umar R, Stochaj U. Oxidative stress inhibits nuclear protein export by multiple mechanisms that target FG nucleoporins and Crm1. Mol Biol Cell. 2009;20:5106–5116. doi: 10.1091/mbc.E09-05-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jones SP, Zachara NE, Ngoh GA, Hill BG, Teshima Y, Bhatnagar A, Hart GW, Marbán E. Cardioprotection by N-acetylglucosamine linkage to cellular proteins. Circulation. 2008;117:1172–1182. doi: 10.1161/CIRCULATIONAHA.107.730515. [DOI] [PubMed] [Google Scholar]
- 83.Love DC, Ghosh S, Mondoux MA, Fukushige T, Wang P, Wilson MA, Iser WB, Wolkow CA, Krause MW, Hanover JA. Dynamic O-GlcNAc cycling at promoters of Caenorhabditis elegans genes regulating longevity, stress, and immunity. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0911857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zou LY, Yang SL, Hu SH, Chaudry IH, Marchase RB, Chatham JC. The protective effects of PUGNAc on cardiac function after trauma-hemorrhage are mediated via increased protein O-GlcNAc levels. Shock. 2007;27:402–408. doi: 10.1097/01.shk.0000245031.31859.29. [DOI] [PubMed] [Google Scholar]
- 85.Ngoh GA, Hamid T, Prabhu SD, Jones SP. O-GlcNAc signaling attenuates ER stress-induced cardiomyocyte death. Amer J Physiol. 2009;297:H1711–H1719. doi: 10.1152/ajpheart.00553.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Not LG, Marchase RB, Fulop N, Brocks CA, Chatham JC. Glucosamine administration improves survival rate after severe hemorrhagic shock combined with trauma in rats. Shock. 2007;28:345–352. doi: 10.1097/shk.0b013e3180487ebb. [DOI] [PubMed] [Google Scholar]
- 87.Not LG, Brocks CA, Vamhidy L, Marchase RB, Chatham JC. Increased O-linked beta-N-acetylglucosamine levels on proteins improves survival, reduces inflammation and organ damage 24 hours after trauma-hemorrhage in rats. Critical Care Medicine. 2010;38:562–571. doi: 10.1097/CCM.0b013e3181cb10b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang SL, Zou LY, Bounelis P, Chaudry I, Chatham JC, Marchase RB. Glucosamine administration during resuscitation improves organ function after trauma hemorrhage. Shock. 2006;25:600–607. doi: 10.1097/01.shk.0000209563.07693.db. [DOI] [PubMed] [Google Scholar]
- 89.Zou LY, Yang SL, Champattanachai V, Hu SH, Chaudry IH, Marchase RB, Chatham JC. Glucosamine improves cardiac function following trauma-hemorrhage by increased protein O-GlcNAcylation and attenuation of NF-kappa B signaling. Amer J Physiol. 2009;296:H515–H523. doi: 10.1152/ajpheart.01025.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu J, Marchase RB, Chatham JC. Increased O-GlcNAc levels during reperfusion lead to improved functional recovery and reduced calpain proteolysis. Amer J Physiol. 2007;293:H1391–H1399. doi: 10.1152/ajpheart.00285.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fulop N, Marchase RB, Chatham JC. Role of protein O-linked N-acetyl-glucosamine in mediating cell function and survival in the cardiovascular system. Cardiovasc Res. 2007;73:288–297. doi: 10.1016/j.cardiores.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fulop N, Zhang ZH, Marchase RB, Chatham JC. Glucosamine cardioprotection in perfused rat hearts associated with increased O-linked N-acetylglucosamine protein modification and altered p38 activation. Amer J Physiol. 2007;292:H2227–H2236. doi: 10.1152/ajpheart.01091.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hwang SY, Shin JH, Hwang JS, Kim SY, Shin JA, Oh ES, Oh S, Kim JB, Lee JK, Han IO. Glucosamine exerts a neuroprotective effect via suppression of inflammation in rat brain ischemia/reperfusion injury. Glia. 2010;58:1881–1892. doi: 10.1002/glia.21058. [DOI] [PubMed] [Google Scholar]
- 94.Liu J, Pang Y, Chang T, Bounelis P, Chatham JC, Marchase RB. Increased hexosamine biosynthesis and protein O-GlcNAc levels associated with myocardial protection against calcium paradox and ischemia. J Mol Cell Cardiol. 2006;40:303–312. doi: 10.1016/j.yjmcc.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 95.Pang Y, Hunton DL, Bounelis P, Marchase RB. Hyperglycemia inhibits capacitative calcium entry and hypertrophy in neonatal cardiomyocytes. Diabetes. 2002;51:3461–3467. doi: 10.2337/diabetes.51.12.3461. [DOI] [PubMed] [Google Scholar]
- 96.Sohn KC, Lee KY, Park JE, Do SI. OGT functions as a catalytic chaperone under heat stress response: a unique defense role of OGT in hyperthermia. Biochem Biophys Res Comm. 2004;322:1045–1051. doi: 10.1016/j.bbrc.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 97.Chatham JC, Marchase RB. The role of protein O-linked beta-N-acetylglucosamine in mediating cardiac stress responses. Biochim Biophys Acta. 2010;1800:57–66. doi: 10.1016/j.bbagen.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fang B, Miller MW. Use of galactosyltransferase to assess the biological function of O-linked N-acetyl-D-glucosamine: A potential role for O-GlcNAc during cell division. Exptl Cell Res. 2001;263:243–253. doi: 10.1006/excr.2000.5110. [DOI] [PubMed] [Google Scholar]
