Abstract
To address limitations of current high-throughput methods for studying cell-cell communication and determining the cell-of-origin of proteins in multicellular environments, we have developed a technique that selectively and continuously labels the proteome of individual cell types in co-culture. Through transgenic expression of exogenous amino acid biosynthesis enzymes, vertebrate cells overcome their dependence on essential amino acids and can be selectively labeled through metabolic incorporation of amino acids produced from heavy isotope-labeled precursors. We have named this method Type specific labeling with Amino acid Precursors (CTAP). Testing CTAP in several human and mouse cell lines, we were able to differentially label the proteome of distinct cell populations in co-culture and determine the relative expression of proteins by quantitative mass spectrometry. In addition, CTAP successfully identified the cell-of-origin of extracellular proteins in co-culture, highlighting its potential use in biomarker discovery for linking secreted factors to their cellular source.
Introduction
The development and maintenance of multicellular environments is dependent on extensive cell-cell communication, and dysregulation of cellular interactions plays a role in many diseases, including cancer1–3. Although widely used for investigating intercellular signal transduction, antibody-based assays are relatively low throughput, vary in specificity, and require preselection of protein readout. While quantitative mass spectrometry-based proteomics4–6 might overcome some of these limitations, studying cell-cell communication with mass spectrometry is hindered by its inability to distinguish between proteins from distinct cell types in multicellular cultures.
Several recent efforts have been made to differentiate the proteome of individual cell populations in co-culture. In one such approach, each distinct cell type is labeled in isolation (e.g., using heavy stable isotope-labeled L-lysine or L-arginine), and the fully labeled cells are subsequently mixed. Peptides identified with liquid chromatography tandem mass spectrometry (LC-MS/MS) can then be assigned a source cell-type from the isotopic label status. Two recent reports demonstrate the feasibility of such an approach for identifying early ephrin signaling responses7 and determining proteins transferred between cell types8. Unfortunately, these labels become rapidly diluted as cells grow and divide in co-culture, making this experimental setup primarily useful for investigating very early signaling events. In a different approach, protein sequence differences between species are used to determine cell-of-origin in cross-species co-cultures and xenografts9,10. Although this approach has the ability to distinguish between proteins from different cell types, the major drawbacks are that only a subset of peptides can be differentiated and the findings from mixed-species models may not be physiologically relevant. Yet another technique utilizes tRNA-synthetases that specifically recognize and incorporate noncanonical amino acids into proteins11–13. This method provides for both proteomic incorporation that is specific to transgenic cells as well as the ability to perform affinity enrichment on chemical moieties (e.g., azides). However, structural differences between noncanonical and canonical amino acids might cause unpredictable functional alterations in mature proteins14. Given the caveats of each of these methods, there is a strong need for a technique that enables continuous cell-specific labeling with canonical amino acids.
In this study, we developed a method for cell-selective proteomic labeling that overcomes the limitations mentioned above. This method utilizes the inability of vertebrate cells to synthesize certain amino acids required for growth and homeostasis. These “essential” amino acids are produced in some plants, bacteria, and lower eukaryotes, and must be supplemented to the media of cultured vertebrate cells or obtained in the diet of animals15. We reasoned that transgenic expression of enzymes that synthesize essential amino acids would allow vertebrate cells to overcome auxotrophy by producing their own amino acids from supplemented precursors. These precursors can be isotopically labeled, allowing cell-of-origin of proteins to be determined by label status identified with MS/MS. We have named this method Cell Type specific labeling with Amino acid Precursors (CTAP) and tested its validity and feasibility using L-lysine, an essential amino acid commonly used in quantitative proteomic methods such as stable isotope labeling by amino acids in cell culture (SILAC)5,16. Using the CTAP method, we were able to continuously and differentially label the proteome of cells in co-culture, determine relative protein expression levels between the cell populations, and identify the cell-of-origin of secreted factors.
Results
Engineering mammalian cells to grow on L-lysine precursors
Several enzymes have been found in bacteria, fungi, and plants that catalyze reactions leading to the production of L-lysine from precursor compounds. We hypothesized that by engineering vertebrate cells to produce their own supply of L-lysine from labeled precursors, we could achieve differential proteomic labeling of specific cell types in co-culture (Fig. 1). We began by identifying a set of precursor-enzyme pairs in which the precursor was readily available and the enzyme had no described orthologs in vertebrate genomes (Supplementary Fig. 1). In a different context, one of the candidate precursor-enzyme pairs had successfully been used to rescue L-lysine auxotrophy when creating a positive selection system for vector incorporation17,18. To investigate the candidate precursors and eliminate those that autonomously rescue L-lysine auxotrophy, we examined growth rates in SILAC media supplemented with L-lysine, various precursors, or in L-lysine-free conditions. With the exception of Nα-acetyl-L-lysine, the tested precursors alone had little or no effect on growth in wild-type cells (Supplementary Fig. 2).
Figure 1. Overview of Cell Type specific labeling with Amino acid Precursors (CTAP).

(a) The CTAP methodology takes advantage of vertebrate cells’ inability to produce essential amino acids, resulting in the requirement that these molecules be supplemented in culture media or diet for cell growth. We focus on one of these amino acids, L-lysine, and the enzymes used to produce it from precursor molecules. By expressing exogenous L-lysine biosynthesis enzymes, transgenic cells produce their own supply of L-lysine and (b) can be labeled selectively by supplementing the media with heavy isotope-labeled forms of the precursors. Expressing distinct L-lysine biosynthesis enzymes in different cell types enables continuous cell-selective proteome labeling with differentially-labeled precursors when grown in media lacking L-lysine. (c) CTAP can be used to investigate direct contact or secreted factor mediated cell-cell communication, relevant for a range of biological phenomena.
