Abstract
This review highlights recent progress in developing DNA aptamers for personalized medicine, with more focus on in vivo studies for potential clinical applications. Examples include design of aptamers in combination with DNA nanostructures, nanomaterials, or microfluidic devices as diagnostic probes or therapeutic agents for cancers and other diseases. The use of aptamers as targeting agents in drug delivery is also covered. The advantages and future directions of such DNA aptamer-based technology for the continued development of personalized medicine are discussed.
Keywords: DNA aptamer, personalized medicine, cancer, diagnosis, therapy, targeted delivery
1. Introduction
1.1 Personalized Medicine
In the last decade we have witnessed an explosive growth of research and development in personalized medicine. The advances in this area promise to improve healthcare while lowering costs of treatment. Despite significant progress, challenges still remain in shifting from traditional medicine to personalized medicine. For instance, clinical tools for the detection of genetic, proteomic, or metabolite biomarkers are required to correctly assess differences in individual conditions, in order to make appropriate medical decisions. Similarly, therapeutic methods for treating diseases need to shift from the administration of broadly acting therapeutic drugs towards the use of more specific drugs or dosages customized to each patient. Among the many approaches for meeting these demands, the use of DNA aptamers are a promising emergent approach, whose growth parallels that of personalized medicine (Figure 1).
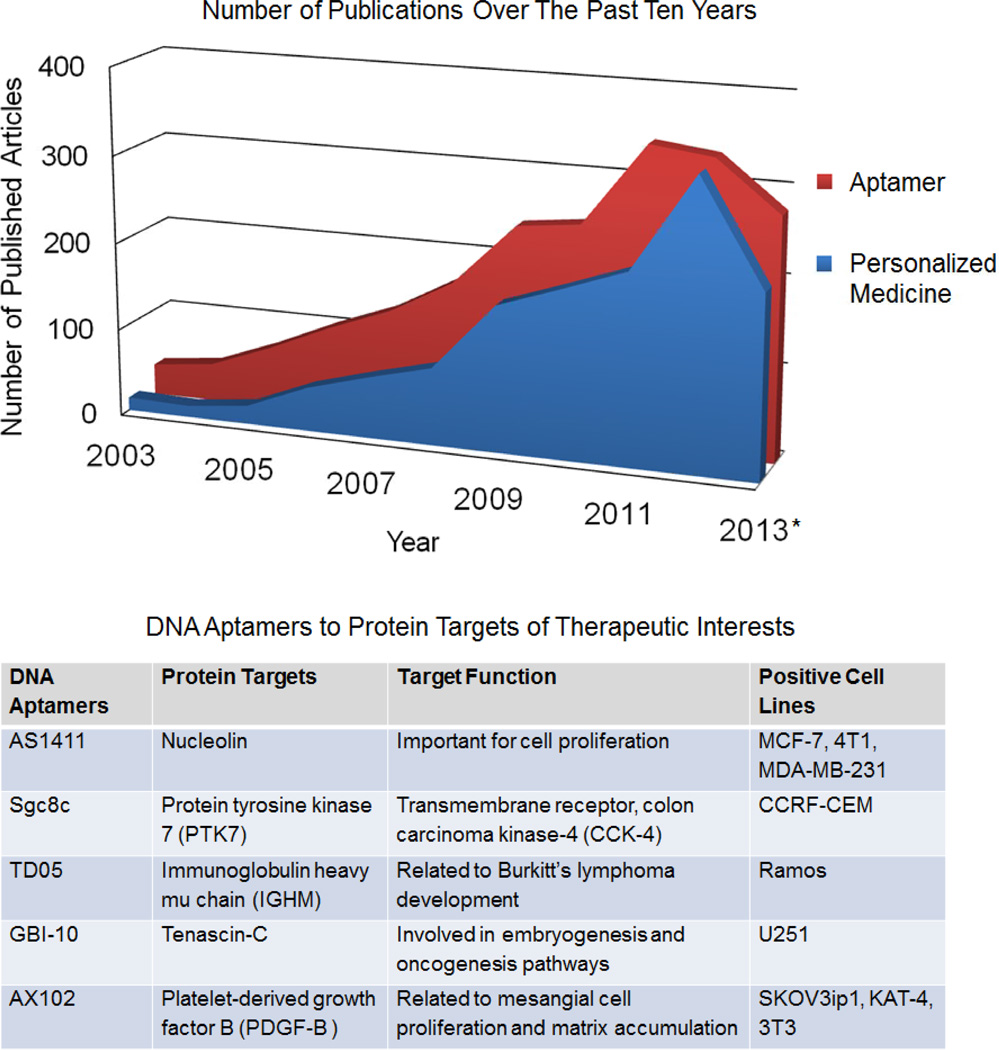
Figure 1.
Top: Graph showing the number of publications over the past ten years with title containing “aptamer” (red) or “personalized medicine” (blue) respectively, according to Web of Science. * The number of publications in 2013 is counted until November. Bottom: Representative DNA aptamers to protein targets of therapeutic interest.
1.2 Overview of DNA Aptamers
A major issue in personalized medicine is the introduction of target specificity or related functionality to diagnostic tools and therapeutic agents. Aptamers, sequences of nucleic acids that are capable of recognizing and binding to a specific target, have become one of the most promising tools for this purpose. Aptamers are selected from pools of oligonucleotides with random sequences in a process called Systematic Evolution of Ligands by EXponential enrichment (SELEX) [1–3]. They can recognize a wide range of biomedically relevant targets including metal ions, small molecules, peptides and proteins, with high affinity and specificity [4–8]. While aptamers may consist of either DNA or RNA, DNA aptamers are more stable against biodegradation than their RNA congeners. Compared with antibodies, aptamers exhibit significant advantages in terms of size, stability, non-immunogenicity, and synthetic accessibility. Furthermore, new aptamers can be selected through a well-established selection process towards any biomedically important target to advance pharmaceutical development, particularly small molecular targets such as organic metabolites, metal ions, or other biomarkers. Owing to these advantages, research in the selection of new aptamers, characterization of their physical properties, as well as application in a variety of biomedical systems has greatly increased over the past decade, and researchers have identified aptamers with high binding affinity for a wide range of clinical targets including kinases, growth factors, and cell-surface receptors (Figure 1) [9–12]. This review highlights recent work on the use of DNA aptamers for diagnosis, targeted therapy of cancers and other diseases, with special focus on those studies that demonstrate in vivo efficacy for potential clinical applications.
