Background: We hypothesize that ER chaperone Cosmc binds specifically to its single client T-synthase.
Results: Using molecular and biochemical approaches, we demonstrate that Cosmc recognizes a specific peptide within unfolded T-synthase.
Conclusion: Cosmc-T-synthase interaction requires unique peptide recognition in the unfolded T-synthase that becomes inaccessible to Cosmc upon final folding.
Significance: This study describes a novel peptide-specific chaperone recognition that regulates protein O-glycosylation.
Keywords: Endoplasmic Reticulum (ER), Glycobiology, Glycoprotein, Glycoprotein Biosynthesis, Glycosylation, Molecular Chaperone, Protein Folding, Cosmc, T-synthase
Abstract
Prior studies suggested that the core 1 β3-galactosyltransferase (T-synthase) is a specific client of the endoplasmic reticulum chaperone Cosmc, whose function is required for T-synthase folding, activity, and consequent synthesis of normal O-glycans in all vertebrate cells. To explore whether the T-synthase encodes a specific recognition motif for Cosmc, we used deletion mutagenesis to identify a cryptic linear and relatively hydrophobic peptide in the N-terminal stem region of the T-synthase that is essential for binding to Cosmc (Cosmc binding region within T-synthase, or CBRT). Using this sequence information, we synthesized a peptide containing CBRT and found that it directly interacts with Cosmc and also inhibits Cosmc-assisted in vitro refolding of denatured T-synthase. Moreover, engineered T-synthase carrying mutations within CBRT exhibited diminished binding to Cosmc that resulted in the formation of inactive T-synthase. To confirm the general recognition of CBRT by Cosmc, we performed a domain swap experiment in which we inserted the stem region of the T-synthase into the human β4GalT1 and found that the CBRT element can confer Cosmc binding onto the β4GalT1 chimera. Thus, CBRT is a unique recognition motif for Cosmc to promote its regulation and formation of active T-synthase and represents the first sequence-specific chaperone recognition system in the ER/Golgi required for normal protein O-glycosylation.
Introduction
Mucin-type O-glycosylation involving modifications of proteins with the core 1 O-glycan Galβ-3GalNAcα1-Ser/Thr and its extensions represent one of the most widespread post-translational modifications of animal cell glycoproteins (1, 2). Mucin-type O-glycosylation is important in many biological processes (3), including platelet functions (4), leukocyte homing (5–7), animal development (8, 9), and microbial interactions (10). However, the overall biosynthesis and regulation of glycans in this pathway are still poorly understood. Many different polypeptide N-acetylgalactosaminyltransferases catalyze the initial step of O-glycosylation by adding GalNAc to Ser/Thr in proteins in the secretory pathway, resulting in Tn antigen expression (11). The Tn antigen then serves within the Golgi apparatus as a substrate for the enzyme core 1 β3-galactosyltransferase (T-synthase),4 which is the only enzyme that catalyzes the addition of galactose onto Tn to generate the core 1 disaccharide, also known as T-antigen (12, 13), which in turn serves as the precursor for many different glycosyltransferases to generate many extended O-glycans in most mammalian cells (3). The Tn antigen can also serve as the substrate for the core 3 GlcNAc-transferase expressed highly in the colon (14, 15).
The formation of active and stable T-synthase requires a specific molecular chaperone termed Cosmc in the endoplasmic reticulum (ER) that directly binds to the T-synthase and limits its aggregation and degradation during folding in the ER (8, 16–18). Thus, expression of Cosmc specifically regulates mucin type O-glycosylation by regulating T-synthase biosynthesis (16–19). Deletion of either Cosmc or T-synthase in mice causes embryonic lethality (8, 9), and acquired mutations, deletion, and hypermethylation of Cosmc are shown to be associated with several human diseases (20–22). Importantly, dysfunctional Cosmc results in the expression of the Tn and sialyl-Tn antigen (23), which are known as tumor-associated carbohydrate antigens present in many cancers such as colon and breast (23, 24). The Tn and sialyl-Tn antigen are also seen in other human diseases including Tn-syndrome, IgA nephropathy, and Henoch-Schönlein purpura (23).
Cosmc promotes the folding of denatured T-synthase both in vivo and in vitro and has properties in that respect that are similar to other classical chaperones (16–19). Cosmc binds directly to non-native T-synthase and promotes the activity of heat/chemically denatured T-synthase independently of other co-chaperones and ATP in vitro (16, 19). The productive interaction of Cosmc with non-native T-synthase results in a relatively stable complex between Cosmc and T-synthase where T-synthase becomes active (19). Active reconstituted T-synthase can be released from the complex in the presence of both non-native and native T-synthase, suggesting a client-driven Cosmc chaperone cycle (19). In vivo, similar to many newly synthesized proteins in eukaryotic cells, T-synthase enters the lumen of the ER where it folds into correct native structure by assistance from Cosmc or forms aggregates in the absence of Cosmc that are subsequently targeted to ER-associated degradation (17, 25, 26). Although a majority of proteins utilize the assistance of a set of conserved promiscuous chaperones (27, 28), T-synthase specifically requires Cosmc to fold correctly and become functional (8, 16–19). The nature of the molecular interactions between Cosmc and T-synthase, however, is not yet understood.
The binding of Cosmc to the denatured but not native T-synthase suggests that a peptide element may be exposed in the denatured T-synthase that we hypothesize is recognized by Cosmc. Here we report our identification of a novel Cosmc binding region within T-synthase (CBRT), which resides within the N-terminal portion of its stem region. Cosmc directly binds to CBRT in the unfolded but not folded T-synthase and thus permits productive folding of the downstream catalytic domain of the T-synthase and prevents unproductive oligomerization of misfolded T-synthase. The unique role of CBRT within the stem region of the T-synthase provides the explanation for the specificity of Cosmc toward a single client protein.
EXPERIMENTAL PROCEDURES
Materials
UDP-Gal and pNP-β-S-GlcNAc were from Sigma-Aldrich and GalNAc-α-4-methylumbelliferone was purchased from Carbosynth Limited (Berkshire, UK). UDP-6-[3H]Gal (40-60 Ci/mmol) was from American Radiolabeled Chemicals, Inc. (St. Louis, MO). Sep-Pak C18 Cartridges were from Waters Corporation (Milford, MA). Insect cells (Hi-5 and Sf-9) were from American Type Culture Collection (Manassas, VA). Restriction enzymes were from New England Biolabs, Inc. (Ipswitch, MA). The baculovirus transfection kit was from BD Biosciences (San Jose, CA). Ni-NTA Superflow beads were from Qiagen. SDS-PAGE gels and Western blot materials were from Bio-Rad. Ultralink beads were from Pierce.
Preparation of Expression Constructs and Peptides
Human soluble HPC4-T-synthase aa 45-363 (HPC4-sTsyn 45-363) and His-sCosmc were prepared as described (17). Using Centricon 10-kDa cut-off membranes, both proteins were concentrated in 10 mm HEPES buffer, 30 mm NaCl, pH 7.8, and stored at −80 °C. C-terminally truncated HPC4-T-syn constructs 45-295, 45-245, 45-196, 45-145, and 45-97 were generated from HPC4-sTsyn 45-363 by introducing stop codons at desired sites using site-directed mutagenesis (Table 1). N-terminal truncated constructs HPC4-sTsyn 83-363, 121-363, and 168-363 were generated by ligating purified larger fragments of pVL1393 XbaI/BamHI and XbaI/BamHI-digested PCR product obtained from the HPC4-T-synthase 45-363 construct (Table 1), as well as a small fragment generated by digesting HPC4-T-synthase 45-363 with BamHI. Human soluble N-terminal HPC4-β4-galactosyltransferase I (HPC4-sβ4GalT1) was synthesized by Genescript. The Emory DNA Custom Cloning Core Facility prepared the following constructs: HPC4-sTsyn-5A, HPC4-sT-synΔCBRT, and HPC4-sβ4GalT1-CBRT. The CBRT-containing peptide (ENTDIAENLYQKVRILCWVMTGPQNLEKKA) with or without N-terminal biotin, the Scrambled CBRT peptide (NYNGKMWNIACTRQLEKVIVLEQAETKDPL), and its N terminus biotinylated version of the peptide (NYNLKMWNIACTRQLEKTIVGEQAEVKDPL) were synthesized by Biomatik (Wilmington, DE). Peptides were dissolved as described by the manufacturer, and scrambled and biotinylated peptides were dissolved in water and stored at −20 °C.
TABLE 1.
