SUMMARY
Tissue homeostasis in metazoans is regulated by transitions of cells between quiescence and proliferation. The hallmark of proliferating populations is progression through the cell cycle, which is driven by cyclin-dependent kinase (CDK) activity. Here, we introduce a live-cell sensor for CDK2 activity and unexpectedly found that proliferating cells bifurcate into two populations as they exit mitosis. Many cells immediately commit to the next cell cycle by building up CDK2 activity from an intermediate level, while other cells lack CDK2 activity and enter a transient state of quiescence. This bifurcation is directly controlled by the CDK inhibitor p21 and is regulated by mitogens during a restriction window at the end of the previous cell cycle. Thus, cells decide at the end of mitosis to either start the next cell cycle by immediately building up CDK2 activity or to enter a transient G0-like state by suppressing CDK2 activity.
INTRODUCTION
Metazoans tightly control the number of cells in each tissue during development and throughout adult life. Imbalances between the creation and removal of cells lead to excessive tissue growth or failure of tissue function. Much of this feat of balanced tissue homeostasis is achieved by switching cells between two different states: proliferative and quiescent. The transitions between proliferation and quiescence are often reversible—cells must be able to switch from a proliferative to a quiescent state (also termed G0) and later re-engage the proliferation machinery from the quiescent state. A better understanding of these transitions is not only important to understand normal development and adult physiology but also to identify better therapeutic approaches for diseases that involve excessive proliferation, such as cancer, or net cell loss, such as aging and neurodegeneration.
Although reduced levels of mitogens, contact inhibition, and various stress conditions are known to promote quiescence, and many molecular regulators of proliferation have been identified, the detailed mechanisms that control the transitions between these two states are still poorly understood. In one prominent model, cells are thought to commit to the cell cycle at a “restriction point” in late G1 (Pardee, 1974). This model was based on experiments in which mitogen-starved cells were restimulated for varying amounts of time to identify a point when the presence of mitogens is no longer necessary to complete the cell cycle. Cells that have crossed the restriction point prior to mitogen removal are committed to completing the cell cycle, whereas cells that have not crossed the restriction point at the time of mitogen withdrawal remain in G0 or G1.
Much is known about the molecular events associated with emergence from a mitogen-starved state. In mitogen-starved cells, CDK activity is off, and the CDK substrate retinoblastoma protein (Rb) is hypophosphorylated, resulting in an inhibition of E2F transcriptional activators. Re-exposure of cells to mitogens triggers CDK4/6-dependent phosphorylation of Rb, which initiates the reactivation of E2F. Active E2F induces expression of cyclin E and other proteins that promote CDK2 activity, leading to further phosphorylation of Rb (Massagué, 2004; Trimarchi and Lees, 2002). This reinforced expression of cell-cycle regulators is thought to engage in G1 a few hours before DNA replication, causing an upregulation of CDK2 activity, full phosphorylation of Rb, and passage through the restriction point (Dou et al., 1993; Weinberg, 1995; Yao et al., 2008; Zetterberg et al., 1995). Ubiquitination and degradation of the CDK inhibitor, p21, is also thought to promote the G1/S transition (Abbas and Dutta, 2009). Despite a significant amount of knowledge about the biochemical processes associated with emergence from quiescence, much less is known about cell-cycle commitment in proliferating cells.
Because cycling cell populations are asynchronous, biochemical analysis of commitment mechanisms cannot readily be performed. Chemical and other synchronization methods can be used to obtain more homogeneous populations, but these procedures can trigger stress responses and may alter the natural behavior of cells. In addition, bulk analysis may mask the existence of distinct sub-states in a population. Even if single-cell methods are used, the lack of accepted molecular markers that distinguish precommitment from postcommitment cells or G0 from G1 cells still leaves challenging problems. For example, there has been a long-standing debate over where between mitosis and S phase G0 ought to be placed (Coller, 2007) (Figure 1A).
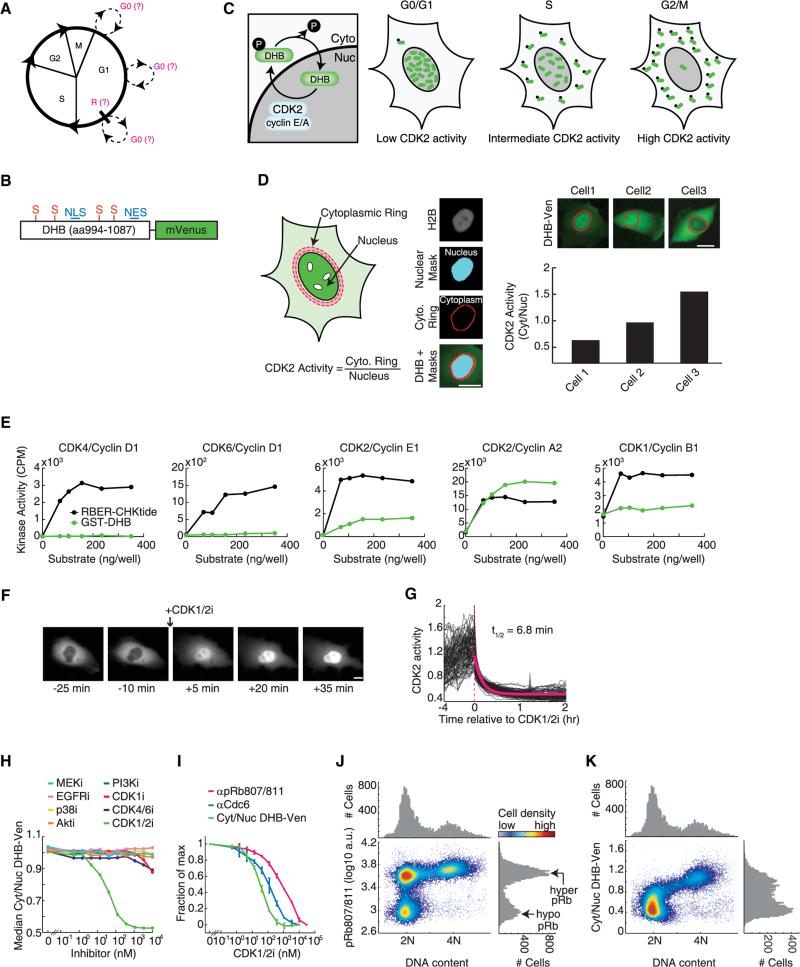
Figure 1. Characterization of a Live-Cell Sensor for CDK2 Activity.
(A) Cell-cycle diagram showing uncertainty about when entry into G0 occurs and where the restriction point (R) is positioned.
(B) Schematic of sensor. NLS, nuclear localization signal; NES, nuclear export signal; S, CDK consensus phosphorylation site on serine.
(C) Schematic of CDK2 phosphorylation-mediated translocation of DHB-Ven.
(D) Method used to obtain the cytoplasmic to nuclear ratio of DHB-Ven. Cell nuclei were identified using fluorescent H2B images to obtain a mask. The nuclear component of DHB-Ven was determined using this mask, whereas the cytoplasmic component of DHB-Ven consisted of a ring around the nuclear mask. See Extended Experimental Procedures.
(E) Purified GST-DHB (aa994–1087) at six different concentrations was used in an in vitro kinase activity assay as a substrate for five different cyclin-CDK complexes. Conditions were selected to be in a range known to work with the established artificial positive control, RBER-CHKtide. Note that CDK1/cyclin B has a high level of basal autophosphorylation. See Extended Experimental Procedures.
(F) Images of DHB-Ven in a single cell before and after treatment with 10 μM CDK1/2 inhibitor.
(G) DHB-Ven traces in individual MCF10A cells before (imaged every 12 min) and after (imaged every 2 min) treatment with 10 μM CDK1/2 inhibitor (red dashed line). The half-life was derived from an exponential fit to the average of individual traces (red curve).
(H) In vivo response of DHB-Ven to titration of eight different kinase inhibitors. Cells were treated with the inhibitors for 30 min in 96-well plate format, fixed, and imaged to obtain the median of single-cell Cyt/Nuc DHB-Ven signals in each well.
(I) Comparison of DHB-Ven and two other CDK2 substrates, Cdc6 and Rb, to a titration of CDK1/2 inhibitor. Cells were treated with the inhibitor for 30 min in 96-well plate format, fixed, stained with antibodies (anti-phosphorylated Rb at serine 807/811 [pRb807/811] or anti-Cdc6; Cdc6 translocates from the nucleus to the cytoplasm in response to CDK2 phosphorylation) and imaged to obtain the median of single-cell Cyt/Nuc DHB-Ven signals, median of single-cell Cyt/Nuc Cdc6 signals or median of single-cell pRb signals in each well. Error bars represent the standard deviation of duplicate wells.
(J and K) Density scatter plot of pRb807/811 versus DNA content (J) or Cyt/Nuc DHB-Ven versus DNA content (K) in single cells obtained by fixed-cell imaging. Single-parameter histograms are shown above and to the right. Scale bars throughout this manuscript are 10 μm.
