Background: H2ABbd is an H2A variant possessing a highly divergent structure. However, the in vivo function of H2ABbd is not understood.
Results: Expression of H2ABbd caused DNA damage, induced apoptotis, and finally led to cell death.
Conclusion: H2ABbd caused apoptosis in caspase 3- and NF-κB-dependent manners.
Significance: This work describes a novel role for human H2ABbd in gene regulation and cell death.
Keywords: Apoptosis, Cell Cycle, Chromatin Histone Modification, DNA Damage, Gene Expression, Histones
Abstract
Histone variants play specific roles in maintenance and regulation of chromatin structures. H2ABbd, an H2A variant, possesses a highly divergent structure compared with canonical H2A and is highly expressed in postmeiotic germ cells, but its functions in the regulation of gene expression are largely unknown. In the present study, we investigated the cellular phenotype associated with enforced H2ABbd expression. Among H2A variants, H2ABbd specifically caused growth defect in human cells and induced apoptosis. H2ABbd expression resulted in degradation of inhibitor of κB-α and translocation of NF-κB into nuclei, indicating the activation of NF-κB. Intriguingly, NF-κB activity was essential for H2ABbd-induced apoptosis. H2ABbd overexpression resulted in DNA damage after release from G1/S, progressed through the S phase slowly, and induced apoptosis. Furthermore, gene expression microarray analysis revealed that expression of H2ABbd activates groups of genes involved in apoptosis and postmeiotic germ cell development, suggesting that H2ABbd might influence transcription. Taken together, our data suggest that H2ABbd may contribute to specific chromatin structures and promote NF-κB activation, which could in turn induce apoptosis in mammalian cells.
Introduction
Genetic information must be stably transmitted during cell divisions through DNA replication followed by rapid packaging of newly synthesized DNA into chromatin. Nucleosomes, the building blocks of chromatin, consist of two pairs of histones, H3-H4 and H2A-H2B, forming an octamer around which DNA is wound (1). Defects in assembly and structure of chromatin may lead to loss of epigenetic information and gross chromosomal rearrangements. However, in order to switch on/off gene expression upon exogenous and endogenous stimuli, remodeling of the chromatin fiber is required to allow/prevent access of transcriptional and epigenetic machineries. One way to achieve this goal is to shuffle canonical histone molecules with histone variants. Previous studies have shown that exchange of canonical histones contributes to a multitude of downstream effects, including alteration in local chromatin organization, chromosome segregation, DNA repair, and transcriptional regulation. This suggests that histone variants might have an important role in chromatin structure and function in general (2).
Histone H2A has several variants with specialized functions. Among them, H2ABbd (H2A Barr body-deficient) is a highly divergent H2A variant (which shares about 48% sequence identity with the canonical H2A) and is found only in mammals (3). Based on its unique structure (3), H2ABbd has been proposed to link specific chromatin states to transcriptional activation (4). Consistent with this notion, recent works showed that H2ABbd was excluded from the inactive X chromosome and colocalized with regions having high levels of acetylated H4 (3). This is further supported by the fact that H2ABbd destabilizes the nucleosome core particles, causing a more relaxed nucleosome core particle conformation (5–8). Indeed, mouse H2ABbd homologue H2AL1 and H2AL2 expressed in postmeiotic cells (9) and H2A.Lap1 are localized at the transcription start sites of active genes (10). Ectopically expressed human H2ABbd in HeLa cells was deposited at intron-exon boundaries, and H2ABbd knockdown cells showed splicing defects, indicating another potential role of this H2A variant in transcriptional regulation through splicing (11).
DNA damage, a potential risk for normal transmission of genetic/epigenetic information, is frequently caused by the collapse of replication forks during DNA replication. To counter such events, mammalian cells are equipped with an elaborate DNA damage response (DDR)4 mechanism, which functions through precise detection of damaged DNA, rapid transmission of signals to downstream effector molecules to facilitate DNA repair, and in extreme cases cell cycle arrest and apoptosis (12). Importantly, it has been reported that incomplete or aberrant DDR leads to diseases such as malignant transformation by causing genetic mutations and genomic instability. Protein kinases ATM (ataxia-telangiectasia, mutated) and ATR (ATM and Rad3-related) are major players in DDR that function through activation of a plethora of downstream targets, including RPA2, H2AX, 53BP1, Chk1, Chk2, and p53. Among these, the p53 and NF-κB transcription factors play key roles in regulation of cell death and cell survival in response to DNA damage, respectively (13). ATM phosphorylates and stabilizes p53, which is the master regulator of apoptosis. In addition, ATM activates NF-κB by cooperating with its essential modulator, NEMO (14). Intriguingly, although it is widely perceived that NF-κB functions as an antiapoptotic molecule, recent studies indicate that NF-κB may also facilitate apoptosis under certain circumstances.
TSEG-1, a rodent version of H2ABbd, has previously been implicated in induction of cell death in cultured spermatocytes (GC-2spd cells) (15). To investigate this possibility further, we expressed H2ABbd in human cells and found that ectopic expression of H2ABbd led to caspase-dependent apoptosis. Intriguingly, treatment with a NF-κB inhibitor blocked H2ABbd-induced apoptosis, suggesting that NF-κB might play a proapoptotic role in H2ABbd-overexpressing cells. To gain insights into the molecular mechanisms linking H2ABbd overexpression with NF-κB-mediated cell death, we performed gene expression microarray analysis and found that indeed ectopic expression of H2ABbd resulted in up-regulation of a set of apoptosis-related genes. Furthermore, gene ontology (GO) tests revealed that genes involved in development/differentiation as well as factors specific to postmeiotic germ cells (i.e. elongating and round spermatids) were preferentially enriched in H2ABbd-expressing cells. Based on these results, we hypothesized that ectopic expression of H2ABbd in somatic cells might cause destabilization of genome integrity, which could potentially lead to activation of the DDR pathway by sensing DNA damage and finally cause cell death by an NF-κB-mediated pathway.
EXPERIMENTAL PROCEDURES
Cell Culture
HeLa cells and MEFs were cultured in DMEM supplemented with 10% FBS RPE cells were cultured in DMEM/F-12 supplemented with 10% FBS. All cells were cultured at 37 °C under 5% CO2.
Construction of Expression Vectors
EGFP-tagged H2A, H2AX, and H2ABbd expression vectors were constructed. We amplified and subcloned human H2ABbd (H2AFB3) (GenBankTM accession number NM_080720), H2A (HIST1H2AM) (GenBankTM accession number NM_003514), and H2AX (H2AFX) (GenBankTM accession number NM_002105) as follows.
EGFP fragments were cut from pEGFP-C1 using NheI and EcoRI restriction endonucleases. Blunt ends were created after NheI single digestion, before EcoRI digestion. pENTR1A-EGFP vector was generated by subcloning the EGFP fragment into pENTR1A (Invitrogen) vector that was already digested with KpnI, blunt-ended, and further cleaved with EcoRI.
pENTR1A-EGFP-H2A and -H2AX vectors were constructed by subcloning human H2A and H2AX genes into pENTR1A-EGFP using EcoRI and EcoRV sites. Human H2A and H2AX were obtained by PCR amplification from total human cDNA library using primers that introduced EcoRI and EcoRV sites on both flanks of the amplified segment.