We next investigated whether transgenic expression of enzymes involved in L-lysine biosynthesis would allow cells to acquire the ability to grow on precursors. The genes encoding the enzymes lysine racemase (lyr) from Proteus mirabilis (optimized for expression and intracellular localization, Supplementary Note 1) and diaminopimelate decarboxylase (DDC) from Arabidopsis thaliana were stably expressed in several cell lines (Supplementary Table 1). DDC-expressing mouse 3T3 and HEK293T cells, along with lyr-expressing human MDA-MB-231 cells, exhibited growth rates in media supplemented with the precursors 2,6-diaminopimelic acid (DAP) and D-lysine, respectively, comparable to those in media containing L-lysine (Fig. 2a, 2b, and Supplementary Fig. 3). Similar observations were found in several additional human and mouse cell lines, although two of the DDC-expressing lines never reached equal growth rates to L-lysine (Supplementary Table 2). The enzyme-precursor pairs were specific, as no growth was observed in the cross enzyme-precursor setup or in empty-vector controls (Fig. 2a and 2b). Furthermore, growth in standard L-lysine conditions with addition of either DAP or D-lysine exhibited little or no growth perturbation (Supplementary Fig. 4). These monoculture results show that transgenic enzyme expression with supplementation of specific precursors is responsible for the growth rescue observed in L-lysine free conditions.
Figure 2. Vertebrate cell lines expressing L-lysine biosynthesis enzymes grow and incorporate L-lysine produced from their precursors.
(a) Mouse fibroblast 3T3 cells that stably express DDC and (b) human breast carcinoma MDA-MB-231 cells that stably express lyr were plated in L-lysine-free media supplemented with 10 mM DAP, 4 mM D-lysine, both precursors, or 0.798 mM L-lysine. Control (empty-vector) cells are shown in the lower panels. Cell growth, assessed with impedance (a correlate of the number of cells) using the xCELLigence system, was normalized to maximum growth. Error bars represent the standard deviation of three biological replicates. (c, d) Molecular incorporation assessed by LC-MS/MS. At the start of the experiment, cell lysates were collected from monocultured (c) DDC-expressing 3T3 cells labeled heavy (H) and (d) lyr-expressing MDA-MB-231 cells labeled light (L) (top panels). Cells were harvested after 10+ days (two passages) in L-lysine-free media containing the indicated precursors (bottom panels). Label status of lysine-containing peptides was assessed by quantitative LC-MS/MS and percent incorporation of heavy label was determined using H/L ratios from MaxQuant analysis. Dashed black line indicates median peptide (percentages indicated). Each histogram depicts one sample processed by LC-MS/MS.
Proteomic incorporation of precursor-based L-lysine
To investigate whether L-lysine is directly produced by enzymatic-turnover of the supplemented precursors, we applied the SILAC principle of exchanging the isotopic label of amino acids from one form to another (e.g., light L-lysine to heavy L-lysine)5. At the beginning of the experiments, DDC-expressing 3T3 cells were labeled with heavy [13C6,15N2]L-lysine (H) and lyr-expressing MDA-MB-231 cells were labeled with light L-lysine (L). These cells were then grown in monoculture for 10+ days (two passages) in L-lysine-free media that contained unlabeled DAP (L), heavy-labeled [2H8]D-lysine (H), or both precursors. Protein from cell lysate was digested with trypsin and/or LysC, submitted to high resolution LC-MS/MS, and the H/L ratio for each peptide was determined by the MaxQuant software package19. These H/L ratios were used to determine the labeling incorporation levels, which we found to be similar to orthogonal enrichment calculations (Supplementary Table 3) and robust to a range of parameter changes, such as peptide score and length (Supplementary Table 4).
In the presence of light-labeled DAP alone, peptides identified in DDC-expressing 3T3 cells switched from being predominantly labeled heavy (94% H, median peptide) to light (97% L) (Fig. 2c). Similarly, the peptides identified in lyr-expressing MDA-MB-231 cells changed from 98% light to 93% heavy in the presence of heavy-labeled D-lysine (Fig. 2d). This level of labeling can be reached after one passage (approximately four doublings, Supplementary Fig. 5) and can be considered near complete as it is similar to the initial samples and levels typically reported in SILAC experiments20,21. To test the amount of unspecific labeling (i.e., cross contamination), cultures were also grown in the presence of both precursors. Supplementing the DDC precursor DAP (L) had little effect on the label switch in lyr-expressing MDA-MB-231 cells, while the presence of D-lysine (H) marginally increased the heavy label in DDC-expressing 3T3 cells (Fig. 2c and 2d, insets). This difference in the 3T3 cells was possibly due to contamination of heavy L-lysine in heavy D-lysine (≥ 95% enantiomeric purity, C/D/N Isotopes) and is expected to be reduced with higher purity. Taken together, these data indicate that lyr and DDC-expressing cells are able to specifically incorporate and grow on L-lysine synthesized directly from their respective precursors.
Limited perturbation to cells growing on precursors
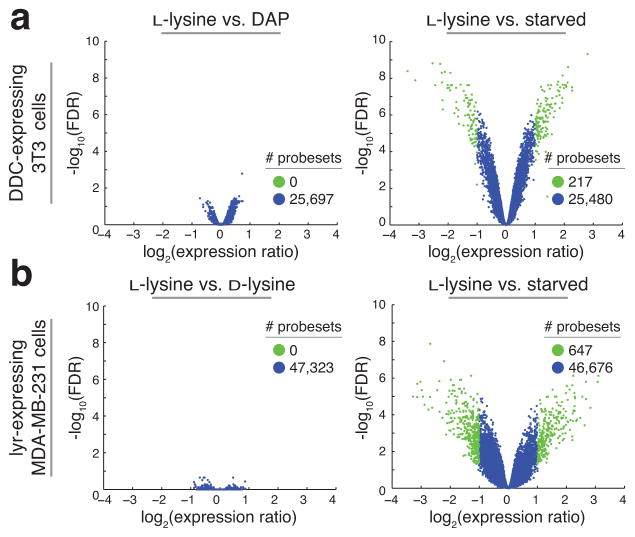
We next investigated whether cells behave similarly when grown on precursors compared to L-lysine. Cells were cultured for three days in media containing L-lysine, precursor, or neither (starved, positive control for perturbed state) and mRNA expression levels were profiled using microarrays (Fig. 3 and Supplementary Fig. 6). Relative to the basal L-lysine condition, no genes changed significantly when enzyme expressing cells were grown on precursor, while hundreds of genes changed in starved conditions (FDR < 0.05 and expression ratio greater than two, Fig. 3). Furthermore, several assays were performed to probe the effects of precursor-based growth, including geneset enrichment analysis (Supplementary Table 5 and 6), measurement of amino acid starvation factors (Supplementary Fig. 7), determination of protein abundance by LC-MS/MS (Supplementary Fig. 8), as well as growth and molecular response to drug perturbation (Supplementary Fig. 9 and 10). Although minor differences exist, overall, these data demonstrate that growing cells on their precursors has little to no effect compared to growth on L-lysine.