All data are from MCF10A cells. See also Figure S1.
Here, we introduce a fluorescent sensor to monitor CDK2 activity in single cycling cells. Using time-lapse microscopy and customized cell tracking, we monitored CDK2 activity over several cell cycles in an immortalized but nontransformed human mammary epithelial cell line, MCF10A. We confirmed our results in primary human foreskin fibroblasts and in a previously used model for cell-cycle progression, murine Swiss 3T3 fibroblasts. By studying unperturbed, asynchronous cultures, we discovered a bifurcation point at mitotic exit where cells choose between two different fates. Many cells exit mitosis with an intermediate level of CDK2 activity, from which they continue to build up CDK2 activity and complete the next cell cycle. The remaining cells enter a transient state of quiescence characterized by low CDK2 activity. We show that cells integrate mitogenic signals during a “restriction window” at the end of the previous cell cycle to decide which of the two paths to take upon completion of mitosis. Finally, using immunofluorescence, genetics, and chemical biology approaches, we show that the bifurcation in cell fate is directly controlled by the CDK inhibitor, p21.
RESULTS
Characterization of a Live-Cell Sensor for CDK2 Activity
We adapted a previously described sensor for the G1/S transition (Hahn et al., 2009) to quantitatively measure changes in CDK activity. The sensor includes amino acids 994–1087 of human DNA helicase B fused to the yellow fluorescent protein mVenus (DHB-Ven; Figure 1B), and contains four CDK consensus phosphorylation sites, a nuclear localization signal, and a nuclear export signal (Gu et al., 2004). We established an MCF10A line stably expressing this construct and found that DHB-Ven localizes to the nucleus in newly born cells. The sensor then translocates progressively out of the nucleus as the cell cycle proceeds and exhibits cytosolic localization at the end of G2 (Figure 1C). We used the cytoplasmic to nuclear ratio (Cyt/Nuc) of DHB-Ven fluorescence intensity as a potential readout for CDK activity. To measure this ratio, we compared the nuclear DHB-Ven signal to the intensity of a region outside, but adjacent to, each nucleus (Figure 1D).
An in vitro kinase activity screen using a panel of CDK/cyclin pairs and amino acids 994–1087 of DHB revealed that DHB is phosphorylated strongly by CDK2/cyclin A, more weakly by CDK2/cyclin E, and is not phosphorylated by CDK1/cyclin B, CDK4/cyclin D, or CDK6/cyclin D (Figure 1E). Using small molecule inhibitors in vivo, we showed that the cytoplasmic localization of DHB-Ven was reversed by addition of a CDK1/2 inhibitor (Figures 1F and 1G) but was not altered by treating cells with selective inhibitors targeting CDK4/6, EGFR, MEK, p38, Akt, or PI3K across a wide range of doses (Figures 1H and S1 available online). The localization of DHB-Ven was also unaffected by addition of a CDK1-selective inhibitor (Figure 1H), arguing that the sensor responds primarily to CDK2 activity, at least in cells where CDK2 is present. Treatment of cells with the CDK1/2 inhibitor caused retro-translocation of DHB-Ven from the cytoplasm to the nucleus with a half-life of 7 min (Figures 1F and 1G), showing that DHB-Ven responds rapidly to changes in CDK2 activity. Furthermore, the rate of translocation of DHBVen did not significantly depend on when during interphase we added the CDK1/2 inhibitor (Figure S1), arguing that the phosphatase activity acting on the sensor is relatively constant during interphase.
We then compared the sensitivity of the response of DHB-Ven to that of two established CDK2 substrates (Cdc6 and Rb) using titration of the CDK1/2 inhibitor (Figure 1I). We found that the three substrates had very different EC50 values, with DHB-Ven being 3.5-fold and 23-fold more sensitive to the inhibitor than Cdc6 and Rb, respectively. Thus, Rb is a highly sensitive CDK2 substrate, meaning it takes very little CDK2 activity to phosphor-ylate Rb, whereas DHB-Ven required a higher level of CDK2 activity to become phosphorylated. When we examined the phosphorylation status of Rb and the localization of DHB-Ven as a function of DNA content in single cells, we found that cells can have hyperphosphorylated Rb already in G1 when they still have a 2N DNA content (Figure 1J). In contrast, DHB-Ven was only partially phosphorylated when cells have a 2N DNA content (Figure 1K). The phosphorylation and consequent cytoplasmic translocation of DHB-Ven then gradually increased as cells progressed through the cell cycle and the DNA content increased from 2N to 4N. This implies that CDK2 activity progressively increases throughout interphase, and that the DHB-Ven sensor has a dynamic range well-suited to monitor changes in CDK2 activity throughout the cell cycle.
Variable Delay in the Activation of CDK2 in Cells Emerging from Mitogen Starvation
To further validate our sensor, we compared single-cell time-lapse measurements of DHB-Ven to earlier bulk experiments in which cells were synchronized by mitogen starvation, restimulated with mitogens, and then lysed for biochemical analysis after varying amounts of time. These earlier studies described an increase in CDK2 activity in late G1 followed by entry into S phase (Dulić et al., 1992; Koff et al., 1992; Ohtsubo et al., 1995) (Figure 2A). Because adherent cells migrate over time, we developed a custom script to track cells and monitor CDK2 activity in thousands of individual cells through several cell cycles (Figure 2B; Extended Experimental Procedures) and made use of a 96-well plate format to test different conditions in the same experiment. In addition to the CDK2 sensor, we monitored the time of entry into S phase in single cells using SCFSkp2-mediated degradation of a fluorescently tagged fragment of Cdt1 (Sakaue-Sawano et al., 2008) (Figures 2C and S2).
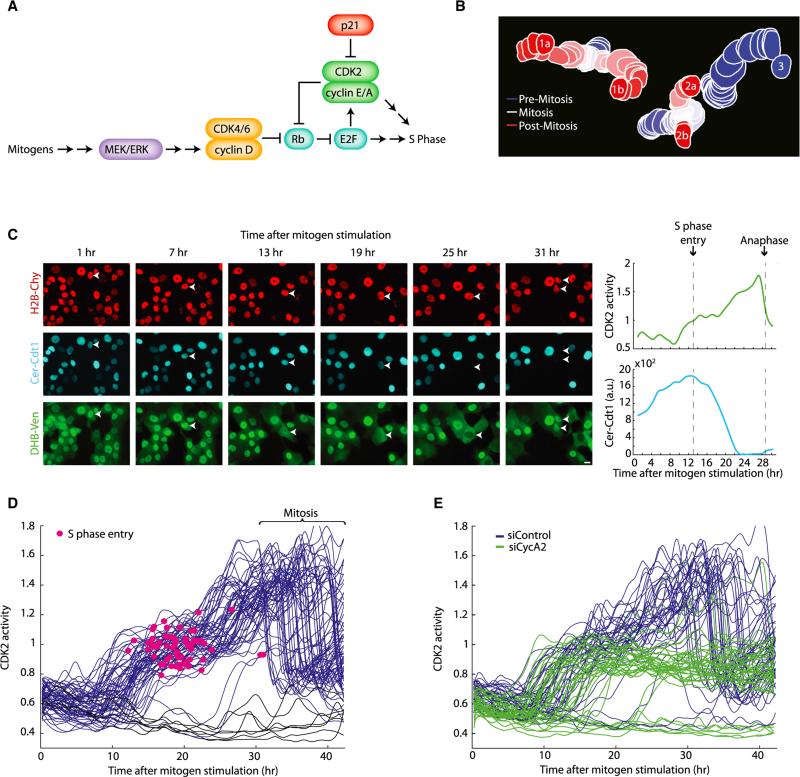
Figure 2. A Variable Delay in CDK2 Activation in Cells Emerging from Mitogen Starvation.
(A) Literature-based schematic of the signaling pathway involved in emergence from mitogen starvation. Mitogens promote upregulation of cyclin D, which binds to CDK4/6 to initiate phosphorylation of Rb. This initiates the release of E2F from its Rb-bound state, freeing it to upregulate cyclin E, cyclin A, CDK2, and other genes needed for S phase. p21 inhibits CDK2.
(B) Tracking of three cells, two of which divide, over 25 frames (5 hr).
(C) Left: images of MCF10A cells emerging from mitogen starvation. An arrow marks the cell tracked in the plot on the right. Top: H2B-Cherry; middle, Cerulean-Cdt1; bottom, DHB-Venus. Right, traces of the cell marked with the arrow in the images. Cer-Cdt1 is degraded at the start of S phase (vertical dashed line) (Sakaue-Sawano et al., 2008); at this time, the Cyt/Nuc ratio of DHB-Ven is about one.
(D) Traces of CDK2 activity in individual cells emerging from 45 hr of mitogen starvation that do (blue) or do not (black) enter S phase during the imaging period. Because there were always a few cells that were not properly starved, only cells with DHB-Ven Cyt/Nuc < 0.8 at t = 0 are included in the plot. A red dot marks the start of S phase (induction of Cer-Cdt1 degradation) for each cell.
