We report on two novel methods of creating complex multiphase tissue constructs from discrete extracellular matrix microenvironments using centrifugation and vacuum molding. In general, the broad field of tissue engineering encompasses a variety of methodologies for creating biological tissues for the replacement or repair of injured or diseased tissues. Modular tissue engineering attempts to create tissues by generating larger structures from repeating subunits, or “modules”.[1] These repeating subunits can be designed to have unique microarchitectural features, and allow for the bottom-up creation of macro-scale tissues. One advantage presented by the modular tissue engineering method is the ability to create complex tissues with precisely designed morphologies and spatially controlled patterning. However, a current limitation of this technology is providing cells with appropriate functional extracellular matrices, which are critical for the development and differentiation of cells into desired tissue types. This paper presents facile techniques by which modular microenvironments comprised of collagen-chitosan microbeads 200–300 μm in diameter can be assembled into larger-scale constructs with defined spatial arrangement. Two methods are presented: vacuum molding and centrifugation. The ability to control the environment surrounding cells in this manner allows for patterning of cellular microenvironments for the study of cell interactions, and this method may be extended to generation of complex tissues.
Assembly of cellular microenvironments into macroscopic, patterned, engineered tissues is a growing area of research. Previously explored methods include cell printing using jet-based 3D printers,[2] self-assembly,[3] bioprinting,[4] robotic 3D printing,[5] microtissue engineering,[6] and cell sheet engineering.[7] The generation of shaped, multilayered tissues containing multiple cell types has applications in developing a wide a variety of complex tissues and organs, with the first efforts being applied to generating vascularized bone tissue and osteochondral interfaces.[1,8] These efforts are progressing as new materials, assembly techniques and cell culture methods are developed. However, there is still a need for more robust approaches to the construction of three dimensional macro-scale tissue constructs with defined architecture, and there is a particular challenge in creating these structures from physiologically relevant extracellular matrix components.
Microscale hydrogels[9] and similar cellular microenvironments have previously been used in the modular tissue engineering approach. Our lab has created hydrogel “microbeads” comprised of cells embedded in spherical modules of defined extracellular matrix proteins and polysaccharides, including collagen, fibrin, agarose, and chitosan.[10–12] Chitosan and collagen are of particular interest, as these naturally derived materials are both generally biodegradable and biocompatible,[13–15] and have been investigated for numerous applications in vascular,[16] skin,[17] ligament,[18] and bone[19] tissue engineering. In general, the function of cells embedded within microbeads can be guided by defining the extracellular matrix composition, incorporating growth factors into the matrix, and by controlling the cell density and mechanical properties of the matrix. In addition, the properties of the microbeads themselves (density, stiffness, cohesiveness) can be modulated to control their assembly into larger scale structures.
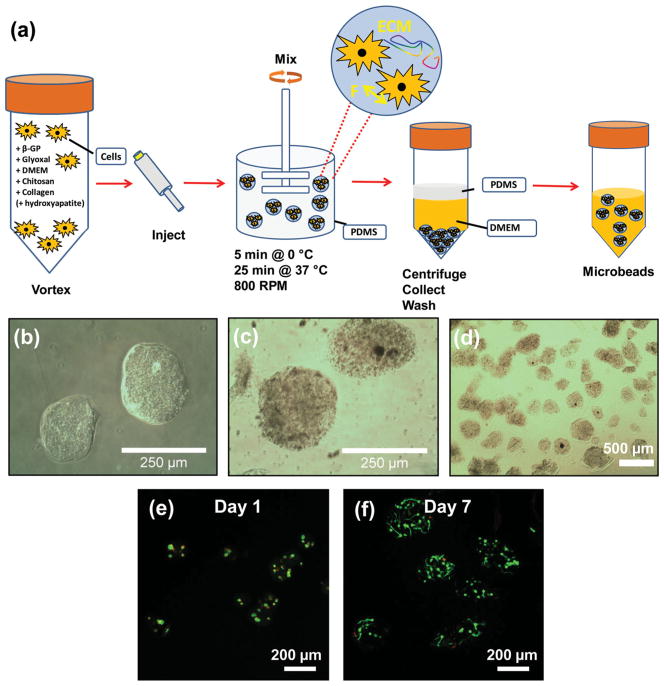