Figure 3. Limited gene expression changes observed when growing cells in precursor versus L-lysine.
(a) DDC-expressing 3T3 cells were plated in SILAC media supplemented with DAP, L-lysine, or neither (starved). After 72 hours, mRNA was harvested and profiled for gene expression levels using the Illumina microarray platform. Expression differences of DAP versus L-lysine (left panel) and starved vs L-lysine (right panel) are plotted as a function of statistical significance (moderated t-statistics adjusted for multiple testing by the Benjamini and Hochberg method). Highlighted genes (green) are more than 2-fold differentially regulated at the level of FDR < 0.05. (b) As in (a) except MDA-MB-231 cells expressing lyr were plated on L-lysine, D-lysine, or in starved conditions. All experiments were performed in triplicate.
Continuous & differential proteome labeling in co-culture
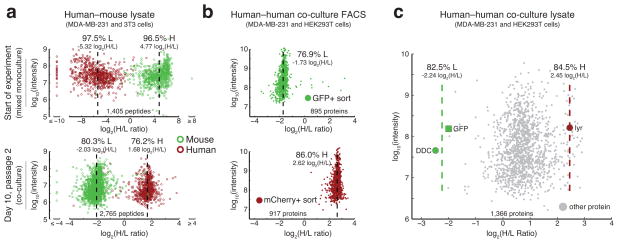
To assess the specificity of labeling in co-culture with each cell population utilizing a distinct enzyme-precursor pair, we took advantage of species-specific sequence differences to compare label status between the enzyme-expressing mouse 3T3 and human MDA-MB-231 cell lines. Labeling each cell type in isolation, the 3T3 cells were initially cultured in heavy L-lysine (H) and the MDA-MB-231 cells in light L-lysine (L). At the start of the experiment, a sample was harvested and combined 1:1 to verify the ability to differentiate label status based on species-specific peptide classification. As expected, labels of mouse-specific and human-specific peptides were confirmed to be primarily heavy and light, respectively (Fig. 4a and Supplementary Fig. 11).
Figure 4. Using two distinct enzyme-precursor pairs, co-cultured cells exhibit precursor-based differential proteome labeling.
(a) DDC-expressing 3T3 cells (mouse) were labeled with heavy L-lysine (H) and lyr-expressing MDA-MB-231 cells (human) with light L-lysine (L) and mixed prior to sample analysis by LC-MS/MS (upper panel). Similarly labeled cells were co-cultured and analyzed after 10 days (two passages) on DAP (L) and D-lysine (H) (lower panel). Peptides unique to the mouse or human proteome are green and red, respectively. Median indicated by dashed black line. (b) GFP+ HEK293T cells expressing DDC were co-cultured with mCherry+ MDA-MB-231 cells expressing lyr in media containing DAP (L) and D-lysine (H) for five days (one passage, approximately four cellular doublings). Sorted GFP+ (upper panel) and mCherry+ (lower panel) cells were lysed, separately subjected to LC-MS/MS, and identified proteins are shown. (c) Proteins derived from unsorted co-culture of cells as in (b). Highlighted are proteins unique to each transgenic cell line (GFP and DDC in HEK293T, lyr in MDA-MB-231 cells). Mean of H/L ratios of the transgenes specific to each cell population (DDC and GFP in HEK293T cells and lyr in MDA-MB-231 cells) are indicated with green and red lines, respectively. Note that although mCherry was detected, it is not included as a distinct protein as it is fused to lyr. Each panel depicts one sample processed by LC-MS/MS.
With the expectation that each cell type would exchange isotopic label, the pre-labeled cells were then combined in co-culture into media containing both light DAP (L) and heavy D-lysine (H). After 10 days (two passages), the two cell types switched labels (Fig. 4a and Supplementary Fig. 12). As expected, the mouse 3T3 peptides became predominantly labeled light (80% or -2.0 log2 H/L) and the human MDA-MB-231 peptides became predominantly labeled heavy (76% or 1.7 log2 H/L). This labeling efficiency is lower than in the mixed monoculture control and could either be due to physiological sharing of L-lysine between cell populations or limitations attributed to the CTAP method. We have optimized CTAP through removal of shared L-lysine from the media with daily media changes and by modifying the lyr sequence to decrease enzyme secretion (e.g., signal peptide removal, Supplementary Fig. 13), and we expect that future development will increase cell type specific label enrichment even further. Despite the fact that the absolute H/L ratios of the species-specific peptides differed between the mixed monoculture control and the CTAP-labeled co-culture, the human and mouse-specific peptides were clearly separable in both experiments. These distinct H/L ratios in species-specific sequences therefore demonstrate the ability to differentially label the proteome across cell types in co-culture.
We next investigated whether the CTAP method could differentiate the proteome of a same-species co-culture system. DDC-expressing GFP+ HEK293T cells were plated together with lyr-expressing mCherry+ MDA-MB-231 cells. After five days (one passage, approximately four cellular doublings) of growth in DAP (L) and D-lysine (H), a co-culture sample was sorted for mCherry+ and GFP+ cells by FACS (Supplementary Fig. 14) and each of the sorted populations was separately subjected to LC-MS/MS. Analysis of protein from the GFP+ and mCherry+ cells of this human-human co-culture showed similar labeling efficiency to that seen in the human-mouse co-culture, with each cell population exhibiting distinct H/L ratios (Fig. 4b).