EGFP-H2ABbd expression vectors were generated in the following way. First pcDNA3.1-H2ABbd-MBD-NLS poly(A) was generated by cutting EGFP from the pcDNA3.1-EGFP-MBD-NLS poly(A) vector (a gift from Dr. Yuki Okada) using HindIII and NotI restriction endonucleases and by subcloning H2ABbd into pcDNA3.1-MBD-NLS poly(A). Human H2ABbd genes (having no introns) were obtained by PCR amplification of human genomic DNA using primers that introduce HindIII and NotI sites at the flanking regions. EGFP fragments with HindIII sites at both ends were religated into pcDNA3.1-H2ABbd-MBD-NLS poly(A), resulting in a pcDNA3.1-EGFP-H2ABbd-MBD-NLS poly(A) vector. Finally, EGFP-H2ABbd fragments were cut from pcDNA3.1-EGFP-H2ABbd-MBD-NLS poly(A), using EcoRI and NotI, and ligated into pENTR1A vector digested with the same enzymes, resulting in a pENTR1A-EGFP-H2ABbd vector. pENTR1A-H2A, H2AX, and H2ABbd vectors were incubated with CSIV-TRE-RfA-UbC-KT vectors and LR Clonase enzyme mix (Invitrogen) for 2 h at 25 °C, which produced CSIV-TRE-RfA-UbC-KT EGFP-H2A, H2AX, and H2ABbd.
Construction of FLAG-HA-tagged histone H2ABbd was as follows. H2ABbd with XhoI and NotI sites was obtained by PCR amplification of pENTR1A-EGFP-H2ABbd. pOZ-FH-N-H2ABbd was generated by subcloning H2ABbd into pOZ-FH-N vector digested with XhoI and NotI.
Next, FLAG-HA-H2ABbd fragments with EcoRI and NotI sites were obtained by PCR amplification of pOZ-FH-N-H2ABbd, digested, and subcloned into pENTR1A that was already cleaved with EcoRI and NotI, producing the pENTR1A-FLAG-HA-H2ABbd construct. The CSIV-TRE-RfA-UbC-KT FLAG-HA-H2ABbd vector was generated as described above.
Lentiviral Transduction
Lentivirus expressing the respective genes was generated by the co-transfection of 293T cells with pCMV-VSV-G-RSV-RevB (a gift from H. Miyoshi), pCAG-HIVgp (also a gift from H. Miyoshi), and the respective CSIV-TRE-RfA-UbC-KT using the calcium phosphate co-precipitation method. Cells infected with viruses were treated with 2 μg/ml puromycin (Sigma-Aldrich) for 2 days. To express the inducible gene, doxycycline (Dox; Sigma-Aldrich) was added to the medium at a concentration of 1 μg/ml.
Immunoblotting
Collected cells were washed with ice-cold PBS, and sample buffer was added to cell pellets. Samples were boiled for 5 min and used as total cell lysate. Chromatin fractionation was performed as described previously (16). Antibodies used in this study are listed in Table 1.
TABLE 1.
Antibodies used in this study
| Antibody | Product number | Company |
|---|---|---|
| GFP | 11814460001 | Roche Applied Science |
| H2ABbd | 06-1319 | Millipore |
| H2A | ab18255 | Abcam |
| H2AX | ab11175 | Abcam |
| H3 | ab1791 | Abcam |
| PP2A | 05-421 | Upstate Biotechnology |
| γ-H2AX | JBW301 | Millipore |
| Caspase3 | 9662 | Cell Signaling |
| Cleaved Caspase3 | 9661 | Cell Signaling |
| actin | ab6276 | Abcam |
| ATM pS1981 | AF1665 | R&D Systems |
| ATM | sc23921 | Santa Cruz Biotechnology |
| 53BP1pS1778 | 2675 | Cell Signaling |
| 53BP1 | sc22760 | Santa Cruz Biotechnology |
| Chk2 pT68 | 2661 | Cell Signaling |
| Chk2 | sc9064 | Santa Cruz Biotechnology |
| RPA2 | 2208 | Cell Signaling |
| Cleaved caspase 6 | 9761 | Cell Signaling |
| IκB-α | sc203 | Santa Cruz Biotechnology |
| p65 | sc372 | Santa Cruz Biotechnology |
| SP1 | sc14027 | Santa Cruz Biotechnology |
Cell Cycle Synchronization
HeLa EGFP-H2A and H2ABbd cells were first synchronized at the G1/S boundary by exposure to 2 mm thymidine for 18 h and then released into S phase by washout of thymidine with PBS and the addition of medium. After 8 h at release, these cells were exposed to 2 mm thymidine for 18 h and released again.
Cell Cycle Analysis
Cells were harvested and fixed with 70% ethanol. These were then washed once with PBS, treated with RNase, and stained with propidium iodide. Flow cytometry was performed using a FACS CANTO2 flow cytometer (BD Biosciences).
DNA Fragmentation Assay
Cells were lysed in 10 mm Tris-HCl (pH 8.0) buffer containing 0.6% SDS and 10 mm EDTA and incubated with 100 μg/ml RNase. After 1 h, NaCl was added to a final concentration of 1 m and incubated for 1 h at 4 °C. Lysates were centrifuged (12,500 rpm, 20 min, 4 °C). The supernatants were extracted with phenol/chloroform. DNA was precipitated for 30 min in 3 volumes of ethanol at −80 °C and centrifuged (17,900 × g, 10 min, 4 °C). The pellet was dissolved in 100 μl of H2O. DNA was resolved by electrophoresis at 100 V for 30 min on 1% agarose gels and stained with 0.1 μg/ml ethidium bromide.
Time Lapse Microscopy
Cells were cultured in 35-mm glass bottom culture dishes (Greiner Bio-one). Medium was changed to a phenol red-free one after doxycycline was added. The dishes were mounted on a fluorescence microscope (Zeiss LSM710) equipped with a video camera. The temperature of the medium was kept constant at 37 °C and CO2 concentration at 5% using a heated stage. Time lapse images were captured every 20 min, and z-stacks of the images were obtained every 2 μm, using a PlanApochromat ×40/1.4 numerical aperture oil lens (Carl Zeiss) as objectives, and were processed by the ZEN 2013 software (Carl Zeiss).
Micrococcal Nuclease (MNase) Assay
Cells were collected after release from thymidine block and permeabilized with 0.01% l-α-lysophosphatidylcholine (Sigma) in 150 mm sucrose, 80 mm KCl, 35 mm HEPES (pH 7.4), 5 mm K2HPO4, 5 mm Mg2Cl, and 0.5 mm CaCl2 for 90 s. Cells were washed and suspended with 20 mm sucrose, 50 mm Tris (pH 7.5), 50 mm NaCl, and 2 mm CaCl2 (MNase buffer). The same amounts of MNase buffer containing MNase (2 units/ml) were added and incubated at room temperature for various times. DNA was isolated and subjected to 0.8% agarose gel electrophoresis.
Real-time PCR
Total RNA was isolated as described previously (17). 50 ng of RNA was reverse-transcribed with random primers using Thermoscript RT-PCR (Invitrogen). Quantitative real-time PCR was performed using Power SYBR Green (Applied Biosystems) and the real time PCR machine ABI7900HT (Applied Biosystems). Expression levels were normalized to GAPDH. Primers used in real-time PCR are listed in Table 2.
TABLE 2.
