Abstract
Oral squamous cell carcinoma (OSCC) is diagnosed in 640,000 patients yearly with a poor (50%) 5-year survival rate that has not changed appreciably in decades. To investigate molecular changes that drive OSCC progression, cDNA microarray analysis was performed using cells that form aggressive poorly differentiated tumors (SCC25-PD) in a murine orthotopic xenograft model compared to cells that produce well-differentiated tumors (SCC25-WD). As this analysis revealed that 59 upregulated genes were NF-κB target genes, the role of NF-κB activation in alteration of the transcriptional profile was evaluated. The mRNA and protein upregulation of a panel NF-κB target genes was validated by real-time qPCR and immunohistochemistry. Additionally, nuclear translocation of RelA was greatly increased in SCC25-PD, increased nuclear RelA was observed in oral tumors initiated with SCC25-PD compared with tumors initiated by SCC25-WD, and nuclear RelA correlated with stage of disease on two human OSCC tissue microarrays. Treatment of SCC25-PD cells with the IKKβ-inhibitor sc-514, that effectively prevents RelA phosphorylation on Ser 536, reversed nuclear-translocation of RelA and strongly inhibited NF-κB gene activation. Furthermore, blocking the phosphorylation of RelA using the MSK1/2 inhibitor SB 747651A significantly reduced the mRNA upregulation of a subset of target genes. Treatment with sc-514 or SB 747651A markedly diminished cellular invasiveness. These studies support a model wherein NF-κB is constitutively active in aggressive OSCC, while blocking the NF-κB pathway reduces NF-κB target gene upregulation and cellular invasiveness.
Keywords: Oral cancer, Immunohistochemistry, Invasion, NF-kappa B (NF-κB), Transcriptional regulation
Introduction
Oral cavity cancer is the 6th most frequently diagnosed cancer, with over 640,000 new diagnoses and more than 200,000 deaths annually worldwide. Oral squamous cell carcinoma (OSCC) is the most common malignancy, with 100 new diagnoses each day in the U.S. alone (1). Approximately 40% of all oral cancers occur in the tongue and this site is more frequently associated with lymph node metastasis due to the rich lymphatic network of the tongue (1). Tongue tumors often remain undetected until advanced stage and patients with tongue tumors have a poorer survival rate relative to other anatomic sites in the oral cavity (1,2). Two-thirds of patients present with locally advanced lesions or regional lymph node involvement (3). Thus, despite advances in chemotherapeutic agents, the survival rate for patients with OSCC has remained at 50% for 35 years and is reduced further to only 29% in patients with cervical lymph node metastases (1). The poor prognosis may reflect a lack of understanding of the molecular mechanisms that regulate growth, differentiation, invasion and metastasis. Therefore, new molecular-based strategies are required to understand and ultimately successfully treat this malignancy.
We have previously modified a well-described orthotopic xenograft model of aggressive invasive tongue OSCC (4) by modulating expression levels of the gene PLAUR, shown to be a component of the OSCC gene signature for molecular stratification of aggressive tumors with poor outcome (5,6). Cells that overexpress PLAUR grow as poorly circumscribed and poorly differentiated SCC (designated SCC25-PD), with infiltrative cords of tumor cells dissecting tongue muscle, high mitotic index, and foci of perineural and vascular invasion (7). In contrast, cells in which PLAUR expression is down-regulated using siRNA grew as well differentiated SCC (designated SCC25-WD) with low mitotic index and numerous keratin aggregates. Using this model system, comparative cDNA microarray analysis revealed that 98 of 151 differentially regulated genes, 73 of which were upregulated, were known or predicted NF-κB target genes. Furthermore, most of the upregulated NF-κB targets were recognized as genes that can be activated through the canonical NF-κB pathway while a majority were genes associated with invasion and metastasis (8-10).
NF-κB is a family of dimeric transcription factors that regulates numerous genes, is constitutively-activated in many cancers, and may play a critical role in transformation, proliferation, aberrant apoptosis and chemoresistance, invasion and metastasis (11). The individual subunits of NF-κB are comprised of RelA (p65), RelB, c-Rel, p50, and p52. All of these subunits contain a Rel homology domain, which facilitates homo- or hetero-dimerization of NF-κB family members as well as DNA binding and the interaction of these dimers with inhibitory IκB proteins (11-13). Homodimers of p50 are abundant in the nuclei of unstimulated cells and while complexed with HDAC1 bind DNA and repress transcription of NF-κB -responsive genes (14). Dimers that contain RelA, RelB and/or c-Rel, including the most abundant of these latent NF-κB dimers the heterodimer RelA/p50, are typically sequestered in the cytoplasm of most unstimulated cells by IκBα (inhibitor of κBα) and other IκB proteins. Upon stimulation by a large variety of substances which activate the canonical pathway, the IκB kinase (IKK) complex, comprised of 2 catalytic subunits IKKα and IKKβ (also known as IKK1 and IKK2) and one regulatory subunit NF-κB essential modulator (NEMO or IKKγ), phosphorylates specific serines of the IκB proteins, triggering their ubiquitination and degradation by the 26S proteasome and release of the NF-κB heterodimer. This exposes the nuclear localization sequence of the NF-κB subunits and results in the nuclear translocation of the NF-κB dimer and subsequent target gene transactivation (14,15). It has been demonstrated that post-translational modification of the RelA subunit is also a requirement for efficient gene transactivation at the promotors of many genes activated by NF-κB signaling (16). Phosphorylation of the RelA subunit at Ser276 or Ser536 is required for the transcription of distinct subsets of NF-κB target genes (17). In the current study, we have examined the role of NF-κB activation in alteration of the transcriptional profile of poorly differentiated OSCC in xenograft tumors and in human OSCC. These studies support a model wherein NF-κB is constitutively active in aggressive OSCC, while blocking the NF-κB pathway reduces NF-κB target gene upregulation and cellular invasiveness.
Materials and Methods
Cell Culture
SCC25-PD and SCC25-WD were created by modulating PLAUR expression in SCC25 parental cells, as previously described (7). Cells that overexpress PLAUR grow as poorly circumscribed and poorly differentiated SCC (SCC25-PD), while cells in which PLAUR expression is down-regulated using siRNA grew as well differentiated SCC (SCC25-WD). Cells were grown in DMEM/F12 (1:1) with 100μg/ml Pen/Strep and 450μg/ml G418 with 10% FBS or without FBS for serum-free medium. Cells were passaged after 50 to 75% confluence.
Reagents and Antibodies
TNF-alpha was obtained from Shenandoah Biotechnology (Warwick, PA.). NF-κB (IKKβ) inhibitor sc-514 was purchased from EMD Millipore Corp. (Billerica, MA). MSK-inhibitor SB 747651A was obtained from Axon MedChem (The Netherlands). Mouse anti-p50, rabbit anti-p50(NLS), rabbit anti-p65 (RelA), rabbit anti-phospho-RelA (S276), and mouse anti-GAPDH, were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA); rabbit anti-phospho-RelA (S536) and rabbit anti-SOD-2 were purchased from Abcam (Cambridge, MA); goat anti-human ICAM-1/CD54 from R&D Systems; mouse anti-Cox-2 was purchased from Cayman Chemical (Ann Arbor Michigan); anti-GAPDH-peroxidase and anti-β-actin-peroxidase were purchased from Sigma-Aldrich (St. Louis, MO). Human oral cancer tissue microarray (OR481 and OR601a) were obtained from US Biomax, Inc. (Rockville, MD.).