Another sample was collected directly from the non-sorted human-human co-cultures and 1,366 proteins were identified with LC-MS/MS. Focusing on the transgenic proteins exclusive to each cell population (GFP and DDC for the HEK293T as well as lyr in MDA-MB-231 cells), we observed the expected H/L ratios corresponding to those determined by FACS (Fig. 4c). This concordance confirms differential labeling in human-human co-culture lysates. When analyzing all identified proteins in this unsorted sample, the H/L ratios exhibited a near-normal distribution with the transgenes lying in the tails. Although these tails contain relatively few members, they likely represent cell type specific proteins (Fig. 4c and Supplementary Fig. 15). This result is consistent with a recent report that found most proteins are ubiquitously expressed across different cell types but at different relative abundance levels22. In summary, these results demonstrate the ability of CTAP to label the proteome in a cell-specific manner and show that label status (H/L ratio) is directly related to the relative protein abundance level between the two cell types.
Linking secreted proteins to their cell-of-origin
To test the potential of the CTAP method to discriminate the cell-of-origin of secreted factors, supernatant was collected from the same human and mouse co-culture setup as in the previous section. Prior to harvesting, the cells were grown for 16 hours in serum-free media to avoid overloading the sample with serum proteins. Secreted proteins were concentrated by ultracentrifugation, precipitated by methanol-chloroform, and subjected to LC-MS/MS. Focusing on species-specific peptides, nearly all could be distinguished by label alone (Fig. 5a and Supplementary Fig. 16). These results demonstrate the ability of the method to determine cell-of-origin for secreted proteins in co-culture.
Figure 5. Application of CTAP for determining cell-of-origin for secreted factors.
(a) DDC-expressing 3T3 cells (mouse) and lyr-expressing MDA-MB-231 cells (human) were co-cultured in DAP (L) and D-lysine (H). Prior to sample collection, cells were grown for 16 hours in serum-free medium and the supernatant (medium) was collected. After concentrating proteins by ultra-centrifugation and methanol-chloroform extraction, the sample was analyzed by LC-MS/MS. Only peptides that are unique to mouse (green) and human (red) are displayed. (b) Similar to (a) except the co-culture consisted of two human cell lines: HEK293T expressing DDC and MDA-MB-231 cells expressing lyr. Colors depict relative protein abundance as determined by SILAC quantitation of mixed, separately labeled monoculture lysates. Uncolored points represent proteins that were not identified in the monoculture sample. Each experiment depicts one sample processed by LC-MS/MS.
Applying a similar approach for analyzing secreted factors in a same-species co-culture, supernatant was collected and subjected to LC-MS/MS from the same co-cultured DDC-expressing HEK293T and lyr-expressing MDA-MB-231 cells as used previously. The H/L ratios of 403 identified proteins spanned a similar range as those detected intracellularly (Fig. 4c and Fig. 5b). Having shown that the H/L ratios are distinct for species-specific proteins in the human and mouse co-culture secretome, the tails of this human-human distribution likely represent cell type specific proteins. Reasoning that a protein secreted primarily by one cell type would also be relatively more abundant intracellularly, we investigated whether CTAP-labeled extracellular protein ratios correlate with SILAC-labeled protein ratios obtained from mixed monoculture lysates. Focusing on the subset of proteins that were common to both samples, good agreement was observed between the H/L ratios from the combined intracellular monocultures and secreted co-culture samples (R2 = 0.57, Fig. 5b and Supplementary Fig. 17). The positive correlation between these conditions indicates that the secreted proteins with the lowest and highest H/L ratios are likely cell type specific as they are most abundant in the HEK293T and MDA-MB-231 cells, respectively. Interestingly, almost half of the putative MDA-MB-231-secreted proteins were not identified intracellularly (47%), highlighting the need for secretome profiling. Taken together with the species-verified secretome analysis, these results establish that the CTAP method can be applied to determine the cell-of-origin of secreted factors in co-culture.
Discussion
Using precursors of the essential amino acid L-lysine and enzymes that catalyze its synthesis, this work shows that the proteome of specific cell types in co-culture can be isotopically labeled by canonical amino acids produced in transgenic cells. Cell types from different tissues of both mouse and human origin successfully overcame L-lysine auxotrophy, and we observed little to no molecular and phenotypic consequences of culturing enzyme-expressing cells with precursors. Mass spectrometry analysis of enzyme-expressing cells in monoculture showed complete molecular labeling by L-lysine derived from precursor. Differential labeling of individual cell types in co-culture was achieved using a dual-enzyme setup with distinctly labeled precursors, allowing the relative expression levels for all identified proteins to be determined in each cell type. In addition, by analyzing the supernatant of co-cultured cells, cell-of-origin of secreted proteins was readily established. Supporting these results, we also found that CTAP was applicable for labeling a specific cell-type of interest in a mixed cell culture system using only one enzyme-precursor pair (Supplementary Fig. 18). Although these are preliminary data, and titrating down the amount of L-lysine in the media was required, a one-enzyme approach may facilitate linking of proteins to a distinct cell population in experimental systems where a two-enzyme approach is either not desirable or not feasible. In addition to DDC and lyr, we also tested and found specific but suboptimal growth rescue with the enzyme CBZcleaver and substrate Z-lysine, supporting the general principle of this method (Supplementary Fig. 19 and Supplementary Note 2). To the best of our knowledge, CTAP is the only method in which the proteome of specific cell populations can be labeled continuously and differentially by canonical amino acids in a complex mixture of cells.