PCR primers used for generating different truncated versions of T-synthase
| Protein | Primers | |
|---|---|---|
| HPC4-sTsyn 83-363 | Forward | 5′-GCGGATCCACTCTATCAGAAAGTTAGAATTCTTTGC-3′ |
| Reverse | 5′-GCTCTAGATCAAGGATTTCCTAACTTCACTTTTGTATC-3′ | |
| HPC4-sTsyn 168-363 | Forward | 5′-GCG GATCCAGCAGATGATGACACGTATG-3′ |
| Reverse | 5′-GCTCTAGATCAAGGATTTCCTAACTTCACTTTTGTATC-3′ | |
| HPC4-sTsyn 121-363 | Forward | 5′-GCGGATCCAATGAGTTCAGAAGAAAATAAAGACTTC-3′ |
| Reverse | 5′-GCTCTAGATCAAGGATTTCCTAACTTCACTTTTGTATC-3′ | |
| HPC4-sTsyn 45-97 | Forward | 5′-GGTTATGACCGGCCCTTAAAACCTAGAGAAAAAGGC-3′ |
| Reverse | 5′-GCCTTTTTCTCTAGGTTTTAAGGGCCGGTCATAACC-3′ | |
| HPC4-sTsyn 45-145 | Forward | 5′-GGCAGAGATCAACTATACTGGTAAACAATTAAAGCTTTTCAG-3′ |
| Reverse | 5′-CTGAAAAGCTTTAATTGTTTACCAGTATAGTTGATCTCTGCC-3′ | |
| HPC4-sTsyn 45-196 | Forward | 5′-GAACCCATTTACTTTGGGAGATGATTTAAGCCTTAT GTAAAGC-3′ |
| Reverse | 5′-GCTTTACATAAGGCTTAAATCATCTCCCAAAGTAAATGGGTTC-3′ | |
| HPC4-sTsyn 45-245 | Forward | 5′-GACTTAGCACTGGGGAGATGAATGGAAATTATGAATGTAGAAGC-3′ |
| Reverse | 5′-GCTTCTACATTCATAATTTCCATTCATCTCCCCAGTGCTAAGTC-3′ | |
| HPC4-sTsyn 45-295 | Forward | 5′-CAACTATTATCCTCCTGTATAGGGTCCTGGTTGCTGCTCTG-3′ |
| Reverse | 5′-CAGAGCAGCAACCAGGACCCTATACAGGAGGATAATAGTTG-3′ |
Pulldown and T-synthase Activity Assay
Hi-5 cells (∼60% confluent) were infected individually with different baculoviruses (both N- and C-terminal deletions as listed above) of HPC4-sT-syn or coinfected with His-sCosmc and incubated at 27 °C for 96 h. The cell suspension (1.5 ml) was pelleted at 600 × g (no. 5415D; Eppendorf, Hauppauge, NY) and lysed in 300 μl of lysis buffer A (50 mm imidazole, 0.5% Triton X-100, 150 mm NaCl, pH 7.8, and protease inhibitors) vortexing periodically for 30 min followed by centrifugation at 18,000 × g for 10 min. Cell lysates (100 μl) from each preparation were mixed with 100 μl of buffer B (50 mm imidazole, 0.1% Triton X-100, 150 mm NaCl, pH 7.8) followed by addition of 50 μl of 50% slurry of Ni-NTA beads equilibrated in buffer B. The mixture was incubated at 4 °C overnight on a rotator at 30 rpm. Beads were pelleted (100 × g) for 1 min and washed five times with 1 ml of buffer B incubating at least 2 min on ice. Elution was carried out by 60 μl of elution buffer containing 300 mm imidazole by incubating for 1 h. of input, unbound, and [1/2.4] of elution was analyzed by SDS-PAGE and Western blot. Each experiment was repeated at least two independent times. A similar approach was used to pull down HPC4-sT-syn-m-5A and control HPC4-sT-syn except that cells were lysed for 15 min and incubated with Ni-NTA beads for 10 min. Cell lysates (5 μl) were directly used to determine T-synthase activity using UDP-Gal as the donor as GalNAc-α-(4-methylumbelliferone) as the acceptor (29).
Pulldowns involving domain swap experiments (HPC4-sβ4GalT1, HPC4-sTsyn, and chimeric versions expressing individually or with His-sCosmc) were carried out as described previously except using half the amount of cell lysate as starting material, 20 μl of 50% Ni-NTA beads and 10 min of incubation. of input in all experiments, except for Cosmc and HPC4-sβ4GalT1 co-expression experiments, unbound was used. [1/5.4] of bound (except for [1/2.7] in Cosmc and HPC4-sβ4GalT1 co-expression experiment) was used to analyze the bound materials. For T-synthase activity assay, the cell lysate was optimized for the expression of HPC4-containing T-syn and β4GalT1; therefore we used 5 μl of cell lysates for T-syn-related experiments, 0.25 μl for HPC4-sβ4GalT1, 20 μl for HPC4-sβ4GalT1 and Cosmc co-expression, and 3 μl for chimeric HPC4-sβ4GalT1 with or without Cosmc; 10 μl of cell lysates were used for Cosmc alone. The same amount of cell lysate that was used to determine activity for each experiment was used respectively to analyze the level of HPC4 protein expression. A similar approach was used to determine β4GalT1 activity and its expression. β4GalT1 activity was determined using UDP-6-[3H]Gal as the donor and pNP-β-S-GlcNAc as the acceptor (30, 31).
Pulldown studies involving direct binding were carried out with 4.3 μg of either biotin WT peptide or biotinylated SC peptide captured on streptavidin beads (40 μl of 50% slurry). To this preparation, 0.2 or 10 μg of His-sCosmc containing 100 μl of buffer C (50 mm Tris-HCl, pH 7.8, containing 150 mm NaCl and 0.1% Triton X-100) was incubated. The mixture was incubated for 30 min, and beads were washed five times with 1 ml of buffer C. Bound materials were eluted by boiling beads on 80 μl of SDS-PAGE sample buffer. of input, unbound, and [1/5.4] of the elution were analyzed by Western blotting against Cosmc.
CBRT-containing Peptide Inhibition of Complex Formation between Cosmc and Reconstituted T-synthase
In vitro refolding experiments were performed as described previously (16). T-synthase (0.45 μg) was used in each experiment, and a constant amount of 45 μl of 50% slurry of Cosmc beads (0.6 μg/μl) or the same amount of control beads was used. Five and 20 μg of peptide containing CBRT and scrambled version were added to 100 μl of reconstitution buffer containing denatured T-synthase prepared by heating at 55 °C for 2 min followed by addition of 45 μl of 50% slurry of Cosmc beads and incubated overnight at 4 °C on a rotator at 35 rpm. Beads were pelleted at 100 × g for 1 min. Beads were washed five times with 450 μl of wash buffer containing 0.1% Triton X-100. Elution was performed by boiling beads for 10 min in SDS-PAGE sample buffer. of input, of unbound, and ⅓ of elution were blotted for HPC4 T-synthase.
In Vitro Refolding Experiment in the Presence of Peptide Containing CBRT and Scrambled Version
In vitro refolding experiments were carried out as described previously (16). HPC4-sT-syn (0.25 μg/20 μl) in buffer (10 mm HEPES, 12 mm MgCl2, 150 mm NaCl, pH 7.8) was used for refolding experiments. The HPC4-sT-syn was heat-denatured at 55 °C for 2 min and allowed to cool to room temperature followed by addition of CBRT-containing peptide or scrambled CBRT peptide. Reconstitution was initiated by the addition of Cosmc (2.67 μg/reaction) for 45 min. T-synthase activity was assayed for 45 min at 37 °C.