Immunofluorescence
Cells were plated on 22-mm2 glass coverslips in 6-well plates to 50-70% confluence, treated with 100μM inhibitor sc-514 or 20μM SB747651A for 25 hours, washed 3X in PBS and serum-starved for 4 hours (in presence of inhibitor). For positive controls, cells were washed with PBS 3X and serum-starved for 4 hours, then treated with 100ng/ml TNFα for 30 min. Cells were washed once with PBS, and then fixed in 4% paraformaldehyde in PBS containing 0.12M sucrose for 20 min. at room temperature and permeabilized for 5 min. in 0.3% Triton in PBS. Cells were rinsed twice in PBS and blocked with 10% BSA in PBS for 1 hr. Primary antibody in 1% BSA was applied for 1 h at room temperature at 1:100 dilution. Cells were washed 3X in PBS followed by application of the secondary antibody (Alexa-fluor 488) for 30 min. at 1:500 dilution. Cells were washed 3X in PBS, rinsed 1X with water and coverslips were allowed to air dry protected from light. Coverslips were inverted onto 10ul of Vectashield with DAPI on glass slides. Fluorescence was examined on inverted microscopes (Olympus IX81with DSU spinning disk confocal microscopy system, Zeiss LSM Meta 510 laser scanning confocal microscope, or the AMG EVOS All-In-One digital microscope). RelA nuclear translocation was scored on a 0-1 scale with 1 = complete nuclear translocation; 0.75 = 75% or greater translocation; 0.5 = nuclear staining intensity equal to cytoplasmic intensity; 0.25 25% some translocation; 0 = no translocation. Cells undergoing mitosis stained more brightly than true positive single cells and were excluded from scoring. Total score divided by the number of cells counted is given as percent of positive RelA nuclear translocation. TNFα treated cells were used as a positive control to confirm RelA nuclear translocation. At least 40 randomly chosen fields (200X magnification) were counted from each cell group. Comparisons between groups were evaluated using the Mann Whitney test (Sigmaplot).
Immunohistochemistry
Orthotopic murine xenograft tumors were generated as previously described (4,7,18). Briefly, three cohorts (8 mice per group) of 6 week old female athymic nu/nu mice were injected into the lateral border of the base of the tongue with SCC25-PD or SCC25-WD cells (6×106 cells in 30 ul sterile PBS). Female mice were used for this study as they exhibit less aggressive biting behavior relative to male mice. At ~9 weeks, mice were sacrificed, the tongue was dissected from each mouse (n=24 per group), fixed in 4% paraformaldehyde (4 C), paraffin embedded, sections (4 um) and stained with H&E or processed for immunohistochemistry as described below. In this model, SCC25-PD grow as poorly circumscribed and poorly differentiated SCC, with infiltrative cords of tumor cells dissecting tongue muscle, high mitotic index, and foci of perineural and vascular invasion, while tumors generated from SCC25-WD grew as well differentiated SCC with low mitotic index, numerous keratin aggregates and no evidence of lymphovascular or perineural invasion (7). Note that institutional ACUC regulations preclude growth of tongue tumors sufficiently large to generate cervical lymph node or distant metastases. For IHC, 4μm formalin-fixed, paraffin-embedded sections of these tumors were deparaffinized with xylene and rehydrated in a series of ethanol washes. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide in methanol for 30 min. Antigen retrieval was enhanced by microwaving in 10 mmol/L sodium citrate (pH 6.0). Nonspecific binding was blocked with 3% normal horse serum in PBS for 30 min. Sections were incubated for 1 h at room temperature (or at 4°C overnight) with primary antibody diluted 1:20 in 1% bovine serum albumin in PBS. Staining was detected using an avidin-biotin horseradish peroxidase system (Vectastain Universal Elite ABC kit, Vector Laboratories), with positive cells staining brown using 3,3′ diaminobenzidine (DAB) chromogen and hydrogen peroxide as substrate (Liquid DAB substrate, Bio-Genex). For quantitation of nuclear RelA, nuclear phospho-RelA, or cytoplasmic staining, the slides were scanned into an Aperio slide-scanner and a macro using a 3 point scoring system was selected and used to analyze annotated tumor cell fields. At least 20 randomly chosen fields (at 200X magnification), or all available fields, were counted from both tumor groups. A minimum of 2800 cells were scored from each group. Results were obtained as a nuclear H-score for nuclear RelA or nuclear phospho-RelA(Ser276), or as a cytoplasmic H-score for all other antigens. To assess nuclear RelA staining in human OSCC progression, a commercial OSCC tissue microarray (TMA) from Biomax (OR481, US Biomax Ltd, Rockville, MD), containing 38 cases of tongue OSCC was analyzed. Results were verified using a second OSCC TMA from Biomax (OR601a) containing 47 cases of tongue SCC. Nuclear RelA staining was performed and quantified as detailed above. In all cases, manual scoring was also performed to verify accuracy of the Aperio macros.
Cell Fractionation and Western Blot Analysis
Cells were washed three times with cold PBS and lysed with a cold hypotonic lysis buffer: 10.0 mM NaCl, 20.0 mM HEPES, pH 7.9, 1.0mM EDTA, 2.0mM MgCl2, 20.0mM β-glycerophosphate, 1.0 mM Na3VO4, 1.0 mM PMSF, 1.0 mM DTT, 200mM sucrose, 0.5% Nonidet P-40, one tablet/10ml of a protease inhibitor cocktail (complete, mini, EDTA-free; Roche Diagnostics, Mannheim, Germany), and a 1:100 dilution of a phosphatase inhibitor cocktail (Halt, Thermo Scientific, Rockford, IL.). Lysate was collected by scraping, passed through a 26-gauge syringe, incubated on ice 5 min., and centrifuged at 13,500g for 5 min. at 4 °C. The cytoplasmic fraction was collected (supernatant), and the pellet was washed twice with hypotonic lysis buffer before treatment with nuclear extraction buffer (420.0 mM NaCl, 20.0 mM HEPES, pH 7.9, 1.0mM EDTA, 2.0mM MgCl2, 20.0mM β-glycerophosphate, 1.0 mM Na3VO4, 1.0 mM PMSF, 1.0 mM DTT, 25% glycerol, one tablet/10ml of a protease inhibitor cocktail (complete, mini, EDTA-free; Roche Diagnostics, Mannheim, Germany), and a 1:100 dilution of a phosphatase inhibitor cocktail (Halt, Thermo Scientific, Rockford, IL.). Following a 15 min. incubation and a 5-min. centrifugation (2,000g at 4 °C), the nuclear fraction (supernatant) was collected. Western blot analysis was done as described previously (30). Each extract lane contained 20 μg of protein as determined using a DC protein assay (Biorad, Hercules, CA.). Western blots were quantified using ImageJ (National Institutes of Health, Bethesda). p values were determined using the t test function (two sample, unequal variance, one-tailed distribution) on Excel (Microsoft Corp., Redmond, WA).
Matrigel Invasion Assays
Cells at 50-70% confluence were untreated or pretreated for 25 hours with inhibitor sc-514 or SB747651A or DMSO control, washed 3X with PBS and serum starved for 4 hours (with or without inhibitor). Invasion chambers (8 micron, 24-well plate pre-treated with 1% collagen) were prepared with 100μl 1mg/ml Matrigel for 1 hour, rinsed with serum-free medium and air-dried. Cells were trypsinized, neutralized with soybean trypsin inhibitor, counted, and reconstituted in SFM with or without inhibitor at a cell density of 500,000 cells/ml. Cells (0.5 ml; 250,000 cells) were added to each invasion chamber in each well (containing 0.75ml SFM). All conditions tested were performed in triplicate. Cells were incubated at 37 C for 64 hours and stained with the Diff-quick kit according to the manufacturer's instructions (Siemens Healthcare Diagnostics). Filters were excised, mounted face down with Permount (Fisher Chemicals), and invading cells enumerated by counting cells in 5 representative fields. Data were evaluated by t-test (Sigmaplot) for significance. Average scores were displayed relative to percent invasion of control SCC25-PD cells (designated 100%).