In this study, microbeads were created from a composite of collagen Type I and chitosan using the process shown in Figure 1. To prepare microbeads, collagen and chitosan were mixed with culture medium, glyoxal, and beta-glycerophosphate (β-GP) to create a 65/35 (mass ratio) collagen/chitosan embedding matrix. Glyoxal is a small aldehyde that is less cytotoxic than glutaraldehyde.[20] β-GP was used to initiate gelation of chitosan/collagen matrix.[14,21] The liquid matrix solution was combined with cells or other components to be embedded, and the mixture was emulsified in a rapidly stirred, ice-cold poly-dimethylsiloxane (PDMS) bath. Emulsification produced spherical droplets 200–300 μm in diameter, which were subsequently gelled into microbeads by raising the temperature (Figure 1a). The gelled microbeads (Figures 1b–1d) were collected and washed for use in experiments, and were shown to maintain the viability of embedded cells (Figures 1e, 1f).
Figure 1.
a) Schematic illustration of microbead fabrication process. b, c) Light microscopy images of 65–35 wt% collagen-chitosan microbeads (panel c show microbeads containing 2.5 mg/mL hydroxyapatite). d) Light microscopy of a population of collagen/chitosan microspheres showing their generally spheroidal morphology. e, f) Fluorescence microscopy of human mesenchymal stem cells embedded in 65–35 wt% collagen-chitosan microbeads at day 1 and day 7 of culture, respectively. The cytoplasm of living cells is stained green, and the nuclei of dead cells are stained red.
The microbead matrix was also augmented at the time of emulsification in selected experiments to facilitate assembly and visualization of macroscale constructs. Hydroxyapatite was added to certain microbead formulations to increase their density, and to thereby facilitate collection from the oil and subsequent use in centrifugation-based assembly. Hydroxyapatite is the mineral component of bone and has been shown to promote osteogenic differentiation of progenitor cells.[22] Fluorescent microspheres were embedded in acellular constructs to allow visualization under visible and UV light. Cellular constructs contained human fibroblasts cells genetically modified to express green and red fluorescent protein for visualization.
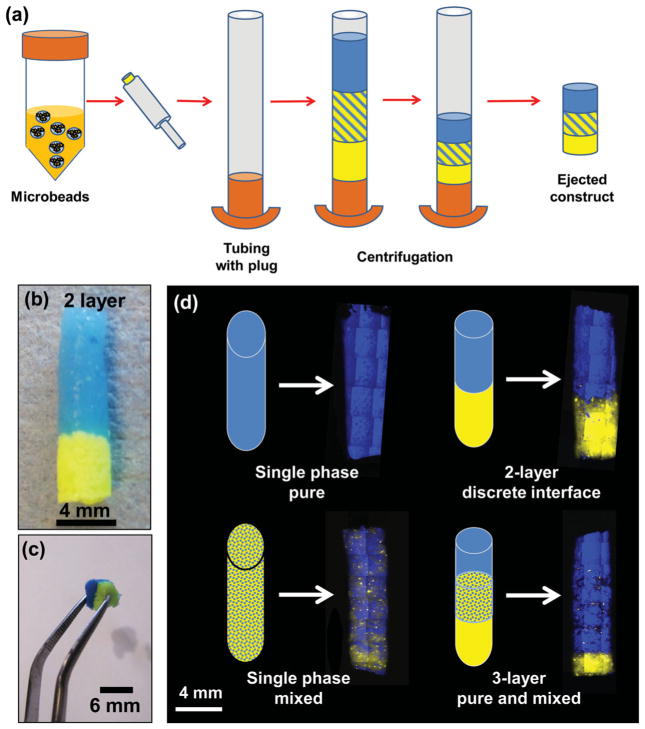
Centrifugation-based assembly of macroscale constructs was accomplished by adding concentrated suspensions of microbeads to a capped tubular container, and then centrifuging to remove the liquid phase, as shown in Figure 2. The centrifugally compacted constructs could then be extruded from the tube to create solid hydrogel plugs with the desired microbead distribution. Figure 2a schematically shows an example of creating a 3-layer construct, and Figure 2b shows an assembled 2-layer construct. The resulting multiphase constructs were cohesive, maintained their shape, and could be easily handled without disrupting their structure. Figure 2 c shows a two-phase construct being lifted with forceps, and constructs could be manipulated and transferred for further processing. This method was used to create several layered architectures (imaged under fluorescence in Figure 2 c), including single phase, two-phase with a discrete interface, two-phase mixed, and three-phase layered constructs. The centrifugation method of assembling constructs is simple and rapid, and has the advantage that it can be done in a closed system to preserve sterility. In addition, we have shown that it can be used to create mixed co-cultures and it may be useful in creating tissue interfaces. However, it is essentially limited to layered construct architectures.