2. DNA Aptamer-Based Techniques for Cancer Diagnosis
Cancer has a major impact on society today. The World Health Organization (WHO) has reported that 7.6 million people die of cancer every year [13]. Identification of cancer cells at the earliest stage is critical to the successful prevention and effective treatment of cancers. Therefore, developing DNA aptamer-based diagnostic tools for cancer cells with high sensitivity and selectivity is important for the continued improvement of clinical cancer management [14].
2.1. Aptamer-containing DNA Nanostructures as Cancer Probes
DNA aptamers capable of recognizing biomarkers or cancer cells can be obtained through in vitro selection or cell-SELEX [15]. When modified with fluorophores, these functional DNA strands can be used as molecular probes for in vivo identification and imaging of cancer cells. The cell-SELEX approach has been adopted to obtain aptamers that specifically bind to and be internalized by glioblastoma (GBM) tumor-initiating cells (TIC). These aptamers were further able to differentiate cells with high tumorigenic potential from GBM xenografts [16]. Another example showed that the use of a DNA aptamer against the A549 lung carcinoma cell line allowed in vivo fluorescence imaging of carcinomas [17].
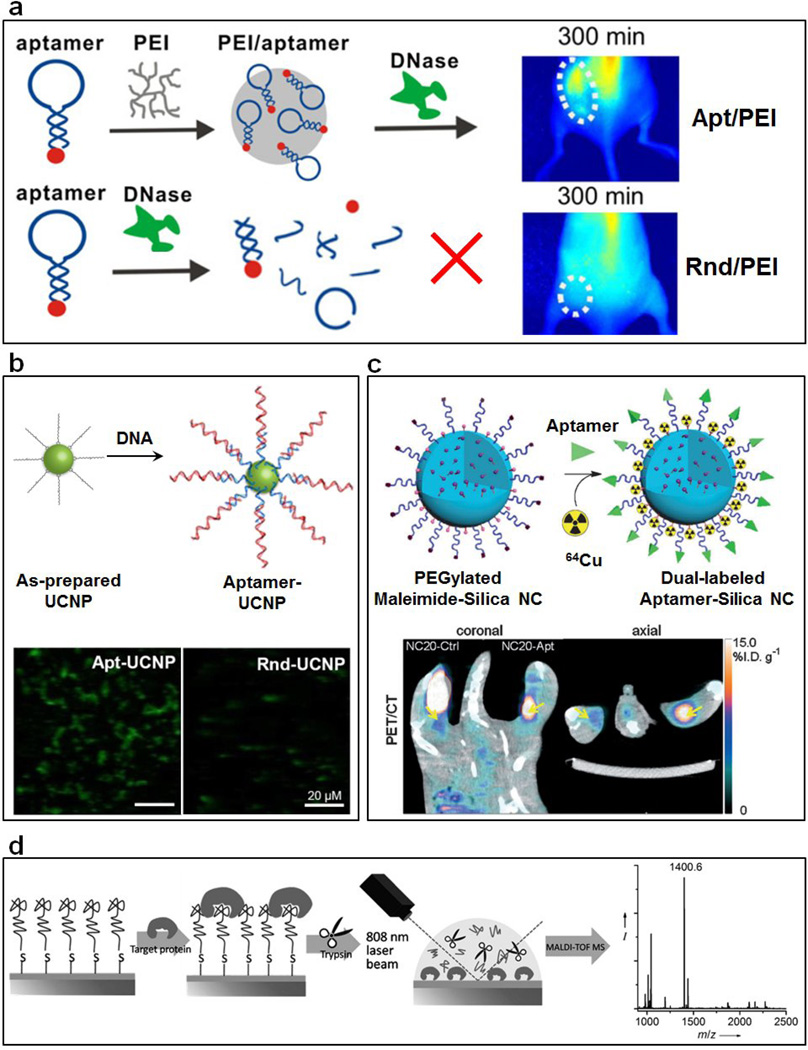
To enhance their performance and functionality, aptamers can be further incorporated into DNA nanostructures. An activatable aptamer probe (AAP) featuring a sgc8 aptamer targeting protein tyrosine kinase-7 (PTK7), a poly-T linker and a short DNA strand can form a molecular beacon structure [18]. Animal studies confirmed that such AAPs could be activated through cell membrane protein-triggered conformational changes, resulting in enhanced fluorescence signals at CCRF-CEM tumor sites. To increase the stability of DNA aptamer probes, branched polyethyleneimine (PEI) was used as a vector to deliver a TD05 aptamer-based probe (Figure 2a) [19]. Such PEI/aptamer probes showed higher stability against DNase degradation and were used for in vivo imaging of a Ramos tumor in mice. Moreover, a more sophisticated DNA-based nanorobot made by DNA origami method was reported for delivery of biologically active payloads for cell-targeting [20]. This stimuli-responsive device was locked with DNA aptamers in a dual-lock mode so that the nanorobot would open and release its payload only in the presence of two different target molecules.
Figure 2.
(a) Schematic illustration of the protection of PEI on DNA and targeted imaging with PEI/aptamer complexes. Adapted from [19]. (b) Schematic view of the synthesis of DNA aptamer-functionalized UCNPs from as-prepared hydrophobic UCNPs and targeted imaging using aptamer-modified UCNPs. Confocal microscopy images of MCF-7 cells treated with aptamer-UCNPs (left) and control-UCNPs (right). Adapted from [21]. (c) Schematic view of the preparation of aptamer-functionalized dual-labeled silica NCs for PET and NIR fluorescence imaging. In vivo whole-body PET/CT imaging of BALB/c mice after hock injection of dual-labeled control-NCs and aptamer-NCs. Adapted from [22]. (d) Design of the aptamer-assisted selective high-throughput detection of biomarkers using MALDI-TOF MS. Adapted from [30].