Real-time quantitative PCR
The following primer sets were employed for comparative quantitation of mRNA: Cox-2 forward: 5’-GCCCAGCACTTCACGCATCAG-3’ and Cox-2 reverse: 5’-AGACCAGGCACCAGACCAAAGACC-3’; ICAM1 forward: 5’-ACGGTGCTGGTGAGGAGAGAT-3’ and ICAM1 reverse: 5’-AGTCGCTGGCAGGACAAAGGT-3’; SOD2 forward: 5’-GGGTTGGCTTGGTTTCAATA-3’ and SOD2 reverse: 5’-ACACATCAATCCCCAGCAGT-3’; IL1A forward: 5’-GTCTCTGAATCAGAAATCCTTCTATC-3’ and IL1A reverse: 5’-CATGTCAAATTTCACTGCTTCATCC-3’; TNFAIP3 forward: 5’-CTTCTCAGTACATGTGGGGCGTTCAGG-3’ and TNFAIP3 reverse: 5’-CCCATTCATCATTCCAGTTCCGAGTATCAT-3’; IL8 forward: 5’-GAGGGTTGTGGAGAAGTTTTTG-3’ and IL8 reverse: 5’-CTGGCATCTTCACTGATTCTTG-3’; endogenous control gene phosphoglycerate kinase (PGK1) forward: 5’-GGGCTGCATCACCATCATAGG-3’ and PGK1 reverse 5’-GAGAGCATCCACCCCAGGAAG-3’. Oligos were custom synthesized by Integrated DNA Technologies (Coralville, Iowa). For inhibitor studies, cells were pretreated for 25 hours with 100μM sc-514 or 20μM SB747651A. RNA was extracted with Trizol Reagent (Invitrogen) and reverse transcription was performed with 5μg of the total RNA from each cell population using the RT2 First Strand Kit (Qiagen, Gaithersburg, Maryland). Real-time PCR was performed with SYBR green Master Mix (Applied Biosystems, Foster City, CA). PCR cycling conditions were 95 °C for 3.5 min. followed by 40 cycles of 95 °C for 15 s and 55°C for 30 s. Melt curve cycling consisted of 81 30 s cycles beginning at 55°C increasing by 0.5 °C to 95 °C. Each sample was analyzed in triplicate for each PCR measurement. Melting curves were checked to ensure specificity. Relative quantification of mRNA expression was calculated using the standard curve method with the endogenous housekeeping gene PGK level as normalizer and control sample as calibrator, or using the [.delta][.delta]CT method where the efficiency of amplification of the target gene was similar (within 10%) to the efficiency of the endogenous control gene (PGK1).
Results
NF-κB target genes are upregulated in invasive OSCC
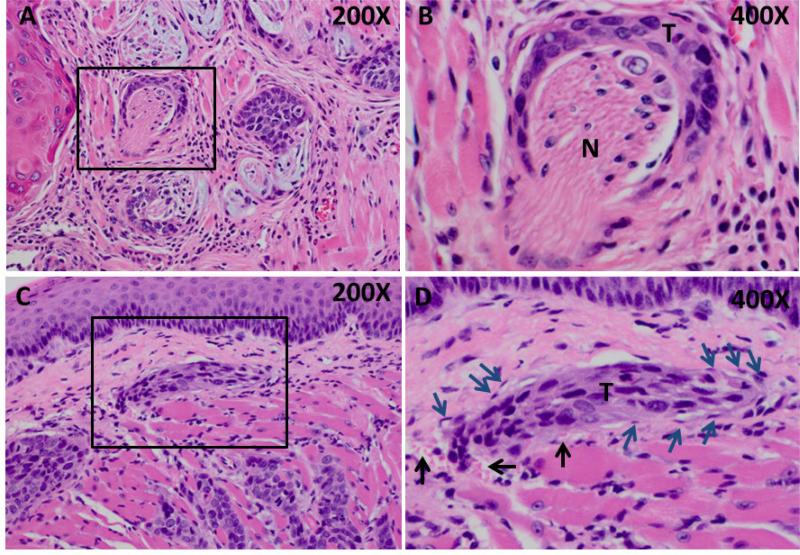
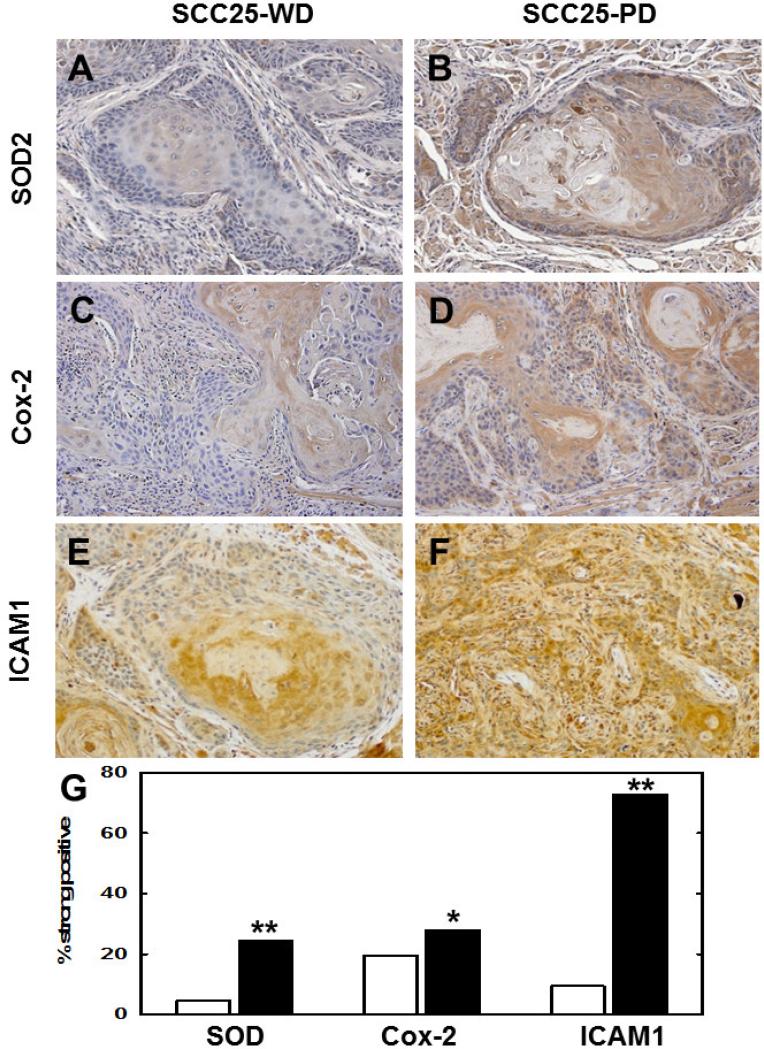
When injected orthotopically into the murine tongue, tumors generated from SCC25-WD grow as well differentiated SCC with low mitotic index, numerous keratin aggregates and no evidence of lymphovascular or perineural invasion (not shown, 7). In contrast, SCC-PD cells develop into poorly differentiated SCC with infiltrative cords of tumor cells exhibiting neural (Fig. 1A,B) and perivascular (Fig. 1C,D) invasion. To identify genes associated with aggressive SCC, comparative cDNA microarray analysis (SCC25-PD vs SCC25-WD) was performed. This analysis identified 151 differentially regulated genes (7), of which 65% were known or predicted NF-κB target genes. Several candidate NF-κB- regulated genes associated with progression, invasion and/or metastasis were validated by real time rtqPCR (Table 1). High level induction of the proinflammatory cytokines IL8 and IL1A as well as the ubiquitin-editing enzyme TNFAIP3 were observed. Additionally, PTGS2, ICAM1, and SOD2 were also significantly upregulated (7-25-fold). Immunohistochemical staining of tumors for SOD2, Cox-2 (PTGS2), and ICAM1 supported the PCR data, showing significantly enhanced staining intensity in SCC25-PD tumors relative to SCC25-WD (Fig. 2A-G, Suppl. Table 1). In addition to these NF-κB target genes, we have previously validated a 3-5-fold upregulation of kallikreins (KLKs) 5, 7, and 10 by qrtPCR and immunohistochemical staining in both murine and human tongue tumors (18).
Figure 1. Perineural and vascular invasion in SCC25-PD tumors.
(A,B) Representative H&E stained section showing tumor cords surrounding nerve bundle (boxed area). (T) - Tumor, (N) - nerve. Panel A – 200X magnification, Panel B – 400X magnification. (C,D) H&E stained section showing vascular invasion of tumor cord. (T) – tumor, black arrows – red blood cells, blue arrows – vascular endothelial cells. Panel C – 200X magnification, Panel D – 400X magnification.
Table 1. Validation of cDNA Microarray Results.
Selected genes showing at least a 2-fold differential expression between SCC25-PD and SCC25-WD cells by cDNA microarray analysis (9) were validated by triplicate real time reverse-transcriptase PCR. Relative expression levels were normalized to the housekeeping gene PGK1. All genes in the panel showed at least a 2-fold upregulation (p<0.05).
| Gene | Fold Change (SCC25-PD vs. SCC25-WD) |
|---|---|
| IL8 | 87.4 +/− 27.0 |
| IL1A | 64.1 +/− 8.0 |
| PTGS2 | 25.2 +/− 3.0 |
| TNFAIP3 | 15.5 +/− 1.0 |
| ICAM1 | 7.8 +/− 3.1 |
| SOD2 | 6.7 +/− 1.9 |
Figure 2. Immunohistochemical validation of upregulated NF-κB target genes.