(E) Traces of CDK2 activity in individual cells emerging from mitogen starvation. Cells were treated for 6 hr with a nontargeting siRNA (blue) or with a pool of four siRNAs against cyclin A2 (green). Cells were then starved for 45 hr then restimulated with full growth media and subjected to time-lapse imaging. Only cells with DHB-Ven Cyt/Nuc < 0.8 at t = 0 are included in the plot.
All data are from MCF10A cells. See also Figure S2 and Movie S1.
When we tracked CDK2 activity in individual cells emerging from mitogen starvation, we observed a period of roughly 8 hr of low CDK2 activity, followed by a broad time window during which individual cells triggered a buildup in CDK2 activity (Figure 2D; Movie S1). These dynamics of CDK2 activity are consistent with results from earlier biochemical experiments. Once the increase in CDK2 activity began in a cell, CDK2 activity continued to increase monotonically. When DHB-Ven reached a Cyt/Nuc ratio of approximately one, cells consistently entered S phase. CDK2 activity then continued to increase for another ~16 hr during S and G2 until the nuclear envelope broke down and the Cyt/Nuc ratio of DHB-Ven became ill-defined until nuclear envelope reformation. Our results also revealed a large cell-to-cell variation in the delay between mitogen restimulation and CDK2 activation, to the extent that some cells in the population failed to build up CDK2 activity at all during the 40 hr observation period. Finally, using siRNA targeting cyclin A, we showed that the initial rise in CDK2 activity (up to a Cyt/Nuc DHB-Ven ratio of one) did not require cyclin A, but the subsequent rise in CDK2 activity did, consistent with a requirement for cyclin A in S phase (Figure 2E).
A Bifurcation in CDK2 Activity at Mitotic Exit in Asynchronously Cycling Cells
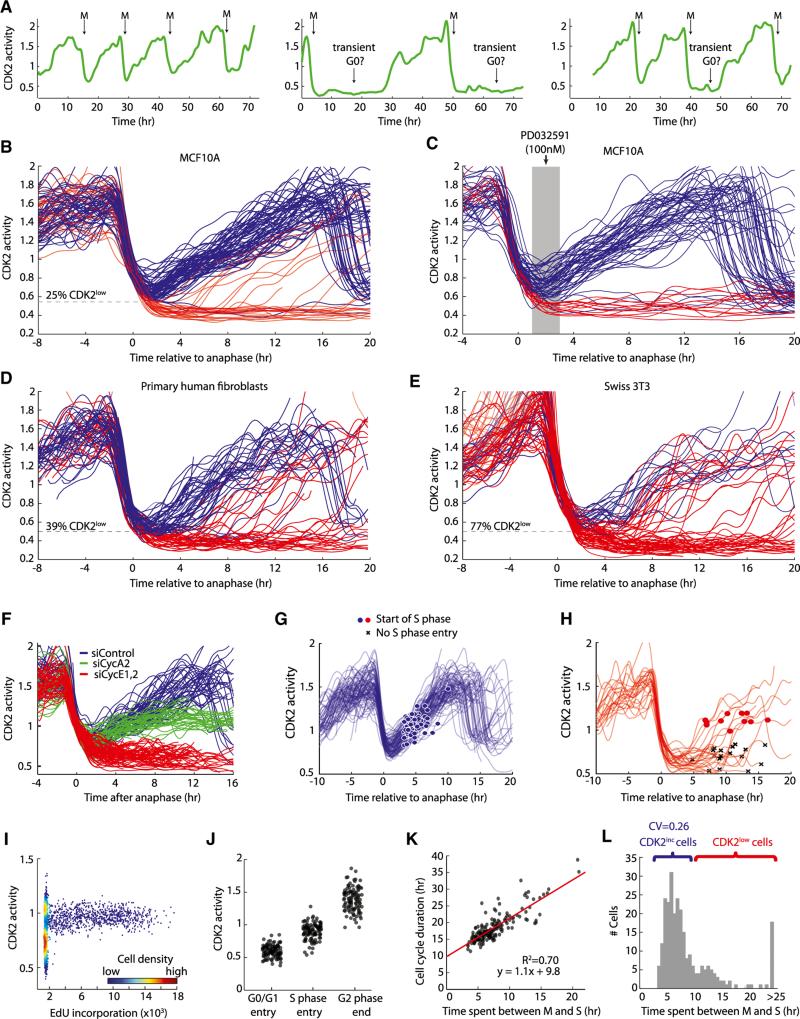
Although the emergence of cells from serum starvation has been extensively studied, it has not previously been possible to study unperturbed cells at a molecular level due to the asynchronous nature of proliferating populations. When we examined CDK2 activity traces from individual cycling MCF10A cells, we noticed two distinct behaviors (Figure 3A and Movie S2), which became even more apparent when we aligned multiple traces of CDK2 activity to the time of anaphase (Figure 3B). In the majority of MCF10A cells (75%), CDK2 activity began to increase immediately after mitosis from an intermediate level (designated CDK2inc; blue, Figure 3B); these cells had short intermitotic times of 16–20 hr. The remaining cells (25%) entered a reversible state of low CDK2 activity (designated CDK2low; red, Figure 3B); these cells had heterogeneous intermitotic times ranging from 20 hr to >50 hr, reminiscent of a G0-like state. These CDK2low cells could later re-enter the proliferative state by increasing CDK2 activity, indicating that these cells remained cell-cycle competent. Indeed, the CDK2inc and CDK2low cells are not fixed subpopulations, but rather could interconvert at mitotic exit (Figures 3A and S3). The two behaviors can be reliably distinguished as early as 2 hr after anaphase, approximately 1 hr after nuclear envelope reformation. The roughly unimodal distribution of CDK2 activity at 2 hr after anaphase quickly evolved into a clearly bimodal distribution by 4 hr after anaphase (Figure S3). We therefore set a threshold in the Cyt/Nuc ratio of DHB-Ven at 2 hr after anaphase and found that the level of CDK2 activity above or below this threshold was highly predictive of whether a cell would enter the CDK2inc or CDK2low state, respectively (Figure S3).
Figure 3. Bifurcation in CDK2 Activity upon Exit from Mitosis.
(A) Single-cell traces of CDK2 activity in proliferating cells. CDK2 activity drops rapidly at mitosis (M) and either immediately builds up again or remains low for a variable amount of time, reminiscent of a G0-like state.
(B–E) Single-cell traces of CDK2 activity aligned computationally to the time of anaphase for MCF10A (B and C), Hs68 primary human fibroblasts (D), or murine Swiss 3T3 fibroblasts (E). Traces were colored red if, 2 hr after anaphase the Cyt/Nuc ratio of DHB-Ven fell below 0.55 (MCF10A) or 0.5 (Hs68); otherwise traces were colored blue. The dashed line marks the cutoff used for the red/blue color scheme. Due to higher noise in Swiss 3T3 traces, we used an expanded rule for these cells in which traces were colored red if the Cyt/Nuc ratio of DHB-Ven fell below 0.5 at 2 hr after anaphase or below 0.6 at 6 hr after anaphase; otherwise traces were colored blue. This red/blue color scheme is used in all subsequent figures. In (C), MCF10A cells were preimaged for 8 hr, and then treated with 100 nM MEKi (PD032591). Only cells that had completed anaphase 1–3 hr prior to addition of MEKi are plotted. MEKi remained in the media for the rest of the imaging period.
(F) Single-cell traces of CDK2 activity aligned computationally to the time of anaphase for cells treated with nontargeting siRNA (blue), siRNAs targeting cyclin A2 (green) or cyclin E1 and E2 (red). Cells were incubated with siRNAs for 6 hr, washed, and immediately subjected to time-lapse imaging.
(G and H) Traces of CDK2 activity aligned computationally to the time of anaphase. The start of S phase (induction of Cer-Cdt1 degradation) is marked with blue or red dots. Cells not yet in S phase at the end of the movie are marked with black Xs.
(I) Density scatter plot of CDK2 activity versus EdU incorporation (a marker for DNA synthesis).
(J) CDK2 activity in single cells at the start of G0/G1 (1 hr after nuclear envelope reformation), at the start of S phase (scored using Cer-Cdt1), or at the end of G2 phase (last frame before nuclear envelope breakdown).
(K) Scatter plot showing the total duration of the cell cycle (intermitotic time) as a function of the M-to-S interval in individual cells (n = 176). The start of S phase was scored using Cer-Cdt1; mitosis was scored using H2B-Chy. Fit was obtained by linear regression; R represents the Pearson correlation coefficient.
(L) Histogram of time spent between mitosis and the start of S phase for individual MCF10A cells (n = 250). Brackets mark CDK2inc cells (blue) or cells that pass through the CDK2low state (red). CV, coefficient of variation.
All data are from MCF10A cells except (D) and (E). See also Figure S3 and Movie S2.