2.2 Aptamer-Conjugated Nanomaterials as Cancer Probes
The conjugation of high-specificity DNA aptamers with nanomaterials featuring unique optical or magnetic properties has resulted in many innovative imaging agents for cancer diagnosis. A prominent example nanomaterial is the luminescent upconversion nanoparticle (UCNP). UCNPs are capable of converting near-infrared (NIR) excitation light into shorter wavelength visible luminescence, which is ideal for deep tissue bioimaging. However, functionalization of such UCNPs for targeting is difficult. Recently, our lab reported a one-step strategy to prepare uniform DNA-modified UCNPs through ligand exchange at the liquid–liquid interface (Figure 2b) [21]. The nucleolin DNA aptamer remained functional on the UCNP surface and enabled specific targeting of MCF-7 cancer cells and cell membrane penetration, with high internalization efficiency.
Besides UCNPs, other aptamer-modified nanomaterials allow different techniques to be used for imaging of tumors. Aptamer-modified, monodisperse silica nanoparticles have been synthesized as probes for multimodal imaging of lymph nodes (Figure 2c) [22]. Positron emission tomography (PET) and NIR fluorescence imaging confirmed that nucleolin aptamer-directed silica nanoparticles accumulated in lymph nodes containing metastatic breast tumors using a 4T1 tumor model. In addition, an AS1411 aptamer-modified cobalt–ferrite nanoparticle was used for targeted multi-model imaging of C6 tumors in mice [23]. Moreover, aptamer-modified nano/micro-sized micelle bubbles [24], quantum dots [25], as well as iron oxide nanoparticles [26] have also been recently reported for cell-specific ultrasound, fluorescence, and magnetic resonance imaging, respectively.
2.3 Aptamers in Combination with Analytical Techniques for Cancer Detection
In addition to conjugation with nanostructures and nanomaterials, DNA aptamers have also been combined with common analytical techniques to enhance sensitivity and specificity. For example, a microfluidic device was combined with the sgc8 aptamer to develop a 3D DNA platform for efficient detection and isolation of CCRF-CEM cancer cells in whole blood samples [27]. In addition, the use of multivalent gold nanoparticle-aptamer conjugates significantly increased the capture of circulating tumor cells in a microfluidic device [28].
In addition, a general approach to the de novo design of electrochemical biosensors with aptamer functionalization has been reported through in vitro selection and functional electrode construction [29]. This approach was able to detect lung cancer biomarker CTAP III/NAP2 with high specificity and sensitivity. Finally, an aptamer microarray was combined with MALDI-TOF MS for high-throughput on-target analysis of protein biomarkers (Figure 2d) [30].
3. DNA Aptamer-based Techniques for Cancer Therapy
Traditional cancer chemotherapy suffers from severe side effects and low therapeutic index. The use of DNA aptamers either alone, as a therapeutic, or in combination with nanomaterials, as a targeting agent, has shown great promise for the improvement of anticancer efficacy while reducing side effects.
3.1 DNA Aptamers as Anticancer Therapeutic Agents
Aptamers can be selected to bind specific protein targets of clinical interest. Among them, some aptamers have been found to have therapeutic effects in a manner similar to monoclonal antibodies, as the binding of aptamers may block the active site of a protein or inhibit ligand-receptor interactions, and thus may function as anticancer therapeutics [14, 31]. The first DNA aptamer-based anticancer drug entering clinical trials was AS1411, which is a 26-mer G-quadruplex oligodeoxynucleotide that binds to nucleolin [9]. Although AS1411 is currently in phase II clinical trials for the treatment of acute myeloid leukaemia, its mechanism of action is not completely clear. A recent study suggests that the anticancer activity of AS1411 might be caused by hyperstimulation of macropinocytosis after binding to nucleolin [32].
Besides AS1411, several new DNA aptamers have been discovered with anticancer properties. For example, a serum-stabilized DNA aptamer for immunotherapy of CD30-expressing lymphoma was generated [33]. In addition, two DNA aptamers which bind to carcinoembryonic antigen (CEA) were selected and used for the pre-treatment of a CEA-expressing tumor model in vivo [34]. Moreover, a DNA aptamer recognizing human epidermal growth factor receptor 2 (ErbB-2/HER2) was recently shown to have antitumor efficacy two-fold higher than a corresponding monoclonal anti-ErbB-2/HER2 antibody, in both human gastric cancer cells and in mice bearing tumor xenographs [35].
3.2 Aptamer-containing DNA Nanostructures for Cancer Therapy
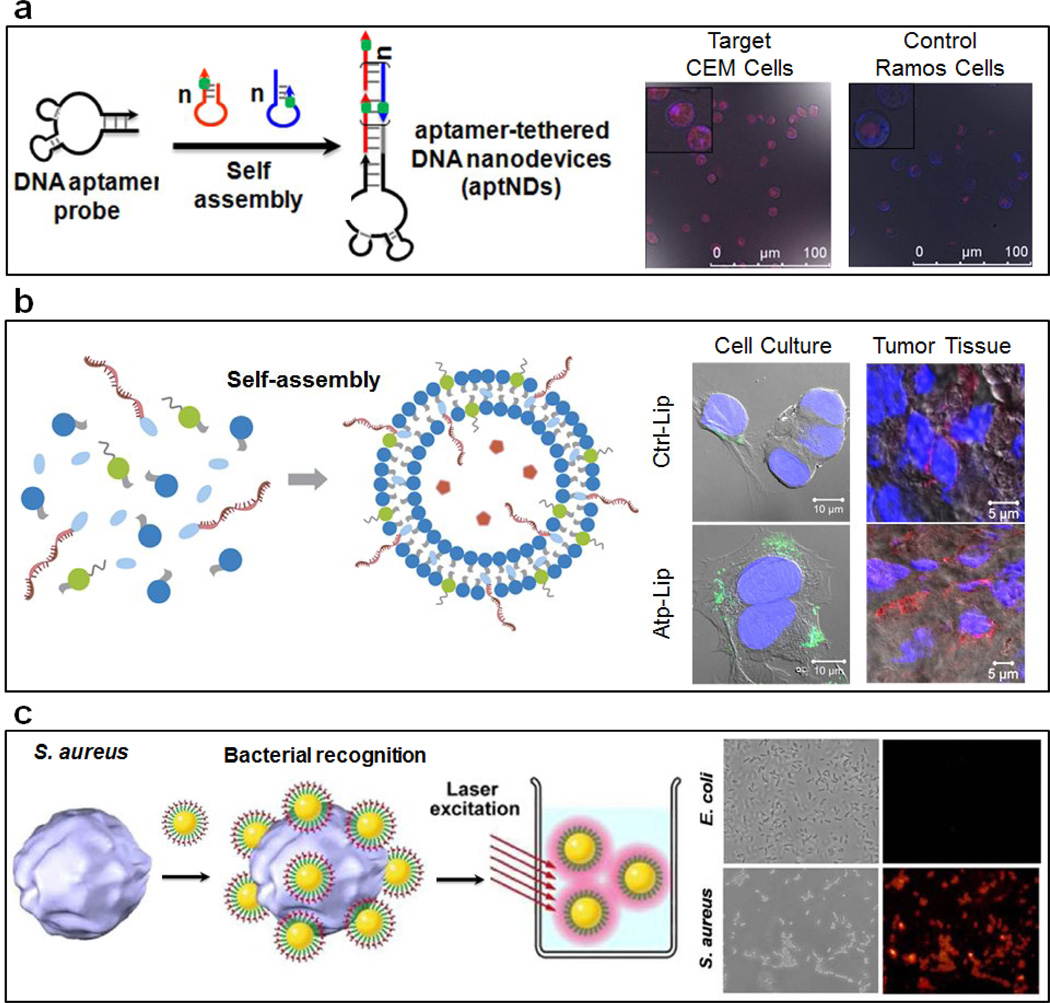