Representative sections from murine orthotopic (tongue) SCC25-WD tumors (left column) and SCC25-PD tumors (right column) were stained with antibodies directed against (A,B) SOD2, (C,D) Cox-2 or (E,F) ICAM1. Magnification 200X. (G) Quantitation of tumor staining for each antigen in SCC25-WD tumors (white bar) and SCC25-PD tumors (black bar). For quantitation of tumor staining, stained slides were scanned on an Aperio ScanScope slide scanner and a minimum of 2500 cells from at least 10 tumor areas were scored on a standard 0 to 3 point scale using algorithm macros developed on the ImageScope software for quantitative analysis according to the manufacturer's instructions.”Strong positive” is defined as 2+ or 3+. (Detailed quantitation results are shown in Supplementary Table 1.) The automated quantitation results were consistent with those obtained by manual counting methods. (*) p<.05; (**) p<.001, relative to the paired SCC25-WD tumor.
RelA nuclear translocation is significantly elevated in aggressive OSCC
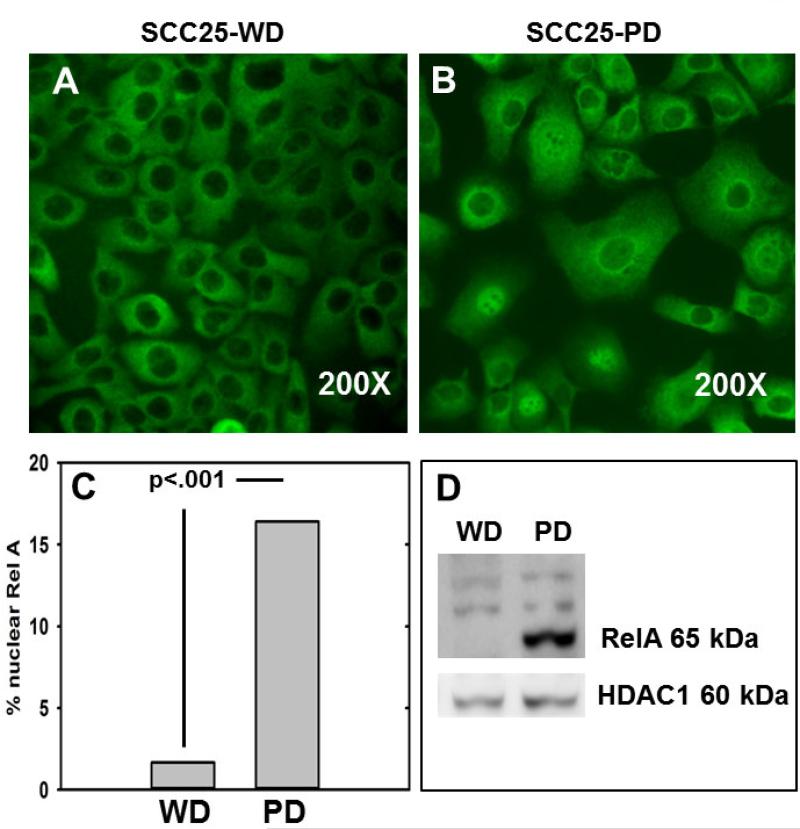
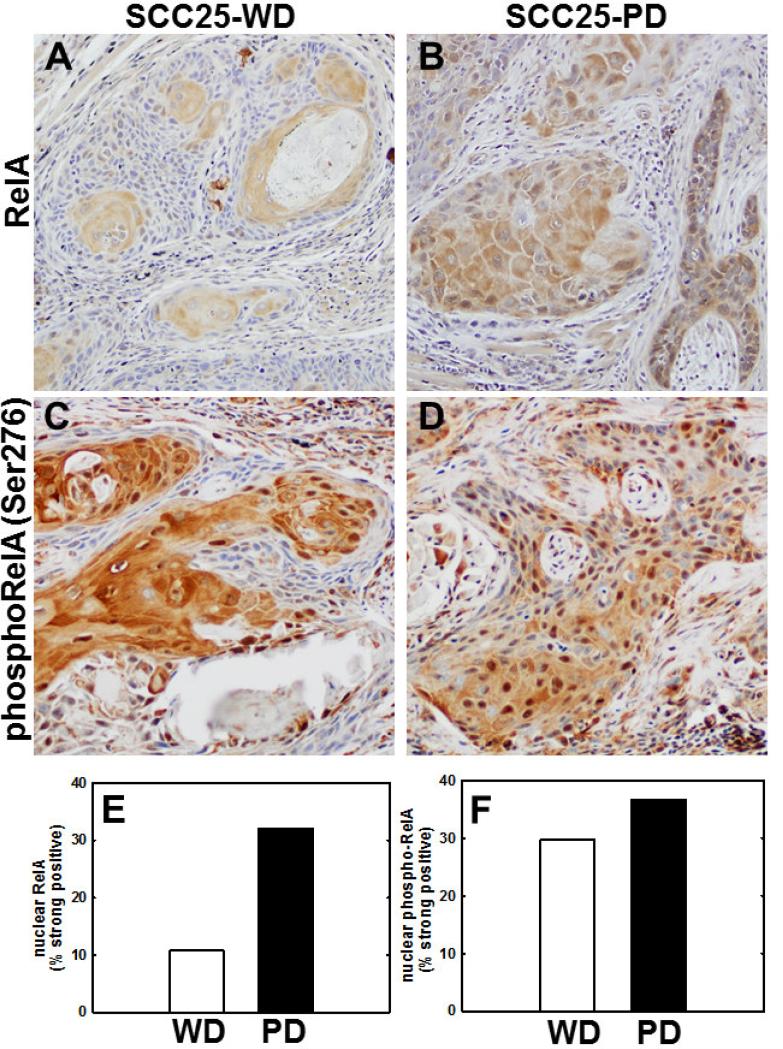
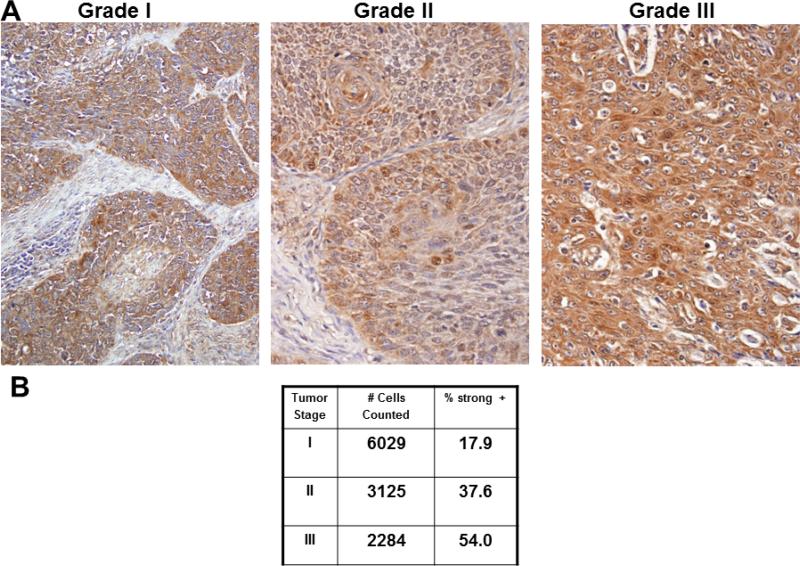
As qrtPCR and immunohistochemistry validated the differential expression of several NF-κB target genes, activation of the NF-κB pathway was assessed in cells and in tumors generated from aggressive SCC25-PD. In initial experiments, localization of p65/RelA was examined in SCC25-PD and SCC25-WD cells by immunofluorescence. While very few nuclei show positive staining for RelA in SCC25-WD cells (Fig. 3A), a significant increase in RelA nuclear occupancy is observed in the more aggressive cell line SCC-PD (Fig. 3B,C). This is confirmed by western blotting of nuclear extracts, showing nuclear localization of RelA in SCC25-PD cells (Fig. 3D). In support of the cellular data, similar results were observed in murine orthotopic tumors, with higher levels of nuclear RelA observed in poorly differentiated tumors (Fig. 4B) relative to well differentiated lesions (Fig. 4A, E). As phosphorylation of RelA is required for transcription of a subset of NFkB target genes (17), tumors were also examined for RelA phosphorylation. Enhanced staining of phospho-Ser276 RelA was also observed in tumors generated from SCC25-PD relative to SCC25-WD (Fig. 4C,D,F; Suppl. Table 2). Human tongue SCCs also exhibit activated NF-κB signaling, as evidenced by enhanced staining of nuclear RelA with increasing tumor stage (Fig. 5A,B; Suppl. Table 3; Suppl. Table 4).