To verify that CDK2inc and CDK2low are really two distinct cell states, we treated asynchronously cycling cells with a MEK inhibitor to test whether MEK inhibition would prevent CDK2low cells from reactivating CDK2 activity. When we examined only those cells that received MEK inhibitor 1–3 hr after anaphase, we found that we were able to block the reactivation of CDK2low cells. In contrast, MEK inhibition had no effect on the already increasing CDK2inc trajectories. Thus, a clear decision is made at the end of mitosis that causes a bifurcation in cell fate in which CDK2inc cells no longer require MEK activity to complete the next cell cycle, whereas CDK2low cells require MEK activity to transition out of the CDK2low state back to the cycling state (Figure 3C).
We then examined two additional cell types, primary human foreskin fibroblasts (Hs68 cells) and a previously used model for cell-cycle progression, murine Swiss 3T3 cells. Both cell types displayed a bifurcation in CDK2 activity at mitotic exit, similar to MCF10A cells. However, the fraction of CDK2low cells was slightly larger in Hs68 cells (39%, Figure 3D) compared to MCF10A cells, and much larger in Swiss 3T3 cells (77%, Figure 3E). For both Hs68 and Swiss 3T3 fibroblasts, cells also tended to persist in the CDK2low state for longer durations than MCF10A cells. This argues that the bifurcation at mitotic exit is likely a general cell-cycle mechanism but that the fraction of cells bifurcating into each state as well as the time cells spend in the CDK2low state vary between cell types, growth factor concentrations, and other experimental conditions (Figure S4).
To determine which cyclin partner of CDK2 was responsible for the immediate build-up of CDK2 activity after mitosis, we treated cycling cells with siRNA against cyclin E and cyclin A. Cells treated with siRNA targeting cyclin E during the previous cell cycle failed to build up CDK2 activity after mitosis and behaved similarly to CDK2low cells (Figure 3F). As in cells emerging from quiescence, cyclin A was required to raise CDK2 activity above a Cyt/Nuc DHB-Ven ratio of one. Thus, the initial rise in DHB-Ven signal requires cyclin E/CDK2 activity, after which cyclin A/CDK2 takes over in S phase. Using Cer-Cdt1 to mark the start of S phase, we found that neither the CDK2inc nor the reactivating CDK2low MCF10A cells entered into S phase until DHB-Ven rose above a Cyt/Nuc ratio of approximately one (Figures 3G and 3H). This was the same ratio that we observed (Figure 2D) for cells emerging from serum starvation. When we used EdU incorporation to monitor DNA replication in individual cells, we found that S phase was restricted to a window of CDK2 activity that corresponds to a DHB-Ven Cyt/Nuc ratio of 0.9–1.1 (Figure 3I). Together with the results from Figure 2, these experiments show that three critical levels of CDK2 activity exist (Figure 3J): (1) CDK2inc cells begin to build up CDK2 activity if they are born with CDK2 activity above a minimal trigger threshold, (2) cells start DNA replication a few hours later after they cross an intermediate CDK2 activity threshold, and (3) cells finish G2 with an even higher level of CDK2 activity.
Variability in cell-cycle duration has long been attributed to variability in the duration of G1 (Prescott, 1968), but the source of this variability was not well understood. We found that the distribution of the time MCF10A cells spend between M and S was long-tailed, with some cells having very long M-to-S intervals (Figures 3K and 3L). The relationship between the duration of the M-to-S interval and the intermitotic time had a slope close to one (Figure 3K), arguing that an increase of 1 hr in the M-to-S interval results in an increase of 1 hr in total cell-cycle time. Moreover, the time that individual cells spent between M and S explained 70% of the variance in the overall cell-cycle duration. Cells that pass through the CDK2low state had long and variable M-to-S intervals, whereas the time spent between M and S for CDK2inc cells was short (5–7 hr) and relatively invariant (Figure 4L; coefficient of variation = 0.26 for CDK2inc cells versus coefficient of variation = 0.42 for all cells with a finite M-to-S interval). Thus, the time cells spend between mitosis and S phase can potentially be described as a G1 phase characterized by steadily increasing CDK2 activity, preceded in a fraction of cells by a transient G0-like state of variable duration characterized by low CDK2 activity.
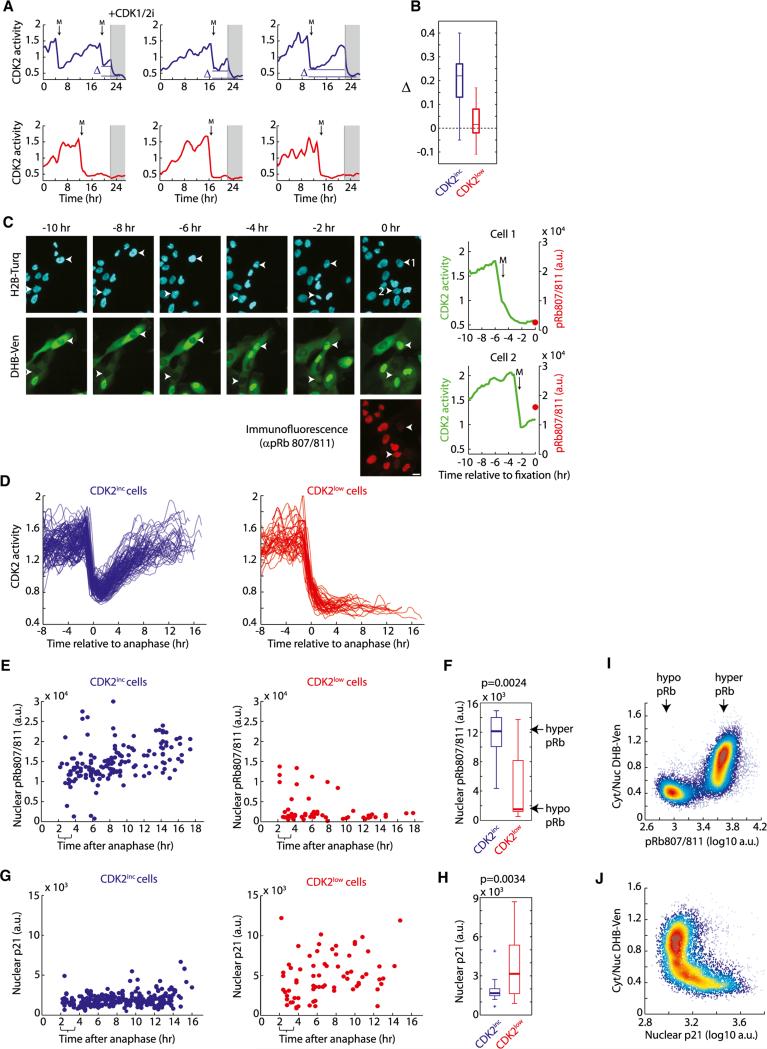
Figure 4. CDK2inc Cells Begin G1 with Residual CDK2 Activity, Hyperphosphorylated Rb, and Low p21.
(A) CDK2 activity in cells treated with 10 μM CDK1/2 inhibitor after 22 hr of imaging. The difference between CDK2 activity at mitotic exit and after drug addition is marked as D.
(B) Boxplot of Δ. Δ is close to zero for CDK2low cells indicating no residual CDK2 activity at mitotic exit.
(C) Time-lapse imaging of CDK2 activity in asynchronous cells was followed by immunofluorescence staining for pRb807/811 (left). The immunofluorescence image was precisely aligned to the time-lapse imaging movie using a custom jitter correction algorithm. This enabled matching of the CDK2 activity trace of each cell to its phospho-Rb level at the end of the movie (right).
(D) Example traces of CDK2 activity for CDK2inc and CDK2low cells used in (E)–(H). For clarity, cells were excluded if they emerged from the CDK2low state or had a second mitosis.
(E and G) Time-lapse imaging of CDK2 activity in asynchronous cells was followed by fixation and immunofluorescence staining for pRb807/811 (E) or p21 (G) as described in (C). Phospho-Rb levels (E) or p21 levels (G) were then reconstructed as a function of time since anaphase for CDK2inc cells (blue dots) and CDK2low cells (red dots). Brackets mark the time window (2–3.5 hr after anaphase) used in the boxplots in (F) and (H).
(F and H) Boxplots comparing phospho-Rb staining (F) or p21 staining (H) in CDK2inc and CDK2low cells selected to be within 2 to 3.5 hr after anaphase. A rank-sum test was used to obtain p values.
(I and J) Density scatter plot of Cyt/Nuc DHB-Ven versus pRb807/811 (I) or versus p21 (J) obtained by fixed-cell imaging in asynchronously cycling cells.
All data are from MCF10A cells. See also Figure S4 and Movie S3.