Primers used in this study
| Sample | Sequence |
|---|---|
| H2A-F | AAAGAATTCAATGTCTGGTCGTGGCAAGCAAG |
| H2A-R | AAAGATATCTCACTTGCCCTTTGCCTTGTG |
| H2AX-F | AAAGAATTCAATGTCGGGCCGCGGCAAGAC |
| H2AX-R | AAAGATATCTTAGTACTCCTGGGAGGCCTG |
| H2ABbd-F (EcoRI) | AAAAGCTTAATGCCGAGGAGGAGGAGACG |
| H2ABbd-R (NotI) | AAAGCGGCCGCCTAGTCCTCGCCAGGGGCC |
| H2ABbd-F (XhoI) | AAACTCGAGATGCCGAGGAGGAGGAGACG |
| FLAG-HA-H2ABbd-F | AAAGAATTCGCCACCATGGACTACAAGGACGACG |
| GAPDH-F | GCCAATCTCAGTCCCTTCC |
| GAPDH-R | TAGTAGCCGGGCCCTACTTT |
| IκB-α-F | CAGCAGCTCACCGAGGA |
| IκB-α-R | GGCCAGGTCTCCCTTCA |
| BIK-F | TCTGCCTGCTGCACTTTG |
| BIK-R | TCTGCCTGCTGCACTTTG |
| NKX3.1-F | TGTGACCCTTGCACCAAA |
| NKX3.1-R | GACAAAGCCACCCCAAGTT |
| p21-F | TCTGCCTGCTGCACTTTG |
| p21-R | TCTGCCTGCTGCACTTTG |
Microarray Experiment
RPE FLAG-HA H2ABbd cells were synchronized at G0 phase with DMEM containing 0% FBS and collected, and total RNA was prepared using ISOGENII (Nippongene). RNA hybridization, wash, and analysis were performed using SurePrint G3 Human GE 8x60k version 2.0 (Agilent Technologies) and GeneSpring GX version 12.6.0 (Agilent Technologies). Microarray expression profiling was performed by Oncomics (Nagoya, Japan).
Microarray and Publicly Available RNA-seq, ChIP-seq Data Analysis
Up-regulated and down-regulated genes were identified by measuring Cy3 and Cy5 probe intensities. Genes that showed elevated or reduced expression (greater or less than 2-fold compared with the control) at the 8 and 16 h time points were listed, and the two groups (8 and 16 h) were compared with identify overlapping/common factors. The odds ratio was calculated as follows, (genes overlapping (common to both 8 and 16 h) × genes not included in either the 8 or 16 h group)/(genes unique for group 1 (8 h) × genes unique for group 2 (16 h)). Representation factor and p value for exact hypergeometric probability were calculated as described previously. RNA-seq data for different staged of male germ cell development was retrieved from the Gene Expression Omnibus (series accession number GSE35005). BedGraph files for control and H2ABbd siRNA knockdown and H2ABbd ChIP-seq experiments were obtained from the data (series accession number GSE38771) deposited by Tolstorukov et al. (11). FPKM values for control and H2ABbd siRNA knockdown experiments were generously provided by Tolstorukov et al.5 University of Toronto BAR (Bio-Analytic Resource) tools were used for heat map analysis. Wig and BedGraph files were uploaded into the IGV Genome Browser (Broad Institute), and images were obtained for respective gene loci.
RESULTS
Overexpression of H2ABbd, but Not H2A and H2AX, Results in Growth Defect
The most recently discovered H2A variant, H2ABbd, possesses an acidic patch smaller than that of canonical H2A and lacks the C-terminal domain for interaction with H3 within the nucleosome (3) (Fig. 1A). To understand the physiological functions of this histone variant, we established cell lines expressing N-terminal EGFP-tagged H2ABbd, H2A, or H2AX that could be induced by Dox. EGFP-H2ABbd, -H2A, or -H2AX was clearly detectable at 24 h after the addition of Dox in nearly all cells, although the expression level was varied (supplemental Fig. 1A).
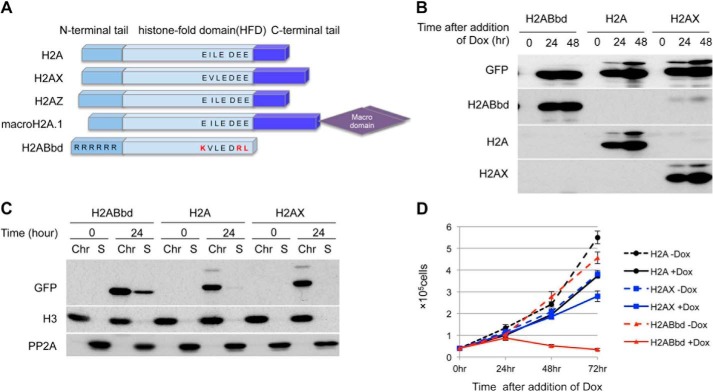
FIGURE 1.
Induction of H2ABbd inhibits proliferation of HeLa cells. A, schematic representation of the histone H2A variants. The C-terminal domain is not conserved among the H2A variants. In particular, H2ABbd does not contain both the acidic patch and the C-terminal region. B, immunoblot analyses showing the time course of EGFP-tagged H2ABbd, H2A, or H2AX induction. HeLa cells transfected with Dox-inducible EGFP-tagged H2ABbd, H2A, or H2AX were cultured in the presence of Dox for the indicated amount of time. Cells were collected, and total cell extracts were subjected to immunoblotting using the respective antibodies indicated in the figure. C, cells were collected as in B, followed by chromatin fractionation and immunoblot analysis. Each type of histone was detected in the chromatin fraction. D, 50,000 HeLa cells were seeded and cultured in the presence of Dox to express EGFP-tagged H2ABbd, H2A, or H2AX. At the indicated time points after the addition of Dox, cell numbers were counted. Data are presented from three independent experiments. Error bars, S.D.
EGFP-tagged H2A or its variants were expressed in almost same level 24 and 48 h after the addition of Dox (Fig. 1B). In this system, the level of ectopically expressed EGFP-H2A was slightly stronger than in endogenous H2A, whereas the EGFP-H2AX level was far higher than in endogenous H2AX. Because H2ABbd is expressed robustly in postmeiotic germ cells (18), endogenous H2ABbd was hardly detectable in HeLa cells. Importantly, subcellular fractionation revealed that ectopically expressed EGFP-H2A and its variants were incorporated into the chromatin fraction in similar levels (Fig. 1C).
HeLa cells expressing EGFP-H2ABbd exhibited marked reduction in the proliferation rate in the presence of Dox (Fig. 1D), showing an increase in the populations of S phase cells at 24 h and sub-G1 at 48 h (supplemental Fig. 1B). In contrast, the effects of EGFP-H2A or EGFP-H2AX expression on cell growth were negligible. Taken together, these results indicate that expression of EGFP-H2ABbd specifically inhibits cell proliferation.
Overexpression of H2ABbd Triggers Caspase 3-dependent Apoptosis
As shown in Fig. 2A, 48 h after induction of EGFP-H2ABbd, most cells became round and detached from culture dishes. These cells were identified as non-mitotic cells by FACS analysis (supplemental Fig. 1B), suggesting that EGFP-H2ABbd expression induced apoptosis. In order to confirm whether the effect of H2ABbd is cell type-specific, we expressed FLAG-HA-tagged H2ABbd in MEFs and RPE cells (Fig. 2A). Similar to HeLa, FLAG-HA-H2ABbd strongly suppressed proliferations of MEFs and RPE cells through induction of cell death. The DNA fragmentation assay revealed that short DNA strands were readily detectable in HeLa and RPE cells expressing EGFP-H2ABbd or FLAG-HA-H2ABbd (Fig. 2B), confirming induction of apoptosis. Further, live cell imaging detected numerous apoptotic cells within 30 h after the induction of EGFP-H2ABbd prior to nuclear envelope breakdown and chromosome separation (Fig. 2C). These results suggest that apoptosis induced by the expression of EGFP-H2ABbd was not caused by mitotic catastrophe. Consistent with this observation, we detected the cleaved form of caspase 3 in EGFP-H2ABbd- or FLAG-HA-H2ABbd-expressing HeLa and RPE cells with a concomitant decrease in the level of procaspase 3 (Fig. 2D). Importantly, low and moderate expression of EGFP-H2ABbd did not trigger apoptosis and activation of procaspase 3. Therefore, the induction of apoptosis appeared dose-dependent (supplemental Fig. 2, A–C) and thus might explain the survival of cells expressing H2ABbd reported previously. In addition, untagged H2ABbd also induced apoptosis as effectively as EGFP-H2ABbd did, eliminating the possibility that apoptosis was not due to an indirect consequence of EGFP tagging into the H2ABbd molecule.