Although the results of this initial study demonstrate the feasibility and functionality of CTAP, there are several avenues for further method development. The co-culture labeling efficiency was lower than expected from mixed monoculture controls (approximately 80% versus 97%, Fig. 4). Incomplete labeling in co-culture can arise from both technical problems and true biological interactions between the two cell populations. During method development, we identified extracellular lyr activity as a potential source of label dilution. Several experimental optimizations, which included removal of a signal peptide, the addition of a mitochondrial targeting sequence, and daily media changes, alleviated much of the label contamination observed in initial experiments and greatly improved method usability (Supplementary Fig. 13). However, even with these changes, extracellular lyr activity likely still plays a role in co-culture label contamination, and future efforts will focus on complete suppression of this activity. Other optimization steps will involve improving enzyme efficacy, decreasing enzyme secretion, increasing precursor uptake17,18 (Supplementary Discussion), and increasing purity of heavy D-lysine (currently only ≥ 95% enantiomeric pure, C/D/N Isotopes). Despite technical improvements, biological exchange of L-lysine between cells in co-culture may prevent cell type specific labeling from reaching isotopic enrichment levels as complete as those of SILAC. Nevertheless, the distinct H/L ratios we observed in each cell population readily enable clear identification of cell type specific factors and relative quantitation of protein expression between the populations.
Several criteria must be met in order for cells to qualify for the use of CTAP to study cell-cell communication. Similar to SILAC, cells must be L-lysine auxotrophic and able to grow in dialyzed FBS or FBS-free media. Currently, the method relies on precursors of L-lysine only, hampering the use of L-arginine containing peptides for protein quantitation of tryptic digests. Additionally, CTAP requires stable expression of exogenous enzymes, growth on amino acid precursors, and investigators should verify that their context-specific phenotype is similar when using this method. Although the H/L ratios of the transgenic proteins themselves can be used to determine the level of labeling in each cell population, it is advisable that the incorporation efficiency be verified through LC-MS/MS analysis of samples sorted by FACS. These levels can be used as cutoffs for assigning the cell-of-origin to individual proteins. Unequal cell number may influence labeling efficiency in co-culture due to sharing of L-lysine from residual extracellular lyr activity or physiological amino acid exchange. This potential issue can be addressed by using different seeding densities of each cell population and daily changes of the culture medium. Despite incomplete labeling, normalizing the H/L ratios (e.g., using z-scores) can enable relative comparison of protein changes between conditions in a cell type specific manner.
Upon overcoming some of the remaining technical challenges, such as competition with endogenous L-lysine, precursor delivery, and enzyme expression, another possible application for CTAP may be identification of disease biomarkers in vivo. Current approaches for biomarker identification are limited by their inability to classify whether a potential marker originates from the diseased tissue itself or from the normal tissue. Using the described technique we can circumvent these limitations, as proteins from specific cell types of interest can in principle be labeled continuously in vivo. Any labeled protein identified in the serum or proximal fluids will have originated from the cell type of interest. As CTAP allows for unbiased and high-throughput LC-MS/MS to differentiate peptides derived from distinct cells in complex cellular environments, we anticipate that CTAP will be an important tool for gaining insight into intercellular signaling in a range of fundamental biological processes.
Methods
Oligonucleotide acquisition
The L-lysine producing enzymes used in this study were DDC, lyr, and CBZcleaver. DDC was directly amplified by PCR from Arabidopsis thaliana cDNA (TAIR id = AT3G14390, primer sequences available in Supplementary Tables 7 and 8). The lyr and CBZcleaver constructs were synthesized by GeneArt with the amino acid sequences specified previously23,24, and nucleotide sequences were optimized for expression in mouse. All sequences are available in supplementary material (Supplementary Note 1). Sequences were verified for all plasmids by the Sanger method of sequencing.
Vector cloning, viral production, and cell line creation
Two MSCV based retroviral vector backbones, one expressing GFP (pMIG) and the other mCherry (pMIC), were used to infect mouse cells. For insert into pMIG, the PCR product of DDC was cloned into the EcoRI site of the vector. CBZcleaver was directly subcloned from the GeneArt supplied vector pMA-RQ into pMIC using EcoRI and XhoI restriction sites. Viral supernatants for pMIG and pMIC were produced by transfecting Phoenix cells with each plasmid and the supernatant was used to infect 3T3 cells 48 hours later as previously described25,26.
The lentiviral backbone pLM was used to infect human cells in this study. Overlapping PCR was performed to generate GFP-DDC and mCherry-lyr constructs that were linked by a P2A peptide preceeded by a Gly-Ser-Gly linker27. The pLM-P2A-enzyme virus was packaged by calcium phosphate transfection of the HEK293T packaging cell line using 10 μg of transfer vector, 6.5 μg of CMVδR8.74, and 3.5 μg of the VSV.G plasmid. MDA-MB-231 and HEK293T cells were then infected with lentiviral supernatant produced from the pLM construct 48 hours post-transfection of the packaging line.
Cellular growth assays
Cell lines were grown in Dulbecco’s modified Eagle’s medium (DMEM) without L-lysine and L-arginine (SILAC-DMEM, Thermo Fisher Scientific) supplemented with 10% dialyzed FBS (Sigma, F0392), antibiotics, and L-glutamine. For monoculture growth assays, 1 mM L-arginine was added to the media and cells were seeded in 200 μL in 96-well plates with 4000 or 5000 cells per well in different concentrations of L-lysine, 2,6-diaminopimelic acid (DAP, Sigma, 33240), D-lysine HCL (Sigma, L5876), Nα-Cbz-L-lysine (Z-lysine, BaChem, C-2200), or Nα-acetyl-L-lysine (N2A, Sigma, A2010). Note that while the substrate of DDC is only the meso- form of DAP, in this work the DAP used contains DD-, LL-, and the meso- form. Cell viability was measured using either the metabolic-activity based Resazurin (Sigma) reagent or the impedance-based xCELLigence system (Roche). For Resazurin experiments, 25 μL of the Resazurin reagent was added to each well and cellular growth was estimated after two to three hours of incubation at 37 °C as described by the manufacturer. For xCELLigence experiments, cells were plated in either 16 or 96-well E-plates, allowed to settle for 30 minutes at room temperature, and then placed in the RTCA DP or RTCA MP analyzer where impedance was measured every 15 minutes for 96–120 hours. At least three replicates were performed for each condition.