Residual CDK2 Activity and Hyperphosphorylation of Rb Immediately after Mitosis
We next investigated whether CDK2inc cells exit mitosis with residual CDK2 activity, as suggested by the intermediate Cyt/Nuc ratio of DHB-Ven in Figures 3B–3E. Indeed, addition of CDK1/2 inhibitor to CDK2inc cells reduced the Cyt/Nuc ratio of DHBVen to a level lower than that observed at mitotic exit and similar to that of CDK2low cells (blue, Figures 4A and 4B), confirming that CDK2inc cells exit mitosis with residual CDK2 activity. In contrast, addition of CDK1/2 inhibitor to CDK2low cells did not further reduce the Cyt/Nuc ratio, indicating that these cells have the minimal measurable level of CDK2 activity (red, Figures 4A and 4B).
We then investigated why some cells exit mitosis with residual CDK2 activity, whereas others do not. We first tested whether local cell density plays a role, but found that the number of neighboring cells was similar in CDK2inc and CDK2low cells under our experimental conditions in which cells remain subconfluent (Figure S4). We also noted that sister cells chose the same CDK2inc or CDK2low fate 98% of the time, suggesting that they share an event in their past that regulates which path a pair of sister cells takes. To examine the process at a molecular level, we compared the endogenous levels of key proteins in single CDK2inc and CDK2low cells immediately after the bifurcation. We established a method in which we fixed cells at the end of a time-lapse imaging series, stained them with antibodies, and used a custom jitter correction script to map each cell in the immunofluorescence image back to the same cell recorded in the CDK2 time-lapse analysis (Figure 4C, Movie S3, and Extended Experimental Procedures). We used this method to monitor a number of potential regulators of the bifurcation including cMyc, cyclin D1, cyclin E, cyclin A2, p21, p27, phospho-Rb, the stress marker phospho-p38, and the DNA damage marker gH2AX. We found that only phospho-Rb and p21 levels were clearly statistically different in CDK2inc and CDK2low cells immediately after the bifurcation (Figures 4D–4H, and S4).
Consistent with our finding that Rb is a highly sensitive CDK2 substrate that requires only minimal CDK2 activity to become phosphorylated (Figure 1I), CDK2inc cells were born with hyper-phosphorylated Rb. In contrast, most cells in the CDK2low state showed low Rb phosphorylation (Figures 4E and 4F). These two distinct sub-populations of phospho-Rb were already evident within the first hour after anaphase (Figure S4). When we examined the relationship between DHB-Ven and phospho-Rb in large numbers of proliferating cells, we found a strong, nonlinear correlation: cells with hypophosphorylated Rb always had a very low Cyt/Nuc DHB-Ven ratio, whereas cells with hyperphosphorylated Rb had intermediate to high Cyt/Nuc DHB-Ven ratios (Figure 4I). Thus, ~75% of MCF10A cells exit mitosis with residual CDK2 activity and hyperphosphorylated Rb.
The Bifurcation in CDK2 Activity Is Controlled by p21
Using our approach of time-lapse imaging followed by immunofluorescence, we also observed a strong inverse relationship between the presence of residual CDK2 activity and endogenous levels of the CDK inhibitor p21 (Figures 4G and 4H). The levels of p21 were elevated in cells born into the CDK2low state, whereas p21 levels remained low in CDK2inc cells throughout the cell cycle. Thus, variation in endogenous p21 levels is correlated with, and perhaps responsible for, the decision made at the bifurcation. When we plotted the DHB-Ven versus p21 signal in single cells, we found that the two are mutually exclusive: cells with high p21 always had low Cyt/Nuc DHB-Ven ratios, whereas cells with low p21 had intermediate to high Cyt/Nuc DHB-Ven ratios (Figure 4J). Because p21 is a direct upstream regulator of CDK2 activity, we decided to investigate the role of p21 in the regulation of the bifurcation.
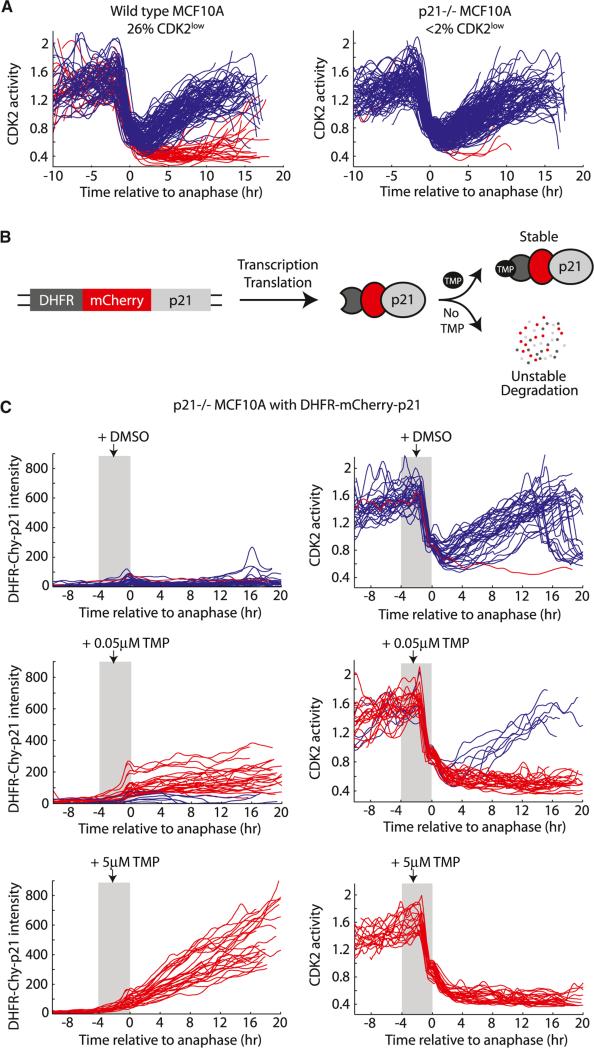
If high levels of p21 were responsible for causing a cell to enter the CDK2low state after mitosis, one would expect that cells lacking p21 would be unable to enter the CDK2low state. We therefore made use of p21–/– MCF10A cells (Bachman et al., 2004), transduced them with DHB-Ven, and performed time-lapse imaging of asynchronous cells as in Figure 3B. Strikingly, when we imaged parental and p21–/– MCF10A cells in parallel, we found that p21–/– MCF10A very rarely entered the CDK2low state (<2% CDK2low for p21–/– cells versus 26% for parental cells) (Figure 5A). The intermitotic times of CDK2inc cells were similar in parental and p21–/– MCF10A cells, arguing that the reported faster population doubling rate of p21–/– cells is primarily due to absence of the CDK2low state.
Figure 5. The Bifurcation in CDK2 Activity Is Controlled by p21.
(A) Single-cell traces of CDK2 activity in parental MCF10A and p21–/– MCF10A aligned to the time of anaphase.
(B) Design of the DHFR-mCherry-p21 construct.
(C) p21–/– MCF10A cells expressing DHFR-mCherry-p21 were preimaged for 6 hr, and then treated with DMSO or with 0.05 μM or 5 μM TMP. Traces were aligned to the time of anaphase and only cells that were treated with TMP 0–4 hr prior to anaphase (gray shaded region) were selected for plotting. Left column, DHFR-mCherry-p21 intensity in single cells over time. Right column, consequent CDK2 activity in the same single cells over time. Traces were colored blue if a buildup of CDK2 activity occurred after mitosis, otherwise traces were colored red.
See also Figure S5.
We then rescued the phenotype of p21–/– MCF10A cells using the dihydrofolate reductase (DHFR)-trimethoprim (TMP) protein stabilization system (Iwamoto et al., 2010) to acutely increase exogenous p21 protein. When the DHFR domain from E. coli is fused to a protein of interest, the entire fusion protein is continuously degraded by the proteasome. Addition of the small molecule ligand TMP rapidly stabilizes the fusion protein (Figure 5B). We transduced p21–/– MCF10A cells with an exogenous copy of p21 tagged with DHFR and mCherry, thereby gaining temporal control of the level of p21 in these cells. Under basal conditions (no TMP), p21 was destabilized and nearly all cells entered the CDK2inc state after mitosis, as expected (Figure 5C, top, and S5). To test whether an increase in p21 protein at the end of the previous cell cycle causes cells to enter the CDK2low state after mitosis, we added TMP to asynchronous cultures and selected for analysis only those cells that were in G2 or M when TMP was added. Remarkably, we found that we were able to control, in a dose-dependent manner, the fraction of CDK2inc versus CDK2low cells simply by regulating the level of p21. At an intermediate dose of TMP, resulting in intermediate levels of p21, a fraction of cells entered the CDK2low state (Figure 5C, middle, red), while the remainder built up CDK2inc activity (Figure 5C, middle, blue; these were invariably the cells with lower p21 levels). At the highest dose of TMP, resulting in high levels of p21, 100% of cells entered the CDK2low state after mitosis (Figure 5C, bottom). Together, our knockout and inducible-rescue experiments demonstrate that, for our experimental conditions, p21 is necessary and sufficient to trigger entry into the CDK2low state at mitotic exit.