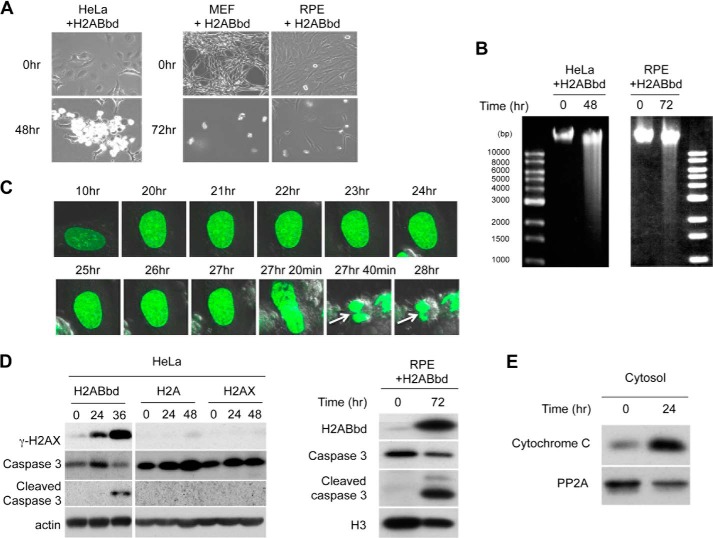
FIGURE 2.
Analysis of H2ABbd-induced apoptosis in HeLa cells. A, EGFP-tagged H2ABbd was induced for 48 or 72 h in HeLa or MEF, respectively. FLAG-HA-H2ABbd was induced for 72 h in RPE cells. Typical differential interference contrast images are shown. B, DNA fragmentation was observed in HeLa cells expressing EGFP-H2ABbd for 48 h or in RPE cells expressing FLAG-HA-H2ABbd for 72 h. Cells were prepared as described in A and lysed, and DNA fragments were analyzed by agarose gel electrophoresis. C, EGFP-H2ABbd was expressed after the addition of Dox in HeLa cells and subjected to time lapse microscopy. Images were captured at the indicated time points. Arrows (27 h 40 min and 28 h time points) show cells starting to undergo apoptosis. D, immunoblot analyses showing the time course of EGFP-tagged H2ABbd, H2A, or H2AX induction. Cells were collected at the indicated times after induction, and total cell extracts were subjected to immunoblotting using the respective antibodies. E, cytosol fraction was prepared from HeLa cells with/without Dox for 24 h and subjected to immunoblot analysis to detect cytochrome c.
We next asked whether H2ABbd overexpression caused cytoplasmic release of cytochrome c, which binds with Apaf1 to activate a series of caspase cascades (19). After induction of H2ABbd, cytochrome c was readily detected in the cytoplasmic fraction (Fig. 2E). In summary, our results indicate that overexpression of EGFP-H2ABbd triggers caspase-dependent apoptosis mediated by cytochrome c release.
Ectopic Expression of H2ABbd Causes DNA Damage
Incorporation of H2ABbd into nucleosomes could result in destabilization of nucleosomes, generating nucleosome-poor regions that easily cause spontaneous DNA damage. In order to investigate whether incorporation of H2ABbd into chromatin actually caused nucleosome-poor regions and whether apoptotic cell death was caused by the activation of DNA damage checkpoints, we synchronized cells at G1/S by double thymidine block and induced expression of EGFP-H2ABbd or -H2A, as shown in Fig. 3A. MNase treatment of cells expressing H2ABbd or H2A revealed that that from cells expressing H2ABbd was far more sensitive than chromatin from cells expressing H2A even at early S phase (Fig. 3B). In addition, the nucleosomal ladder became degraded in H2ABbd-expressing cells but not in H2A-expressing cells, showing increased signal between the nucleosomal bands. These destabilizations were also observed in cells expressing H2ABbd even at early S phase (Fig. 3C). Taken together, these results strongly suggested that incorporation of H2ABbd into chromatin probably resulted in destabilizing nucleosomes. FACS analysis revealed that cells expressing EGFP-H2A normally progressed through S phase and completed DNA replication within 8 h (Fig. 3D). In contrast, cells expressing EGFP-H2ABbd showed a severe defect in S phase progression and a marked increase in sub-G1 population prior to the completion of S phase. As demonstrated in Fig. 3E, a strong signal of γ-H2AX was detected in HeLa cells expressing H2ABbd 4 h after release. Given that apoptosis was induced 12 h after release, as judged by cleaved caspase 3, this γ-H2AX signal was probably due to DNA damage but not DNA fragmentation as a consequence of apoptosis. Indeed, two-color flow cytometric analysis using propidium iodide and antibodies to γ-H2AX revealed that γ-H2AX-positive cells were detected in S phase, but not sub-G1 phase cells (supplemental Fig. 3). Further, association of DNA damage in cells expressing EGFP-H2ABbd with DNA replication suggested the activation of the intra-S phase checkpoint. Therefore, we examined the activation of target proteins of DNA damage checkpoint kinases. As exhibited in Fig. 3E, hyperphosphorylated RPA2 and phosphorylation of ATM at Ser-1981 (a marker of ATM activation) were significantly up-regulated in cells expressing EGFP-H2ABbd. ATM activation was also confirmed by phosphorylations of 53BP1 and Chk2. DNA replication-induced DNA damage and activation of intra-S phase checkpoint were not detected in cells expressing EGFP-H2A. We went on to examine whether apoptosis induced by the expression of H2ABbd was caused by aberrant DNA replication. RPE cells expressing FLAG-HA-H2ABbd were synchronized at quiescent state by serum starvation. Surprisingly, ectopic expression of EGFP-H2ABbd induced cell death in the quiescent cells, showing that H2ABbd-mediated apoptosis could occur without DNA replication (Fig. 3F). In particular, accumulation of γ-H2AX was also observed (Fig. 3G), further confirming that H2ABbd is associated with DNA damage. Collectively, these observations suggest that H2ABbd overexpression-mediated cell death is not restricted to DNA damage during S phase.
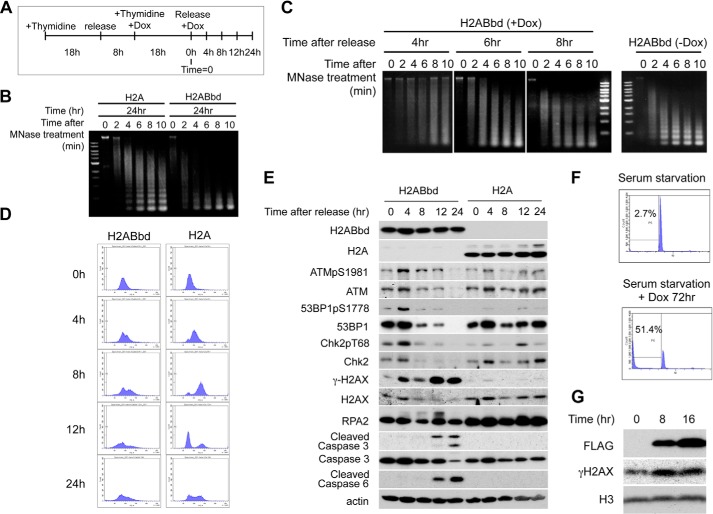
FIGURE 3.