Figure 2.
a) Schematic illustration of centrifugation method for construct assembly. b) Image of 2-layer construct created using blue microbeads and yellow microbeads with hydroxyapatite. c) Image of vacuum-molded multiphase construct being handled with forceps. d) Fluorescence imaging of single phase and layered multiphase constructs. Schematic shows intended pattern of microbead distribution and mosaic reconstruction of fluorescent images shows the construct created.
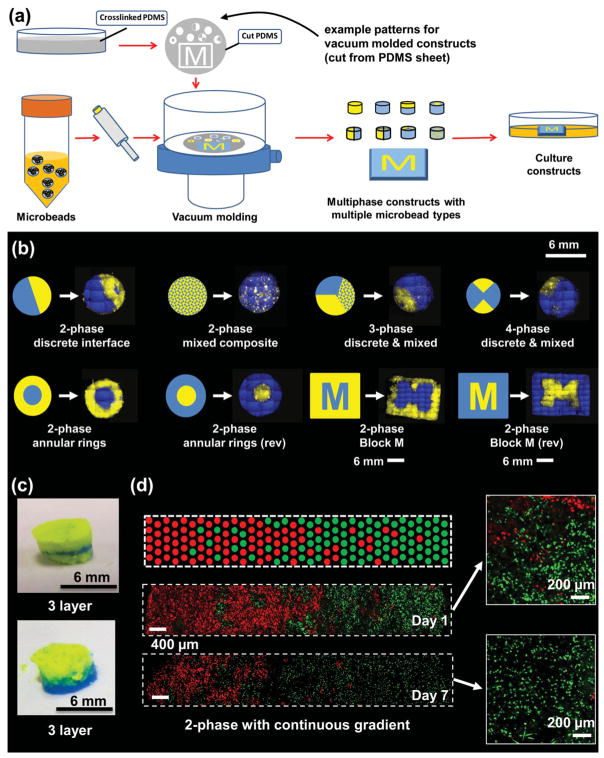
Vacuum molding was also used to create multiphase microbead constructs, as shown in Figure 3. PDMS elastomer was used as a molding template because wells of the desired shape could be easily cut from this material, which is also biologically inert and maintains its form after deformation.[23–25] Wells were constructed in a variety of shapes, including multi-part disks, annular rings, as well as the Michigan “Block M” logo. A PDMS sheet containing these patterns was overlaid on a membrane filter with aspiration from below. To create constructs, microbeads were suspended in solution and then transferred into the molds with mild suction applied from below (Figure 3a). Removal of the interstitial liquid between microbeads resulted in cohesive constructs that retained their shape upon removal from the wells. Multiphase constructs could be formed by sequential disassembly of the PDMS mold to fill in only the portions desired.
Figure 3.