RESULTS
T-synthase Contains a Novel Cosmc Binding Region within its N-terminal Stem Region
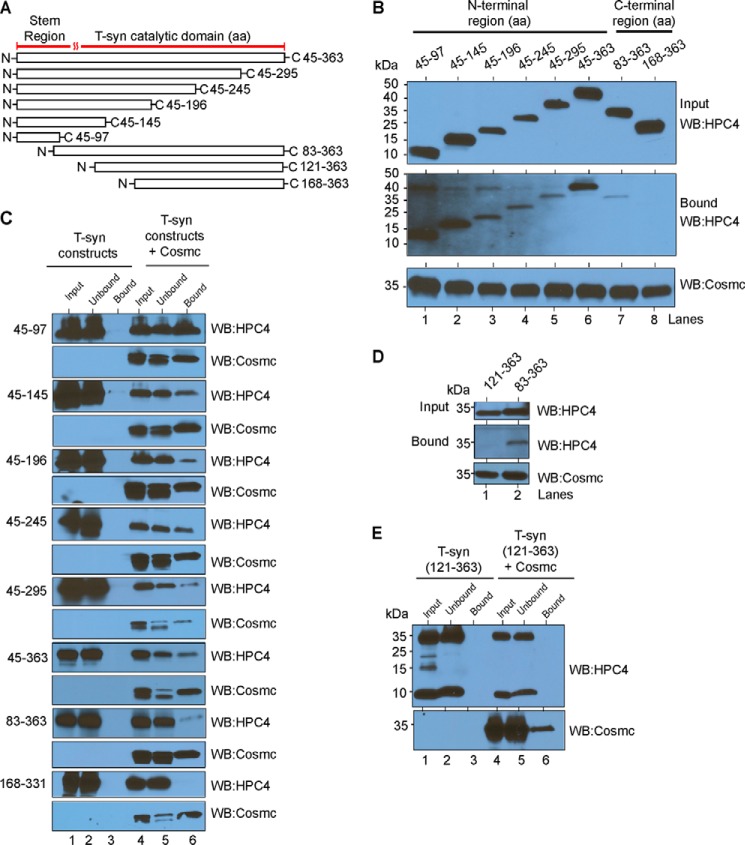
In our initial approach to define the interaction between Cosmc and T-synthase, we engineered a wide range of truncated versions of HPC4-tagged T-synthase (HPC4-sT-syn) with various N-terminal or C-terminal deletions (Fig. 1A and Table 2). We expressed these constructs individually or co-expressed each of them with His-tagged soluble Cosmc (His-sCosmc) in an insect cell system that constitutively lacks Cosmc. We first explored the interaction between the truncated T-syn and Cosmc using a pulldown approach with Ni-NTA beads. Whole cell lysates expressing various HPC4-sT-syn with or without His-sCosmc were incubated with Ni-NTA beads followed by Western blot analysis of pulled down material. We observed that all of the expressed HPC4-sT-syn constructs associated with His-sCosmc (Fig. 1B, lanes 1–7), except for the N-terminal deleted HPC4-sT-syn 168-363 (Fig. 1B, lane 8). In control studies, none of the individually expressed HPC4-sT-syn constructs bound to beads alone (Fig. 1C, lane 3). Because we see interaction with construct 83-363 but not 168-363, the results indicate that the binding region is lost in the 168-363 truncation and therefore must be contained between residues 83 and 168. Given that the 45-97 construct is bound to Cosmc (Fig. 1B), the results indicate that the binding domain for Cosmc is contained within residues 83-97. To test this prediction, we successfully expressed the HPC4-sT-syn 121-363 construct (Fig. 1A), which is 38 aa shorter from the N-terminal stem region than HPC4-sT-syn 83-363. We expressed both constructs either alone or in co-expression with His-sCosmc. Interestingly, HPC4-sT-syn 83-363 (Fig. 1, B, lane 7, and D, lane 2), but not HPC4-sTsyn 121-363, associated with His-sCosmc (Fig. 1, D, lane 1, and E, lane 6). Together, these data suggest that the Cosmc binding region of the T-synthase, which we have termed CBRT, resides within its N-terminal stem region and requires residues 83-97. Interestingly, this region is relatively conserved within T-synthase found in vertebrates but not in invertebrates (Fig. 2), which do not have an ortholog for Cosmc.
FIGURE 1.
Identification and functional characterization of a novel Cosmc binding region in human T-synthase, the CBRT. A, schematics depicting the truncated soluble versions of HPC4-T-synthase. All constructs contain a signal sequence and HPC4 tag followed by different lengths of lumenal T-synthase as indicated. B and C, cosmc binds to the N-terminal stem region of T-synthase. B, input and bound fractions from the pulldown experiment were immunoblotted using monoclonal antibody against HPC4 for T-synthase, and the His-sCosmc used for pulldown was detected by monoclonal antibody against Cosmc. C, whole cell lysates from Hi-5 cells expressing various constructs of the human N-terminal HPC4 epitope-tagged soluble T-synthase (HPC4-sT-syn) expressed individually or co-expressed with human N-terminal His-tagged soluble Cosmc (His-sCosmc) were incubated with Ni-NTA beads followed by pulldown experiments. Input, unbound, and bound fractions were detected by antibody against HPC4 for T-synthase (top panels). Similarly, the amount of Cosmc used in the pulldown experiment was detected by antibody against Cosmc (bottom panels). D, pulldown experiment demonstrating that aa 83-121 in the N-terminal stem region of T-synthase are essential for binding to Cosmc. Input, bound, and amount of Cosmc used in the pulldown experiment were analyzed as described in B. E, pulldown experiment showing HPC4-sT-syn 121-363 does not bind to Cosmc or beads alone. A representative example of two independent experiments is shown. The data in B and D were repeated at least two independent times, and a representative example is shown. WB, Western blot.
TABLE 2.
List of truncated T-synthase proteins
Refer to Fig. 2 for the complete sequences.
| Protein | Amino acid sequence |
|---|---|
| HPC4-sTsyn 45-363 | 45DPHAR. . . . . .LGNP363 |
| HPC4-sTsyn 45-295 | 45DPHAR. . . . . .YPPV295 |
| HPC4-sTsyn 45-245 | 45DPHAR. . . . . .ALGR245 |
| HPC4-sTsyn 45-196 | 45DPHAR. . . . . .YFGR196 |
| HPC4-sTsyn 45-145 | 45DPHAR. . . . . .QLYW145 |
| HPC4-sTsyn 45-97 | 45DPHAR. . . . . .MTGP97 |
| HPC4-sTsyn 83-363 | 83LYQK . . . . . .LGNP363 |
| HPC4-sTsyn 121-363 | 121MSSE. . . . . .LGNP363 |
| HPC4-sTsyn 168-363 | 168ADDD. . . . . .LGNP363 |
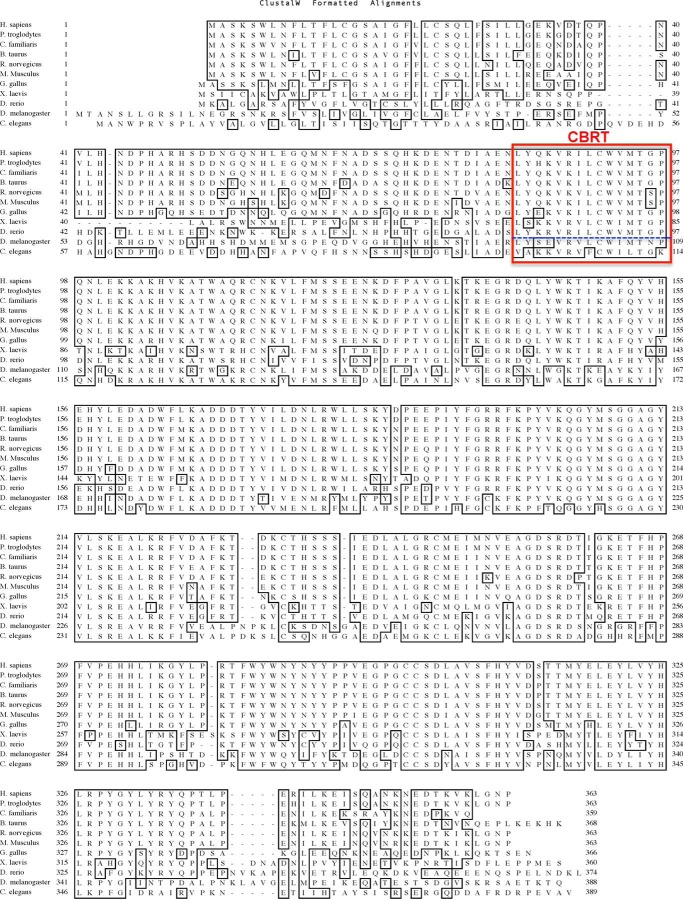
FIGURE 2.
Comparison of amino acid sequences of T-synthase from different species showing CBRT within vertebrates is conserved (67). The CBRT region as determined in Fig. 1 is boxed in red, and vertebrate CBRT and invertebrate CBRT are separated by blue dashes. No significant homology of this CBRT region was found with other peptide sequences in other proteins upon BLAST searches.
Cosmc Binding to CBRT Is Necessary for T-synthase Function
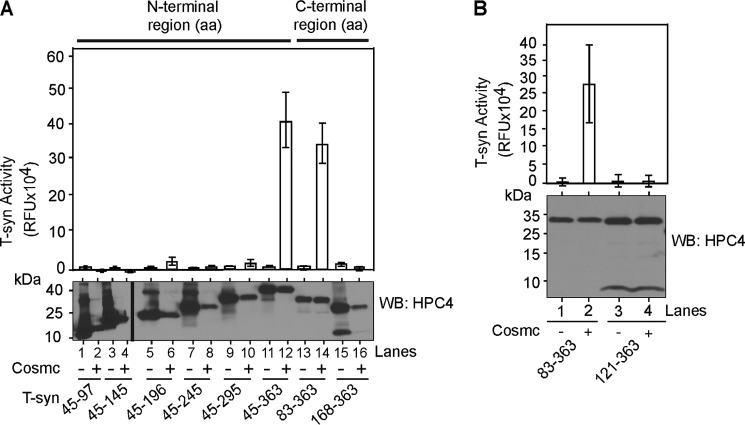
We measured T-synthase activity as another approach to access the productive interaction of T-synthase with Cosmc using the whole cell lysates from above. The positive control HPC4-sTsyn 45-363 was highly active when co-expressed with His-sCosmc (Fig. 3A). HPC4-sTsyn 83-363, which interacts with His-sCosmc (Fig. 1, B, lane 7, and D, lane 2), also had significant activity in the presence of His-sCosmc co-expression (Fig. 3A). However, for the HPC4-sTsyn 45-196 (a 167-aa C-terminal deletion) construct, which interacts with His-sCosmc (Fig. 1B, lane 3), we detected only a small amount of T-synthase activity in co-expression with His-sCosmc (Fig. 3A, lane 6). We also did not detect significant activity of the other C-terminal deletion constructs (Fig. 3A, lanes 8 and 10), even though they contain a DXD motif and interact with His-sCosmc (Fig. 1B, lanes 4 and 5), thus suggesting that they may lack an active site for the acceptor. Importantly, HPC4-sTsyn 121-363 construct, which does not associate with Cosmc (Fig. 1D, lane 1), did not demonstrate any detectable activity with or without co-expressed Cosmc (Fig. 3B, lanes 3 and 4). These data indicate that the N-terminal stem region of T-synthase containing aa 83-121 encompasses the majority of information required for binding to Cosmc and that this region, along with other sequence elements, is important for the biosynthesis of functional and active T-synthase.
