Background: Sj7170 is a new peptide from scorpion venom.
Results: Sj7170 specifically inhibits the activity of α-chymotrypsin. Sj7170 promotes glioblastoma tumorigenesis and metastasis by up-regulating cyclin D1 and Snail.
Conclusion: Sj7170 not only specifically inhibits α-chymotrypsin activity but also promotes the tumorigenesis and metastasis of glioblastoma.
Significance: Sj7170 is a unique dual-function peptide with a protease inhibitory activity and a potent tumor-activating effect.
Keywords: Glioblastoma, Metastasis, Peptides, Proliferation, Protease Inhibitor
Abstract
A new peptide precursor, termed Sj7170, was characterized from the venomous gland cDNA library of the scorpion Scorpiops jendeki. Sj7170 was deduced to be a 62-amino acid peptide cross-linked by five disulfide bridges. The recombinant Sj7170 peptide (rSj7170) with chromatographic purity was produced by a prokaryotic expression system. Enzyme inhibition assay in vitro and in vivo showed that rSj7170 specifically inhibited the activity of α-chymotrypsin at micromole concentrations. In addition, Sj7170 not only promoted cell proliferation and colony formation by up-regulating the expression of cyclin D1 in vitro but also enhanced tumor growth in nude mice. Finally, Sj7170 accelerated cellular migration and invasion by increasing the expression of the transcription factor Snail and then inducing the epithelial-mesenchymal transition. Moreover, Sj7170 changed cell morphology and cytoskeleton of U87 cells by the GTPase pathway. Taken together, Sj7170 is a unique dual-function peptide, i.e. a specific α-chymotrypsin inhibitor and a potent tumorigenesis/metastasis activator. Our work not only opens an avenue of developing new modulators of tumorigenesis/metastasis from serine protease inhibitors but also strengthens the functional link between protease inhibitors and tumor activators.
Introduction
Human glioblastoma is the most frequent primary malignant glioma brain tumor in adults and is one of the most aggressive malignancies. Approximately half of the newly diagnosed glioblastoma patients are in the late stages and have a poor prognosis. Current therapy with surgery, radiation, and chemotherapy, if ever, rarely cures the disease and infrequently prolongs life for more than 1 year. The median survival rate for patients with glioblastoma is less than 1 year, and the average life expectancy remains ∼14–18 months (1).
Glioblastomas have the following four characteristics: rapidly dividing cells, higher increasing vascularity, invasion into normal brain tissue, and a strong resistance to death-inducing effects. The diffusely infiltrative nature of glioblastoma is one of the major obstacles to its successful surgical control. The specific molecular mechanisms underlying the invasive behavior of glioblastomas remain largely unclear (2). Therefore, the development of effective modulators (activators or inhibitors) against glioblastomas will help to reveal the specific molecular mechanism of tumorigenesis, which certainly contributes to finding methods and cures for glioblastoma diagnosis and therapy.
Serine protease inhibitors (SPIs)4 are widely found in a variety of organisms in nature, not only in the bodies of various animals but also in the secretions of parasites, hematophagous invertebrates, amphibian skins, and the venom glands of poisonous animals. SPIs are very fundamental tools because they act as modulators by playing key roles in regulating the activities of numerous target serine proteases, blocking these in emergency cases, and signaling receptor interactions or clearance.
SPIs are classified into several families based on sequence homology, location of reactive site, structural characteristics, and mechanism of action. Generally, they are grouped into the Kunitz family, potato 1 family, and potato 2 family, Kazal family, Bowman-Birk family, and Ascaris family. They control complement activation and a variety of other physiological functions, such as blood coagulation, fibrinolysis, inflammation, tumor cell metastasis, apoptosis, and others. Several serine protease inhibitors from human or other organisms have evolved functions that do not involve protease inhibition (3). For example, TFPI-2, a Kunitz-type serine protease inhibitor, has been described as a tumor suppressor gene in several types of cancers, including glioma. Its expression was absent in five of the nine investigated high grade glioma cell lines (4). SPIs are often overexpressed in different tumor types, suggesting that the overexpression of these inhibitors might favor tumor progression (5).
Indeed, it has been demonstrated that the overexpression of a number of SPIs from the Serpin and Kunitz families results in the enhancement of cancer cell malignancy. However, all of these SPIs are secreted by endogenous human cells, and none of chymotrypsin-inhibitory peptides or Ascaris-type SPIs were found to promote malignancy of cancer cells, especially glioma cells.