Overexpression of H2ABbd induces DNA damage during S phase. A, the experimental strategy to synchronize HeLa cells in G1/S phase by double thymidine block and induce H2ABbd or H2A expression after the addition of Dox is shown. Cells were released from the thymidine block at 0 h, and samples were taken. B, cells expressing EGFP-H2ABbd and EGFP-H2A were collected at 24 h after release and subjected to MNase analysis. DNA was isolated from cells after MNase treatment for the indicated time (min) and subjected to agarose gel electrophoresis. C, EGFP-H2ABbd-expressing cells were collected at the indicated times after release and subjected to MNase analysis (left). As a control, the data from HeLa cells without induction of H2ABbd are also shown (right). D, cells prepared as in A were collected at the indicated times after thymidine block for FACS analysis. E, total cell extracts were subjected to immunoblotting using the indicated antibodies. F, RPE cells were cultured in medium without FBS for 5 days to synchronize in G0 (left) or further incubated in the serum-starved condition with Dox for 72 h (right) to express FLAG-HA-tagged H2ABbd. G, RPE cells were cultured in medium without FBS for 5 days and further incubated in the presence of Dox. At 8 or 16 h after the addition of Dox, cells were collected, and total cell extracts were subjected to immunoblotting using antibodies as indicated in the figure.
NF-κB Is Activated by H2ABbd Expression
Given that NF-κB plays a role in the induction of apoptosis, we examined whether activation of NF-κB occurred in cells expressing EGFP-H2ABbd. Indeed, we found that IκB-α was markedly decreased in cells expressing EGFP-H2ABbd (Fig. 4A). Reduced IκB was presumably caused by enhanced degradation because the amount of IκB-α transcript did not change despite the reduction in the protein level (Fig. 4B). Consistent with this, we found that a fraction of p65 and p50 proteins translocated into the nucleus when EGFP-H2ABbd was expressed (Fig. 4, C and D). Small fractions of p50 and p65 already existed in non-treated cells; however, this level was increased upon H2ABbd induction. In short, these results indicate that H2ABbd expression activated the NF-κB signaling pathway.
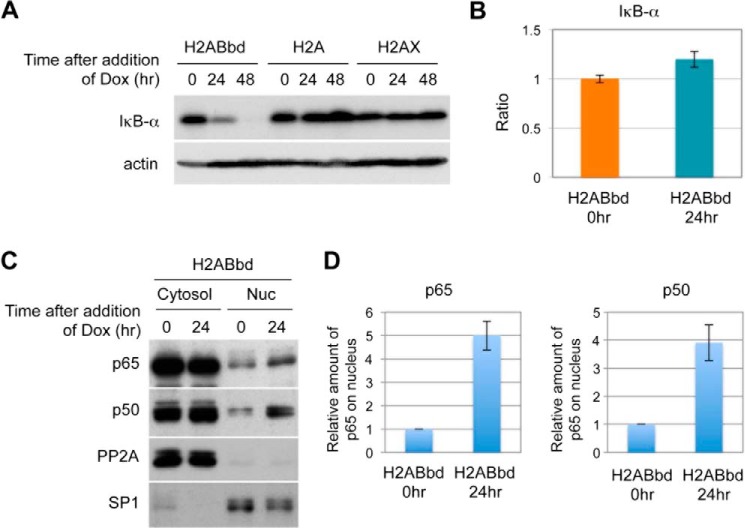
FIGURE 4.
NF-κB signaling cascade was activated upon overexpression of H2ABbd. A, degradation of inhibitor of NF-κB, IκB-α, was observed in HeLa cells overexpressing EGFP-H2ABbd. B, IκB-α mRNA was not decreased upon H2ABbd expression. Total RNAs were prepared from asynchronous HeLa cells (72 h after the addition of Dox). Quantitative real-time RT-PCR was performed using a set of primers for the IκB-α transcript. Relative mRNA expression was normalized to GAPDH. C, NF-κB activation was observed in H2ABbd-expressing HeLa cells. Cells were treated with Dox for 24 h to induce EGFP-H2ABbd or EGFP-H2A, collected for subcellular fractionation, and subjected to immunoblotting. PP2A and SP1 were used as markers of the cytosol fraction and nuclear fraction, respectively. D, p65 and p50 were accumulated in nucleus upon the addition of Dox. The signal intensity was analyzed by ImageJ software and normalized with SP1. Data are presented from three independent experiments. Error bars, S.D.
Apoptosis Induced by the Expression of H2ABbd Is Dependent on the Activation of Caspase 3 and NF-κB
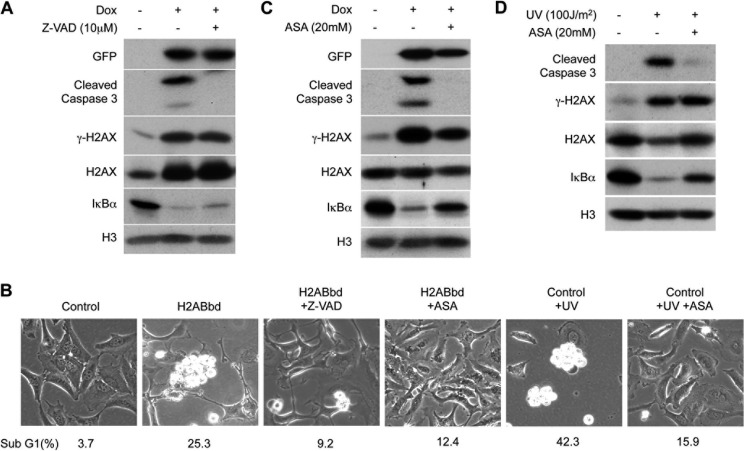
We examined whether apoptosis induced by H2ABbd was dependent on caspase 3 activation. Treatment with pan-caspase inhibitor benzyloxycarbonyl-VAD-fluoromethyl ketone (Z-VAD) for 48 h inhibited EGFP-H2ABbd-induced activation of caspase 3 (Fig. 5A) and suppressed apoptosis, as well as showing significant reduction in the sub-G1 population (25.3% versus 9.2%) (Fig. 5B). Intriguingly, treatment with inhibitors of NF-κB, aminosalicylic acid (ASA) (20) and ammonium pyrrolidine dithiocarbamate (21), markedly suppressed degradation of IκB-α, and the activation of caspase 3 (Fig. 5C and supplemental Fig. 4A). Suppression of EGFP-H2ABbd-dependent apoptosis was further confirmed by the observation that ASA or ammonium pyrrolidine dithiocarbamate treatment reduced sub-G1 fraction cells (25.3% versus 12.4%, 22.4% versus 3.6%) (Fig. 5B and supplemental Fig. 4B). As a control, we treated HeLa cells to induce apoptosis under NF-κB inhibitors. Consistent with previous reports, inhibition of NF-κB could suppress UV-induced apoptosis (Fig. 5, B and D) and UV-, 4NQO-, and actinomycin D-induced apoptosis (supplemental Fig. 4, C and D).