FIGURE 3.
Cosmc binding to CBRT is necessary for T-synthase function. A, Cosmc binding to CBRT is necessary for T-synthase function. A portion of whole cell lysates from Hi-5 cells expressing various constructs of HPC4-sT-syn as depicted in Fig. 1A expressed individually or co-expressed with His-sCosmc were directly assayed for the activity (top panel). Half of the other portion of lysate was directly used to determine the amount HPC4-sT-syn expression (bottom panel), using mAb against HPC4 for T-synthase constructs. B, T-synthase activity demonstrating 83-121 aa is necessary for the biosynthesis of functional T-synthase. Similar to A, T-synthase activity assay (top panel) and the amount of T-synthase present in those samples were determined by Western blotting against HPC4-sT-syn. Each assay was performed in duplicate, three replicate experiments were performed, and the data represent the averages of all experiments. Error bars, ± S.D. from the average. WB, Western blot.
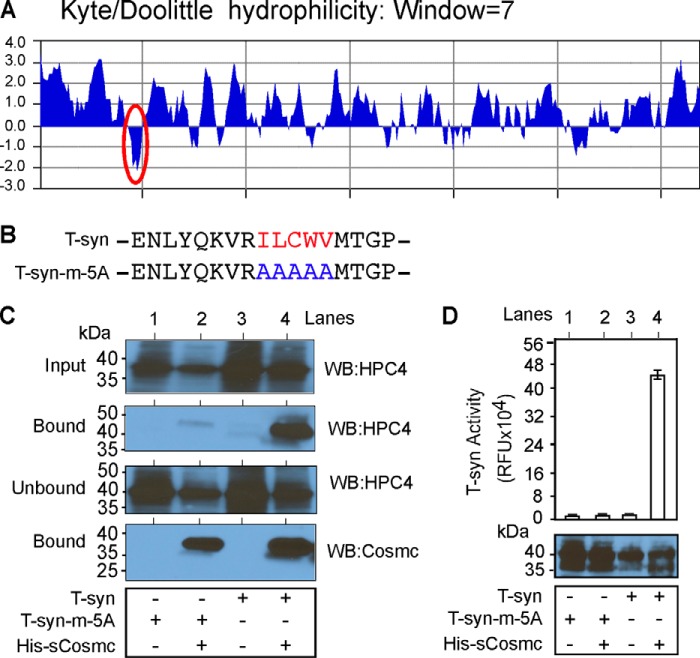
Cosmc Binds to a Hydrophobic Sequence within CBRT
Analysis using a Kyte/Doolittle hydropathy plot shows that the CBRT domain resides within a relatively hydrophobic region within the stem domain of T-synthase (Fig. 4A). To explore whether hydrophobic amino acids in that region are important to Cosmc binding, we mutated those amino acid residues to alanines to generate the construct T-syn-m-5A (Fig. 4B). In the pulldown experiments, we found that T-syn-m-5A, although well expressed in insect cells, did not bind significantly to His-sCosmc (Fig. 4C, lane 2) as compared with wild-type T-syn (Fig. 4C, lane 4). The bead control shows that neither T-syn-m-5A nor T-syn bind nonspecifically to beads (Fig. 4C, lanes 1 and 3). Furthermore we did not detect activity of T-syn-m-5A with or without Cosmc co-expression (Fig. 4D, lanes 1 and 2), whereas we observed robust activity of T-syn in the presence of His-sCosmc (Fig. 4D, lane 4). These data demonstrate that the small hydrophobic region within the CBRT is required for the interaction with Cosmc and acquisition of functional enzyme activity.
FIGURE 4.
Cosmc binds to hydrophobic amino acids of CBRT to make functional T-synthase. A, Kyte/Doolittle hydrophobicity analysis of the lumenal domain of T-synthase. The hydrophobic area in the stem region is highlighted with a red oval. B, primary aa sequence showing clustered hydrophobic sequence (red) of HPC4-sT-syn (T-syn) and the primary aa sequence where the hydrophobic sequence is substituted by five alanines (blue) of HPC4-sT-syn-m-5A (T-syn-m-5A). C, the hydrophobic region of CBRT is important for Cosmc binding. Input, unbound, and bound fractions from the pulldown experiment were detected by monoclonal antibody against HPC4 for T-synthase. Similarly, the amount of Cosmc used in the pulldown experiment was detected by monoclonal antibody against Cosmc. D, the hydrophobic region of CBRT is important for T-synthase function. Whole cell lysates from Hi-5 cells expressing T-syn or Tsyn-m-5A individually or co-expressed with His-sCosmc were directly assayed for T-synthase activity and the level of T-synthase expression by Western blotting against HPC4. The data in C were repeated three independent times, and a representative example is shown. In D, each assay was performed in duplicate, three replicate experiments were performed, and the data represent the averages of all experiments. Error bars, ± S.D. from the average. WB, Western blot.
Cosmc Interacts Directly with the Synthetic Peptides Containing CBRT
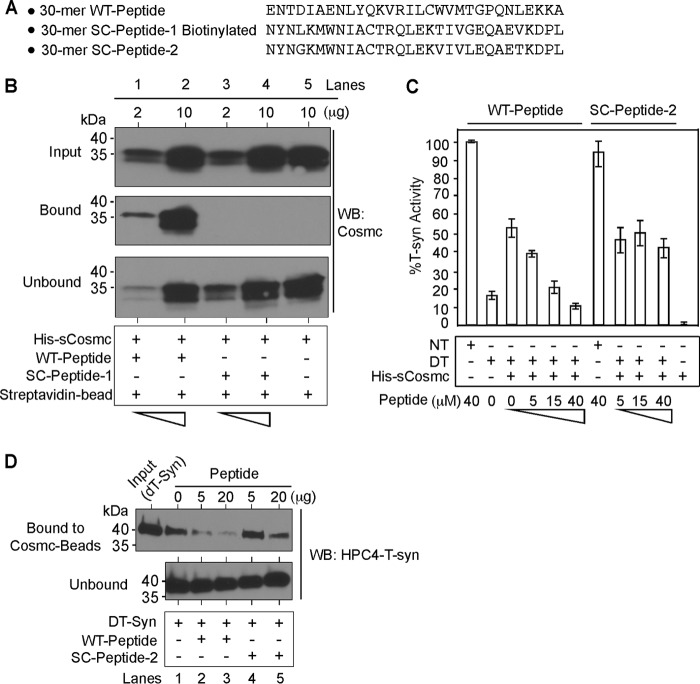
To directly explore the interaction of Cosmc to the CBRT domain, we synthesized two polypeptides: one was a biotinylated 30-mer peptide (Fig. 5A, WT-Peptide) representing key residues within CBRT, and the other was a biotinylated, scrambled version of the 30-mer peptide (Fig. 5A, SC-Peptide-1). We also expressed and purified the recombinant His-sCosmc in an insect cell system. We immobilized both peptides onto streptavidin-Sepharose and incubated them with His-sCosmc followed by pulldown experiments and Western blotting against Cosmc. Although the biotinylated WT peptide was effective in pulling down His-sCosmc in a dose-dependent manner (Fig. 5B, lanes 1 and 2), the SC peptide 1 was much less effective (Fig. 5B, lanes 3 and 4), indicating that this interaction with the WT peptide is specific and sequence dependent and does not simply reflect binding to a hydrophobic domain.
FIGURE 5.
Cosmc directly associates with the CBRT to assist T-synthase folding. A, three peptides were synthesized representing aa of the CBRT region and designated as shown. B, synthetic biotinylated peptide containing CBRT directly interacts with Cosmc in a dose-dependent manner but not with scrambled biotinylated peptide. Approximately similar amounts of biotinylated CBRT-containing peptide (WT-Peptide) or scrambled peptide (SC-Peptide-1) were immobilized on streptavidin beads and incubated with varied concentrations of Cosmc as indicated followed by pulldown. Input, bound, and unbound materials were analyzed by Western blotting against Cosmc. C, the WT peptide, but not the SC peptide 2, inhibits His-sCosmc assisted refolding of heat-denatured T-synthase (DT-syn) in a dose-dependent manner. Reconstitution of heat-denatured HPC4-sT-syn, which was heated for 2 min at 55 °C, was initiated by the addition of His-sCosmc, and the percentage of restored T-synthase activity was determined in the presence of varied concentrations of WT peptide and SC-peptide. D, WT peptide, but not the SC peptide 2, significantly inhibits the formation of Cosmc-T-synthase complex in a dose-dependent manner. Input, bound, and unbound materials from the pulldown experiment were analyzed by Western blotting against HPC4. In B and D, the experiment was repeated three independent times, and a representative example is shown. Biotinylated CBRT-containing peptide (WT-Peptide) or biotinylated scrambled peptide (SC-Peptide-1) were only used in B, and nonbiotinylated forms were used in C and D. In C, the data show averages of three independent experiments. Error bars, ± S.D. from the average. DT, heat-denatured T-synthase; NT, native T-synthase; WB, Western blot.