Here, we describe a new Ascaris-type serine protease inhibitor that was first discovered from the venom of the scorpion Scorpiops jendeki. We also demonstrate that the recombinant peptide Sj7170 (rSj7170) effectively promoted the proliferation of glioma U87 cells in vitro and tumor growth in vivo. This effect could be suppressed by knockdown of the expression of cyclin D1, indicating that the proliferation triggered by Sj7170 occurs through the cyclin D1-Rb-E2F pathway. In addition, Sj7170 also enhanced the migration and invasion of U87 cells by inducing cellular EMT progress. The cell motility induced by Sj7170 also could be inhibited by knockdown of the expression of the EMT transcription factor Snail. Finally, we confirmed that Sj7170 changed morphology of U87 cells and rearranged cytoskeleton by GTPase pathway. Overall, this study describes a new tumor modulator of serine protease inhibitor from animal venom and provided a potential molecular tool for cancer research.
EXPERIMENTAL PROCEDURES
cDNA Library Construction and Screening
The venomous gland cDNA library of the scorpion S. jendeki was constructed as described in our previous work (6, 7). Some new and randomly selected colonies were sequenced using the ABI 3730 automated sequencer (Applied Biosystems, Foster City, CA). Sequences were identified for open reading frames using the ORF finder program (www.ncbi.nlm.nih.gov). Signal peptide was removed using the SignalP 4.0 Server. Sequences of Ascaris-type toxins were obtained by searching the data of this venomous gland cDNA library and the GenBankTM National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov) using the Basic Local Alignment Search Tool algorithm. Sequence alignment was performed using Clustal_X 1.83 software followed by manual adjustment.
Construction of Sj7170 Expression Vector
Expression vector pET-28a was used to produce the recombinant Sj7170 (rSj7170) in Escherichia coli. The cDNA sequence of Sj7170 from the venom gland cDNA library of the scorpion S. jendeki was used as the template for the generation of fragments using PCR. The PCR product of Sj7170 cDNA was digested with NcoI and XhoI and inserted into the NcoI-XhoI cutoff pET-28a expression vector. After confirmation by sequencing, the recombinant plasmid pET-28a-Sj7170 was transformed into E. coli Rosetta (DE3) cells for expression.
Expression and Purification of Sj7170
Transformed cells containing the expression plasmid pET-28a-Sj7170 were cultured at 37 °C in Luria-Bertani (LB) medium with 30 μg/ml kanamycin. Protein synthesis was induced by the addition of 10 mm isopropyl β-d-thiogalactoside (IPTG) when the absorbance at 600 nm reached 0.8–1.0. After 4 h of continued growth at 37 °C, cells from 1 liter of culture were harvested by centrifugation. The cell pellet was resuspended in phosphate-buffered saline (PBS) buffer and lysed by sonication on ice. rSj7170 was exclusively accumulated in inclusion bodies. The insoluble inclusion bodies were washed twice with washing buffer (1–2% Triton X-100 in PBS) and denatured in 2.5 ml of denaturation solution (6 m guanidinium HCl, 0.1 m Tris-HCl, pH 8.0, 1 mm EDTA, 30 mm reduced glutathione). The rSj7170 was then reactivated by 100-fold dilution in renaturation solutions (0.2 m ammonium acetate at pH 8.0 containing 0.2 mm oxidized glutathione and 0.5 m arginine) at 16 °C for 24 h. The soluble material was then desalted and enriched using centrifugal filter devices (Sartorius Stedim Biotech, Germany, cutoff value >3 kDa). The renatured peptide was finally purified by HPLC on a C18 column (10 mm × 250 mm, 5 μm; Elite-HPLC, China) with a constant flow rate of 5 ml/min. Peaks were detected at a wavelength of 230 nm. The fraction containing rSj7170 was collected manually and lyophilized immediately. The molecular mass of the purified rSj7170 was further analyzed by MALDI-TOF-MS (Voyager-DESTR; Applied Biosystems). The secondary structures of Sj7170 and its mutants were analyzed by CD spectropolarimetry. All purified peptides were dissolved in water at a concentration of 0.2 mg/ml. Spectra from 250 to 190 nm were recorded at 25 °C with a scan rate of 50 nm/min on a Jasco-810 spectropolarimeter. The final CD spectra were obtained by averaging three scans and subtracting the signal from a water blank.
Serine Protease Inhibition Activity Assays in Vitro
The inhibitory activity of rSj7170 was tested by measuring the hydrolysis of synthetic chromogenic substrates in the presence of serine proteases. Trypsin (bovine pancreatic trypsin; EC 3.4.21.4), chymotrypsin (bovine pancreatic A-chymotrypsin; EC 3.4.21.1), elastase (porcine pancreatic elastase; EC 3.4.21.36), and the chromogenic substrates Nα-benzoyl-l-arginine 4-nitroanilide hydrochloride, N-succinyl-Ala-Ala-Pro-Phe p-nitroanilide, and N-succinyl-Ala-Ala-Ala-p-nitroanilide were purchased from Sigma. The initial rates of every reaction was monitored continuously at 405 nm for 5 min at 25 °C. Lineweaver-Burk plots (1/(V versus 1/[S]) were used to determine the 1/V versus [I] values of chymotrypsin activity on N-succinyl-Ala-Ala-Pro-Phe p-nitroanilide in the presence of different inhibitor concentrations. The slopes (1/V versus [I]) of curves were plotted against the concentration of inhibitor. The inhibitory constant (Ki) of the chymotrypsin-inhibitor complex can be determined from the intercept point of the secondary plot on the x axis. Inhibitory tests for trypsin and elastase were carried out in the same manner as for chymotrypsin.