Location of the Restriction Point
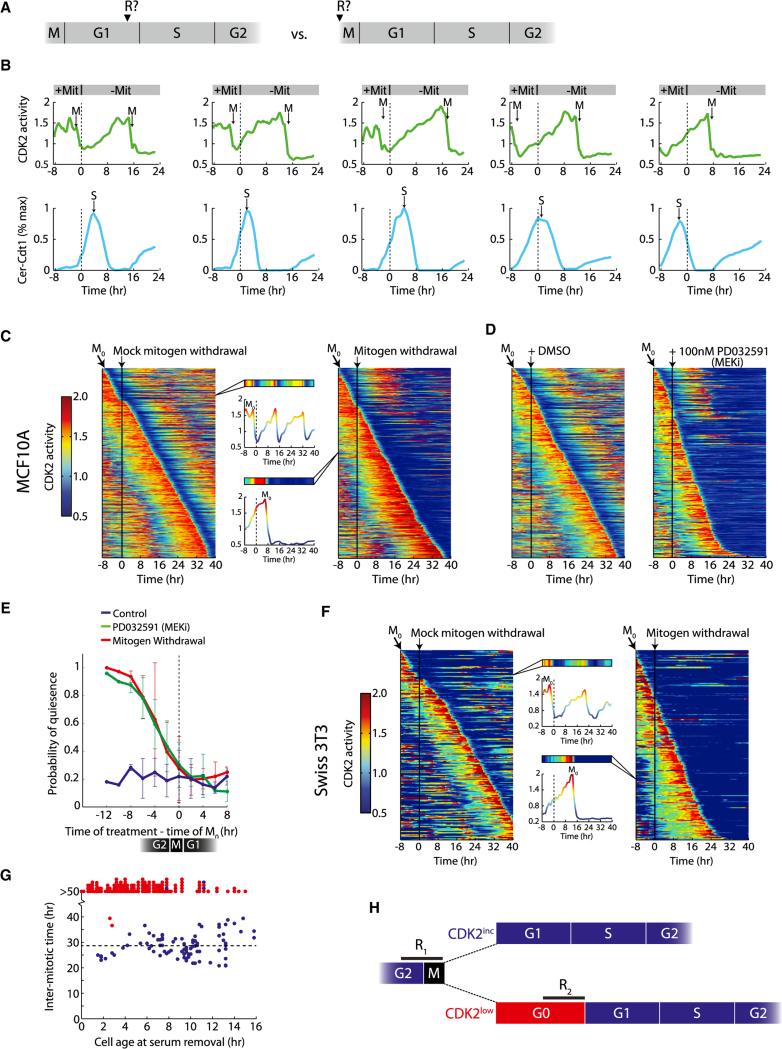
Nearly every cell that initiated a buildup of CDK2 activity in early G1 continued to increase CDK2 activity until the subsequent mitosis (Figures 3B–3E). This suggests that the time of bifurcation represents a cell-cycle commitment for CDK2inc cells and that the quiescence-proliferation decision window in these cells might occur at the end of the previous cell cycle (Figure 6A, right). However, this idea conflicts with the classic model of the restriction point in which cell-cycle commitment occurs in late G1 (Figure 6A, left). To directly test where the restriction point is positioned in cycling cells, we preimaged asynchronous cultures to obtain cell-cycle phase information, then removed mitogens and followed the cellular response. Every MCF10A cell that initiated a buildup in CDK2 activity continued on its trajectory and completed mitosis even if mitogens were withdrawn at any time within G1, S, or G2. Mitogen withdrawal did not significantly alter the duration of the ongoing cell cycle but did promote entry into the CDK2low state after the subsequent mitosis (Figures 6B–6E). We obtained similar results in Hs68 and Swiss 3T3 cells (Figure S6).
Figure 6. Mitogenic Stimuli during the Previous Cell Cycle Control CDK2 Activation and Commitment to the Next Cell Cycle.
(A) The following experiments are aimed at determining whether cell-cycle commitment occurs in late G1 as posited by the restriction point model (left) or at the end of the previous cell cycle (right).
(B) Cells were preimaged in full growth media, washed three times, and then transferred to media without mitogens after 8 hr (dashed line at t = 0). CDK2 activity builds up even when mitogens are removed as early as 25 min after anaphase (leftmost cell). Top, single-cell traces of CDK2 activity. Bottom, corresponding normalized traces of Cer-Cdt1 used to determine the start of S phase. Mit, mitogens.
(C and F) Heatmap of CDK2 activity in MCF10A cells (C) or Swiss 3T3 cells (F) sorted computationally by the time of mitosis (M0). Each row in the heatmap represents the CDK2 activity trace of a single cell over time. After 8 hr of preimaging, cells were washed and put back in full growth media (mock, left, vertical black line) or washed and put in starvation media (right, vertical black line).
(D) Same as (C), except instead of removing mitogens after 8 hr of preimaging, MCF10A cells were treated with DMSO (left, vertical black line), or treated with 100 nM MEKi (PD032591) in full growth media.
(E) Probability of quiescence following M0, for the case of mitogen withdrawal (red), addition of 100 nM MEKi (green), or control (blue). A cell was considered quiescent if, 6 hr after M0, the DHB-Ven Cyt/Nuc ratio was less than one. Error bars are standard deviation from duplicate experiments.
(G) For each Swiss 3T3 cell, the time between the last mitosis and the time serum was removed was recorded (cell age at serum removal), as well as the length of the subsequent cell cycle (intermitotic time). Points are red if CDK2 activity was not yet building up when mitogens were removed; points are blue if CDK2 activity was already building up when mitogens were removed.
(H) Schematic of key decision points in cell-cycle commitment based on our data. Cells integrate mitogenic stimuli during a restriction window (R1) at the end of the previous cell cycle and choose between CDK2inc and CDK2low fates upon completion of mitosis. CDK2low cells face a second decision point (R2) in which they can re-enter the cell cycle by building up CDK2 activity.
All data are from MCF10A cells except (F) and (G). See also Figure S6 and Movie S4.
To determine the precise timing of these events in MCF10A cells, we scaled up our analysis and used a heatmap representation to quantify the results (Figure 6C). It then became apparent that if mitogens were removed more than 8 hr before mitosis, cells would enter the CDK2low state after mitosis and become quiescent. If mitogens were removed in a window 0–8 hr prior to mitosis, cells showed a decreasing probability of entering the proliferative state with a midpoint approximately 3 hr prior to anaphase (Figure 6E). If mitogens were withdrawn after mitosis, cells entered the CDK2inc state with the same probability as control cells and completed one additional cell cycle before becoming quiescent. We then considered that we may not have removed all growth factors or that growth factor signaling may have continued from internalized growth factor receptors. We therefore suppressed growth factor signaling by adding a MEK inhibitor to cycling cells and obtained similar results (Figures 6D and 6E and Movie S4).
We then scaled up our mitogen withdrawal experiments in cycling Swiss 3T3 cells to more directly compare our findings to studies by Zetterberg (Zetterberg and Larsson, 1985) who investigated the restriction point in cycling Swiss 3T3 cells using bright-field time-lapse microscopy. In their experiments, serum withdrawal within the first 4 hr after mitosis led to an immediate lengthening of G1 within the same cell cycle, in apparent conflict with our findings. As in MCF10A, we found that CDK2inc Swiss 3T3 cells (only 20% of cells) were immune to serum withdrawal at any time in G1, S, or G2, but would enter into the CDK2low state after completing the next mitosis (Figures 6F and 6G). However, a majority (80%) of Swiss 3T3 cells naturally enter the CDK2low state after mitosis (Figure 3E) and therefore remain sensitive to serum withdrawal as long as they have low CDK2 activity (Figures 6F–6G and S6). Thus, Zetterberg and Larsson (1985) likely described a restriction point (serum-sensitivity window) in mid to late G1 because they used a cell type in which the dominant behavior is entry into the CDK2low state after mitosis.
These results suggest that cells integrate mitogenic inputs in G2/M of the previous cell cycle during a first restriction window (R1), and then bifurcate into either a CDK2inc or CDK2low cell fate after completing mitosis. One consequence of the location of R1 is that sister cells typically share the same fate upon completion of mitosis. Once cells enter the CDK2inc state, they are committed to completing the cell cycle whether mitogenic stimuli and MEK activity are present or not. Cells in the CDK2low state face another decision point that is mitogen sensitive (R2) in which they can build up CDK2 activity and reenter the cell cycle. If mitogens are withdrawn or MEK is inhibited while cells are in the CDK2low state, cells will persist in this state (Figure 6H).