While aptamers can be used directly as therapeutic agents, they require a relatively large dosage in order to be effective. Since delivery of the highly negatively charged DNA is a major hurdle, an alternate approach using DNA as a targeting agent in combination with other drugs may be more promising, as this approach takes advantage of the high selectivity of aptamer with the strong therapeutic potential of known drugs; even if relatively few DNA strands are delivered to the target, the high loading of the drug may still allow the combined system to be effective. Along these lines, it has been shown that DNA nanostructures can be used as a delivery system with antitumor drugs intercalated within strands of double-stranded DNA (dsDNA) [36]. Together with cancer cell-specific aptamers, different DNA nanostructures have been demonstrated as targeting vectors for the delivery of anticancer agents. As an example, an aptamer-tethered, long linear dsDNA structure, dubbed a “DNA nanotrain” (aptNTr) has been developed to deliver anticancer drugs and bioimaging agents to tumor cells (Figure 3a) [37, 38]. The long dsDNA chain serves as a carrier in which doxorubicin may be loaded with very high density. The enhanced antitumor efficacy and reduced side effects of drugs delivered by aptNTrs were demonstrated in a xenograft tumor model in vivo. Similarly, a poly-aptamer-drug system was constructed by rolling circle amplification using a leukemia cell-binding aptamer and doxorubicin-loaded strands. Enhanced targeting effects and improved cellular uptake were observed as compared to the monovalent counterpart, due to aptamer multivalency [39].
Figure 3.
(a) Schematic of the self-assembly of aptamer-tethered DNA nanotrains (aptNTrs) and the illustration of the drug transportation process via aptNTrs to target cells. Adapted from [37]. (b) Schematic illustration of the assembly of aptamer-conjugated liposomes with encapsulated cargos and confocal microscope images of MCF-7 cells treated with non-aptamer-conjugated liposomes and aptamer-conjugated liposomes in cell culture (left) and in tumor tissue sections (right). Adapted from [40]. (c) Schematic view of the light-scattering detection of S. aureus cells using aptamer-conjugated gold nanoparticles. Adapted from [47].
3.3 Aptamers as Targeting Agents in Drug Delivery
While aptamers themselves have the potential to be therapeutic agents, their successful clinical application requires a large amount of aptamers in order to bind and suppress their targets. Given the difficulty in delivering sufficient quantities of DNA aptamers into cells or the human body, it may be difficult to achieve the desired therapeutic effect in patients. On the other hand, by using aptamers as targeting agents in conjunction with known drugs or drug delivery vehicles with high potency but less selectivity, one can achieve better efficacy without requiring the delivery of as much aptamer to the patient. One example is the conjugation of aptamers to drug-containing liposomes, which are one of the most successful drug delivery platforms in clinical use today. To confer selectivity upon this system, the AS1411 aptamer was conjugated to a 200 nm liposome loaded with doxorubicin (Figure 3b) [40]. Cellular and animal studies demonstrated that these highly stable liposomes were able to target MCF-7 cells specifically and improve the inhibition of tumor growth, attributable to enhanced tumor tissue penetration. More importantly, because the DNA aptamer sequence is known and the aptamer can bind its complementary DNA sequence strongly through hybridization, the effectiveness of this system can be tuned with different concentrations of the complementary DNA strand of the aptamer as an antidote [41].
Other delivery systems, such as micelles and polymer nanoparticles, have also been used in combination with cancer-specific DNA aptamers. For example, PEG-PLGA nanoparticles have been functionalized with the AS1411 nucleolin aptamer for targeted delivery of paclitaxel (PTX) to mice bearing C6 gliomas [42]. The biodistribution test showed enhanced tumor accumulation for the aptamer-functionalized nanoparticles compared to either the nanoparticles or drug alone. Prolonged circulation time and the targeting effect of the system facilitated tumor inhibition. Moreover, a lipid tail was attached onto the end of the TDO5 aptamer, forming an aptamermicelle with high binding affinity to Ramos cells and extremely low binding off rate (10−5–10−6 s−1) [43]. Finally, the aptamer-conjugated micelle could be used not only for the targeted imaging of cancer cells but also specific destruction of the same, via ultrasound-mediated acoustic droplet vaporization [44].
4. DNA Aptamers for Diagnostic and Therapeutic Applications of Other Diseases
While much effort has been devoted towards the detection and treatment of cancer, DNA aptamers have also been applied to other diseases. We highlight here a few such applications, again emphasizing those that have been applied in vivo or which are undergoing clinical trials.