Chymotrypsin Activity Assay in Vivo
Clear-bottom, white-walled 96-well microtiter cell culture plates (Greiner Bio-One North America Inc., Monroe, NC) were seeded with U87 cells as indicated and treated with epoxomicin or rSj7170. Proteasome inhibition was measured using the proteasome Glo reagent according to the manufacturer's instructions (Promega). In brief, tumor cells were treated with epoxomicin or rSj7170 at different concentrations and incubated for 2 h. After incubation, cells were incubated for 15 min with 100 μl of proteasome Glo reagent, which permeabilizes cells and contains the bioluminescent substrates succinyl-LLVY-amino luciferin to determine the chymotrypsin-like activities, respectively. The luminescence was measured with a Dynex MLX luminometer.
Cell Line and Cell Culture
The human glioblastoma cell line U87 was purchased from American Type Culture Collection (ATCC). The cell was cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin (1000 units/ml), and streptomycin (1000 μg/ml) in a humidified incubator at 37 °C with 5% CO2. Cell culture dishes and plates were obtained from Wuxi NEST Biotechnology Co. Ltd. (Wuxi, China).
Cell Proliferation Assay
The effects of rSj7170 on cell proliferation were characterized by cell counting. To assess cellular proliferation, a growth curve was generated by seeding 5 × 104 cells/well into a 24-well tissue culture plate. Six groups of cells were treated with 0, 2, 4, 6, 8, and 10 μm rSj7170 for 48 h and counted using a hemocytometer. The seventh group of cells was treated with 10 μm rSj7170 and counted daily over a 3-day period.
Plate Colony Formation Assay
For the plate colony formation assay, the cells were plated into a 6-well tissue culture plate (500 cells per well) and incubated at 37 °C for 14 days. The resulting colonies were rinsed with 1× PBS, fixed with methanol for 10 min, and stained with crystal violet. The colonies were photographed and counted for three independent experiments. The colony index of rSj7170-treated cells was derived using the following formula: colony index (%) = (the number of colonies in rSj7170-treated group)/(the number of colonies in the untreated group) × 100. As for the untreated U87 cells, the colony index was regarded as 100%.
Cell Cycle Analysis
The cells were plated into a 6-well tissue culture plate at concentrations determined to yield 70% confluence within 48 h. The cells were harvested using 0.25% trypsin and subsequently pelleted by centrifugation at 2000 rpm for 5 min. The pellet was fixed in 70% ethanol at 4 °C overnight. Next, the cells were washed with 1× PBS, resuspended in propidium iodide solution (50 μg/ml) containing RNase, and incubated at room temperature in the dark for 30 min. The cell suspensions were filtered through a 50-μm nylon mesh, and the cell cycle profiles were analyzed using a BD FACSCalibur flow cytometer (BD Biosciences). The cell cycle distribution (G1, G2/M, or S phase) was determined using FlowJo software (Tree Star, Ashland, OR).
Western Blot Analysis
The following antibodies were used for the Western blot analyses: anti-Sj7170 was prepared from Beijing Perfect Biotechnology, Ltd.; anti-cyclin D1, anti-cyclin D2, anti-cyclin D3, anti-CDK2, anti-CDK4, anti-CDK6, anti-cyclinE1, anti-cyclinE2, anti-MMP2 and anti-MMP9 were purchased from ProteinTech Group Inc.; anti-Rb2/p130, anti-E2F1, and anti-E2F5 were purchased from Abgent (China); anti-P21, anti-P27, anti-E-cadherin, anti-vimentin, and anti-Snail were purchased from Cell Signaling Technology; and anti-β-actin antibody was purchased from Santa Cruz Biotechnology. Briefly, the cells were collected, washed in 1× PBS, and resuspended on ice in RIPA buffer (50 mm Tris-HCl, pH 7.2; 250 mm NaCl; 2% Nonidet P-40; 2.5 mm EDTA; 0.1% SDS; 0.5% sodium deoxycholate; and protease inhibitor mixture (Roche Applied Science)). The protein concentration was determined using the Bradford assay (Bio-Rad). The proteins were separated by 8–15% SDS-PAGE and electroblotted using a semidry transfer system (Bio-Rad) onto nitrocellulose filter membranes (Millipore Corp., Billerica, MA). The membranes were blocked with nonfat milk and probed with primary antibody. Next, the membranes were washed three times for 5 min in PBS/Tween (0.1%), incubated with a horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology) for 1 h at room temperature, and then washed another three times for 5 min in PBS/Tween (0.1%). The blots were visualized using the SuperSignal chemiluminescent detection module (Pierce).