(a) Annotated schematic illustration of vacuum molding method for construct assembly. b) Fluorescence imaging of patterned multiphase constructs using mixed and pure microbead preparations. Schematic shows intended pattern and mosaic reconstruction of fluorescent images shows the construct created. c) Images of layered constructs created using vacuum molding. d) Fluorescent imaging of graded microbead constructs containing red- and green-labeled fibroblast cells and day 1 and day 7 in culture. Schematic shows approximation of intended gradient across construct. Insets show higher magnification view of embedded cells.
Constructs with various geometries were created using the vacuum molding method. Figure 3B shows fluorescently illuminated examples of multiphase constructs containing both blue and yellow microbeads. Pure phases could be easily assembled in patterns with discrete interfaces, in these examples either as pie-shaped wedges in a disk or as annular rings. Microbeads could also be combined into uniformly mixed composite phases, which in turn could be assembled with pure phases. Finally, a Block M logo was used to demonstrate the ability to create arbitrary patterns using the same molding technique. It should be noted that the patterns were not perfect, and displayed some distortion from the intended pattern. This variation is partially a result of imperfect addition of microbeads to the wells, as well as the “resolution” of the constructs, which was determined by the size of the microbeads (in this case 200–300 μm). Layered constructs could also be made using this method by sequential addition of microbead preparations (Figure 3c). In all cases, collagen-chitosan constructs were robust enough to be handled with forceps and could easily be cultured as cohesive 3D macro-scale structures.
Fluorescently labeled, living fibroblast cells were also incorporated into microbeads and were patterned to create a gradient across a 3D rectangular construct (Figure 3d) by sequentially adding preparations with changing ratios of red and green cells. The cells remained viable over the 7 day culture period, showing that removal of interstitial fluid in the constructs did not substantially harm the cells. The gradient produced was not ideal, but there was a clear and gradual shift in the predominance of red- or green-labeled cells across the construct. Closer examination of the embedded cells (insets in Figure 3d) showed that they were more elongated at day 7, which suggests that the fibroblasts were attached to and spread within the protein matrix.
The mechanical properties of the constructs depend on the interactions between the constituent microbeads. The collagen-chitosan microenvironments used in the present study formed cohesive pastes in which microbeads presumably adhered to each other through electrostatic protein-protein and/or protein-polysaccharide interactions. The constructs created were cohesive enough to be routinely handled and transferred using standard instruments, and their properties could be modulated by controlling the amount of water that was removed (data not shown). However, the desired mechanical properties will depend on the intended application, and different materials can be used to enhance or diminish interactions of the microbeads. Furthermore, multiphase tissues created using these techniques can be expected to mature under the appropriate conditions in vitro or in vivo, which will lead to matrix deposition, remodeling, and alteration of the mechanical properties.
Our studies have demonstrated that protein-polysaccharide microbeads can be assembled into larger-scale constructs with prescribed architecture. Centrifugation-based assembly is facile and requires little handing of the microbeads, but is limited to layered geometries. Vacuum molding is also relatively simple and rapid, and allows more complex multiphase constructs to be assembled. The patterns that can be produced are limited only by the ability to create the appropriate molds, and we demonstrated millimeter scale resolution in these initial studies. Furthermore, we showed that microbeads support the viability of cells embedded within them, and that cells remain viable after assembly of larger-scale constructs. These methods may have applications in patterning of cells and microenvironments into tissue-like structures. Such models could be used to study cell-cell communication and the effects of defined co-cultures on cell function. In addition, the assembly of microenvironments into prescribed architectures could have utility in creating more complex engineered tissues, in which the spatial composition can be tailored to direct cell function and achieve improved tissue performance.