4.1 Aptamer-Enabled Diagnosis of Other Diseases
One major area of application of aptamers is bacteria and virus detection. The generalizability of aptamer selection allows the aptamer to be tailored not only to a specific cell, bacteria, or viral target, but even to a particular protein or protein domain on the surface of these species. For example, five ssDNA aptamers capable of binding Staphylococcus aureus were identified, each aptamer binding different protein targets. They were used either individually or together for detection of S. aureus in clinical samples of pyogenic fluid taken from burn victims [45]. Further conjugation of these aptamers to the surface of single-walled carbon nanotubes (SWCNTs) allowed for detection of S. aureus via real-time potentiometry, down to a concentration of 800 colony forming units per mL; such sensitivity made it possible to detect bacteria from a sample of contaminated pig skin [46]. Similarly, gold nanoparticle-conjugated DNA aptamers were used for detection of S. aureus down to the single-cell level by means of light scattering measurement (Figure 3c) [47]. Another example involved the selection of DNA aptamers for the H5N1 an influenza virus. These aptamers were used for detection of the virus in bird swab samples using surface plasmon resonance [48].
4.2 Aptamer-Based Treatment for Other Diseases
The potential application of aptamers as therapeutic agents for other diseases has also been investigated. For example, an aptamer for an envelope protein of the Hepatitis C virus, identified through whole-cell SELEX, was shown to inhibit the interaction between the virus and the CD81 receptor of human cells [49]. Similar results were obtained using an aptamer identified from SELEX using a purified viral protein [50]. Both sets of aptamers demonstrated inhibition of HCV infection in cell culture models.
Another disease that has been investigated for aptamer-based therapy includes thrombosis, or obstruction of blood flow due to clotting. DNA aptamers have been selected for von Willebrand Factor (vWF), a glycoprotein implicated in thrombosis formation due to cardiovascular injury [51]. ARC1779 is an aptamer targeting vWF that has been studied for use as an inhibitor to platelet function, as a potential replacement of current anti-platelet treatments, which carry the significant drawback of hemorrhage [52]. Phase 1 and 2 studies have been carried out on this aptamer, which have indicated efficacy in treating a number of cardiovascular diseases (thrombocytopenia, type 2B von Willebrand disease) [53].
5. Summary and Perspectives
Much progress has been made in developing DNA aptamer-based techniques for diagnosis and therapy of cancers such as leukemias, lymphomas, gliomas, gastric and breast cancers, as well as other diseases including S. aureus infection, influenza, Hepatitis C, and thrombosis. This review focuses more on work that has demonstrated in vivo efficacy. Despite this progress, aptamer research in the laboratory has been slow to reach clinical applications in the hospital. So far, only one aptamer-based drug has been approved for marketing by the US Food and Drug Administration (FDA), with several others currently under clinical evaluation [54]. While many DNA aptamers have been well characterized and show promise, until now most work has only demonstrated proof-of-concept models, with a few showing application in animal models. With a highly negatively charged phosphate backbone, delivery of these DNA aptamers into cells and and the human body is still a challenge. Therefore, their effectiveness in vivo still requires further investigation in order to be used in future personalized medicine.
In addition, even though a number of aptamers for targets of interest in medicine have been identified, aptamers for many other targets of clinical significance have yet to be selected, such as small molecules that serve as disease markers in the blood or urine [55]. Another potential class of target may be metal ions, which are critical in many aspects of health and disease, including cancer [56]. The ability to obtain DNAzymes selective for a specific metal ion has already been demonstrated and will likely serve as the basis for such future work [57].
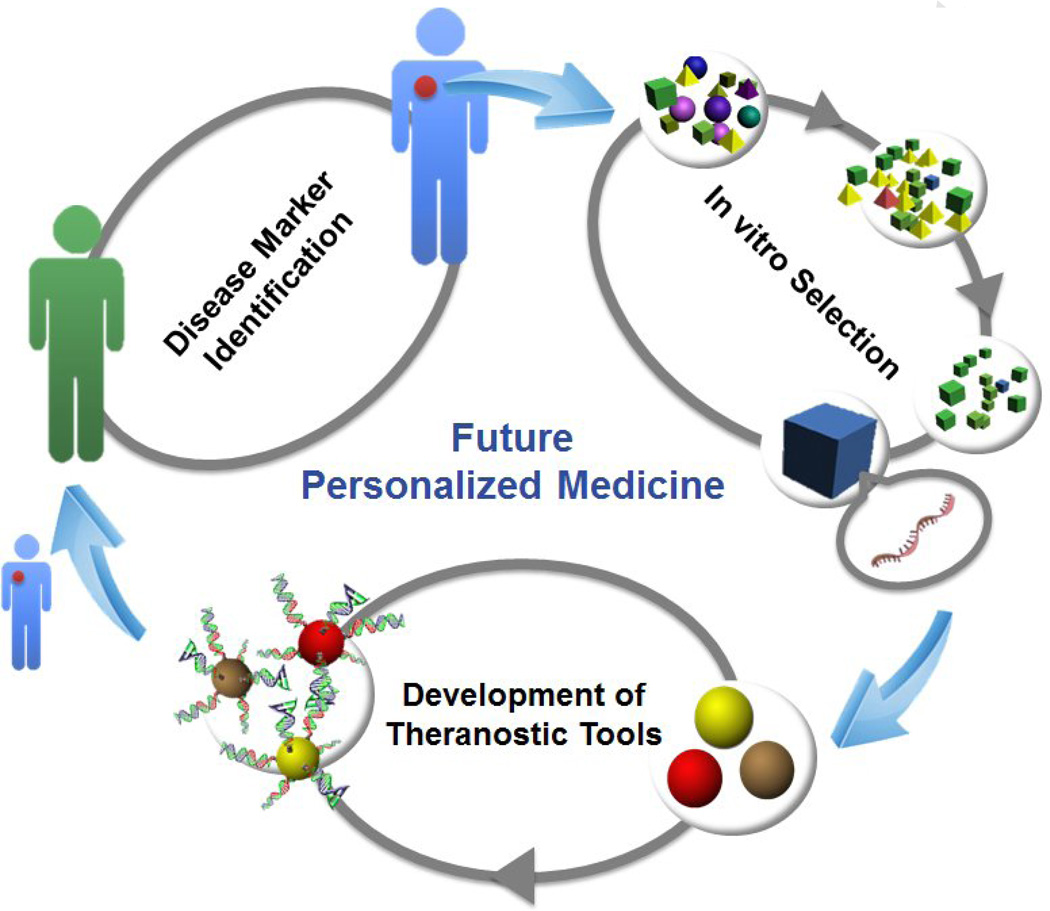
To realize the full potential of personalized medicine, diagnostic tests must become more portable, using simple devices for real-time and on-site detection and monitoring, instead of the laborious and time-consuming diagnostic tests currently available only in clinical labs. The incorporation of DNA aptamers into the personal glucose meter and dipsticks provides a general method for sensing a broad range of targets using simple devices [58, 59]. For therapy, there is still huge untapped potential in the combination of the target recognition ability of aptamers with exquisitely designed nanomaterials that can be used as effective drug delivery platforms. Many nanomaterials, such as liposomes, polymer vesicles and silica nanoparticles, combined with DNA aptamers, have shown feasibility for use in in vivo targeted drug delivery. In all therapeutic approaches, a general drug delivery platform with physiochemical stability, stimuli-responsiveness, controlled release profile and desired in vivo biodistribution will be needed to incorporate with specific aptamers selected from individuals. Finally, the combination of diagnosis and therapy in one system (theranostics) holds the key to the future success of personal medicine. To achieve such a goal, a development loop is required, in which the development of medicines will start from identification of disease markers from an individual patient, proceed through the expedited in vitro selection of DNA aptamers for these markers, and finally facilitate the production of theranostic tools specifically designed for that patient (Figure 4). Successful implementation of this development loop will make DNA nanostructures and aptamers key drivers for the future development of personalized medicine.