shRNA Preparation and Cell Transfection
shRNA sequences of cyclin D1 and Snail were designed by the software of siRNA Sequence-Selector and then synthesized (Table 1). The fragments digested by endonuclease BbsI and BamHI were inserted into the vector pGPU/GFP/Neo (Shanghai GenePharma Co. Ltd. China) to generate pGPU/GFP/Neo-cyclin D1 and pGPU/GFP/Neo-Snail. Transient transfection was performed at ∼80% confluency in 6-well plates (NEST) using FuGENE HD (Roche Applied Science) according to the manufacturer's instructions. Briefly, a total of 5 × 105 cells were seeded into each well in DMEM containing 10% fetal bovine serum the day before transfection. Four micrograms of the purified shRNA expression vectors (shcyclin D1–326, shcyclin D1–526, shcyclin D1–719, or shcyclin D1–901) were transfected into U87 cells with 10 μl of FuGENE HD reagent. After 48 h of transfection, RT-PCR and Western blotting were done to assess the efficiency of cyclin D1 knockdown. G418 is an analog of neomycin. For stable transfection, the optimal concentration of G418 for selection was determined by titration before transfection. About 2 × 104 cells were seeded into each well of a 24-well dish (NEST). After 24 h, cells were cultured with G418 concentrations of 0, 200, 400, 600, 800, 1000, 1200, and 1400 μg/ml. The lowest concentration of G418 initiated cell death in ∼7–9 days and killed all cells within 2 weeks. After 24–48 h transfection as described above, cells were cultured with 10% FBS at the optimal concentration of G418. After 2 weeks, resistant cell clones were picked and transferred to 24-well plates and gradually expanded to 6-well plates and 1- cm dishes. At 90% confluence, cyclin D1 expression was measured with Western blotting.
TABLE 1.
Insert sequence of cyclin D1 shRNA fragments
| Name | Sequence |
|---|---|
| shcyclin D1–326 | 5′-CACCGCCCTCGGTGTCCTACTTCAATTCAAGAGATTGAAGTAGGACACCGAGGGCTTTTTTG-3′ |
| shcyclin D1–526 | 5′-CACCGCATGTTCGTGGCCTCTAAGATTCAAGAGATCTTAGAGGCCACGAACATGCTTTTTTG-3′ |
| shcyclin D1–719 | 5′-CACCGGCGGAGGAGAACAAACAGATTTCAAGAGAATCTGTTTGTTCTCCTCCGCCTTTTTTG-3′ |
| shcyclin D1–901 | 5′-CACCGCTTCCTCTCCAGAGTGATCATTCAAGAGATGATCACTCTGGAGAGGAAGCTTTTTTG-3′ |
| shsnail-142 | 5′-GAGCGAGCTGCAGGACTCTAATTTCAAGAGAATTAGAGTCCTGCAGCTCGCTT-3′ |
| shsnail-771 | 5′-GCCACTCAGATGTCAAGAAGTATTCAAGAGATACTTCTTGACATCTGAGTGGTT-3′ |
| shsnail-822 | 5′-GCCGAATGTCCCTGCTCCACAATTCAAGAGATTGTGGAGCAGGGACATTCGGTT-3′ |
Real Time Quantitative PCR
Total RNA was reverse-transcribed into cDNA using SuperScript II (Invitrogen). Real time quantitative PCR was performed using an ABI 7500 real time PCR system (Applied Biosystems, Foster City, CA) and the SYBR Green real time PCR Master Mix (TOYOBO, Osaka, Japan). Primers used for real time quantitative PCR are shown in Table 2. Each PCR was performed in triplex tubes, with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) being used as an endogenous control to standardize the amount of sample RNA.
TABLE 2.