Experimental Section
Preparation of 65-35 Collagen-Collagen Microbeads: Collagen-chitosan microbeads were prepared using a water-in-oil emulsion procedure. Chitosan (2.0% wt/vol, dissolved in 0.1 N acetic acid) and Type I collagen (4.0 mg/mL dissolved in 0.02 N acetic acid) were mixed, and fluorescent microspheres were added to facilitate visualization of the formed microbeads. Beta-glycerophosphate (β-GP) and 1.0 mM glyoxal were mixed separately. A third solution of DMEM culture medium was also prepared, and contained the cellular component and/or the hydroxyapatite component, as appropriate for the desired microbead formulation. The fibroblasts were previously labeled using a retroviral expression system as previously described[26] and bead preparations were fabricated at 1.0 × 106 cells/mL. To fabricate microbeads, the three solutions were combined and the mixture was injected into a bath of ice-chilled liquid PDMS stirred using an impeller. Once the microbead matrix was fully emulsified, the mixture was transferred to a 37 °C water bath to induce cogelation of the collagen and chitosan phases. Formed microbeads were separated from the oil phase by centrifugation and washing with phosphate buffered saline (PBS) supplemented with Pluronic L101 surfactant.
Assembly of 3D constructs by centrifugation: A 5 cm section of polyvinyl chloride (PVC) tubing with a 4 mm internal diameter and plugged at one end with a metal screw was used as a vessel to hold microbead preparations for centrifugation. Microbeads were injected directly into the tube using a micropipetter, and the tube was placed into a 15 mL centrifuge tube and centrifuged at 200g for 5 minutes. For constructs with multiple layers, this procedure was performed multiple times to arrive at the desired geometry. The formed construct could then be extruded from the tubing by gently plunging using a 4 mm diameter rod.
Assembly of 3D constructs by vacuum molding: PDMS molds were prepared by mixing liquid PDMS (Sylgard) with curing agent in a 10:1 ratio by weight. The mixture was poured into Petri dishes and allowed to set for 2 days to achieve full solidification, and was sterilized under UV light. Molding wells could be made either by putting a positive cast into the PDMS during curing, or by punching the desired shape out of the cured sheet. In this study, a Michigan Block M and rectangular casts were used for positive casting, and circular wells were made using biopsy punches. For creating multiphase constructs, portions of the PDMS were removed from the wells sequentially as microbead preparations were added. In order to create 3D constructs, the PDMS mold was placed on top of a 0.22 μm filter, and a vacuum was pulled through the filter. Microbeads suspended in PBS were injected into the mold with a micropipette and the interstitial liquid was aspirated to waste.
For microbeads containing cells, red and green fluorescently labeled human fibroblasts were grown in culture and then embedded in the microbeads during fabrication. The 3D macroscale constructs produced from the cell-laden microbeads were cultured in fibroblast cell medium in a 37 °C incubator, with medium changes every two days. Images of microbeads and constructs were taken either by visible light microscopy or using fluorescence microscopy. Macroscale fluorescence images visualized the embedded fluorescent microspheres, whereas laser scanning confocal microscopy was used to image inside the cell-laded constructs to visualize the cells. Image processing was performed in ImageJ.
Acknowledgments
This work was supported in part by a National Science Foundation Graduate Research Fellowship (to RRR), and by a grant from the AO Foundation Large Bone Defect Healing Consortium (to JPS). The authors would like to thank Jake Ceccarelli for his assistance in creating fluorescently labeled cells.