FIGURE 5.
H2ABbd-mediated apoptosis was blocked by caspase inhibitor and NF-κB inhibitor. A, HeLa cells were treated with or without Dox and benzyloxycarbonyl-VAD-fluoromethyl ketone (Z-VAD) (10 μm), a pancaspase inhibitor, for 48 h. Total cell extracts were prepared for immunoblotting. B, HeLa cells were treated with or without Dox, benzyloxycarbonyl-VAD-fluoromethyl ketone (10 μm), or ASA (20 mm), an inhibitor of NF-κB, for 48 h. As a control, HeLa cells were irradiated with UV (100 J/m2) and cultured with or without ASA (20 mm) for 48 h. Typical differential interference-contrast images and sub-G1 DNA contents (percentages) analyzed by FACS are shown. C, HeLa cells were treated with or without Dox and ASA (20 mm) for 48 h. Total cell extracts were prepared for immunoblotting. D, HeLa cells were irradiated with UV (100 J/m2) and cultured with or without ASA for 48 h, and total cell extracts were prepared and analyzed by immunoblotting.
Expression of H2ABbd Perturbs Gene Transcription Profiles
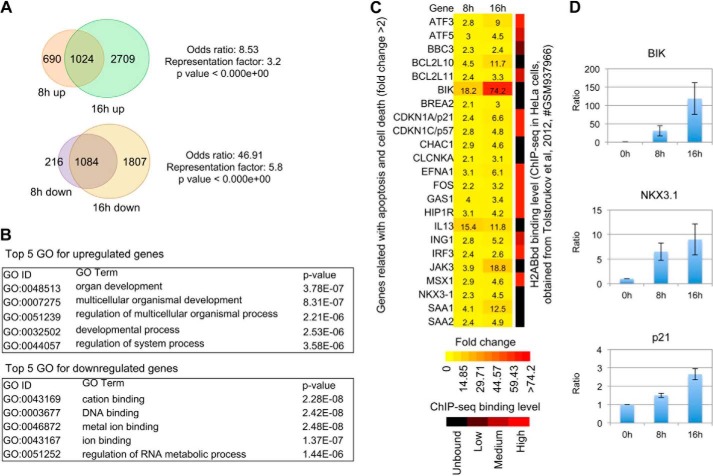
To gain further insight into the changes in the transcriptional network upon ectopic expression of H2ABbd in RPE cells, we performed gene expression microarray analysis in control (0 h) and H2ABbd-induced (8 and 16 h) cells. In order to avoid indirect effects of H2ABbd expression, we chose relatively early time points after H2ABbd expression. 1714 and 3733 genes were found to be up-regulated (>2-fold) after 8 and 16 h, respectively (Fig. 6A; full list given in supplemental Table 1). Comparison of the 8 and 16 h gene sets showed significant overlap, with 1024 genes in common. Further, statistical analyses revealed that the odds ratio and representation factor for overlap were 8.5 (>2 is generally significant) and 3.2 (>1 is generally significant), respectively (Fig. 6A). The p value for association was nearly equal to zero, suggesting that the overlap between the 8 and 16 h groups was not random. Previous studies have investigated the roles of H2ABbd in human cells by gene knockdown experiments (10). Interestingly, some of the genes, such as ARC, BLK, and ICAM5, that are repressed upon H2ABbd knockdown exhibited transcriptional up-regulation upon enforced H2ABbd expression in our study (supplemental Fig. 6A). These results further supported our hypothesis that there is a functional link between H2ABbd and regulation of gene transcription.
FIGURE 6.
Genes up-regulated and down-regulated upon enforced H2ABbd expression. A, up-regulated and down-regulated genes upon enforced H2ABbd expression are summarized in Venn diagrams. 1024 genes were found to be activated (>2-fold) at both the 8 and 16 h time points after H2ABbd expression. In contrast, 1084 genes were repressed (<2 fold) after 8 and 16 h of enforced H2ABbd expression. The odds ratio, representation factor, and p value are shown beside the diagrams. B, top five GO categories for up-regulated and down-regulated genes, selected by statistical significances determined with p value, are shown within their respective boxes. C, a group of apoptosis- and cell death-related genes were up-regulated in H2ABbd-overexpressing cells. -Fold changes in gene expression (>2-fold) are demonstrated in a heat map (left). The color scale for the heat map is given below. The ratio of up-regulation (-fold change) for each gene is shown within its respective box inside the heat map. Right, summary of the H2ABbd deposition pattern obtained from previously published ChIP-seq data in HeLa cells for each gene. The level of enrichment was judged by eye from the BedGraph data visualized by IGV and for simplification categorized into four groups: unbound, low, medium, and high, respectively (demonstrated in a black to red color scale at the bottom). D, quantitative real-time PCR results for three randomly picked loci, namely BIK, NKX3-1, and p21, from the up-regulated gene list. The relative ratio of activation for these genes, compared with the control (0 h, value = 1), is demonstrated in bar plots. Error bars, S.D. (n = 3). The relative mRNA expression was normalized to GAPDH.
As for the down-regulated loci, we found that the expression of 1300 and 2891 genes was reduced by more than 2-fold after 8 and 16 h of H2ABbd induction, respectively. This association was also highly significant, with 1084 genes in common. The odds ratio for overlap was ∼47, the representation factor was 5.8, and the p value was nearly zero (Fig. 6A).
Up-regulated Genes upon H2ABbd Expression Exhibit Association with Reproductive Functions
To categorize up-regulated and down-regulated genes in H2ABbd-overexpressing cells, we carried out GO tests. GO analysis revealed that development- and differentiation-associated genes were predominantly enriched among common up-regulated genes (Fig. 6B). It is interesting to note that H2ABbd is expressed in differentiated germ cells, such as elongating and round spermatids (Fig. 7A). Thus, selective activation of differentiation-associated genes might reflect the biological function of H2ABbd to regulate such genes during germ cell development. To examine this point further, we carried out functional clustering of the up-regulated genes by using the publicly available DAVID (Database for Annotation, Visualization, and Integrated Discovery) Functional Classification Tool. Indeed, this analysis revealed that the up-regulated genes were enriched in a functional cluster (cluster 14) enriched for germ cell- and reproductive function-associated GO terms, such as male genitalia development (GO:0030539) and spermatogenesis (GO:0007283). The association between the genes and functional terms in this group is shown in a two-dimensional view heat map (supplemental Fig. 7A). A positive gene term association that passes the statistical filter assigned by DAVID is given a value of 1 (shown in red), and an association that has not been reported yet is denoted with 0 (shown in black). The enrichment score of for this functional category was 2.1, which is statistically significant (in general, scores of >1 are accepted as significant).
FIGURE 7.