Primers used for quantitative real time PCR
| Gene | Sequence | |
|---|---|---|
| Cyclin D1 | Forward | 5′-CTGGCCATGAACTACCTGGA-3′ |
| Reverse | 5′-GTCACACTTGATCACTCTGG-3′ | |
| E-cadherin | Forward | 5′-ACCAGAATAAAGACCAAGTGACCA-3′ |
| Reverse | 5′-AGCAAGAGCAGCAGAATCAGAAT-3′ | |
| Vimentin | Forward | 5′-AATGGCTCGTCA CCTTCGTGAAT-3′ |
| Reverse | 5′-CAGATTAGTTTCCCTCAGGTTCAG-3′ | |
| Snail | Forward | 5′-TTGGATACAGCTGCTTTGAG-3′ |
| Reverse | 5′-ATTGCATAGTTAGTCACACCTC-3′ | |
| Twist | Forward | 5′-GAAAATGGACAGTCTAGAGACTCTG3′ |
| Reverse | 5′-GTGGCTGATTGGCAAGACCTCTTG-3′ | |
| MMP2 | Forward | 5′-ATAACCTGGATGCCGTCGT-3′ |
| Reverse | 5′-AGGCACCCTTGAAGAAGTAGC-3′ | |
| MMP9 | Forward | 5′-GAACCAATCTCACCGACAGG-3′ |
| Reverse | 5′-GCCACCCGAGTGTAACCATA-3′ |
In Vivo Xenograft Tumor Study
Male BALB/c nude mice (6 weeks old) were obtained from the Disease Prevention Center of Hubei Province, China. Each mouse was inoculated in the right axilla with 1 × 106 U87 cells suspended in 0.1 ml of PBS. At 1 week later, mice with tumors reached 200 ± 100 mm3 in the rSj7170 group (n = 4). Tumor volumes and body weights were measured every day. All mice were maintained for 30 days before they were sacrificed to collect tumors. Tumor volume was calculated using the following formula: tumor volume = 1/2 × (longer diameter) × (shorter diameter)2. All animal experiments were approved by the Animal Research Ethics Board of Wuhan University in China and were in compliance with institutional guidelines on the care of experimental animals.
Immunohistochemistry (IHC) of Tumor Xenografts
To assess cyclin D1 expression in xenograft tumors, rabbit monoclonal anti-cyclin D1 antibody (diluted to 1:50, Cell Signaling Technology) was used in IHC. Specimens of U87 xenografts were fixed in 4% paraformaldehyde and embedded with paraffin. All immunohistochemistry staining assays were performed by Hubei BIOS Biological Technology Co., Ltd. (Wuhan, China), following a standard procedure. The expression signal in tumor tissue was observed and analyzed under microscopy.
Cell Migration and Invasion Assay
The cell migration assay was performed using a Transwell Assay Chamber (PET track-etched membrane; BD Biosciences). For the invasion assays, the cells were seeded into BioCoat Matrigel Invasion Chambers (BD Biosciences). The untreated U87 clones (5 × 104 cells) maintained in DMEM supplemented with 0.1% FBS were seeded in the top chamber, and DMEM supplemented with 10% FBS was added to the bottom chamber. For both assays, the cells were allowed to migrate or to invade. After incubation for 24 h at 37 °C, the cells on the top side of the membranes were removed by cotton swab, and those on the bottom side were fixed and stained with crystal violet. Migrated and invaded cells were fixed and stained after 24 h. Six randomly selected fields per well were photographed, and the numbers of migrated or invaded cells were counted. The extent of cell migration was expressed as a migration index. The migration index of rSj7170-treated cells was calculated using the following formula: migration index (%) = (number of cells that migrated through the insert membrane in rSj7170-treated group)/(number of cells that migrated through the insert membrane in the untreated group) × 100. As for the untreated cells, the migration index was regarded as 100%. The invasion indices of the rSj7170-treated cells and the untreated cells were calculated in the same way.
Immunofluorescent Staining
Cells were grown on sterile coverslips up to 50% confluence. Coverslip-grown cells were washed twice with 1× PBS and fixed with 4% paraformaldehyde for 15 min. The cells were first incubated with primary antibodies at a 1:100 dilution in 0.05% Triton X-100 and 1% BSA in 1× PBS at 4 °C overnight. After thorough washing, the cells were incubated for 1 h with fluorescence-conjugated secondary antibodies (TRITC-conjugated sheep anti-rabbit). The nuclei were labeled blue with 4′,6-diamidino-2-phenylindole (DAPI). The coverslips were mounted using Vectashield mounting medium (Vector Laboratories, Burlingame, CA). The fluorescence signals were visualized using confocal laser microscopy (Fluoview FV1000; Olympus, Tokyo, Japan).
Statistical Analysis
Mean values were calculated from at least three independent experiments and expressed as mean ± S.D. Student's t test was used to evaluate statistically significant differences, which were considered at p < 0.05 (two-sided).