Because Cosmc assists in refolding heat-denatured T-synthase in vitro independently of other factors (16), we hypothesized that the CBRT-containing peptide could compete with heat-denatured T-synthase for binding to Cosmc, which might cause inhibition of Cosmc-assisted refolding of denatured T-synthase. We expressed and purified recombinant HPC4-T-synthase from an insect cell system and tested the WT peptide and SC peptide in refolding experiments. The WT peptide inhibited the His-sCosmc-assisted refolding of heat-denatured HPC4-T-synthase, as measured by T-synthase activity, in a dose-dependent manner (Fig. 5C, lanes 1–6), whereas SC peptide 2 had no significant effect (Fig. 5C, lanes 7–11). Additionally, we performed pulldown experiments with heat-denatured HPC4-T-synthase toward His-sCosmc in the presence of varying concentration of WT peptide and its scrambled version. We prepared active Cosmc covalently conjugated to beads and the denatured version of HPC4-T-synthase as described (19). We incubated Cosmc-conjugated beads with heat-denatured T-synthase in the presence of varying concentrations of the CBRT containing WT peptide and the SC peptide 2. Although the WT peptide inhibited binding of heat-denatured T-synthase in a dose-dependent manner (Fig. 5D, lanes 2 and 3), the SC peptide 2 was less effective (Fig. 5D, lanes 4 and 5). Together, these data demonstrate that Cosmc binds directly to the CBRT sequence within the N-terminal stem region of T-synthase to promote its productive folding.
Cosmc Interacts with Chimeric β4GalT1 Containing CBRT
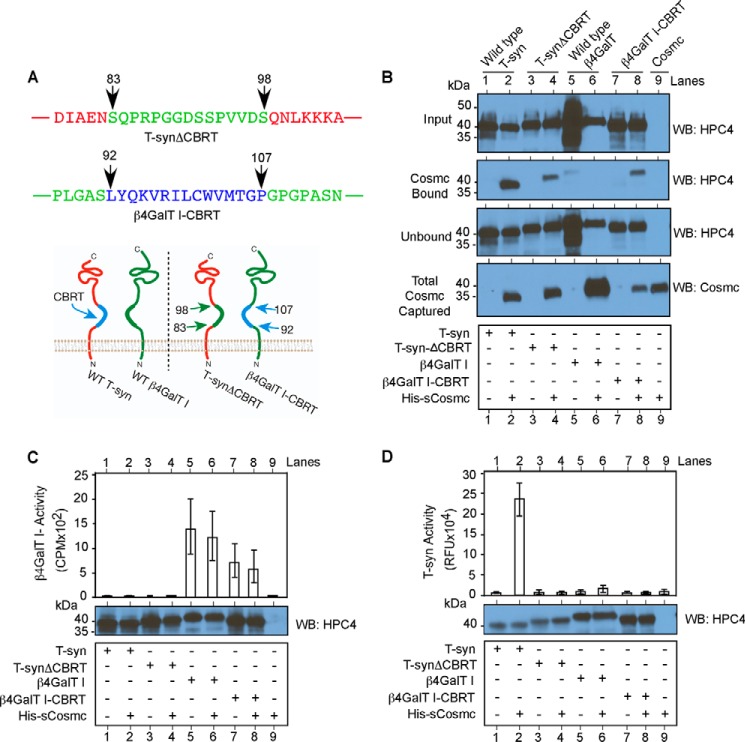
As a robust test to further determine the predicted role of the CBRT in binding to Cosmc, we prepared a chimeric glycosyltransferase, in which we swapped the 15 residues within CBRT of T-synthase (residues 83-98) (shown in blue text in Fig. 6A) with the stem region of β4GalT1 (residues 92–107) (shown in green text in Fig. 6A), which is also a type II transmembrane protein and a β-galactosyltransferase (32) (Fig. 6A). This change generated the chimeras T-synΔCBRT and β4GalT1-CBRT, respectively. We expressed HPC4-sT-syn and HPC4-sβ4GalT1, as well as their chimeric constructs, individually or co-expressed with His-sCosmc in an insect cell system. Whole cell lysates from each of the experiments were pulled down by Ni-NTA beads followed by Western blotting against HPC4-sT-syn or Cosmc. Although wild-type T-syn bound Cosmc, the T-synΔCBRT was less efficiently bound (Fig. 6B, lanes 2 and 4, respectively). Conversely, whereas the wild-type β4GalT1 was not bound by His-Cosmc, the chimeric HPC4-sβ4GalT1-CBRT was bound (Fig. 6B, lanes 6 and 8, respectively). Control beads alone did not bind to either versions of HPC4-sβ4GalT1 (Fig. 6B, lanes 5 and 7). We also measured β4GalT1 activity, and although, as expected, HPC4-sTsyn and its chimeric version lack β4GalT1 activity (Fig. 6C, lanes 1–4), we also did not observe any promotion of β4GalT1 activity of chimeric HPC4-sβ4GalT1-CBRT with or without His-sCosmc as compared with HPC4-sβ4GalT1 (Fig. 6C, lane 7 versus lane 8 and lane 5 versus lane 6, respectively). In addition, although T-syn was enzymatically active only when co-expressed with Cosmc, as expected (Fig. 6D, lanes 1 versus lane 2), there was no significant activity observed for the chimeric T-synthase in the absence of CBRT and also with or without His-sCosmc (Fig. 6D, lane 3 versus lane 4). In control studies, we did not detect T-synthase activity with either version of HPC4-sβ4GalT1 (Fig. 6D, lanes 5–8). These data demonstrate the important role of the CBRT in T-synthase biosynthesis and that this domain can independently confer binding of a chimeric protein to Cosmc and that in the absence of this domain, T-synthase is inactive.
FIGURE 6.
Characterization of domain swapped chimeric HPC4-sTsyn and HPC4-sβ4GalT1. A, primary aa sequence showing CBRT swapped HPC4-sTsyn (red) and HPC4-sβ4GalT1 (green) and schematic showing chimeric HPC4-sTsyn and HPC4-sβ4GalT1. B, pulldown experiment demonstrating significant loss of HPC4-sT-synΔCBRT binding to Cosmc and gain of HPC4-sβ4GalT1-CBRT binding to Cosmc. Input, unbound, and bound to Cosmc fractions from the pulldown experiment were analyzed by antibody against HPC4 for T-synthase/β4GalT1. Similarly, the amount of Cosmc used in the pulldown experiment was analyzed by monoclonal antibody against Cosmc (bottom panel). C, β4GalT1 activity was determined from whole cell lysates from Hi-5 cells expressing HPC4-sβ4GalT1 or HPC4-sT-syn or chimeric versions expressed individually or co-expressed with His-sCosmc (top panel). The data show averages of three independent experiments. Error bars, ± S.D. from the average. HPC4 containing either T-syn or β4GalT1 or their respective chimera were detected by Western blotting against HPC4 (bottom panel), and a representative Western blot from three is shown. D, similar to C, T-synthase activity and HPC4 containing proteins were determined. The data show averages of three independent experiments performed in duplicate. A representative Western blot from three independent analyses is shown. Error bars, ± S.D. from the average. WB, Western blot.
DISCUSSION
Although the linear amino acid sequence of a protein determines its most thermodynamically stable native conformation (33), protein folding in vivo commonly requires the assistance of a cellular molecular chaperone system (28). A number of chaperone binding regions within protein substrates have been defined in great detail, but in many cases, the chaperone-binding sites within the non-native substrate contain hydrophobic sequences normally buried inside the core of the native protein or otherwise inaccessible (34), such as for BiP (GRP78), an HSP70 molecular chaperone (35), and GroEL, the bacterial chaperonin (36). These are examples of multiclient chaperones that recognize peptide features of a broad clientele. However, there is growing awareness of client-specific chaperones and those that have a very restricted set of clients. In the former category is HSP47, which exhibits collagen-specific binding whereby the trimeric HSP47 binds specific triple helical features containing the specific Xaa-Arg-Gly triplet sequence (37–41). An example of the latter category of client-restricted chaperones is gp96/GRP94, which interacts with Toll-like receptors and integrins, although the exact mechanism of its specificity is unknown (42–44).