Figure 3. Enhanced RelA nuclear translocation in SCC25-PD.
(A,B) Immunofluorescence evaluation of RelA in (A) SCC25-WD cells and (B) SCC25-PD cells. (C) Quantitation of nuclear RelA staining. (D) Representative western blot of nuclear extracts from SCC25-WD (WD) and SCC25-PD (PD), as indicated, probed with antibodies directed against RelA (upper panel) and HDAC1 (lower panel). Experiment was performed in triplicate.
Figure 4. Nuclear staining for RelA and phospho-RelA(Ser276) is enhanced in SCC25-PD tumors relative to SCC25-WD tumors.
(A,B) Immunohistochemical analysis of tumor section from orthotopic (tongue) mouse xenograft model initiated with (A) SCC25-WD cells or (B) SCC25-PD cells stained with antibodies directed against RelA. (C,D) Immunohistochemical analysis of tumor section from orthotopic murine xenograft initiated with (C) SCC25-WD cells and (D) SCC25-PD cells stained with antibodies directed against phospho-RelA(Ser276). Magnification – 200X. (E) Quantitation of nuclear RelA staining. (F) Quantitation of nuclear phospho-RelA(Ser276) staining. (Detailed quantitation results are shown in Supplementary Table 2).
Figure 5. Immunohistochemical analysis of RelA staining in human tongue tumors.
(A) Representative image fields from staged human tongue SCC tissue microarray stained with antibodies directed against RelA. (B) Quantitation of staining as described in Materials and Methods. %strong positive is the sum of the percentage of cells staining 2+ and 3+.
Inhibition of NF-κB signaling alters nuclear localization, invasion and target gene expression
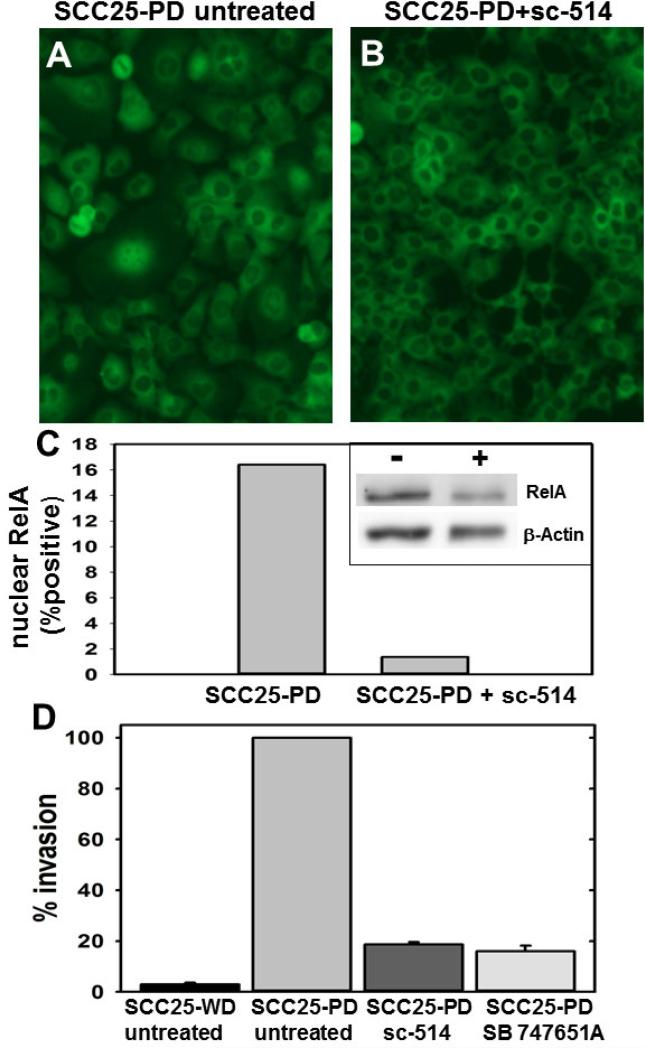
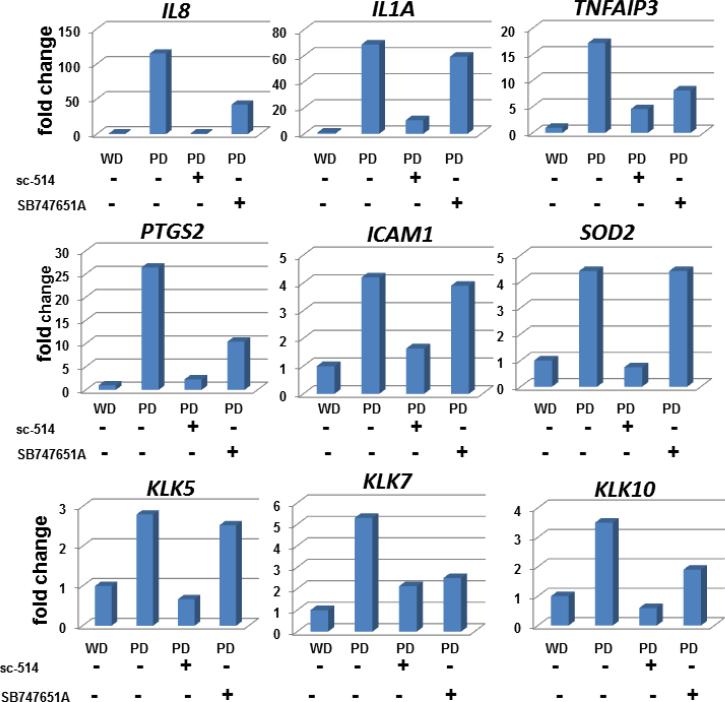
It has been demonstrated that post-translational modification of the RelA subunit is required for efficient gene transactivation at the promoters of many genes activated by NF-κB signaling (16). Phosphorylation of the RelA subunit at Ser276 or Ser536 is required for the transcription of distinct subsets of NF-κB target genes (17). In control experiments, SCC25-WD cells treated with TNFα and the IKKβ-inhibitor sc-514 completely abrogated the TNFα-induced nuclear translocation of RelA (Suppl. Fig. 1). A similar response was observed with SCC25-PD, demonstrating significantly diminished constitutive RelA nuclear translocation (Fig.6A-C). The decrease in nuclear RelA was confirmed by western blotting of nuclear extracts (Fig. 6C, inset). Orthotopic tumors formed from SCC25-PD show invasive behavior in vivo, exhibiting both perineural and lymphovascular invasion (Fig. 1). To examine the effect of inhibition of NF-κB signaling on invasive behavior, the ability of SCC25-PD cells to invade Matrigel was examined in the presence or absence of inhibitors. Treatment of SCC25-PD cells with either the IKKβ-inhibitor sc-514 or the MSK1/2 inhibitor SB 74751A greatly diminished the invasive character of SCC25-PD cells (Fig.6D). Furthermore, NF-κB target gene upregulation was significantly reduced in SCC25-WD cells for all the genes studied after treatment with the IKKβ inhibitor sc-514 (Fig.7). Expression levels of some mRNAs were reduced below the levels seen in control SCC25-WD cells. Treatment of SCC25-PD with SB 747651A, which has no effect on RelA nuclear translocation but prevents MSK1-mediated RelA phosphorylation at Ser276, also significantly diminished the mRNA upregulation of 6 of the 9 genes studied (Fig. 7).
Figure 6. Inhibitors reverse nuclear translocation of RelA and block invasion.
(A,B) Cells were treated as indicated and processed for immuno-fluorescence microscopy with antibodies directed against RelA. Experiment was performed in triplicate. (A) SCC25-PD cells, untreated. (B) SCC25-PD cells treated with sc-514 (10 μM, 25 h). (C) Graph showing percentage of cells with positive nuclear RelA in control and sc-514-treated SCC25-PD cells, as indicated. (inset) Western blot of nuclear extracts of control (-) and sc-514-treated (+) SCC25-PD cells probed with antibodies directed against RelA (upper panel) and β-actin (lower panel). (D) Analysis of invasion. Cells were untreated or treated, as indicated, prior to addition to a Boyden chamber containing Matrigel as described in Experimental Procedures. Invading cells were enumerated and results shows as % of invasion relative to untreated SCC25-PD cells (designated 100%). (sc-514, 10 uM; SB 747651A, 20 uM)
Figure 7. Effect of NF-κB pathway inhibitors on target gene upregulation.