RESULTS
Sequence Analysis of Sj7170, a Member of the Ascaris-type Toxin Family
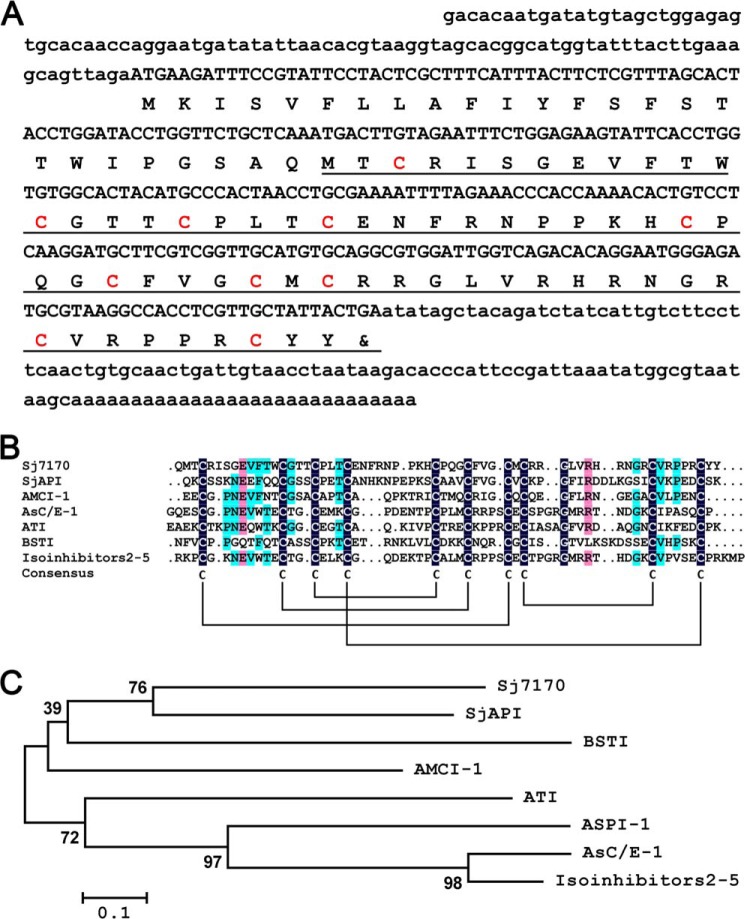
Using random screening and bioinformatics analysis of the S. jendeki venom gland cDNA library, a number of new scorpion venom peptides were characterized. After searching for homologs in the GenBankTM NCBI database, a new venom peptide termed Sj7170 (GenBankTM accession number GH547578) was found. Sj7170 has a precursor nucleotide sequence of 459 nucleotides including three parts as follows: 5′-untranslated region (UTR), open reading frame (ORF), and 3′-UTR. The lengths of 5′- and 3′-UTR parts are 95 and 96 nucleotides long, respectively. The ORF region of 261 nucleotides encodes a precursor polypeptide of 86 amino acid residues, including a 24-residue signal peptide and a 62-residue mature peptide, with the unique feature of 10 cysteine residues (Fig. 1A).
FIGURE 1.
Sequence analysis of Sj7170 from the scorpion S. jendeki. A, predicted protein sequence is shown below the nucleotide sequence. The 5′- and 3′-UTR regions are written in lowercase letters. The signal peptide is not underlined; the mature peptide is underlined, and the cysteine residues are highlighted in red color. B, sequence alignment of Sj7170 and other Ascaris-type peptides. Identical and similar residues are highlighted by different colors. Predicted disulfide connections are shown with lines. C, minimum evolution tree of representative Ascaris-type proteins based on the multiple sequence alignment.
Multiple sequence alignments showed that the mature Sj7170 peptide shares homology with typical Ascaris-type venom peptides, including SjAPI from S. jendeki (8), AMCI-1 from Apis mellifera (9), AsC/E-1 from Ascaris suum (10), ATI from Ascaris lumbricoides var. suum (11), BSTI from Bombina bombina (12), and isoinhibitors 2–5 isolated from A. lumbricoide (Fig. 1B) (13). The mature Sj7170 peptide possesses the classical Ascaris-type cysteine framework reticulated by five disulfide bridges, which is conserved in classical Ascaris-type peptides but significantly different from all known protease inhibitors from venomous animals. Phylogenetic tree analysis indicated that seven Ascaris-type peptides (AsC/E-1, isoinhibitors 2–5, ATI, AMCI-1, Sj7170, SjAPI, and BSTI) were assigned to a single group among the representative Ascaris-type peptides, whereas a membrane-associated Kunitz-type serine protease inhibitor HAI-1 from human belongs to another group (Fig. 1C), suggesting similarities between peptide toxins from animal venom glands.