Cosmc is an example of a client-specific chaperone that uniquely recognizes a single protein substrate, as evidenced by a variety of in vitro and in vivo studies (4, 8, 17, 18, 20, 45), but the molecular explanation for this specificity was unknown previously. Here we used a variety of molecular and biochemical approaches to define key aspects of the molecular basis for the specific recognition of the T-synthase by Cosmc and its role as a regulator of T-synthase folding. Our results demonstrate that Cosmc, an ER resident protein (46), directly binds to a unique hydrophobic region within the N-terminal stem region of T-synthase, which we have termed CBRT. Furthermore, in the absence of Cosmc in cells, the T-synthase cannot fold to become active, and thus any hydrophobic sequences within the T-synthase that might contribute to binding by other chaperones do not assist in correct folding of the enzyme in the absence of Cosmc.
We showed previously that Cosmc selectively binds to non-native T-synthase but not the native T-synthase (19). Moreover, interaction of Cosmc with T-synthase creates a relatively stable complex representing an intermediate form of T-synthase with enzyme activity, thus promoting downstream folding of the catalytic domain independently of other factors. The folded active T-synthase is released from Cosmc in a client-dependent manner, and we postulated that this represents a maturation exchange or cycle, in which newly synthesized T-synthase, once folded into an intermediate state through interactions with Cosmc, is then processed into a dimeric form that cannot rebind to Cosmc (19). Such folding intermediates are common for protein folding pathways and are critical to the path of maturation (47). The CBRT of T-synthase is unique and is not found in non-T-synthase proteins in the secretory pathway (Table 3), consistent with the specificity of interactions of T-synthase with Cosmc.
TABLE 3.
Result showing the top nine proteins from Homo sapiens using NCBI/BLAST/blastp for the CBRT sequence suggesting that the CBRT is unique
| Accession/description | Max score | Total score | Query coverage | E value | Identity | Sequence alignment |
|---|---|---|---|---|---|---|
| % | % | |||||
| NP_064541.1/Glycoprotein-N-acetylgalactosamine 3β-galactosyltransferase 1 | 55.8 | 55.8 | 100 | 2E−10 | 100 | 83LYQKVRILCWVMTGP97 |
| XP_005249869.1/predicted: glycoprotein-N-acetylgalactosamine 3β-galactosyltransferase 1 isoform X1 | 55.8 | 55.8 | 100 | 2E−10 | 100 | 91LYQKVRILCWVMTGP105 |
| NP_001005239.1/olfactory receptor 11H1 | 26.5 | 40.7 | 93 | 0.91 | 73 | 155LY*K**ILCWV165 |
| 75 | 150*MTG153 | |||||
| NP_001184216.1/olfactory receptor 11H2 | 26.5 | 40.7 | 93 | 0.91 | 73 | 155LY*K**ILCWV165 |
| 75 | 150*MTG153 | |||||
| XP_005271646.1/predicted: SID1 transmembrane family member 2 isoform X3 | 25.2 | 42.0 | 60 | 2.4 | 80 | 108LYQKV*R*LC117 |
| 100 | 591LYQK594 | |||||
| NP_001035545.1/SID1 transmembrane family member 2 precursor | 25.2 | 42.0 | 60 | 2.4 | 80 | 108LYQKV*R*LC117 |
| 100 | 594LYQK597 | |||||
| XP_005271645.1/predicted: SID1 transmembrane family member 2 isoform X2 | 25.2 | 42.0 | 60 | 2.4 | 80 | 108LYQKV*R*LC117 |
| 100 | 594LYQK597 | |||||
| XP_005271644.1/predicted: SID1 transmembrane family member 2 isoform X1 | 25.2 | 42.0 | 60 | 2.4 | 80 | 108LYQKV*R*LC117 |
| 100 | 598LYQK601 | |||||
| NP_653280.1/AP-4 complex accessory subunit tepsin | 23.1 | 23.1 | 40 | 12 | 100 | 116LYQKVR121 |
The ER is a very specialized environment, and folding of newly synthesized proteins and glycoproteins must occur efficiently or quality control systems cause their removal from the ER and degradation (48–50). In this regard it is important to note that the T-synthase is not glycosylated and lacks N-glycosylation sequons, so it cannot participate in glycoprotein-dependent folding pathways. It is possible that Cosmc, an ER resident protein, may scan newly synthesized T-synthase starting from the N terminus in the ER seeking the hydrophobic CBRT region, which is consistent with evidence that Cosmc is needed for co-translational biosynthesis of T-synthase (45). This binding may allow Cosmc to sequester T-synthase to promote its folding and prevent T-synthase from forming dead-end oligomers. A model depicting the proposed biosynthetic steps is shown in Fig. 7. The T-synthase within the ER is predicted to form a heterocomplex with Cosmc through hydrophobic interaction with the exposed hydrophobic sequences of the stem region of the newly translocating T-synthase, thus promoting downstream folding of the catalytic domain. The hydrophobic sequence of T-synthase that was involved in the interaction with Cosmc may be further buried inside the protein after being released from Cosmc as part of the folding process toward native confirmation, or the hydrophobic sequence may be engaged for Cosmc-assisted homodimerization of T-synthase, suggesting a cryptic homodimeric hydrophobic interface between dimeric T-synthase. Future crystallization studies of T-synthase will be needed to understand in greater detail the molecular interactions of these hydrophobic residues within the CBRT that are directly involved in binding to Cosmc, as well as the peptide determinants within the T-synthase that promote its homodimerization. The finding that Cosmc directly interacts with the N-terminal stem region of T-synthase in the unfolded but not native conformation greatly aids our understanding of the specific mechanism of Cosmc function and has important implications for understanding molecular chaperone functions in general and the specific roles of Cosmc and T-synthase in biosynthetic regulation of O-glycosylation and glycoprotein generation.
FIGURE 7.
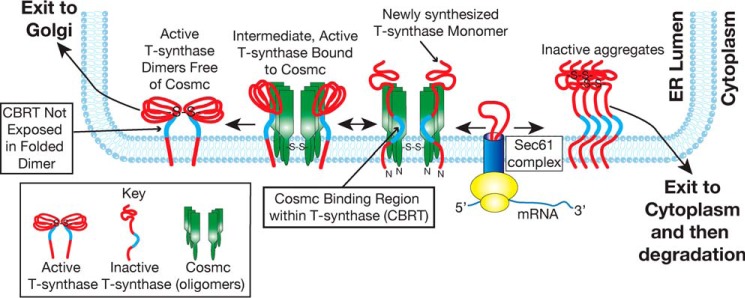
Proposed model of Cosmc function. In this model newly synthesized T-synthase co-translationally translocates into the lumen of the ER and directly interacts with Cosmc through the CBRT region (blue). Such interaction prevents the formation of an inactive oligomeric complex of T-synthase, which is removed to the cytoplasm and degraded. Binding of T-synthase to Cosmc promotes the formation of a transient complex, resulting in the folding of T-synthase. Active T-synthase is released from the complex when Cosmc interacts with other co-translationally translocated non-native T-synthase for another possible cycle of refolding (19). The CBRT region with the active and released T-synthase is no longer accessible to binding by Cosmc.
The discovery here of the molecular nature of the specific and single client activity of Cosmc recognition of T-synthase is consistent with the physiological consequence of losing Cosmc function, as we first identified in studies of individuals with Tn syndrome (20). Those individuals express dysfunctional T-synthase in some blood cell lineages caused by acquired loss of function mutation in Cosmc, an X-linked gene. Subsequent studies including Cosmc and T-synthase knock-out mice show many abnormalities correlated with defects in mucin type O-glycosylation that are related to several human diseases (8, 9) and physiological processes, including platelet formation (4), lymphangiogenesis (51, 52), and cancer (21–23, 53–56). In all such studies, the phenotypes of Cosmc deficiencies mirror those for T-synthase deficiencies (2, 57, 58), further supporting the evidence that Cosmc is a specific molecular chaperone for a single client T-synthase.
The T-synthase, like most glycosyltransferases, is a type II single-pass transmembrane protein with a short N-terminal cytoplasmic domain and a short region known as the stem that separates the C-terminal and lumenal catalytic domain from the transmembrane domain (59). The general functions of the stem regions of glycosyltransferases are not well understood, but there is growing evidence that this region may be important in many aspects of glycosyltransferase maturation and function. These include roles of the stem region in enzyme dimerization (60), in Golgi retention (61) and localization (59), in acceptor recognition (62), and as a site of proteolysis for release of soluble enzyme (63). The stem region of some enzymes such as the β4GalT1, when expressed independently of the catalytic domain of the enzyme, may act in a chaperone-like fashion to promote formation of the active enzyme refolded from bacterially expressed inclusion bodies (64). Finally, the short but specific stem region of β4GalNAcTB is required for its binding to GABPI (65, 66), a polytopic ER protein that may act as a type of pilot protein for targeting of β4GalNAcTB to the Golgi apparatus. Thus, expanding knowledge of the stem region functions of glycosyltransferases indicate they are important to enzyme function and localization. The evidence here in regard to the CBRT domain of the T-synthase provides for the first time key primary sequence functions of a glycosyltransferase stem region in protein folding and interaction with a molecular chaperone.