Quantitative rtPCR was used to determine mRNA levels for the indicated genes in (column 1) untreated SCC25-WD cells, (column 2) untreated SCC25-PD cells, (column 3) SCC25-PD cells treated with sc-514 (10 μM, 25 h), and (column 4) SCC25-PD cells treated with SB747651A (20μM, 25 h). Graphs show fold change relative to untreated SCC25-WD cells (designated as ‘1’). Experiments were performed in triplicate.
Discussion
In addition to the effects of NF-κB pathway deregulation on malignant transformation, anti-apoptosis, promotion of cell growth and acquired chemoresistance, the link between NF-κB activation and invasion and metastasis has been investigated in several different cancers (17). In OSCC, overexpression of RelA is sufficient to induce EMT (19) and constitutive activation of NF-κB in oral cancer has also been reported (3,20-22). It should be noted, however, that some authors define “constitutive activation of NF-κB” as nuclear translocation of NF-κB dimers (3,21), while others refer to an increase in cytoplasmic or whole cell NF-κB subunits (20,23-24), or an increase in nuclear or cytoplasmic phospho-RelA(Ser276) or phospho-RelA(Ser536) as evidence of NF-κB activation (22). For the purposes of this study, activation is defined as an upregulation in nuclear RelA.
In a related study in oral cancer, nuclear NF-κB staining was detected in 52% of primary head and neck squamous cell carcinoma (HNSCC) tumors with positive nodes compared to only 23% of primary HNSCC tumors without positive nodes and blockade of NF-κB decreased invasion and metastasis (3,25). The results presented here are consistent with published data, as activation of NF-κB in aggressive SCC25-PD cells correlated with significantly enhanced invasive potential which was attenuated more than 80% by NF-κB inhibitors. Although ethical and institutional ACUC regulations preclude growth of tongue tumors in this model of sufficient size to generate cervical lymph node or distant metastases, the tumors initiated with these cells in the xenograft mouse model exhibited perineural and vascular invasion, which has been shown to be associated with lymph node metastasis and poor prognosis in human OSCC (26-27). Furthermore, human tumors exhibiting the cord-like and single cell type pattern of invasion seen in the murine SCC25-PD tumors are characterized by poor survival and lymph node metastasis (28-29).
Highly aggressive OSCC tumors generated by SCC25-PD demonstrate upregulation of a large number of NF-κB target genes, prompting an investigation of NF-κB regulation. Our results indicate that nuclear RelA is significantly increased in aggressive SCC25-PD cells, correlating with enhanced invasiveness and upregulation of 59 invasion and metastasis genes (7). Invasiveness was diminished by inhibiting the NF-κB pathway with sc-514. This inhibition also resulted in a reversal of RelA nuclear translocation and a significant decrease in NF-κB target gene transcription. Interestingly, most target genes in our panel required long-term treatment with inhibitor before a significant decrease in transcription was obtained. This could be due to the fact that constitutive activation of NF-κB can lead to prolonged IKK activation and repeated cycles of RelA nuclear occupancy (30), since SCC25-PD RelA nuclear occupancy was also undiminished at earlier time points (data not shown). The specific IKKβ-inhibitor sc-514 delays IκBα phosphorylation, slows and decreases RelA nuclear import, increases the rate of RelA nuclear export, and prevents IKKβ-mediated RelA phosphorylation on Ser536 (31). In the NF-κB constitutively active cell line SCC25-PD, sc-514 was a very effective inhibitor of gene activation for all 10 of the mRNAs studied.
In unstimulated cells, p50 homodimers associated with the histone deacetylase HDAC-1 repress transcription of NF-κB target genes (14). Upon stimulation of the canonical pathway, RelA-containing dimers enter the nucleus and displace these complexes and activate transcription. However, RelA phosphorylation is required for full transactivation of many NF-κB target genes. For these genes, phosphorylation on Ser536 and/or Ser276 is required for RNA polymerase II recruitment, enhanceosome formation and maximal gene transactivation (11,13,14,16,17,32-34). Since IKKβ is the primary kinase that phosphorylates RelA on Ser536 in the canonical pathway (15,35-37), and the IKKβ inhibitor sc-514 prevents this modification, we elected to also include an inhibitor of RelA-Ser276 phosphorylation in our investigation. Treatment with the inhibitor SB 747651A, which does not affect RelA nuclear translocation but does prevent MSK1-mediated RelA phosphorylation at Ser276 in the nucleus, significantly diminished mRNA expression of 6 of the 9 tested NF-κB target genes and was also an effective inhibitor of SCC25-PD cell invasiveness. This is consistent with published data showing that RelA-Ser276 phosphorylation results in the upregulation of a subset of NF-κB target genes and is critical for the TNF- and IL1-mediated inflammatory response. Phosphorylation on Ser276 is required for recruitment of CREB binding protein (CBP)/p300 and the positive transcription elongation factor b on some NF-κB targets which facilitates recruitment of polymerase II and drives transcription (14,39-40). Also, phosphorylation of RelA/p65 at Ser276 prevents its constitutive degradation by ubiquitin-mediated proteolysis in the nucleus (41). Under the current experimental conditions, no evidence of increased apoptosis or decreased growth rate was observed using either inhibitor.
MSK1 and the catalytic subunit of PKA (PKAc) are both known to phosphorylate RelA at Ser276 (14,42), dependent on the cell type and stimulus. There is strong evidence that a ROS-dependent PKAc pathway phosphorylates Ser276 through an NF-κB canonical pathway in some cell types (40). However, even when the same stimulus is investigated, there is some disagreement in the literature on which kinase is involved (43). In fact, a third kinase Pim1 has also been shown to phosphorylate RelA at Ser276 in response to TNFα in some cell lines (41). We performed preliminary experiments to examine the potential ROS-related NF-κB activation by abrogating the pathway using DMSO (0.5, 1, and 2%) but found no inhibition of target gene activation (data not shown). Since this pathway generally involves PKAc as the primary kinase that phosphorylates RelA at Ser276 (14,40), we chose an inhibitor that was specific to the other major kinase known to phosphorylate RelA at Ser276, the nuclear kinase MSK1 (33,43,44). The specific MSK inhibitor SB 747651A does not affect IκBα phosphorylation and degradation, or nuclear translocation of RelA-containing dimers (45). The only known molecular effect on NF-κB signaling is the prevention of RelA phosphorylation on Ser276. However, SB 747651A was an effective inhibitor of mRNA transcription for 6 of the 9 genes studied, which include genes previously shown to be activated by phospho-RelA(Ser276)-containing dimers such as IL-8, Cox-2 and TNFAIP3 (16,39,46).
Although the upstream causes of NF-κB activation in OSCC are varied and unknown in most clinical cases, inhibitors of IKKβ such as sc-514 represent a promising group of therapeutic agents. IKKβ null mice closely resemble RelA null mice phenotypically, and most activators of NF-κB require functional IKKβ (17). In the canonical pathway, IKKβ is also the kinase that typically phosphorylates RelA on Ser536, a post-translational modification that is required for full transactivation of some NF-κB targets. Also, inhibition of the transcription factor NF-κB is attractive since NF-κB is now known to be involved in numerous stages in carcinoma progression. NF-κB activation in human cancer is not limited to the tumor cells themselves, as several types of tumor-associated macrophages are NF-κB-dependent and release cytokines which orchestrate angiogenesis and further activate NF-κB signaling in tumor cells (47). Furthermore, the anti-cancer effects of IKKβ inhibition extend to other pathways. Phosphorylation of TSC-1 by IKKβ activates the mTOR pathway and enhances angiogenesis, and phosphorylation of FOXO3a induces its degradation, leading to enhanced cell proliferation and survival (48). The inhibitor sc-514 has been shown to suppress TNFα-promoted metastasis of murine colon adenocarcinoma cells (49). The administration of NF-κB pathway inhibitors in cancer is not without challenges. IKKβ-inhibitors are unlikely to be effective for cancers activated by the non-canonical pathway, which is characterized by signaling through IKKα. Normal B lymphocytes are constitutively active for NF-κB, which is required for their survival, and NF-κB is required for T and B cell development and immune response (50). The administration of any IKKβ inhibitor will have cell-type and dose-specific effects. Inhibition of IKKβ by sc-514 does not completely abrogate the canonical NF-κB pathway, and the successful clinical employment of sc-514 will likely depend on establishment of a dose-escalation endpoint that achieves sufficient NF-κB inhibition while maintaining immune function. Successful use of these compounds will necessitate further elucidation of the molecular details of NF-κB pathway activation in OSCC.