Expression and Purification of rSj7170
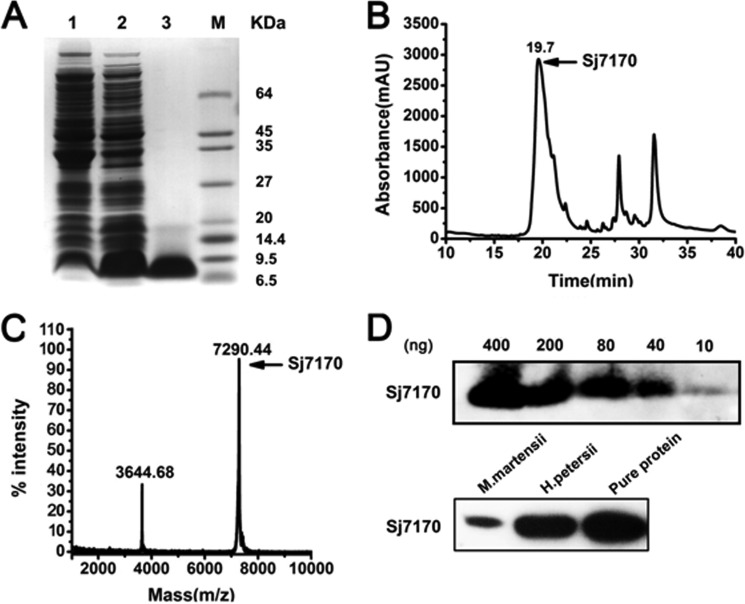
The nucleotide sequence encoding mature Sj7170 peptide was inserted the NcoI-XhoI cut of the vector pET-28a (+) to generate the recombinant expression vector pET-28a/Sj7170. After induction of the E. coli Rosetta (DE3) cell culture with IPTG, the rSj7170 peptide was found exclusively in inclusion bodies. Using a refolding protocol described under “Experimental Procedures,” soluble rSj7170 peptide was recovered (Fig. 2A). After concentration, the soluble material was separated by reverse phase (RP)-HPLC. The peak eluting at 19.5 min corresponding to rSj7170 peptide was collected (Fig. 2B) and identified by MALDI-TOF-MS. MALDI-TOF-MS showed a doubly charged ion at m/z 3644.68 and a singly charged ion at m/z 7290.44, all corresponding to the same peptide with an average mass of 7241.51 Da, consistent with the calculated value (Fig. 2C). Both MALDI-TOF-MS and SDS-PAGE analyses showed that rSj7170 was expressed and purified successfully. Subsequently, the purified rSj7170 peptide was quantified by the BCA protein assay kit (Thermo Fisher Scientific) and stored at −20 °C after freeze-drying. The rSj7170 peptide yield was 6 mg/ml LB media. We prepared the polyclonal antibody of Sj7170 in Beijing Perfect Biotechnology, Ltd. (China). The titer of antibody was tested by Western blot (Fig. 2D) because the numbers of scorpion S. jendeki are very scarce. So the venom of S. jendeki is also too hard to obtain. Thus, we chose other two homologous species Mesobuthus martensii and Heterometrus petersii, which were obtained easily. We used Western blot to detect whether Sj7170 or homolog existed in the venom of these two species of scorpion. The results are shown in Fig. 2D; Sj7170 peptide actually exists in scorpion venom.
FIGURE 2.
Expression and purification of the recombinant Sj7170 peptide. A, Tricine-SDS-PAGE analysis of the recombinant Sj7170 peptide. Lanes 1 and 2 are the pellet fractions of non-IPTG-induced and IPTG-induced Rosetta (DE3) cells containing the expression plasmid pET-28a-Sj7170, respectively. Lane 3 is refolded Sj7170 after desalting and enrichment and HPLC. B, purification of the recombinant Sj7170 by RP-HPLC. The fractions containing Sj7170 are indicated by arrows. C, MALDI-TOF-MS mass spectrum of the recombinant Sj7170. D, detection of the titer of Sj7170 antibody and the expression of Sj7170 in the venom of two species of scorpion venom. M, mass.
Serine Protease Inhibitory Activity of Sj7170 in Vitro and in Vivo
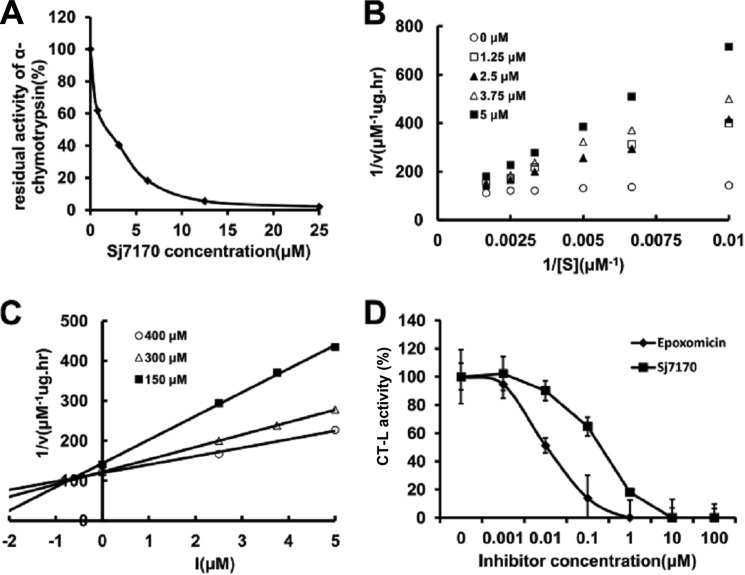
The purified rSj7170 peptide was assayed for inhibitory activity against chymotrypsin, trypsin, and elastase by measuring the inhibition of hydrolysis of synthetic chromogenic substrates by serine proteases. The results showed that the rSj7170 peptide inhibited α-chymotrypsin with a 1:1 stoichiometric ratio (Fig. 3A), but it exhibited no inhibitory effect on trypsin and elastase even at a high concentration (data not shown). Furthermore, the inhibitory constant (Ki) of the chymotrypsin-rSj7170 complex was determined by Lineweaver-Burk plots and further slope replotting, yielding a Ki value of 1.0 × 10−7 m (Fig. 3, B and C).