- 99.Laurell E, Kutay U. Dismantling the NPC permeability barrier at the onset of mitosis. Cell Cycle. 2011;10:2243–2245. doi: 10.4161/cc.10.14.16195. [DOI] [PubMed] [Google Scholar]
- 100.Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, Aebersold R, Antonin W, Kutay U. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell. 2011;144:539–550. doi: 10.1016/j.cell.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 101.Nuclear Transport: A Switch for the Oxidative Stress-Signaling Circuit? J Signal Transduct. 2012;2012 doi: 10.1155/2012/208650. Article ID 208650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mizuguchi-Hata C, Ogawa Y, Oka M, Yoneda Y. Quantitative regulation of nuclear pore complex proteins by O-GlcNAcylation. Biochim Biophys Acta. 2013;1833:2682–2689. doi: 10.1016/j.bbamcr.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 103.Yu S-H, Boyce M, Wands AM, Bond MR, Bertozzi CR, Kohler JJ. Metabolic labeling enables selective photocrosslinking of O-GlcNAc-modified proteins to their binding partners. Proc Natl Acad Sci U S A. 2012;109:4834–4839. doi: 10.1073/pnas.1114356109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pham N, Parker R, Kohler J. Photocrosslinking approaches to interactome mapping. Curr Opin Chem Biol. 2013;17:90–101. doi: 10.1016/j.cbpa.2012.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rout MP, Aitchison JD, Magnasco MO, Chait BT. Virtual gating and nuclear transport: the hole picture. Trends Cell Biol. 2003;13:622–628. doi: 10.1016/j.tcb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 106.Denning DP, Patel SS, Uversky V, Fink AL, Rexach M. Disorder in the nuclear pore complex: The FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci U S A. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Frey S, Görlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130:512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 108.Labokha AA, Gradmann S, Frey S, Hulsmann BB, Urlaub H, Baldus M, Gorlich D. Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 2013;32:204–218. doi: 10.1038/emboj.2012.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- 110.Arib G, Akhtar A. Multiple facets of nuclear periphery in gene expression control. Curr Opin Cell Biol. 2011;23:346–353. doi: 10.1016/j.ceb.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 111.Blobel G. Gene gating: a hypothesis. Proc Natl Acad Sci U S A. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 113.Hou C, Corces VG. Nups take leave of the nuclear envelope to regulate transcription. Cell. 2010;140:306–308. doi: 10.1016/j.cell.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 116.Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins Nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 2010;6:e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood. 2011;118:6247–6257. doi: 10.1182/blood-2011-07-328880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jackson SP, Tjian R. Purification and analysis of RNA polymerase II transcription factors by using wheat germ agglutinin affinity chromatography. Proc Natl Acad Sci U S A. 1989;86:1781–1785. doi: 10.1073/pnas.86.6.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zachara N, Molina H, Wong K, Pandey A, Hart G. The dynamic stress-induced “O-GlcNAc-ome” highlights functions for O-GlcNAc in regulating DNA damage/repair and other cellular pathways. Amino Acids. 2011;40:793–808. doi: 10.1007/s00726-010-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang Z, Pandey A, Hart GW. Dynamic interplay between O-linked N-acetylglucosaminylation and glycogen synthase kinase-3-dependent phosphorylation. Mol Cell Proteom. 2007;6:1365–1379. doi: 10.1074/mcp.M600453-MCP200. [DOI] [PubMed] [Google Scholar]
- 121.Sprung R, Nandi A, Chen Y, Kim SC, Barma D, Falck JR, Zhao Y. Tagging-via-substrate strategy for probing O-GlcNAc modified proteins. J Proteome Res. 2005;4:950–957. doi: 10.1021/pr050033j. [DOI] [PubMed] [Google Scholar]
- 122.Hahne H, Sobotzki N, Nyberg T, Helm D, Borodkin VS, van Aalten DMF, Agnew B, Kuster B. Proteome wide purification and identification of O-GlcNAc-modified proteins using click chemistry and mass spectrometry. J Proteome Res. 2013;12:927–936. doi: 10.1021/pr300967y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Roquemore EP, Chou T-Y, Hart GW. Detection of O-Linked N-acetylglucosamine (O-GlcNAc) on cytoplasmic and nuclear proteins. In: William J, Lennarz GWH, editors. Methods in Enzymology. Academic Press; 1994. pp. 443–460. [DOI] [PubMed] [Google Scholar]
- 124.Tai H-C, Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Parallel Identification of O-GlcNAc-Modified Proteins from Cell Lysates. Journal of the American Chemical Society. 2004;126:10500–10501. doi: 10.1021/ja047872b. [DOI] [PubMed] [Google Scholar]