Figure 4.
A scheme showing how DNA aptamer technology powers future personalized medicine.
Highlights.
The development of DNA aptamers parallels that of personalized nanomedicine
Both aptamer-functionalized nanomaterials and DNA nanostructures have been developed
They have been used for detection, diagnosis, and treatment of cancer and other diseases
The review focuses more on studies that demonstrate in vivo efficacies
Acknowledgements
We thank the US National Institute of Health (ES016865) and the National Science Foundation (DMR-0117792, CTS-0120978 and DMI-0328162) for financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L. Systematic Evolution of Ligands by Exponential Enrichment - RNA ligands to bacteriophage-T4 DNA-polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Beaudry AA, Joyce GF. Directed evolution of an RNA enzyme. Science. 1992;257:635–641. doi: 10.1126/science.1496376. [DOI] [PubMed] [Google Scholar]
- 4. Willner I, Zayats M. Electronic aptamer-based sensors. Angew Chem Int Ed. 2007;46:6408–6418. doi: 10.1002/anie.200604524. * A comprehensive review about functional nucleic acid sensors, including nucleic acid enzyme, aptamer, and aptazyme -based sensors.
- 5.Liu JW, Cao ZH, Lu Y. Functional nucleic acid sensors. Chem Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li D, Song SP, Fan CH. Target-responsive structural switching for nucleic acid-based sensors. Acc Chem Res. 2010;43:631–641. doi: 10.1021/ar900245u. [DOI] [PubMed] [Google Scholar]
- 7. Xing H, Wong NY, Xiang Y, Lu Y. DNA aptamer functionalized nanomaterials for intracellular analysis, cancer cell imaging and drug delivery. Curr Opin Chem Biol. 2012;16:429–435. doi: 10.1016/j.cbpa.2012.03.016. ** A review highlights the using of different nanomaterials with DNA aptamer functionalization for the applications of intracellular analysis, targeted cancer imaging and therapy.
- 8. Tan WH, Donovan MJ, Jiang JH. Aptamers from cell-based selection for bioanalytical applications. Chem Rev. 2013;113:2842–2862. doi: 10.1021/cr300468w. ** A recently published thorough review on the development of cell-based SELEX for aptamers discovery and their bioanalytical applications.
- 9.Soundararajan S, Chen WW, Spicer EK, Courtenay-Luck N, Fernandes DJ. The nucleolin targeting aptamer AS1411 destabilizes bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008;68:2358–2365. doi: 10.1158/0008-5472.CAN-07-5723. [DOI] [PubMed] [Google Scholar]
- 10.Mallikaratchy P, Tang ZW, Kwame S, Meng L, Shangguan DH, Tan WH. Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt's lymphoma cells. Mol Cell Proteomics. 2007;6:2230–2238. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. A tenascin-C aptamer identified by tumor cell SELEX: Systematic evolution of ligands by exponential enrichment. Proc Natl Acad Sci U S A. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sennino B, Falcon BL, McCauley D, Le T, McCauley T, Kurz JC, Haskell A, Epstein DM, McDonald DM. Sequential loss of tumor vessel pericytes and endothelial cells after inhibition of platelet-derived growth factor B by selective aptamer AX102. Cancer Res. 2007;67:7358–7367. doi: 10.1158/0008-5472.CAN-07-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. Plos Medicine. 2006;3 doi: 10.1371/journal.pmed.0030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ireson CR, Kelland LR. Discovery and development of anticancer aptamers. Mol Cancer Ther. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 15.Fang XH, Tan WH. Aptamers generated from Cell-SELEX for molecular medicine: a chemical biology approach. Acc Chem Res. 2010;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim Y, Wu Q, Hamerlik P, Hitomi M, Sloan AE, Barnett GH, Weil RJ, Leahy P, Hjelmeland AB, Rich JN. Aptamer identification of brain tumor-initiating cells. Cancer Res. 2013;73:4923–4936. doi: 10.1158/0008-5472.CAN-12-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi H, Cui WS, He XX, Guo QP, Wang KM, Ye XS, Tang JL. Whole cell-SELEX aptamers for highly specific fluorescence molecular imaging of carcinomas in vivo. PLoS One. 2013;8 doi: 10.1371/journal.pone.0070476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi H, He XX, Wang KM, Wu X, Ye XS, Guo QP, Tan WH, Qing ZH, Yang XH, Zhou B. Activatable aptamer probe for contrast-enhanced in vivo cancer imaging based on cell membrane protein-triggered conformation alteration. Proc Natl Acad Sci U S A. 2011;108:3900–3905. doi: 10.1073/pnas.1016197108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong P, Shi BH, Zheng MB, Wang B, Zhang PF, Hu DH, Gao DY, Sheng ZH, Zheng CF, Ma YF, et al. PEI protected aptamer molecular probes for contrast-enhanced in vivo cancer imaging. Biomaterials. 2012;33:7810–7817. doi: 10.1016/j.biomaterials.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 20.Douglas SM, Bachelet I, Church GM. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 21. Li LL, Wu PW, Hwang K, Lu Y. An Exceptionally Simple Strategy for DNA-Functionalized Up-Conversion Nanoparticles as Biocompatible Agents for Nanoassembly, DNA Delivery, and Imaging. J Am Chem Soc. 2013;135:2411–2414. doi: 10.1021/ja310432u. * The authors developed a one-step ligand-exchange method for the functionalization of upconversion nanoparticles with DNA aptamers and demonstrated the aptamer-UCNPs for cancer cell targeted imaging.