Measuring the percentage of mCherry+ and GFP+ cells in co-culture was performed by either flow cytometry (BD LSR II) or Tali image-based cytometry (Invitrogen). For flow cytometric assays, 25,000 cells from each cell line were seeded together in 6-well plates in 3–4 mL media supplemented with different concentrations of L-lysine and/or L-lysine precursors. After 72 hours, cells were trypsinized, washed, and resuspended in 200 μL PBS containing 2% dialyzed FBS and 0.1% NaN3. 20 μL was used for estimating total cell numbers using the ViaCount assay (Millipore) as described by the manufacturer. The remaining 180 μL was mixed with an equal volume of 2% paraformaldehyde. The percentage of GFP+ and mCherry+ cells in each sample was analyzed by flow cytometry. At least two replicates were performed for each condition. For Tali assays, cells were trypsinized, resuspended in media, 25 μL of co-culture cell suspension was used to determine the percentage of GFP+ and RFP+ cells in biological triplicate.
Stable isotope labeling and cell passaging
For exchange-of-label experiments (all monocultures, all human and mouse co-cultures, and Supplementary Fig. 19), cells were first metabolically labeled by growth for at least 10 cellular doublings (10+ days) in L-arginine and 10% dialyzed FBS-containing SILAC DMEM supplemented with 798 μM light L-lysine (L), medium [2H4]L-lysine (M, +4 Daltons), or heavy [13C6,15N2]L-lysine (H, +8 Daltons) (Cambridge Isotopes). Cells were then seeded in mono- or co-culture with 10 mM light DAP (L, Sigma), 2.5 mM or 4 mM heavy [2H8]D-lysine (H, +8 Daltons, C/D/N Isotopes, 3,3,4,4,5,5,6,6-d8), 2.5 mM heavy labeled [13C6,15N2]Z-lysine (H, +8 Daltons, Supplementary Fig. 19), or both DAP (L) and D-lysine (H). For experiments that maintained label (all human-human co-cultures), cells were initially grown for at least 10 cellular doublings (10+ days) in their respective precursors: DDC-expressing in DAP (L) and lyr-expressing in D-lysine (H). Populations were then combined in 10 mM DAP (L) and 1 mM D-lysine (H) and grown together for five days (approximately four cellular doublings) in co-culture. Co-cultures were seeded at ratios in which an equal number of cells were expected at the end of the experiment. All cell lines were passaged 1:10–1:15 at 95% confluence.
mRNA microarray expression profiling
Cells were seeded at equal densities into L-arginine and 10% dialyzed FBS-containing SILAC media supplemented with 798 μM L-lysine, 798 μM L-lysine (M), 4 mM D-lysine HCl, or 10 mM DAP. After 72 hours, cells were washed, trypsinized, pelleted, and frozen at −80 °C. RNA was extracted using the RNeasy mini kit (Qiagen), labeled, and hybridized to Illumina Mouseref-8 or Human HT-12 microarrays. After median centering the probe intensities for each array, moderated t-statistics and false discovery rate calculations for multiple hypothesis correction were performed using the eBayes method provided in LIMMA28,29.
Drug Perturbation Assays
Cells were seeded in 96-well plates (2000 cells/well) and grown to 40% confluence in SILAC media containing 0.798 mM K0 or 10 mM DAP DMEM with 10% dialyzed fetal bovine serum (FBS). Cells were then inhibited with eight different drug concentrations (2 fold dilution) in eight replicates. Drugs used were Stattic (STAT3 inhibitor), PI3K-IV (PI3K inhibitor), AKT-VIII (AKT inhibitor), and SL327 (MEK inhibitor). After 48 hours drug treatment cell viability was measured by Resazurin (Sigma) as described by manufacturer. Cell viability relative to untreated cells was calculated to obtain dose-response curves.
Western Blotting
Frozen cell pellets were thawed and lysed for 20 min with NP40 lysis buffer, which contained 1% Nonident P-40, 1 mM sodium orthovanadate, and Complete protease inhibitors (Roche Diagnostics) in PBS. Protein concentrations were determined by the Bradford assay (BioRad) and adjusted to 1–1.5 mg/mL. Protein was then denatured in 2% SDS for 5 minutes at 95 °C. Approximately 20 μg of each sample was then separated by SDS-PAGE, transferred to PVDF membrane, and immunoblotted using primary and secondary antibodies. All antibodies were from Cell Signaling, except anti-ATF4 which was from Abcam. Chemoluminescence visualization was performed on Kodak or HyBlotCL films and films were scanned by a microTEK scanner at 600 d.p.i. in gray scale. The membranes were stripped and reprobed with anti-GAPDH (Cell Signaling) to test for protein loading.
Gene Set Enrichment Analysis (GSEA)
Data from Illumina Mouseref-8 or Human HT-12 microarray experiments were analyzed with the GSEA software package version 2.0730. The KEGG pathway (v2.5) was selected as gene sets database and the experimental treatment conditions (L-lysine vs. precursor, or L-lysine vs. starved) were used as phenotype vector. Default input parameters were applied. Default significance levels were used to report perturbed KEGG pathway ontologies (Nominal p-value < 0.01 and FDR < 0.25).
Mass spectrometry sample preparation
For harvesting of cell lysate, cells were trypsinized, resuspended in SILAC DMEM, washed three times in ice cold PBS, and cell pellets frozen at −80 °C. For FACS samples, co-cultures of GFP+ and mCherry+ cells were trypsinized, washed, and resuspended in PBS with 20% media (2% FBS) to a concentration of approximately 2×107 cells/mL Cells were then sorted into single GFP+ and mCherry+ populations on a MoFlo cell sorter (Dako), washed twice with ice cold PBS, and cell pellets were stored at −80 °C for further analysis. A small aliquot of each sorted population was immediately reanalyzed to determine purity. For cultured media samples, cells were washed three times with PBS and supplied with serum-free SILAC DMEM 24 hours prior to supernatant sample collection. Media was collected, filtered with a 0.22 μm filter, and proteins were concentrated to around 1 mg/mL using a 3 kDa Amicon Ultra Centrifuge filter (Millipore) as described by the manufacturer.