References
- 1.Nichol JW, Khademhosseini A. Soft Matter. 2009;5:1312. doi: 10.1039/b814285h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mironov V, Boland T, Trusk T, Forgacs G, Markwald RR. Trends Biotechnol. 2003;21:157. doi: 10.1016/S0167-7799(03)00033-7. [DOI] [PubMed] [Google Scholar]
- 3.Tejavibulya N, Yousseff Jj, Bao B, Ferruccio TM, Morgan JR. Biofabrication. 2011;3:034110. doi: 10.1088/1758-5082/3/3/034110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jakab K, Norotte C, Marga F, Murphy K, Vunjak-Novakovic G, Forgacs G. Biofabrication. 2010;2:022001. doi: 10.1088/1758-5082/2/2/022001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsunaga YT, Morimoto Y, Takeuchi S. Adv Mater. 2011;23:H90. doi: 10.1002/adma.201004375. [DOI] [PubMed] [Google Scholar]
- 6.Kelm JM, Fussenegger M. Adv Drug Deliv Rev. 2010;62:753. doi: 10.1016/j.addr.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Elloumi-hannachi I, Yamato M, Okano T. J Intern Med. 2010;267:54. doi: 10.1111/j.1365-2796.2009.02185.x. [DOI] [PubMed] [Google Scholar]
- 8.Cheng HW, Luk KD, Cheung KM, Chan BP. Biomaterials. 2011;32:1526. doi: 10.1016/j.biomaterials.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 9.Tekin H, Tsinman T, Sanchez JG, Jones BJ, Camci-Unal G, Nichol JW, Langer R, Khademhosseini A. J Am Chem Soc. 2011;133:12944. doi: 10.1021/ja204266a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batorsky A, Liao J, Lund AW, Plopper GE, Stegemann JP. Biotechnol Bioeng. 2005;92:492. doi: 10.1002/bit.20614. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z, Wang L, Stegemann JP. J Microencapsul. 2011;28:344. doi: 10.3109/02652048.2011.569764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lund AW, Bush JA, Plopper GE, Stegemann JP. J Biomed Mater Res B Appl Biomater. 2008;87:213. doi: 10.1002/jbm.b.31098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan BP, Hui TY, Yeung CW, Li J, Mo I, Chan GC. Biomaterials. 2007;28:4652. doi: 10.1016/j.biomaterials.2007.07.041. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Stegemann JP. Biomaterials. 2010;31:3976. doi: 10.1016/j.biomaterials.2010.01.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubbell JA. Nat Biotechnol. 1995;13:565. [Google Scholar]
- 16.Zhu C, Fan D, Duan Z, Xue W, Shang L, Chen F, Luo Y. J Biomed Mater Res A. 2009;89:829. doi: 10.1002/jbm.a.32256. [DOI] [PubMed] [Google Scholar]
- 17.Ma L, Gao C, Mao Z, Zhou J, Shen J, Hu X, Han C. Biomaterials. 2003;24:4833. doi: 10.1016/s0142-9612(03)00374-0. [DOI] [PubMed] [Google Scholar]
- 18.Peng L, Cheng XR, Wang JW, Xu DX, Wang G. J Bioact Compat Pol. 2006;21:207. [Google Scholar]
- 19.Arpornmaeklong P, Suwatwirote N, Pripatnanont P, Oungbho K. Int J Oral Maxillofac Surg. 2007;36:328. doi: 10.1016/j.ijom.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 20.Kieć-Swierczyńska M, Krecisz B, Krysiak B, Kuchowicz E, Rydzyński K. Int J Occup Med Environ Health. 1998;11:349. [PubMed] [Google Scholar]
- 21.Song K, Qiao M, Jiang B, Macedo HM, Ma X, Cui Z. J Mater Sci Mater Res. 2010;21:2835. doi: 10.1007/s10856-010-4131-4. [DOI] [PubMed] [Google Scholar]
- 22.Kim HW, Gu HJ, Lee HH. Tissue Eng. 2007;13:965. doi: 10.1089/ten.2006.0299. [DOI] [PubMed] [Google Scholar]
- 23.Belanger MC, Marois Y. J Biomed Mater Res. 2001;58:467. doi: 10.1002/jbm.1043. [DOI] [PubMed] [Google Scholar]
- 24.Regehr KJ, Domenech M, Koepsel JT, Carver KC, Ellison Zelski SJ. Lab Chip. 2008;9:2132. doi: 10.1039/b903043c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy KC, Leach JK. BMC Res Notes. 2012;5:423. doi: 10.1186/1756-0500-5-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao RR, Peterson AW, Ceccarelli J, Putnam AJ, Stegemann JP. Angiogenesis. 2012;15:253. doi: 10.1007/s10456-012-9257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]