- 22. Tang L, Yang XJ, Dobrucki LW, Chaudhury I, Yin Q, Yao C, Lezmi S, Helferich WG, Fan TM, Cheng JJ. Aptamer-Functionalized, Ultra-Small, Monodisperse Silica Nanoconjugates for Targeted Dual-Modal Imaging of Lymph Nodes with Metastatic Tumors. Angew Chem Int Ed. 2012;51:12721–12726. doi: 10.1002/anie.201205271. * Using DNA aptamer-functionalized silica nanoconjugates loaded with both NIR dye and 64Cu, the authors demonstrated the in vivo PET and NIR fluoresce imaging of lymph nodes in mice.
- 23.Hwang DW, Ko HY, Lee JH, Kang H, Ryu SH, Song IC, Lee DS, Kim S. A Nucleolin-Targeted Multimodal Nanoparticle Imaging Probe for Tracking Cancer Cells Using an Aptamer. J Nucl Med. 2010;51:98–105. doi: 10.2967/jnumed.109.069880. [DOI] [PubMed] [Google Scholar]
- 24.Wang CH, Huang YF, Yeh CK. Aptamer-Conjugated Nanobubbles for Targeted Ultrasound Molecular Imaging. Langmuir. 2011;27:6971–6976. doi: 10.1021/la2011259. [DOI] [PubMed] [Google Scholar]
- 25.Zhang CL, Ji XH, Zhang Y, Zhou GH, Ke XL, Wang HZ, Tinnefeld P, He ZK. One-Pot Synthesized Aptamer-Functionalized CdTe:Zn2+ Quantum Dots for Tumor-Targeted Fluorescence Imaging in Vitro and in Vivo. Anal Chem. 2013;85:5843–5849. doi: 10.1021/ac400606e. [DOI] [PubMed] [Google Scholar]
- 26.Rosen JE, Chan L, Shieh DB, Gu FX. Iron oxide nanoparticles for targeted cancer imaging and diagnostics. Nanomedicine. 2012;8:275–290. doi: 10.1016/j.nano.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 27.Zhao WA, Cui CH, Bose S, Guo DG, Shen C, Wong WP, Halvorsen K, Farokhzad OC, Teo GSL, Phillips JA, et al. Bioinspired multivalent DNA network for capture and release of cells. Proc Natl Acad Sci U S A. 2012;109:19626–19631. doi: 10.1073/pnas.1211234109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheng WA, Chen T, Tan WH, Fan ZH. Multivalent DNA Nanospheres for Enhanced Capture of Cancer Cells in Microfluidic Devices. ACS Nano. 2013;7:7067–7076. doi: 10.1021/nn4023747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas JM, Chakraborty B, Sen D, Yu HZ. Analyte-Driven Switching of DNA Charge Transport: De Novo Creation of Electronic Sensors for an Early Lung Cancer Biomarker. J Am Chem Soc. 2012;134:13823–13833. doi: 10.1021/ja305458u. [DOI] [PubMed] [Google Scholar]
- 30.Zhang XY, Zhu SC, Xiong Y, Deng CH, Zhang XM. Development of a MALDI-TOF MS Strategy for the High-Throughput Analysis of Biomarkers: On-Target Aptamer Immobilization and Laser-Accelerated Proteolysis. Angew Chem Int Ed. 2013;52:6055–6058. doi: 10.1002/anie.201300566. [DOI] [PubMed] [Google Scholar]
- 31. Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discovery. 2010;9:537–550. doi: 10.1038/nrd3141. ** A comprehensive review and perspective on the therapeutic applications of DNA and RNA aptamers.
- 32.Reyes-Reyes EM, Teng Y, Bates PJ. A New Paradigm for Aptamer Therapeutic AS1411 Action: Uptake by Macropinocytosis and Its Stimulation by a Nucleolin-Dependent Mechanism. Cancer Res. 2010;70:8617–8629. doi: 10.1158/0008-5472.CAN-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parekh P, Kamble S, Zhao NX, Portier BP, Zu YL. Immunotherapy of CD30-expressing lymphoma using a highly stable ssDNA aptamer. Biomaterials. 2013;34:8909–8917. doi: 10.1016/j.biomaterials.2013.07.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orava EW, Abdul-Wahid A, Huang EHB, Mallick AI, Gariepy J. Blocking the attachment of cancer cells in vivo with DNA aptamers displaying anti-adhesive properties against the carcinoembryonic antigen. Mol Oncol. 2013;7:799–811. doi: 10.1016/j.molonc.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahlknecht G, Maron R, Mancini M, Schechter B, Sela M, Yarden Y. Aptamer to ErbB-2/HER2 enhances degradation of the target and inhibits tumorigenic growth. Proc Natl Acad Sci U S A. 2013;110:8170–8175. doi: 10.1073/pnas.1302594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li N, Ma Y, Yang C, Guo LP, Yang XR. Interaction of anticancer drug mitoxantrone with DNA analyzed by electrochemical and spectroscopic methods. Biophys Chem. 2005;116:199–205. doi: 10.1016/j.bpc.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 37. Zhu GZ, Zheng J, Song EQ, Donovan M, Zhang KJ, Liu C, Tan WH. Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc Natl Acad Sci U S A. 2013;110:7998–8003. doi: 10.1073/pnas.1220817110. ** The authors showed the assembly of sgc8 aptamer-tethered DNA nanotrains and demonstrated the targeted delivery of therapeutic agents against CEM xenograft mouse tumor model with enhanced antitumor efficacy in vivo.