DISCUSSION
A Live-Cell Sensor for CDK2 Activity that Marks a G0-Like State and Tracks Interphase Progression
The ability to monitor events at the molecular level in unperturbed single cells marks an improvement over previous methods of studying the G0 and G1 phases of the cell cycle. Although there may be many forms of quiescence (O'Farrell, 2011), a unifying feature of quiescence is a lack of progression through the cell cycle, which is readily detectable with our CDK2 sensor. It is tempting to use this new capability to monitor CDK2 activity in single cells to revisit the nomenclature of G0 and G1 phases of the cell cycle. For cells emerging from a starvation-induced G0 state, the CDK2low state could be interpreted as a continued G0 state that persists for at least 8 hr after mitogen addition, and the initial activation of CDK2 could be interpreted as the beginning of the G1 phase. We therefore propose that the CDK2low state could serve as a marker for the currently imprecisely defined G0 phase, and that the initiation of CDK2 activation could serve as a marker for the start of G1. Alternatively, the CDK2low state could be described as early G1 and entry into the CDK2inc state as late G1, but this nomenclature does not distinguish if or when a switch from G0 to early G1 occurs, nor does it reflect the fact that a fraction of the CDK2low cells remain permanently in this state. The stereotypical increase in CDK2 activity throughout G1, S, and G2 demonstrates that DHB-Ven also provides a long sought-after molecular timer for progression through interphase. Finally, previous classifications of committed versus uncommitted cells required a perturbation (mitogen withdrawal) followed by observation of future events (mitosis) in order to retrospectively determine whether a cell had crossed the restriction point at the time of mitogen withdrawal. In contrast, our CDK2 sensor allows one to determine a cell's commitment to the cell cycle without needing to perturb it or monitor its future behavior.
Unification of Our Results and Previous Observations
There are several discrepancies between our results and previous publications. First, we were initially surprised to observe a gradual increase in CDK2 activity beginning shortly after nuclear envelope reformation, as it is commonly believed that CDK2 activity only increases at the end of G1 (Hochegger et al., 2008; Massagué, 2004). Second, the presence of hyperphosphorylated Rb in newly born CDK2inc cells in our studies was unexpected because previous studies found Rb to be fully dephosphorylated during mitosis and only fully rephosphorylated in late G1 (Buchkovich et al., 1989; Chen et al., 1989; Ludlow et al., 1993; Ludlow et al., 1990; Mihara et al., 1989; Weinberg, 1995). Third, we initially expected to see a boost in CDK2 activity at the G1/S transition in cycling cells as CDK2 overcomes inhibition by p21 and promotes p21 degradation (Abbas and Dutta, 2009). Instead, we observe a steady ramp of CDK2 activity in CDK2inc cells that is unaffected by the rapid induction of ubiquitin ligase activity at the G1/S transition. This is consistent with a fourth unanticipated result, in which we find that p21 levels remain uniformly low in CDK2inc cells, starting from birth.
Our use of single-cell analysis in cycling cells can explain the discrepancies with past studies in which synchronization and bulk analysis were used. The previously observed multihour delay in the phosphorylation of Rb and the activation of CDK2 can be attributed to the use of synchronization via mitogen starvation, which sets cells far back into G0. Measurements of the extent of Rb dephosphorylation in mitosis were previously performed in cells synchronized by nocodazole, which triggers a stress response and increases p21 (Lanni and Jacks, 1998; Uetake and Sluder, 2010), and we show in our experiments that increased p21 during G2 causes entry into the CDK2low and hypophosphorylated Rb states in the next cell cycle. The lack of a noticeable increase in CDK2 activity at the G1/S transition can be explained by the fact that CDK2inc cells start the cell cycle with already low p21. Thus, we believe that our results represent an internally consistent set of observations.
Identification of a Bifurcation in CDK2 activity at the End of Mitosis That Is Controlled by p21
Our study shows that cycling cells cross a bifurcation point at the end of mitosis where they either immediately build up CDK2 activity and start the next cell cycle or enter a transient G0 state characterized by low CDK2 activity (Figure 7A). Independent of whether cells enter the cell cycle directly from mitosis or out of a G0-like state, we identified a requirement for a threshold level of CDK2 activity for DNA replication to begin. This notion is consistent with previous work in S. pombe demonstrating that S phase is triggered at an intermediate threshold level of CDK activity (Coudreuse and Nurse, 2010).
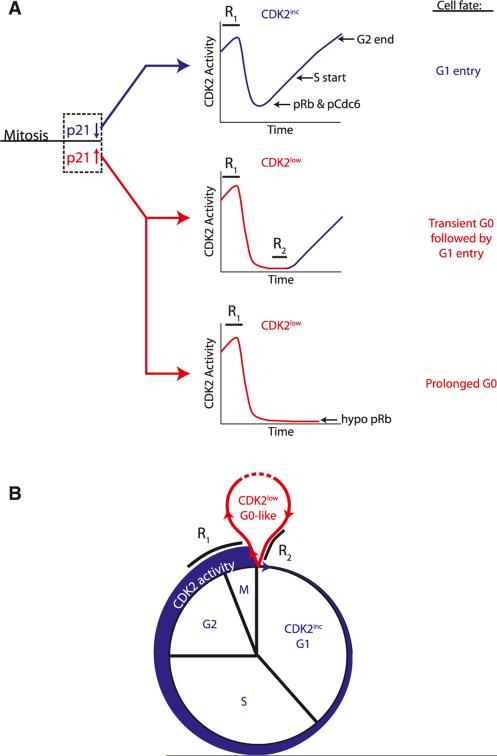
Figure 7. Model for Cell-Cycle Commitment and Transitions between G0 and G1.
(A) Cells choose between CDK2inc and CDK2low fates during a restriction window (R1) at the end of the previous cell cycle. Suppression of p21 protein during this period sends cells into the CDK2inc state upon completion of mitosis. These cells are born into G1, committed to the cell cycle, with low p21, residual CDK2 activity, hyperphosphorylated Rb, and enter S phase in 5–7 hr. Elevation of p21 protein at the end of the previous cycle sends cells into the CDK2low state upon completion of mitosis. These cells are born into a transient or prolonged G0-like state with high p21, no residual CDK2 activity, and hypophosphorylated Rb. These cells remain sensitive to mitogen withdrawal and are not committed to the cell cycle until they pass a second restriction window (R2) prior to the buildup of CDK2 activity.
(B) Upon exit from mitosis, cells execute a quiescence-proliferation decision by choosing between a CDK2low, G0-like state (red), and a CDK2inc, G1 state (blue).
The fraction of cells that bifurcate into the CDK2inc or CDK2 low state after mitosis varies among different cell types and is regulated by the strength of mitogen stimulation. CDK2inc cells have a relatively short and invariant G1 phase of approximately 5–7 hr duration. In contrast, CDK2low cells spend variable amounts of time in this transient G0-like state that lengthens the total cell-cycle time and explains most of the observed variability in intermitotic time. Thus, the fraction of cells that pass through a G0-like state and the amount of time they remain in this state appear to be critical determinants of the population doubling time.
Under optimal proliferation conditions, the path a cell takes in the bifurcation is directly controlled by p21. We demonstrate the central role of p21 by showing (1) that the CDK2inc versus CDK2low decision closely correlates with the endogenous level of p21 in single cells, (2) that p21-null MCF10A cells rarely enter the CDK2low state, and (3) that rapid induction of p21 protein immediately prior to mitosis causes cells to enter the CDK2low state upon completion of mitosis. Together, these results show that, for our experimental conditions, p21 is necessary and sufficient to control the bifurcation in CDK2 activity at mitotic exit. There are two potential benefits to the tunability of this system. At a cell-intrinsic level, even healthy cycling cells need time to deal with metabolic imbalances or errors arising from DNA replication and mitosis and the CDK2low state may represent a pause for cells to deal with these challenges. At a tissue level, the ability to regulate the fraction of cells entering a proliferative or a quiescent state allows for control of tissue homeostasis.
Evidence for Two Restriction Windows in the Cell Cycle
The restriction point was originally defined as a point in late G1 after which cells would continue through the cell cycle even if mitogens were withdrawn (Pardee, 1974). It has also been proposed that commitment to the next cell cycle is made at the end of the preceding cycle (Chassot et al., 2008; Hitomi and Stacey, 1999). Our study argues that these apparently contradictory models can be explained by taking into account the two types of cell behaviors that we have observed within the same cell population. Our results argue that cells integrate mitogenic and potentially other inputs during a restriction window at the end of the previous cell cycle (R1) to regulate the bifurcation into the CDK2inc or CDK2low state upon completion of mitosis. Only the cells in the CDK2low state experience a second restriction window (R2) in which mitogens are needed to re-enter the cell cycle and build up CDK2 activity. Given the different fractions of CDK2low cells we found in the cell types we tested, the relative importance of R1 versus R2 will vary depending on the cell type, strength of mitogen stimuli and other conditions. Although the two restriction windows R1 and R2 cover different phases of the cell cycle, they reflect an underlying principle that cells require the continued presence of mitogens for several hours before they commit to building up CDK2 activity (Figure 7B). Finally, the relationship between CDK2 activity and cell-cycle commitment that we describe here may be integral to other cell fate decisions such as the senescence of somatic cells, the differentiation of stem cells, or the progression of cancer cells.
EXPERIMENTAL PROCEDURES
Cell Lines
MCF10A human mammary epithelial cells were obtained from the laboratory of Dr. Joan Brugge and cultured as described previously in MCF10A full growth media (Debnath et al., 2003). Hs68 primary human foreskin fibroblasts (#CRL-1635) and Swiss 3T3 murine fibroblasts (#CCL-92) were obtained from ATCC and cultured in DMEM plus 10% FBS and penicillin/streptomycin. Hs68 cells were used for experiments up to passage 16 after receipt of the cells.