Acknowledgments
We thank members of the Cummings Lab for critical reading of the manuscript; Oskar Laur (Emory DNA Custom Cloning Core Facility) for helping to generate HPC4-sT-syn-m-5A, chimeric HPC4-sT-syn containing β4GalT1, and HPC4-sβ4GalT1-CBRT; and Dr. Jamie Heimburg-Molinaro for reviewing and editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants DK0808760 (to T. J.) and GM068559 (to R. D. C.).
- T-synthase or T-syn
- core 1 β3-galactosyltransferase
- Cosmc
- Core 1 β3-galactosyltransferase specific molecular chaperone
- CBRT
- Cosmc binding region within T-synthase
- ER
- endoplasmic reticulum
- Ni-NTA
- nickel-nitrilotriacetic acid
- aa
- amino acid(s).
REFERENCES
- 1. Tian E., Ten Hagen K. G. (2009) Recent insights into the biological roles of mucin-type O-glycosylation. Glycoconj. J. 26, 325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Steentoft C., Vakhrushev S. Y., Joshi H. J., Kong Y., Vester-Christensen M. B., Schjoldager K. T., Lavrsen K., Dabelsteen S., Pedersen N. B., Marcos-Silva L., Gupta R., Bennett E. P., Mandel U., Brunak S., Wandall H. H., Levery S. B., Clausen H. (2013) Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hang H. C., Bertozzi C. R. (2005) The chemistry and biology of mucin-type O-linked glycosylation. Bioorg. Med. Chem. 13, 5021–5034 [DOI] [PubMed] [Google Scholar]
- 4. Wang Y., Jobe S. M., Ding X., Choo H., Archer D. R., Mi R., Ju T., Cummings R. D. (2012) Platelet biogenesis and functions require correct protein O-glycosylation. Proc. Natl. Acad. Sci. U.S.A. 109, 16143–16148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rosen S. D. (2004) Ligands for l-selectin: homing, inflammation, and beyond. Annu. Rev. Immunol. 22, 129–156 [DOI] [PubMed] [Google Scholar]
- 6. McEver R. P., Cummings R. D. (1997) Perspectives series: cell adhesion in vascular biology: role of PSGL-1 binding to selectins in leukocyte recruitment. J. Clin. Invest. 100, 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McEver R. P., Cummings R. D. (1997) Role of PSGL-1 binding to selectins in leukocyte recruitment. J. Clin. Invest. 100, S97–S103 [PubMed] [Google Scholar]
- 8. Wang Y., Ju T., Ding X., Xia B., Wang W., Xia L., He M., Cummings R. D. (2010) Cosmc is an essential chaperone for correct protein O-glycosylation. Proc. Natl. Acad. Sci. U.S.A. 107, 9228–9233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xia L., Ju T., Westmuckett A., An G., Ivanciu L., McDaniel J. M., Lupu F., Cummings R. D., McEver R. P. (2004) Defective angiogenesis and fatal embryonic hemorrhage in mice lacking core 1-derived O-glycans. J. Cell Biol. 164, 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johansson M. E., Larsson J. M., Hansson G. C. (2011) The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. U.S.A. 108, 4659–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ten Hagen K. G., Fritz T. A., Tabak L. A. (2003). All in the family: the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology 13, 1R–16R [DOI] [PubMed] [Google Scholar]
- 12. Ju T., Brewer K., D'Souza A., Cummings R. D., Canfield W. M. (2002) Cloning and expression of human core 1 β1,3-galactosyltransferase. J. Biol. Chem. 277, 178–186 [DOI] [PubMed] [Google Scholar]
- 13. Ju T., Cummings R. D., Canfield W. M. (2002) Purification, characterization, and subunit structure of rat core 1 β1,3-galactosyltransferase. J. Biol. Chem. 277, 169–177 [DOI] [PubMed] [Google Scholar]
- 14. Brockhausen I., Matta K. L., Orr J., Schachter H. (1985) Mucin synthesis: UDP-GlcNAc:GalNAc-R β3-N-acetylglucosaminyltransferase and UDP-GlcNAc:GlcNAc β1–3GalNAc-R (GlcNAc to GalNAc) β6-N-acetylglucosaminyltransferase from pig and rat colon mucosa. Biochemistry 24, 1866–1874 [DOI] [PubMed] [Google Scholar]
- 15. Iwai T., Inaba N., Naundorf A., Zhang Y., Gotoh M., Iwasaki H., Kudo T., Togayachi A., Ishizuka Y., Nakanishi H., Narimatsu H. (2002) Molecular cloning and characterization of a novel UDP-GlcNAc:GalNAc-peptide β1,3-N-acetylglucosaminyltransferase (β3Gn-T6), an enzyme synthesizing the core 3 structure of O-glycans. J. Biol. Chem. 277, 12802–12809 [DOI] [PubMed] [Google Scholar]
- 16. Aryal R. P., Ju T., Cummings R. D. (2010) The endoplasmic reticulum chaperone Cosmc directly promotes in vitro folding of T-synthase. J. Biol. Chem. 285, 2456–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ju T., Aryal R. P., Stowell C. J., Cummings R. D. (2008) Regulation of protein O-glycosylation by the endoplasmic reticulum-localized molecular chaperone Cosmc. J. Cell Biol. 182, 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ju T., Cummings R. D. (2002) A unique molecular chaperone Cosmc required for activity of the mammalian core 1 β3-galactosyltransferase. Proc. Natl. Acad. Sci. U.S.A. 99, 16613–16618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aryal R. P., Ju T., Cummings R. D. (2012) Tight complex formation between Cosmc chaperone and its specific client non-native T-synthase leads to enzyme activity and client-driven dissociation. J. Biol. Chem. 287, 15317–15329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ju T., Cummings R. D. (2005) Protein glycosylation: chaperone mutation in Tn syndrome. Nature 437, 1252. [DOI] [PubMed] [Google Scholar]
- 21. Ju T., Lanneau G. S., Gautam T., Wang Y., Xia B., Stowell S. R., Willard M. T., Wang W., Xia J. Y., Zuna R. E., Laszik Z., Benbrook D. M., Hanigan M. H., Cummings R. D. (2008) Human tumor antigens Tn and sialyl Tn arise from mutations in Cosmc. Cancer Res. 68, 1636–1646 [DOI] [PubMed] [Google Scholar]
- 22. Mi R., Song L., Wang Y., Ding X., Zeng J., Lehoux S., Aryal R. P., Wang J., Crew V. K., van Die I., Chapman A. B., Cummings R. D., Ju T. (2012) Epigenetic silencing of the chaperone Cosmc in human leukocytes expressing Tn antigen. J. Biol. Chem. 287, 41523–41533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ju T., Otto V. I., Cummings R. D. (2011) The Tn antigen-structural simplicity and biological complexity. Angew. Chem. Int. Ed. Engl. 50, 1770–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Springer G. F. (1984) T and Tn, general carcinoma autoantigens. Science 224, 1198–1206 [DOI] [PubMed] [Google Scholar]
- 25. Stevens F. J., Argon Y. (1999) Protein folding in the ER. Semin. Cell Dev. Biol. 10, 443–454 [DOI] [PubMed] [Google Scholar]
- 26. Hartl F. U., Bracher A., Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 [DOI] [PubMed] [Google Scholar]
- 27. Fink A. L. (1999) Chaperone-mediated protein folding. Physiol. Rev. 79, 425–449 [DOI] [PubMed] [Google Scholar]
- 28. Hartl F. U. (1996) Molecular chaperones in cellular protein folding. Nature 381, 571–579 [DOI] [PubMed] [Google Scholar]
- 29. Ju T., Xia B., Aryal R. P., Wang W., Wang Y., Ding X., Mi R., He M., Cummings R. D. (2011) A novel fluorescent assay for T-synthase activity. Glycobiology 21, 352–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kawar Z. S., Van Die I., Cummings R. D. (2002) Molecular cloning and enzymatic characterization of a UDP-GalNAc:GlcNAc(β)-R β1,4-N-acetylgalactosaminyltransferase from Caenorhabditis elegans. J. Biol. Chem. 277, 34924–34932 [DOI] [PubMed] [Google Scholar]
- 31. van Die I., van Tetering A., Schiphorst W. E., Sato T., Furukawa K., van den Eijnden D. H. (1999) The acceptor substrate specificity of human β4-galactosyltransferase V indicates its potential function in O-glycosylation. FEBS Lett. 450, 52–56 [DOI] [PubMed] [Google Scholar]
- 32. Qasba P. K., Ramakrishnan B., Boeggeman E. (2008) Structure and function of β-1,4-galactosyltransferase. Curr Drug Targets 9, 292–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Anfinsen C. B. (1973) Principles that govern the folding of protein chains. Science 181, 223–230 [DOI] [PubMed] [Google Scholar]
- 34. Kim Y. E., Hipp M. S., Bracher A., Hayer-Hartl M., Hartl F. U. (2013) Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82, 323–355 [DOI] [PubMed] [Google Scholar]
- 35. Blond-Elguindi S., Cwirla S. E., Dower W. J., Lipshutz R. J., Sprang S. R., Sambrook J. F., Gething M. J. (1993) Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell 75, 717–728 [DOI] [PubMed] [Google Scholar]