- 38.Zhu GZ, Zhang SF, Song EQ, Zheng J, Hu R, Fang XH, Tan WH. Building Fluorescent DNA Nanodevices on Target Living Cell Surfaces. Angew Chem Int Ed. 2013;52:5490–5496. doi: 10.1002/anie.201301439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Z, Ali MM, Eckert MA, Kang DK, Chen YY, Sender LS, Fruman DA, Zhao W. A polyvalent aptamer system for targeted drug delivery. Biomaterials. 2013;34:9728–9735. doi: 10.1016/j.biomaterials.2013.08.079. [DOI] [PubMed] [Google Scholar]
- 40. Xing H, Tang L, Yang XJ, Hwang K, Wang WD, Yin Q, Wong NY, Dobrucki LW, Yasui N, Katzenellenbogen JA, et al. Selective delivery of an anticancer drug with aptamer-functionalized liposomes to breast cancer cells in vitro and in vivo. J Mater Chem B. 2013;1:5288–5297. doi: 10.1039/C3TB20412J. ** The authors developed a nucleolin aptamer-functionalized liposome as targeted delivery system. The system showed targeting effects to MCF-7 breast cancer cells and improved antitumor efficacy against xenograft MCF-7 tumors in athymic nude mice.
- 41.Cao ZH, Tong R, Mishra A, Xu WC, Wong GCL, Cheng JJ, Lu Y. Reversible Cell-Specific Drug Delivery with Aptamer-Functionalized Liposomes. Angew Chem Int Ed. 2009;48:6494–6498. doi: 10.1002/anie.200901452. [DOI] [PubMed] [Google Scholar]
- 42. Guo JW, Gao XL, Su LN, Xia HM, Gu GZ, Pang ZQ, Jiang XG, Yao L, Chen J, Chen HZ. Aptamer-functionalized PEG-PLGA nanoparticles for enhanced anti-glioma drug delivery. Biomaterials. 2011;32:8010–8020. doi: 10.1016/j.biomaterials.2011.07.004. * The authors demonstrated the anti-glioma targeted paclitaxel delivery by sgc8 aptamer-functionalized PEG-PLGA nanoparticles. The biodistribution results of the aptamer-PLGA nanoparticles in mouse were reported.
- 43.Wu YR, Sefah K, Liu HP, Wang RW, Tan WH. DNA aptamer-micelle as an efficient detection/delivery vehicle toward cancer cells. Proc Natl Acad Sci U S A. 2010;107:5–10. doi: 10.1073/pnas.0909611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang CH, Kang ST, Lee YH, Luo YL, Huang YF, Yeh CK. Aptamer-conjugated and drug-loaded acoustic droplets for ultrasound theranosis. Biomaterials. 2012;33:1939–1947. doi: 10.1016/j.biomaterials.2011.11.036. [DOI] [PubMed] [Google Scholar]
- 45. Cao XX, Li SH, Chen LC, Ding HM, Xu H, Huang YP, Li J, Liu NL, Cao WH, Zhu YJ, et al. Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus. Nucleic Acids Res. 2009;37:4621–4628. doi: 10.1093/nar/gkp489. * The authors presented of bacterium-based selection of DNA aptamer strands for the recognition and detection of S. aureus in clinical samples of pyogenic fluid taken from burn victims
- 46.Zelada-Guillen GA, Sebastian-Avila JL, Blondeau P, Riu J, Rius FX. Label-free detection of Staphylococcus aureus in skin using real-time potentiometric biosensors based on carbon nanotubes and aptamers. Biosens Bioelectron. 2012;31:226–232. doi: 10.1016/j.bios.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 47.Chang YC, Yang CY, Sun RL, Cheng YF, Kao WC, Yang PC. Rapid single cell detection of Staphylococcus aureus by aptamer- conjugated gold nanoparticles. Sci Rep. 2013;3 doi: 10.1038/srep01863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang RH, Zhao JJ, Jiang TS, Kwon YM, Lu HG, Jiao PR, Liao M, Li YB. Selection and characterization of DNA aptamers for use in detection of avian influenza virus H5N1. J Virol Methods. 2013;189:362–369. doi: 10.1016/j.jviromet.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 49.Chen F, Hu YL, Li DQ, Chen HD, Zhang XL. CS-SELEX Generates High-Affinity ssDNA Aptamers as Molecular Probes for Hepatitis C Virus Envelope Glycoprotein E2. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang DR, Meng XH, Yu QQ, Xu L, Long Y, Liu B, Fang XH, Zhu HZ. Inhibition of Hepatitis C Virus Infection by DNA Aptamer against Envelope Protein. Antimicrob Agents Chemother. 2013;57:4937–4944. doi: 10.1128/AAC.00897-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jilma-Stohlawetz P, Knobl P, Gilbert JC, Jilma B. The anti-von Willebrand factor aptamer ARC1779 increases von Willebrand factor levels and platelet counts in patients with type 2B von Willebrand disease. Thromb Haemostasis. 2012;108:284–290. doi: 10.1160/TH11-12-0889. [DOI] [PubMed] [Google Scholar]
- 52.Bae ON. Targeting von Willebrand factor as a novel anti-platelet therapy; Application of ARC1779, an Anti-vWF aptamer, against thrombotic risk. Arch Pharmacal Res. 2012;35:1693–1699. doi: 10.1007/s12272-012-1000-3. [DOI] [PubMed] [Google Scholar]
- 53.Wang P, Yang Y, Hong H, Zhang Y, Cai W, Fang D. Aptamers as Therapeutics in Cardiovascular Diseases. Curr Med Chem. 2011;18:4169–4174. doi: 10.2174/092986711797189673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu MD, Zhang KH. The application of aptamers in cancer research: an up-to-date review. Future Oncol. 2013;9:369–376. doi: 10.2217/fon.12.201. [DOI] [PubMed] [Google Scholar]
- 55.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 56.Kolenko V, Teper E, Kutikov A, Uzzo R. Zinc and zinc transporters in prostate carcinogenesis. Nat Rev Urol. 2013;10:219–226. doi: 10.1038/nrurol.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB, Lu Y. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc Natl Acad Sci U S A. 2007;104:2056–2061. doi: 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiang Y, Lu Y. Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nature Chem. 2011;3:697–703. doi: 10.1038/nchem.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu JW, Mazumdar D, Lu Y. A simple and sensitive "dipstick" test in serum based on lateral flow separation of aptamer-linked nanostructures". Angew Chem Int Ed. 2006;45:7955–7959. doi: 10.1002/anie.200603106. [DOI] [PubMed] [Google Scholar]