Constructs
cDNAs for Histone 2B fused to mCherry or mCerulean, Cdt1 (aa30–120) (Sakaue-Sawano et al., 2008) fused to mCerulean, and DHB (aa994–1087) (Hahn et al., 2009) fused to mVenus were cloned into the CSII-EF lentiviral vector. Transduced cells were sorted on a Becton Dickinson Influx to obtain pure populations expressing the desired fluorescent reporters. cDNAs for DHFRR12Y/G67S/Y100I (Iwamoto et al., 2010), mCherry, and human p21 were cloned into the pCru5 MoMLV retroviral vector and selected with 1 μg/ml puromycin for 3 days.
Immunofluorescence
Cells were fixed in 4% formaldehyde, washed twice with PBS, permeabilized with 0.2% triton, and stained overnight at 4°C with αpRb807/811 (Cell Signaling Technology #9308) or ap21 (BD #556430). Primary antibodies were visualized using a secondary antibody conjugated to Alexa Fluor-647 and imaged with a far red filter. For Figure 3I, cells expressing DHB-Ven were treated with 10 μM EdU for 15 min and fixed and processed according to manufacturer's instructions (Invitrogen # C10356).
Inhibitors
The inhibitors used in this study were: PD0325901 (MEK inhibitor), Gefitinib (EGFR inhibitor), SB 203580 (p38 inhibitor), MK-2206 (Akt inhibitor), GDC-0941 (PI3K inhibitor), VX-680 (Aurora kinase inhibitor), RO3306 (CDK1 inhibitor), PD0332991 (CDK4/6 inhibitor), and EMD Biosciences #217714 (CDK1/2 inhibitor).
siRNA Transfection
MCF10A cells were transfected using Dharmafect 1 or 3 (Thermo Scientific) according to the manufacturer's instructions. The following siRNAs were used: control siRNA (nontargeting #2), siGenome pooled set of four siRNAs for cyclin A2, or siGenome pooled set of eight siRNAs for cyclin E1 and E2 (Dharmacon) at final concentrations of 20–25 nM.
In Vitro Kinase Assays
Kinase assays were performed by ProQinase GmbH. See Extended Experimental Procedures.
Time-Lapse Microscopy and Image Processing
Images were taken on an ImagExpress5000A or on an IXMicro microscope (Molecular Devices) and image processing was performed in MATLAB (MathWorks). See Extended Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jia-Yun Chen, Sean Collins, James Ferrell, Xuedong Liu, Julien Sage, Matthew Scott, and John Albeck for helpful discussions; Matthew Clutter, Garry Nolan, Ben Ho Park, Kyuho Han, and Akiko Seki for reagents; and the Stanford Shared FACS Facility for cell sorting. S.L.S and S.D.C were supported by the Damon Runyon Cancer Research Foundation (DRG-[2043-10] and DRG-[2141-12]). S.L.S was also supported by an American Cancer Society Robert and Mary Ann Forsland Postdoctoral Fellowship (PF-13-304-01-CCG). T.M. was supported by NIH grants CA120732 and MH095087.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, six figures, one table, and four movies and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.08.062.
REFERENCES
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman KE, Blair BG, Brenner K, Bardelli A, Arena S, Zhou S, Hicks J, De Marzo AM, Argani P, Park BH. p21(WAF1/CIP1) mediates the growth response to TGF-beta in human epithelial cells. Cancer Biol. Ther. 2004;3:221–225. doi: 10.4161/cbt.3.2.666. [DOI] [PubMed] [Google Scholar]
- Buchkovich K, Duffy LA, Harlow E. The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell. 1989;58:1097–1105. doi: 10.1016/0092-8674(89)90508-4. [DOI] [PubMed] [Google Scholar]
- Chassot AA, Lossaint G, Turchi L, Meneguzzi G, Fisher D, Ponzio G, Dulic V. Confluence-induced cell cycle exit involves pre-mitotic CDK inhibition by p27(Kip1) and cyclin D1 downregulation. Cell Cycle. 2008;7:2038–2046. doi: 10.4161/cc.7.13.6233. [DOI] [PubMed] [Google Scholar]
- Chen PL, Scully P, Shew JY, Wang JY, Lee WH. Phosphor-ylation of the retinoblastoma gene product is modulated during the cell cycle and cellular differentiation. Cell. 1989;58:1193–1198. doi: 10.1016/0092-8674(89)90517-5. [DOI] [PubMed] [Google Scholar]
- Coller HA. What's taking so long? S-phase entry from quiescence versus proliferation. Nat. Rev. Mol. Cell Biol. 2007;8:667–670. doi: 10.1038/nrm2223. [DOI] [PubMed] [Google Scholar]
- Coudreuse D, Nurse P. Driving the cell cycle with a minimal CDK control network. Nature. 2010;468:1074–1079. doi: 10.1038/nature09543. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Dou QP, Levin AH, Zhao S, Pardee AB. Cyclin E and cyclin A as candidates for the restriction point protein. Cancer Res. 1993;53:1493–1497. [PubMed] [Google Scholar]
- Dulić V, Lees E, Reed SI. Association of human cyclin E with a periodic G1-S phase protein kinase. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- Gu J, Xia X, Yan P, Liu H, Podust VN, Reynolds AB, Fanning E. Cell cycle-dependent regulation of a human DNA helicase that localizes in DNA damage foci. Mol. Biol. Cell. 2004;15:3320–3332. doi: 10.1091/mbc.E04-03-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn AT, Jones JT, Meyer T. Quantitative analysis of cell cycle phase durations and PC12 differentiation using fluorescent biosensors. Cell Cycle. 2009;8:1044–1052. doi: 10.4161/cc.8.7.8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi M, Stacey DW. Cellular ras and cyclin D1 are required during different cell cycle periods in cycling NIH 3T3 cells. Mol. Cell. Biol. 1999;19:4623–4632. doi: 10.1128/mcb.19.7.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochegger H, Takeda S, Hunt T. Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nat. Rev. Mol. Cell Biol. 2008;9:910–916. doi: 10.1038/nrm2510. [DOI] [PubMed] [Google Scholar]
- Iwamoto M, Björklund T, Lundberg C, Kirik D, Wandless TJ. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem. Biol. 2010;17:981–988. doi: 10.1016/j.chembiol.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koff A, Giordano A, Desai D, Yamashita K, Harper JW, Elledge S, Nishimoto T, Morgan DO, Franza BR, Roberts JM. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- Lanni JS, Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol. Cell. Biol. 1998;18:1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow JW, Shon J, Pipas JM, Livingston DM, DeCaprio JA. The retinoblastoma susceptibility gene product undergoes cell cycle-dependent dephosphorylation and binding to and release from SV40 large T. Cell. 1990;60:387–396. doi: 10.1016/0092-8674(90)90590-b. [DOI] [PubMed] [Google Scholar]
- Ludlow JW, Glendening CL, Livingston DM, DeCarprio JA. Specific enzymatic dephosphorylation of the retinoblastoma protein. Mol. Cell. Biol. 1993;13:367–372. doi: 10.1128/mcb.13.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. G1 cell-cycle control and cancer. Nature. 2004;432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- Mihara K, Cao XR, Yen A, Chandler S, Driscoll B, Murphree AL, T'Ang A, Fung YK. Cell cycle-dependent regulation of phosphorylation of the human retinoblastoma gene product. Science. 1989;246:1300–1303. doi: 10.1126/science.2588006. [DOI] [PubMed] [Google Scholar]
- O'Farrell PH. Quiescence: early evolutionary origins and universality do not imply uniformity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011;366:3498–3507. doi: 10.1098/rstb.2011.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo M, Theodoras AM, Schumacher J, Roberts JM, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol. Cell. Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee AB. A restriction point for control of normal animal cell proliferation. Proc. Natl. Acad. Sci. USA. 1974;71:1286–1290. doi: 10.1073/pnas.71.4.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM. Regulation of cell reproduction. Cancer Res. 1968;28:1815–1820. [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr. Biol. 2010;20:1666–1671. doi: 10.1016/j.cub.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- Yao G, Lee TJ, Mori S, Nevins JR, You L. A bistable Rb-E2F switch underlies the restriction point. Nat. Cell Biol. 2008;10:476–482. doi: 10.1038/ncb1711. [DOI] [PubMed] [Google Scholar]
- Zetterberg A, Larsson O. Kinetic analysis of regulatory events in G1 leading to proliferation or quiescence of Swiss 3T3 cells. Proc. Natl. Acad. Sci. USA. 1985;82:5365–5369. doi: 10.1073/pnas.82.16.5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterberg A, Larsson O, Wiman KG. What is the restriction point? Curr. Opin. Cell Biol. 1995;7:835–842. doi: 10.1016/0955-0674(95)80067-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.