- 36. Raineri E., Ribeca P., Serrano L., Maier T. (2010) A more precise characterization of chaperonin substrates. Bioinformatics 26, 1685–1689 [DOI] [PubMed] [Google Scholar]
- 37. Yagi-Utsumi M., Yoshikawa S., Yamaguchi Y., Nishi Y., Kurimoto E., Ishida Y., Homma T., Hoseki J., Nishikawa Y., Koide T., Nagata K., Kato K. (2012) NMR and mutational identification of the collagen-binding site of the chaperone Hsp47. PLoS One 7, e45930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dafforn T. R., Della M., Miller A. D. (2001) The molecular interactions of heat shock protein 47 (Hsp47) and their implications for collagen biosynthesis. J. Biol. Chem. 276, 49310–49319 [DOI] [PubMed] [Google Scholar]
- 39. Koide T., Takahara Y., Asada S., Nagata K. (2002) Xaa-Arg-Gly triplets in the collagen triple helix are dominant binding sites for the molecular chaperone HSP47. J. Biol. Chem. 277, 6178–6182 [DOI] [PubMed] [Google Scholar]
- 40. Koide T., Asada S., Takahara Y., Nishikawa Y., Nagata K., Kitagawa K. (2006) Specific recognition of the collagen triple helix by chaperone HSP47: minimal structural requirement and spatial molecular orientation. J. Biol. Chem. 281, 3432–3438 [DOI] [PubMed] [Google Scholar]
- 41. Widmer C., Gebauer J. M., Brunstein E., Rosenbaum S., Zaucke F., Drögemüller C., Leeb T., Baumann U. (2012) Molecular basis for the action of the collagen-specific chaperone Hsp47/SERPINH1 and its structure-specific client recognition. Proc. Natl. Acad. Sci. U.S.A. 109, 13243–13247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Randow F., Seed B. (2001) Endoplasmic reticulum chaperone gp96 is required for innate immunity but not cell viability. Nat. Cell Biol. 3, 891–896 [DOI] [PubMed] [Google Scholar]
- 43. Yang Y., Liu B., Dai J., Srivastava P. K., Zammit D. J., Lefrançois L., Li Z. (2007) Heat shock protein gp96 is a master chaperone for Toll-like receptors and is important in the innate function of macrophages. Immunity 26, 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu S., Hong F., Gewirth D., Guo B., Liu B., Li Z. (2012) The molecular chaperone gp96/GRP94 interacts with Toll-like receptors and integrins via its C-terminal hydrophobic domain. J. Biol. Chem. 287, 6735–6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Narimatsu Y., Kubota T., Furukawa S., Shimojima M., Iwasaki H., Tozawa Y., Tachibana K., Narimatsu H. (2011) Co-translational function of Cosmc, core 1 synthase specific molecular chaperone, revealed by a cell-free translation system. FEBS Lett. 585, 1276–1280 [DOI] [PubMed] [Google Scholar]
- 46. Sun Q., Ju T., Cummings R. D. (2011) The transmembrane domain of the molecular chaperone Cosmc directs its localization to the endoplasmic reticulum. J. Biol. Chem. 286, 11529–11542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Merulla J., Fasana E., Soldà T., Molinari M. (2013) Specificity and regulation of the endoplasmic reticulum-associated degradation machinery. Traffic 14, 767–777 [DOI] [PubMed] [Google Scholar]
- 48. Frydman J., Hartl F. U. (1996) Principles of chaperone-assisted protein folding: differences between in vitro and in vivo mechanisms. Science 272, 1497–1502 [DOI] [PubMed] [Google Scholar]
- 49. Siegers K., Waldmann T., Leroux M. R., Grein K., Shevchenko A., Schiebel E., Hartl F. U. (1999) Compartmentation of protein folding in vivo: sequestration of non-native polypeptide by the chaperonin-GimC system. EMBO J. 18, 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thulasiraman V., Yang C. F., Frydman J. (1999) In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO J. 18, 85–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Herzog B. H., Fu J., Wilson S. J., Hess P. R., Sen A., McDaniel J. M., Pan Y., Sheng M., Yago T., Silasi-Mansat R., McGee S., May F., Nieswandt B., Morris A. J., Lupu F., Coughlin S. R., McEver R. P., Chen H., Kahn M. L., Xia L. (2013). Podoplanin maintains high endothelial venule integrity by interacting with platelet CLEC-2. Nature 502, 105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fu J., Gerhardt H., McDaniel J. M., Xia B., Liu X., Ivanciu L., Ny A., Hermans K., Silasi-Mansat R., McGee S., Nye E., Ju T., Ramirez M. I., Carmeliet P., Cummings R. D., Lupu F., Xia L. (2008) Endothelial cell O-glycan deficiency causes blood/lymphatic misconnections and consequent fatty liver disease in mice. J. Clin. Invest. 118, 3725–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Madsen C. B., Lavrsen K., Steentoft C., Vester-Christensen M. B., Clausen H., Wandall H. H., Pedersen A. E. (2013) Glycan elongation beyond the mucin associated Tn antigen protects tumor cells from immune-mediated killing. PLoS One 8, e72413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schietinger A., Philip M., Yoshida B. A., Azadi P., Liu H., Meredith S. C., Schreiber H. (2006) A mutant chaperone converts a wild-type protein into a tumor-specific antigen. Science 314, 304–308 [DOI] [PubMed] [Google Scholar]
- 55. Lee J., Jr., Chen C. H., Chen Y. H., Huang M. J., Huang J., Hung J. S., Chen M. T., Huang M. C. (2013) COSMC is overexpressed in proliferating infantile hemangioma and enhances endothelial cell growth via VEGFR2. PLoS One 8, e56211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huang J., Che M. I., Lin N. Y., Hung J. S., Huang Y. T., Lin W. C., Huang H. C., Lee P. H., Liang J. T., Huang M. C. (2014) The molecular chaperone Cosmc enhances malignant behaviors of colon cancer cells via activation of Akt and ERK. Mol. Carcinog. 53, E62–E71 [DOI] [PubMed] [Google Scholar]
- 57. Bergstrom K. S., Xia L. (2013) Mucin-type O-glycans and their roles in intestinal homeostasis. Glycobiology 23, 1026–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ju T., Wang Y., Aryal R. P., Lehoux S. D., Ding X., Kudelka M. R., Cutler C., Zeng J., Wang J., Sun X., Heimburg-Molinaro J., Smith D. F., Cummings R. D. (2013). Tn and sialyl-Tn antigens, aberrant O-glycomics as human disease markers. Proteomics Clin. Appl., 7, 618–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Colley K. J. (1997) Golgi localization of glycosyltransferases: more questions than answers. Glycobiology 7, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee H. J., Barry C. H., Borisova S. N., Seto N. O., Zheng R. B., Blancher A., Evans S. V., Palcic M. M. (2005) Structural basis for the inactivity of human blood group O2 glycosyltransferase. J. Biol. Chem. 280, 525–529 [DOI] [PubMed] [Google Scholar]
- 61. Kitazume-Kawaguchi S., Dohmae N., Takio K., Tsuji S., Colley K. J. (1999) The relationship between ST6Gal I Golgi retention and its cleavage-secretion. Glycobiology 9, 1397–1406 [DOI] [PubMed] [Google Scholar]
- 62. Legaigneur P., Breton C., El Battari A., Guillemot J. C., Auge C., Malissard M., Berger E. G., Ronin C. (2001) Exploring the acceptor substrate recognition of the human β-galactoside α2,6-sialyltransferase. J. Biol. Chem. 276, 21608–21617 [DOI] [PubMed] [Google Scholar]
- 63. Cho S. K., Cummings R. D. (1997) A soluble form of α1,3-galactosyltransferase functions within cells to galactosylate glycoproteins. J. Biol. Chem. 272, 13622–13628 [DOI] [PubMed] [Google Scholar]
- 64. Boeggeman E. E., Ramakrishnan B., Qasba P. K. (2003) The N-terminal stem region of bovine and human β1,4-galactosyltransferase I increases the in vitro folding efficiency of their catalytic domain from inclusion bodies. Protein Expr. Purif. 30, 219–229 [DOI] [PubMed] [Google Scholar]
- 65. Kraft B., Johswich A., Kauczor G., Scharenberg M., Gerardy-Schahn R., Bakker H. (2011) “Add-on” domains of Drosophila β1,4-N-acetylgalactosaminyltransferase B in the stem region and its pilot protein. Cell Mol. Life Sci. 68, 4091–4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Johswich A., Kraft B., Wuhrer M., Berger M., Deelder A. M., Hokke C. H., Gerardy-Schahn R., Bakker H. (2009) Golgi targeting of Drosophila melanogaster β4GalNAcTB requires a DHHC protein family-related protein as a pilot. J. Cell Biol. 184, 173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ju T., Zheng Q., Cummings R. D. (2006) Identification of core 1 O-glycan T-synthase from Caenorhabditis elegans. Glycobiology 16, 947–958 [DOI] [PubMed] [Google Scholar]