Protein extraction and digestion
Cell pellets were resuspended with Denaturation buffer (6 M Urea and 2 M thio Urea in 10 mM Tris), 1 μL of benzonase was added, followed by incubation for 10 minutes at room temperature. Cellular debris was removed by centrifugation at 4000 g for 30 min. For the supernatant samples, the secreted proteins were precipitated by chloroform-methanol extraction. Protein concentration was assessed by the Bradford assay (Bio-Rad). Crude protein extracts were subjected to either in-gel or in-solution digest. For the in-gel digestion, protein extracts were cleaned on a 10 cm, 4–12% gradient SDS-PAGE gel (Novex). The resulting lane was cut from the gel and subjected to in-gel digestion with trypsin and/or LysC as described previously31. Upon gel extraction, peptides were cleaned using Stage-tips and analyzed by nano-LC-MS/MS. For in-solution digestion, proteins from the crude extract were reduced with 1 mM dithiothreitol (DTT), alkylated with 5 mM iodoacetamide, predigested with the endoproteinase LysC (Wako) for 3 h, and further digested with LysC or trypsin overnight32. The resulting peptide mixture was cleaned using Stage-tips33 and subjected to nano-LC-MS/MS without prior peptide separation.
LC-MS/MS analysis
All samples were analyzed by online nanoflow LC-MS/MS as previously described34 with a few modifications. Briefly, nano-LC-MS/MS experiments were performed on an EASY-nLC system (Proxeon Biosystems, Odense, Denmark) connected to an LTQ-Orbitrap XL or LTQ-Orbitrap Elite (Thermo Scientific, Bremen, Germany) through a nanoelectrospray ion source. Peptides were auto-sampled directly onto the 15 cm long 75 mm-inner diameter analytical column packed with reversed-phase C18 Reprosil AQUA-Pur 3 mm particles at a flow rate of 500 nl/min. The flow rate was reduced to 250 nl/min after loading, and the peptides were separated with a segmented linear gradient of acetonitrile from 5–50% in 0.5% acetic acid for either 100, 150, or 240 minutes. Eluted peptides from the column were directly electrosprayed into the mass spectrometer. For the LTQ-Orbitrap XL analyses, the instrument was operated in positive ion mode, with the following acquisition cycle: a full scan recorded in the orbitrap analyzer at resolution R 60,000 was followed by MS/MS (CID) of the top 10 most intense peptide ions in the LTQ analyzer. The total acquisition time was either 150 or 240 minutes. For LTQ-Orbitrap Elite data acquisition the instrument was operated in the positive ion mode, with the following acquisition cycle: a full scan recorded in the orbitrap analyzer at resolution R 120,000 was followed by MS/MS (CID Rapid Scan Rate) of the 20 most intense peptide ions in the LTQ analyzer. The total acquisition time was either 100 or 240 minutes depending on the method of sample preparation. Mono-enzyme co-culture samples were measured on the LTQ-Orbitrap XL with slight modifications: a full scan recorded in the orbitrap analyzer at resolution R 60,000 was followed by MS/MS (CID) of the top 5 most intense peptide ions, with a total acquisition time of 95 minutes.
Processing of mass spectrometry data
The MaxQuant software package with the Andromeda search engine was used to identify and quantify proteins in cellular lysates and media19,35. MaxQuant version 1.2.2.5 was used to analyze all samples expect for comparing protein intensities across differentially labeled samples (Supplementary Fig. 8), where version 1.3.0.5 was required to select multiple groups with different labels. Mouse and human IPI protein databases (both version 3.84, http://www.ebi.ac.uk/IPI/) plus common contaminants and CTAP transgenes were used. With the exception of “second peptides”, which was deselected, default parameters were selected. Detection and quantitation of L-lysine containing peptides was specified as light L-lysine, medium L-lysine, and heavy L-lysine. For L-lysine derived from precursors DAP, Z-lysine (H), and D-lysine (H), variable labels were specified as light L-lysine, heavy L-lysine (lys8), and a custom modification (8 deuterium atoms for L-lysine), respectively. Only L-lysine-containing peptides were used for label quantitation. Default parameters in MaxQuant were used; MS/MS mass tolerance of 20 ppm, 1% FDR for peptide and protein identification, Re-quantify on, carbamethylation of cysteine (fixed modification), and oxidation of methionine and N-terminal protein acetylation (variable modifications).
Peptide and protein statistics (e.g., sequences, H/L ratios, intensities) were extracted from MaxQuant exported peptides.txt and proteingroups.txt, respectively. Entries that MaxQuant classified as contaminants were removed. Unless otherwise stated, no other filters or normalizations were applied to the H/L ratios. Peptides were determined to be species-specific if they appeared exclusively in one of the human or mouse IPI protein databases. For the species-specific sequence determination an exact peptide sequence match to the protein database was required, except Isoleucine (I) and Leucine (L) were used interchangeably. Percent heavy label was calculated from the H/L ratio (r) as = 100 *r/(r + 1). All peptide data presented in this manuscript were filtered for human and mouse specific sequences, irrespective of whether the samples were from mono- or co-cultures. No organism-specific filtering was performed for any protein data presented. At least two peptides (razor and unique) were required for protein identification. For displaying histograms of H/L ratios, 50 bins were used and a nonparametric kernel-smoothing function was applied to fit the distribution of the histogram.
Supplementary Material
Acknowledgments
We gratefully acknowledge E. Larsson, Y. Gruber, D.S. Marks, A. Arvey, J. Joyce, and A. Koff for helpful discussions. H. Erdjument-Bromage for pilot MS/MS investigation. A.N. Miller, J. Cross, X. Jing for technical help. E. Larsson, J. Gauthier, J. Joyce, and A.M. Miller for helpful comments on the manuscript. This work was funded in part by US National Cancer Institute grant U54 CA148967.
Footnotes
Storage of primary data
The raw LC-MS/MS data are available on the CTAP website (http://www.ctap.ms). Illumina microarray experiments are uploaded to GEO (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE43894 and GSE43895.