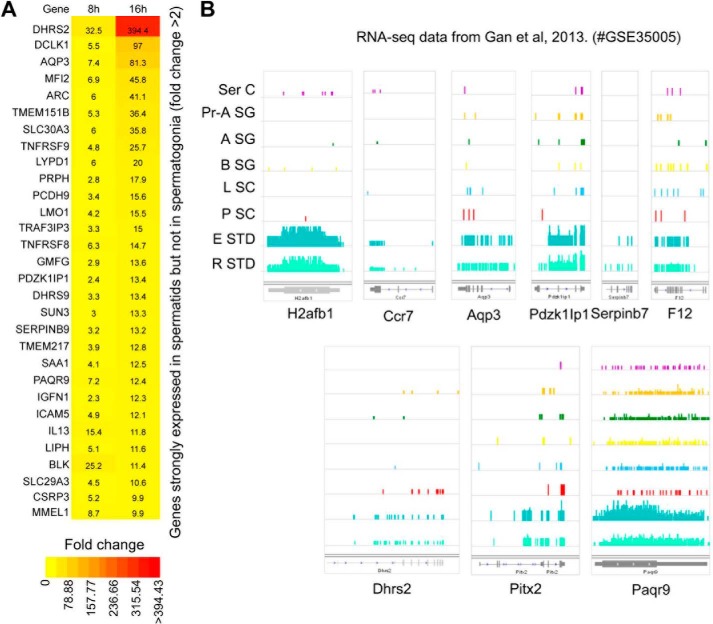
An in vivo link between postmeiotic germ cell-specific gene expression and H2ABbd. A, genes up-regulated upon H2ABbd expression in RPE cells included factors that are enriched in postmeiotic germ cells (e.g. in elongating and round spermatids). -Fold enrichment of such genes after 8 and 16 h are shown in a heat map. Similar to Fig. 6C, the ratio of up-regulation (>2-fold) is shown within the respective boxes for each gene, and the color scale is given below. B, comprehensive RNA-seq data for sertoli cells (Ser C, somatic cells in testis), primitive type-A spermatogonia (Pr-A SG), type-A spermatogonia (A SG), type-B spermatogonia (B SG), leptotene spermatocytes (L SC), pachytene spermatocytes (P SC), elongating spermatids (E STD), and round spermatids (R STD) were retrieved from the publicly available Gene Expression Omnibus database (18) (series accession number GSE35005). The expression of the murine H2ABbd homolog, H2afb1, is demonstrated on the left. The transcription levels for four other factors, namely Dhrs2, Aqp3, Pdzk1Ip1, and Paqr9, all of which are expressed specifically in the elongation and round spermatid cells, similar to H2afb1, are shown on the right.
Previous studies have reported that H2ABbd is expressed robustly in postmeiotic germ cells. To further examine this point, we retrieved comprehensive RNA sequencing (RNA-seq) data for different stages of murine male germ cell development from the publicly available Gene Expression Omnibus database (18). Transcription of H2afb1, the murine homolog of human H2ABbd, could be detected only in the postmeiotic elongating and round spermatids, but not in spermatogonia (Type A and B) or spermatocytes (leptotene, pachytene stages) (Fig. 7B). We asked whether the factors up-regulated by enforced H2ABbd expression in RPE cells included genes that are normally transcribed in postmeiotic germ cells. To this end, we checked the wig files provided in this data set by eye and selected ∼100 genes that were up-regulated upon H2ABbd overexpression as well as demonstrating robust expression patterns in spermatid but not in progenitor cells, such as DHRS2 (dehydrogenase/reductase member 2), PDZK1IP1 (PDZK1-interacting protein 1), PAQR9 (progestin and adipoQ receptor family member IX), and AQP3 (aquaporin 3). These genes were classified as putative “spermatid-enriched genes” (Fig. 7, A and B, and supplemental Table 3).
In contrast, GOs enriched for common down-regulated genes (1084 annotations) were associated with cellular processes, such as cation binding, metal ion binding, and DNA binding. The full list of GOs is given in supplemental Table 2.
Overexpression of H2ABbd Activates Apoptosis-related Genes
Given the fact that overexpression of H2ABbd induces apoptosis, we wondered whether cell death-related genes were up-regulated after 8 and 16 h. Indeed, around 80 genes were found to be included in this category (representative genes shown by heat map in Fig. 6C; full list given in supplemental Fig. 5). Interestingly, well known proapoptotic factors, such as BIK (Bcl-2-interacting killer) and BCL2L10 and BCL2L11 (Bcl2-like protein 10 and 11), were activated at the 8 and 16 h time points (Fig. 6C). We performed quantitative real-time PCR analysis for three randomly picked genes, BIK, NKX3-1, and p21, and found that these were up-regulated by nearly 100-, 10-, and 3-fold in 16 h samples (Fig. 6D, results shown for 8 and 16 h). These results were highly similar to our microarray data, suggesting the reproducibility of the findings. In addition, functional tests using DAVID tools demonstrated that up-regulated genes could be clustered in an apoptosis-related group (functional cluster 43), showing association with terms such as regulation of caspase activity (GO:0006919) and apoptosis regulator Bcl2, BH (IPR000712) (supplemental Fig. 7B). The enrichment factor for this cluster was >1 (1.09).
The ChIP-seq deposition pattern of H2ABbd in human (HeLa) cells has been already reported (10). We wondered whether apoptosis-related genes, identified from our study, demonstrated H2ABbd enrichment in HeLa cells. Indeed, we found that many of these loci, such as CDKN1A, MSX1, and ATF3, showed H2ABbd deposition around the promoter and/or gene body regions. The level of H2ABbd enrichment (strong, moderate, and low) for apoptosis-associated factors is shown at the right side of the gene expression heat map (Fig. 6C and supplemental Fig. 6B). Collectively, these results suggest that at least some of the apoptosis-related loci could be marked by a basal level of H2ABbd enrichment in wild type cells. Upon increased H2ABbd binding due to some cellular cues/signals, these genes could be switched on in turn and contribute to the observed apoptotic phenotype in our study.
DISCUSSION
Previous studies examining the role of the murine homolog of H2ABbd have suggested that this histone variant plays a role in spermatogenesis via promoting apoptosis of male germ cells. However, the mechanisms underlying H2ABbd-mediated apoptosis are not fully understood. In the present study, we found that ectopic expression of human H2ABbd specifically induced NF-κB-dependent apoptosis in a dose-dependent manner, showing that cell death during spermatogenesis may be in part dependent on NF-κB signaling.
Dysregulation of histone expression has been linked with increased mitotic chromosome loss and DNA damage sensitivity, resulting in genomic instability (22, 23), depending on the types or contexts of the histones (22). In this study, we noted that enforced expression of H2A or H2AX did not profoundly affect any cell function, although a slight delay of cell proliferation was observed (Fig. 1D). However, ectopic expression of H2ABbd led to drastic consequences, namely apoptosis.
Impairment of histone incorporation into chromatin leads to defects in chromatin assembly, causing stalled replication fork and leading to DNA replication-dependent DNA damage that activates ATM-dependent DDR pathways (24). Indeed, ectopic expression of H2ABbd caused DNA replication-dependent DNA damage, activating the intra-S phase checkpoint (Fig. 3, C and D). Surprisingly, ectopic expression of H2ABbd caused DNA damage even in quiescent cells (Fig. 3, F and G), suggesting that incorporation of H2ABbd into nucleosomes resulted in severe destabilization of nucleosomes and generation of anucleosome regions that might consist of fragile DNA sites.
Microarray analysis revealed that H2ABbd overexpression activates postmeiotic germ cell-enriched genes and apoptosis-related genes, although physiological implication of H2ABbd in sperm cell differentiation or other developmental pathways remains elusive. This raises the possibility that H2ABbd could be selectively incorporated into specific gene loci to regulate transcription. However, any histone chaperone responsible for incorporation of H2ABbd into chromatin remains to be identified. We speculate that nucleosomal assembly of H2ABbd might be regulated by cooperation of histone chaperones and transcription factors that control development and differentiation of postmeiotic germ cells. Recently, it has been reported that human H2ABbd was deposited effectively at intron-exon boundaries in HeLa cells. Moreover, knockdown of H2ABbd resulted in splicing defect of mRNA, indicating that H2ABbd might be linked with regulation of splicing (11). Summarizing these possibilities, it could be speculated that expression of genes involved in development and/or postmeiotic germ cells could be mediated by the regulation of splicing of specific gene transcripts. In addition, recent genome-wide analysis showed that human H2ABbd was localized in both active and repressed chromatin domains (25). Thus, it should be important to determine the mechanism of how H2ABbd regulates the expression of genes involved in development and differentiation.