Supplementary Material
Acknowledgement
This work was supported by grant RO1 CA085870 to M.S.S. from the National Institutes of Health/National Cancer Institute.
List of abbreviations
- NF-κB
nuclear factor kappa B
- OSCC
oral squamous cell carcinoma
- HNSCC
head and neck squamous cell carcinoma
- SCC25-PD
subline of squamous cell carcinoma 25 cell line that forms poorly differentiated tumors in vivo
- SCC25-WD
subline of squamous cell carcinoma 25 cell line that forms well differentiated tumors in vivo
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors have no conflict of interest to disclose.
References
- 1.Sano D, Myers JN. Metastasis of squamous cell carcinoma of the oral tongue. Can. Met. Rev. 2007;26(3-4):645–62. doi: 10.1007/s10555-007-9082-y. [DOI] [PubMed] [Google Scholar]
- 2.Rusthoven K, Ballonoff A, Raben D, Chen C. Poor prognosis in patients with stage I and II oral tongue squamous cell carcinoma. Cancer. 2008;112(2):345–51. doi: 10.1002/cncr.23183. [DOI] [PubMed] [Google Scholar]
- 3.Yan M, Xu Q, Zhang P, Zhou XJ, Zhang ZY, Chen WT. Correlation of NF-κB signal pathway with tumor metastasis of human head and neck squamous cell carcinoma. BMC Cancer. 2010;10:437. doi: 10.1186/1471-2407-10-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Myers JN, Holsinger FC, Jasser SA, Bekele BN, Fidler IJ. An orthotopic nude mouse model of oral tongue squamous cell carcinoma. Clin Cancer Res. 2002;8:293–8. [PubMed] [Google Scholar]
- 5.Ziober AF, Patel KR, Alawi F, Gimotty P, Weber RS, Feldman MM, Chalian AA, Weinstein GS, Hunt J, Ziober BL. Identification of a gene signature for rapid screening of oral squamous cell carcinoma. Clin Cancer Res. 2006;12(20 Pt 1):5960–71. doi: 10.1158/1078-0432.CCR-06-0535. [DOI] [PubMed] [Google Scholar]
- 6.Nagata M, Fujita H, Ida H, Hoshina H, Inoue T, Seki Y. Identification of potential biomarkers of lymph node metastasis in oral squamous cell carcinoma by cDNA microarray analysis. Int J Cancer. 2003;106(5):683–9. doi: 10.1002/ijc.11283. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh S, Koblinski J, Johnson J, Liu Y, Ericsson A, Davis JW, Shi Z, Ravosa MJ, Crawford S, Frazier S, Stack MS. Urinary-type plasminogen activator receptor/alpha 3 beta 1 integrin signaling, altered gene expression, and oral tumor progression. Mol Cancer Res. 2010;8(2):145–58. doi: 10.1158/1541-7786.MCR-09-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.NF-KB Transcription Factors Home/The Gilmore Lab. Boston University; www.bu.edu/NF-KB [Internet]. Available at www.bu.edu/NF-KB/gene-resources/target-genes/ [Google Scholar]
- 9.Chung CH, Parker JS, Karaca G, Wu J, Funkhouser WK, Moore D, Butterfoss D, Xiang D, Zanation A, Yin X, Shockley WW, Weissler MC, Dressler LG, Shores CG, Yarbrough WG, Perou CM. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5(5):489–500. doi: 10.1016/s1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, Su L, Pirani AA, Wu H, Zhang H, Shin DM, Gernert KM, Chen ZG. Understanding metastatic SCCHN cells from unique genotypes to phenotypes with the aid of an animal model and DNA microarray analysis. Clin Exp Metastasis. 2006;23(3-4):209–22. doi: 10.1007/s10585-006-9031-0. [DOI] [PubMed] [Google Scholar]
- 11.Perkins ND. The diverse and complex roles of NF- B subunits in cancer. Nat Rev Cancer. 2012;12(2):121–32. doi: 10.1038/nrc3204. [DOI] [PubMed] [Google Scholar]
- 12.Chaturvedi MM, Sung B, Yadav VR, Kannappan R, Aggarwal BB. NF-κB addiction and its role in cancer: ‘one size does not fit all’. Oncogene. 2011;30(14):1615–30. doi: 10.1038/onc.2010.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oeckinghaus A, Ghosh S. The NF-κB Family of Transcription Factors and Its Regulation. Cold Spring Harb. Perspect. Biol. 2009;1(4):a000034. doi: 10.1101/cshperspect.a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong H, May MJ, Jimi E, Ghosh S. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol Cell. 2002;9(3):625–36. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- 15.Perkins ND. Integrating cell-signaling pathways with NF-κB and IKK function. Nat Rev Mol Cell Biol. 2007;8(1):49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 16.Arun P, Brown MS, Ehsanian R, Chen Z, Van Waes C. Nuclear NF-kappaB p65 phosphorylation at serine 276 by protein kinase A contributes to the malignant phenotype of head and neck cancer. Clin Cancer Res. 2009;15(19):5974–84. doi: 10.1158/1078-0432.CCR-09-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayden MS, Ghosh S. Shared Principles in NF-κB Signaling. Cell. 2008;132(3):344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 18.Pettus JR, Johnson JJ, Shi Z, Davis JW, Koblinski J, Ghosh S, Liu Y, Ravosa MJ, Frazier S, Stack MS. Multiple kallikrein (KLK 5, 7, 8, and 10) expression in squamous cell carcinoma of the oral cavity. Histol Histopathol. 2009;24(2):197–207. doi: 10.14670/hh-24.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Julien S, Puig I, Caretti E, Bonaventure J, Nelles L, van Roy F, Dargemont C, de Herreros AG, Bellacosa A, Larue L. Activation of NF-kappaB by Akt upregulates Snail expression and induces epithelium mesenchyme transition. Oncogene. 2007;26(53):7445–56. doi: 10.1038/sj.onc.1210546. [DOI] [PubMed] [Google Scholar]
- 20.Nakayama H, Ikebe T, Beppu M, Shirasuna K. High expression levels of nuclear factor kappaB, IkappaB kinase alpha and Akt kinase in squamous cell carcinoma of the oral cavity. Cancer. 2001;92(12):3037–44. doi: 10.1002/1097-0142(20011215)92:12<3037::aid-cncr10171>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 21.Español A, Dasso M, Cella M, Goren N, Sales ME. Muscarinic regulation of SCA-9 cell proliferation via nitric oxide synthases, arginases and cyclooxygenases. Role of the nuclear translocation factor-κB. Eur J Pharmacol. 2012;683(1-3):43–53. doi: 10.1016/j.ejphar.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Furuta H, Osawa K, Shin M, Ishikawa A, Matsuo K, Khan M, Aoki K, Ohya K, Okamoto M, Tominaga K, Takahashi T, Nakanishi O, Jimi E. Selective inhibition of NF-κB suppresses bone invasion by oral squamous cell carcinoma in vivo. Int J Cancer. 2012;131(5):E625–35. doi: 10.1002/ijc.27435. [DOI] [PubMed] [Google Scholar]
- 23.Carbone C, Melisi D. NF-κB as a target for pancreatic cancer therapy. Expert Opin Ther Targets. 2012;16(Suppl 2):S1–10. doi: 10.1517/14728222.2011.645806. [DOI] [PubMed] [Google Scholar]
- 24.Lun M, Zhang PL, Pellitteri PK, Law A, Kennedy TL, Brown RE. Nuclear factor-kappaB pathway as a therapeutic target in head and neck squamous cell carcinoma: pharmaceutical and molecular validation in human cell lines using Velcade and siRNA/NF-kappaB. Ann Clin Lab Sci. 2005;35(3):251–8. [PubMed] [Google Scholar]