Accession codes
Oligonucleotide sequences for inserts are uploaded to GenBank (KC962560, KC962561, KC962562, and KC962563) and are available in Supplementary Note 1.
Author contributions
N.P.G. and M.L.M. designed, performed, and analyzed the experiments. W.E.W. generated reagents. B.S., K.J.M., and V.A.P. contributed with experiments. N.P.G. and M.L.M. wrote the manuscript. B.S., W.E.W., B.M., K.J.M., V.A.P., D.Y.G., and C.S. contributed to discussions and editing the manuscript. N.P.G. conceived the hypothesis. N.P.G., C.S., and M.L.M. developed the concept and managed the project.
Competing financial interests
The authors declare no competing financial interests. A provisional patent application relating to the use of exogenous enzymes for proteomic labeling in multicellular culture has been filed by Memorial Sloan-Kettering Cancer Center.
References
- 1.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–52. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mcmillin DW, et al. Tumor cell-specific bioluminescence platform to identify stroma-induced changes to anticancer drug activity. Nat Med. 2010;16:483–9. doi: 10.1038/nm.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olumi AF, et al. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59:5002–11. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gygi SP, et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–9. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 5.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Molecular & Cellular Proteomics. 2002;1:376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 6.Ross PL, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Molecular & Cellular Proteomics. 2004;3:1154–69. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Jørgensen C, et al. Cell-specific information processing in segregating populations of Eph receptor ephrin-expressing cells. Science. 2009;326:1502–9. doi: 10.1126/science.1176615. [DOI] [PubMed] [Google Scholar]
- 8.Rechavi O, et al. Trans-SILAC: sorting out the non-cell-autonomous proteome. Nat Methods. 2010;7:923–7. doi: 10.1038/nmeth.1513. [DOI] [PubMed] [Google Scholar]
- 9.Naba A, et al. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol Cell Proteomics. 2012;11:M111.014647. doi: 10.1074/mcp.M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van den Bemd GJCM, et al. Mass spectrometric identification of human prostate cancer-derived proteins in serum of xenograft-bearing mice. Mol Cell Proteomics. 2006;5:1830–9. doi: 10.1074/mcp.M500371-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Ngo JT, et al. Cell-selective metabolic labeling of proteins. Nat Chem Biol. 2009;5:715–7. doi: 10.1038/nchembio.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Truong F, Yoo TH, Lampo TJ, Tirrell DA. Two-strain, cell-selective protein labeling in mixed bacterial cultures. Journal of the American Chemical Society. 2012;134:8551–6. doi: 10.1021/ja3004667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ngo JT, Schuman EM, Tirrell DA. Mutant methionyl-tRNA synthetase from bacteria enables site-selective N-terminal labeling of proteins expressed in mammalian cells. Proc Natl Acad Sci USA. 2013;110:4992–4997. doi: 10.1073/pnas.1216375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annual review of biochemistry. 2010;79:413–44. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 15.Xu H, Andi B, Qian J, West AH, Cook PF. The alpha-aminoadipate pathway for lysine biosynthesis in fungi. Cell Biochem Biophys. 2006;46:43–64. doi: 10.1385/CBB:46:1:43. [DOI] [PubMed] [Google Scholar]
- 16.Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2006;1:2650–60. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 17.Saqib KM, Hay SM, Rees WD. The expression of Escherichia coli diaminopimelate decarboxylase in mouse 3T3 cells. Biochim Biophys Acta. 1994;1219:398–404. doi: 10.1016/0167-4781(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 18.Jouanneau J, Stragier P, Bouvier J, Patte JC, Yaniv M. Expression in mammalian cells of the diaminopimelic acid decarboxylase of Escherichia coli permits cell growth in lysine-free medium. Eur J Biochem. 1985;146:173–8. doi: 10.1111/j.1432-1033.1985.tb08635.x. [DOI] [PubMed] [Google Scholar]
- 19.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 20.Sury MD, Chen JX, Selbach M. The SILAC fly allows for accurate protein quantification in vivo. Mol Cell Proteomics. 2010;9:2173–83. doi: 10.1074/mcp.M110.000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spellman DS, Deinhardt K, Darie CC, Chao MV, Neubert TA. Stable isotopic labeling by amino acids in cultured primary neurons: application to brain-derived neurotrophic factor-dependent phosphotyrosine-associated signaling. Mol Cell Proteomics. 2008;7:1067–76. doi: 10.1074/mcp.M700387-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geiger T, Wehner A, Schaab C, Cox J, Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol Cell Proteomics. 2012;11:M111.014050. doi: 10.1074/mcp.M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuan YC, et al. Biochemical characterization of a novel lysine racemase from Proteus mirabilis BCRC10725. Process Biochemistry. 2011;46:1914–1920. [Google Scholar]
- 24.Nanduri V, Goldberg S, Johnston R, Patel R. Cloning and expression of a novel enantioselective N-carbobenzyloxy-cleaving enzyme. Enzyme and Microbial Technology. 2004;34:304–312. [Google Scholar]
- 25.Mavrakis KJ, et al. Tumorigenic activity and therapeutic inhibition of Rheb GTPase. Genes & Development. 2008;22:2178–88. doi: 10.1101/gad.1690808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swift S, Lorens J, Achacoso P, Nolan GP. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. Curr Protoc Immunol. 2001;Chapter 10(Unit 10.17C) doi: 10.1002/0471142735.im1017cs31. [DOI] [PubMed] [Google Scholar]
- 27.Szymczak AL, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–94. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 28.Smyth G. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and computational biology solutions using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 30.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–60. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 32.Macek B, et al. Phosphorylation of the human full-length protein kinase Ciota. J Proteome Res. 2008;7:2928–35. doi: 10.1021/pr800052z. [DOI] [PubMed] [Google Scholar]
- 33.Ishihama Y, Rappsilber J, Mann M. Modular stop and go extraction tips with stacked disks for parallel and multidimensional Peptide fractionation in proteomics. J Proteome Res. 2006;5:988–94. doi: 10.1021/pr050385q. [DOI] [PubMed] [Google Scholar]
- 34.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–48. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 35.Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.