Supplementary Material
Acknowledgments
We thank Yoshie Chiba and Dr. Chisato Yamada for technical assistance and Dr. Mireille Delhase for critical reading of the manuscript. We also thank Drs. Peter J. Park and Michael Y. Tolstorukov (Massachusetts General Hospital, Boston, MA) for generously providing the RNA-seq FPKM data for control and H2Abbd knockdown experiments previously reported by Tolstorukov et al. (11).
This work was supported by grants from the NEXT program (LS105) and the Naito Foundation (to M. S.).

This article contains supplemental Tables 1–3 and Figs. 1–7.
M. Y. Tolstorukov, J. A. Goldman, C. Gilbert, V. Ogryzko, R. E. Kingston, and P. J. Park, personal communication.
- DDR
- DNA damage response
- GO
- gene ontology
- MEF
- mouse embryo fibroblast
- EGFP
- enhanced green fluorescent protein
- Dox
- doxycycline
- ASA
- aminosalicylic acid
- MNase
- micrococcal nuclease.
REFERENCES
- 1. Lorch Y., Zhang M., Kornberg R. D. (1999) Histone octamer transfer by a chromatin-remodeling complex. Cell 96, 389–392 [DOI] [PubMed] [Google Scholar]
- 2. Talbert P. B., Henikoff S. (2010) Histone variants: ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 11, 264–275 [DOI] [PubMed] [Google Scholar]
- 3. Chadwick B. P., Willard H. F. (2001) A novel chromatin protein, distantly related to histone H2A, is largely excluded from the inactive X chromosome. J. Cell Biol. 152, 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhou J., Fan J. Y., Rangasamy D., Tremethick D. J. (2007) The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat. Struct. Mol. Biol. 14, 1070–1076 [DOI] [PubMed] [Google Scholar]
- 5. Bao Y., Konesky K., Park Y. J., Rosu S., Dyer P. N., Rangasamy D., Tremethick D. J., Laybourn P. J., Luger K. (2004) Nucleosomes containing the histone variant H2A. Bbd organize only 118 base pairs of DNA. EMBO J. 23, 3314–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gautier T., Abbott D. W., Molla A., Verdel A., Ausio J., Dimitrov S. (2004) Histone variant H2ABbd confers lower stability to the nucleosome. EMBO Rep. 5, 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Angelov D., Verdel A., An W., Bondarenko V., Hans F., Doyen C. M., Studitsky V. M., Hamiche A., Roeder R. G., Bouvet P., Dimitrov S. (2004) SWI/SNF remodeling and p300-dependent transcription of histone variant H2ABbd nucleosomal arrays. EMBO J. 23, 3815–3824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Montel F., Fontaine E., St-Jean P., Castelnovo M., Faivre-Moskalenko C. (2007) Atomic force microscopy imaging of SWI/SNF action: mapping the nucleosome remodeling and sliding. Biophys. J. 93, 566–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Govin J., Escoffier E., Rousseaux S., Kuhn L., Ferro M., Thévenon J., Catena R., Davidson I., Garin J., Khochbin S., Caron C. (2007) Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J. Cell Biol. 176, 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Soboleva T. A., Nekrasov M., Pahwa A., Williams R., Huttley G. A., Tremethick D. J. (2012) A unique H2A histone variant occupies the transcriptional start site of active genes. Nat. Struct. Mol. Biol. 19, 25–30 [DOI] [PubMed] [Google Scholar]
- 11. Tolstorukov M. Y., Goldman J. A., Gilbert C., Ogryzko V., Kingston R. E., Park P. J. (2012) Histone variant H2A. Bbd is associated with active transcription and mRNA processing in human cells. Mol. Cell 47, 596–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shimada M., Nakanishi M. (2006) DNA damage checkpoints and cancer. J. Mol. Histol. 37, 253–260 [DOI] [PubMed] [Google Scholar]
- 13. Nowsheen S., Yang E. S. (2012) The intersection between DNA damage response and cell death pathways. Exp. Oncol. 34, 243–254 [PMC free article] [PubMed] [Google Scholar]
- 14. Wu Z. H., Shi Y., Tibbetts R. S., Miyamoto S. (2006) Molecular linkage between the kinase ATM and NF-κB signaling in response to genotoxic stimuli. Science 311, 1141–1146 [DOI] [PubMed] [Google Scholar]
- 15. Gu C., Tong Q., Zheng L., Liang Z., Pu J., Mei H., Hu T., Du Z., Tian F., Zeng F. (2010) TSEG-1, a novel member of histone H2A variants, participates in spermatogenesis via promoting apoptosis of spermatogenic cells. Genomics 95, 278–289 [DOI] [PubMed] [Google Scholar]
- 16. Shimada M., Niida H., Zineldeen D. H., Tagami H., Tanaka M., Saito H., Nakanishi M. (2008) Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell 132, 221–232 [DOI] [PubMed] [Google Scholar]
- 17. Shimada M., Haruta M., Niida H., Sawamoto K., Nakanishi M. (2010) Protein phosphatase 1gamma is responsible for dephosphorylation of histone H3 at Thr 11 after DNA damage. EMBO Rep. 11, 883–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gan H., Wen L., Liao S., Lin X., Ma T., Liu J., Song C. X., Wang M., He C., Han C., Tang F. (2013) Dynamics of 5-hydroxymethylcytosine during mouse spermatogenesis. Nat. Commun. 4, 1995. [DOI] [PubMed] [Google Scholar]
- 19. Danial N. N., Korsmeyer S. J. (2004) Cell death: critical control points. Cell 116, 205–219 [DOI] [PubMed] [Google Scholar]
- 20. Kopp E., Ghosh S. (1994) Inhibition of NF-κB by sodium salicylate and aspirin. Science 265, 956–959 [DOI] [PubMed] [Google Scholar]
- 21. Bessho R., Matsubara K., Kubota M., Kuwakado K., Hirota H., Wakazono Y., Lin Y. W., Okuda A., Kawai M., Nishikomori R. (1994) Pyrrolidine dithiocarbamate, a potent inhibitor of nuclear factor κB (NF-κB) activation, prevents apoptosis in human promyelocytic leukemia HL-60 cells and thymocytes. Biochem. Pharmacol. 48, 1883–1889 [DOI] [PubMed] [Google Scholar]
- 22. Meeks-Wagner D., Hartwell L. H. (1986) Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell 44, 43–52 [DOI] [PubMed] [Google Scholar]
- 23. Gunjan A., Verreault A. (2003) A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115, 537–549 [DOI] [PubMed] [Google Scholar]
- 24. Ye X., Franco A. A., Santos H., Nelson D. M., Kaufman P. D., Adams P. D. (2003) Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol. Cell 11, 341–351 [DOI] [PubMed] [Google Scholar]
- 25. Ioudinkova E. S., Barat A., Pichugin A., Markova E., Sklyar I., Pirozhkova I., Robin C., Lipinski M., Ogryzko V., Vassetzky Y. S., Razin S. V. (2012) Distinct distribution of ectopically expressed histone variants H2A. Bbd and MacroH2A in open and closed chromatin domains. PloS One 7, e47157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.