- 25.Tanaka T, Nakayama H, Yoshitake Y, Irie A, Nagata M, Kawahara K, Takamune Y, Yoshida R, Nakagawa Y, Ogi H, Shinriki S, Ota K, Hiraki A, Ikebe T, Nishimura Y, Shinohara M. Selective inhibition of nuclear factor-κB by nuclear factor-κB essential modulator-binding domain peptide suppresses the metastasis of highly metastatic oral squamous cell carcinoma. Cancer Sci. 2012;103(3):455–63. doi: 10.1111/j.1349-7006.2011.02174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dissanayaka WL, Pitiyage G, Kumarasiri PV, Liyanage RL, Dias KD, Tilakaratne WM. Clinical and histopathologic parameters in survival of oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol. 2012;113(4):518–25. doi: 10.1016/j.oooo.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Jerjes W, Upile T, Petrie A, Riskalla A, Hamdoon Z, Vourvachis M, Karavidas K, Jay A, Sandison A, Thomas GJ, Kalavrezos N, Hopper C. Clinicopathological parameters, recurrence, Head Neck locoregional and distant metastasis in 115 T1-T2 oral squamous cell carcinoma patients. Oncol. 2010;2:9. doi: 10.1186/1758-3284-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondoh N, Ishikawa T, Ohkura S, Arai M, Hada A, Yamazaki Y, Kitagawa Y, Shindoh M, Takahashi M, Ando T, Sato Y, Izumo T, Hitomi K, Yamamoto M. Gene expression signatures that classify the mode of invasion of primary oral squamous cell carcinomas. Mol Carcinog. 2008;(10):744–56. doi: 10.1002/mc.20431. [DOI] [PubMed] [Google Scholar]
- 29.Kaihara T, Kusaka T, Kawamata H, Oda Y, Fujii S, Morita K, Imura J, Fujimori T. Decreased expression of E-cadherin and Yamamoto-Kohama's mode of invasion highly correlates with lymph node metastasis in esophageal squamous cell carcinoma. Pathobiology. 2001;69(3):172–8. doi: 10.1159/000048772. [DOI] [PubMed] [Google Scholar]
- 30.Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, Edwards SW, McDowell HP, Unitt JF, Sullivan E, Grimley R, Benson N, Broomhead D, Kell DB, White MR. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306(5696):704–8. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 31.Kishore N, Sommers C, Mathialagan S, Guzova J, Yao M, Hauser S, Huynh K, Bonar S, Mielke C, Albee L, Weier R, Graneto M, Hanau C, Perry T, Tripp CS. A selective IKK-2 inhibitor blocks NF-kappa B-dependent gene expression in interleukin-1 beta-stimulated synovial fibroblasts. J Biol Chem. 2003;278(35):32861–71. doi: 10.1074/jbc.M211439200. [DOI] [PubMed] [Google Scholar]
- 32.Hayden MS, Ghosh S. Signaling to NF-κB. Genes Dev. 2004;18(18):2195–224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 33.Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene. 2006;25(51):6717–30. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 34.Sasaki CY, Slemenda CF, Ghosh P, Barberi TJ, Longo DL. Traf1 Induction and Protection from Tumor Necrosis Factor by Nuclear Factor-κB p65 Is Independent of Serine 536 Phosphorylation. Cancer Res. 2007;67(23):11218–25. doi: 10.1158/0008-5472.CAN-07-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madrid LV, Mayo MW, Reuther JY, Baldwin AS., Jr Akt stimulates the transactivation potential of the RelA/p65 Subunit of NF-kappa B through utilization of the I kappa B kinase and activation of the mitogen-activated protein kinase p38. J Biol Chem. 2001;276(22):18934–40. doi: 10.1074/jbc.M101103200. [DOI] [PubMed] [Google Scholar]
- 36.Haller D, Russo MP, Sartor RB, Jobin C. IKK beta and phosphatidylinositol 3-kinase/Akt participate in non-pathogenic Gram-negative enteric bacteria-induced RelA phosphorylation and NF-kappa B activation in both primary and intestinal epithelial cell lines. J Biol Chem. 2002;277(41):38168–78. doi: 10.1074/jbc.M205737200. [DOI] [PubMed] [Google Scholar]
- 37.Jeong SJ, Pise-Masison CA, Radonovich MF, Park HU, Brady JN. Activated AKT regulates NF-kappaB activation, p53 inhibition and cell survival in HTLV-1-transformed cells. Oncogene. 2005;24(44):6719–28. doi: 10.1038/sj.onc.1208825. [DOI] [PubMed] [Google Scholar]
- 38.Okazaki T, Sakon S, Sasazuki T, Sakurai H, Doi T, Yagita H, Okumura K, Nakano H. Phosphorylation of serine 276 is essential for p65 NF-κB subunit-dependent cellular responses. Biochem Biophys Res Commun. 2003;300(4):807–12. doi: 10.1016/s0006-291x(02)02932-7. [DOI] [PubMed] [Google Scholar]
- 39.Nowak DE, Tian B, Jamaluddin M, Boldogh I, Vergara LA, Choudhary S, Brasier AR. RelA Ser276 phosphorylation is required for activation of a subset of NF-kappaB-dependent genes by recruiting cyclin-dependent kinase 9/cyclin T1 complexes. Mol Cell Biol. 2008;28(11):3623–38. doi: 10.1128/MCB.01152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jamaluddin M, Wang S, Boldogh I, Tian B, Brasier AR. TNF-α-induced NF-κB/RelA Ser276 phosphorylation and enhanceosome formation is mediated by an ROS-dependent PKAc pathway. Cell Signal. 2007;19(7):1419–33. doi: 10.1016/j.cellsig.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 41.Nihira K, Ando Y, Yamaguchi T, Kagami Y, Miki Y, Yoshida K. Pim-1 controls NF-kappaB signalling by stabilizing RelA/p65. Cell Death Differ. 2010;17(4):689–98. doi: 10.1038/cdd.2009.174. [DOI] [PubMed] [Google Scholar]
- 42.Reber L, Vermeulen L, Haegeman G, Frossard N. Ser276 phosphorylation of NF-κB p65 by MSK1 controls SCF expression in inflammation. PLoS One. 2009;4(2):e4393. doi: 10.1371/journal.pone.0004393. doi: 10.1371/journal.pone.0004393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moynagh PN. The NF-kappaB pathway. J Cell Sci. 2005;118(Pt 20):4589–92. doi: 10.1242/jcs.02579. [DOI] [PubMed] [Google Scholar]
- 44.Vermeulen L, De Wilde G, Van Damme P, Vanden Berghe W, Haegeman G. Transcriptional activation of the NF-κB p65 subunit by mitogen- and stress-activated protein kinase-1(MSK1). EMBO J. 2003;22(6):1313–24. doi: 10.1093/emboj/cdg139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naqvi S, Macdonald A, McCoy CE, Darragh J, Reith AD, Arthur JS. Characterization of the cellular action of the MSK inhibitor SB-747651A. Biochem J. 2012;441(1):347–57. doi: 10.1042/BJ20110970. [DOI] [PubMed] [Google Scholar]
- 46.Sundar IK, Chung S, Hwang JW, Lapek JD, Jr, Bulger M, Friedman AE, Yao H, Davie JR, Rahman I. Mitogen- and stress-activated kinase 1 (MSK1) regulates cigarette smoke-induced histone modifications on NF-κB-dependent genes. PLoS One. 2012;7(2):e31378. doi: 10.1371/journal.pone.0031378. doi: 10.1371/journal.pone.0031378. Epub 2012 Feb 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bassères DS, Baldwin AS. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25(51):6817–30. doi: 10.1038/sj.onc.1209942. [DOI] [PubMed] [Google Scholar]
- 48.Israel A. The IKK Complex, A Central Regulator of NF-κB Activation. Cold Spring Harb Perspect Biol. 2010;2(3):a000158. doi: 10.1101/cshperspect.a000158. doi: 10.1101/cshperspect.a000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choo MK, Sakurai H, Kim DH, Saiki I. A ginseng saponin metabolite suppresses tumor necrosis factor-alpha-promoted metastasis by suppressing nuclear factor-kappaB signaling in murine colon cancer cells. Oncol Rep. 2008;19(3):595–600. [PubMed] [Google Scholar]
- 50.Olivier S, Robe P, Bours V. Can NF-κB be a target for novel and efficient anti-cancer agents? Biochem Pharmacol. 2006;72(9):1054–68. doi: 10.1016/j.bcp.2006.07.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.