FIGURE 3.
Inhibitory activity of rSj7170 against α-chymotrypsin. A, concentration-dependence curve of rSj7170 inhibitory activity on α-chymotrypsin in vitro. α-Chymotrypsin (final concentration 100 nm) was incubated with different concentrations of rSj7170 for 30 min. B, Lineweaver-Burk plots of chymotrypsin reaction rate reciprocal (1/V) to different concentrations of substrate reciprocal (1/[S]) in the different concentrations of 1.25, 2.5, 3.75, and 5 μm Sj7170, respectively. C, Dixon plots of chymotrypsin reaction rate reciprocal (1/V) to different concentrations of rSj7170 ([I]) in the different concentrations of 150, 300, and 400 μm substrate, respectively. The inhibitory constant (Ki) was determined from the intercept point on the x axis. D, effect of rSj7170 on the cell-based chymotrypsin-like activity of U87. U87 cells were treated individually over a period of 2 h with epoxomicin or rSj7170 at different concentrations. The natural product epoxomicin was used as the positive control. Data normalized to controls represent the mean of three independent experiments, and each experiment was performed in duplicate (n = 3). Bars show the standard deviation.
To investigate further the chymotrypsin-like activity of rSj7170 in vivo, we tested the effectiveness of rSj7170 in a cell culture-based system. As shown in Fig. 3D, rSj7170 inhibited the chymotrypsin-like activity in a dose-dependent manner. Epoxomicin as a positive control showed a stronger inhibitory effect, compared with rSj7170. These results suggest the conclusion that rSj7170 was confirmed to be a potent α-chymotrypsin inhibitor in vitro and in vivo.
Sj7170 Promotes Cell Proliferation and Colony Formation
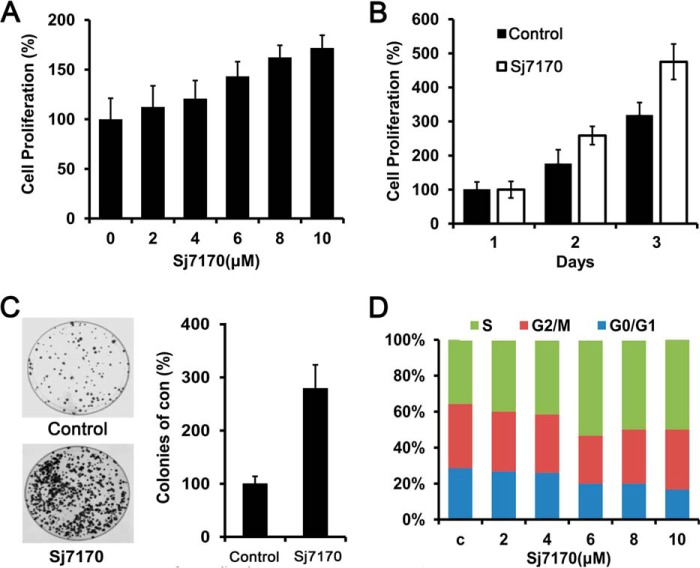
To investigate the cellular effects of Sj7170 on glioma U87 cells, we treated U87 cells using dose- and time-dependent assays. U87 cells were treated with rSj7170 at various concentrations (0, 2, 4, 6, 8, and 10 μm) for 48 h and with 10 μm for various timings (24, 48, and 72 h). The results showed that rSj7170 significantly increased cell proliferative capacity in a dose- and time-dependent manner, compared with control cells (Fig. 4, A and B). Furthermore, in vitro colony formation assays demonstrated that the frequencies of colony formation of the rSj7170-treated U87 cells are markedly higher than those of control cells (p < 0.01; Fig. 4C). To further probe the molecular mechanism of its proliferative effect on U87 cells, cell cycle analysis was performed. The results indicated that the treatment of rSj7170 peptide resulted in an increased S phase population and a concomitantly decreased G1 population (Fig. 4D).
FIGURE 4.
Effects of Sj7170 on cell proliferation and colony formation in vitro. A, dose-dependent effect of Sj7170 on the proliferation of U87 cells. Results are means ± S.D. B, time-dependent effect of Sj7170 on the proliferation of U87 cells. Results are means ± S.D. C, effect of Sj7170 on tumor cell growth by monolayer colony formation assay. Left panel, representative plates for the colony formation assay. Right panel, statistical results. D, cell cycle analysis of U87 treated by Sj7170 using flow cytometry. U87 cells were treated with different concentrations of Sj7170 for 48 h. c, control.
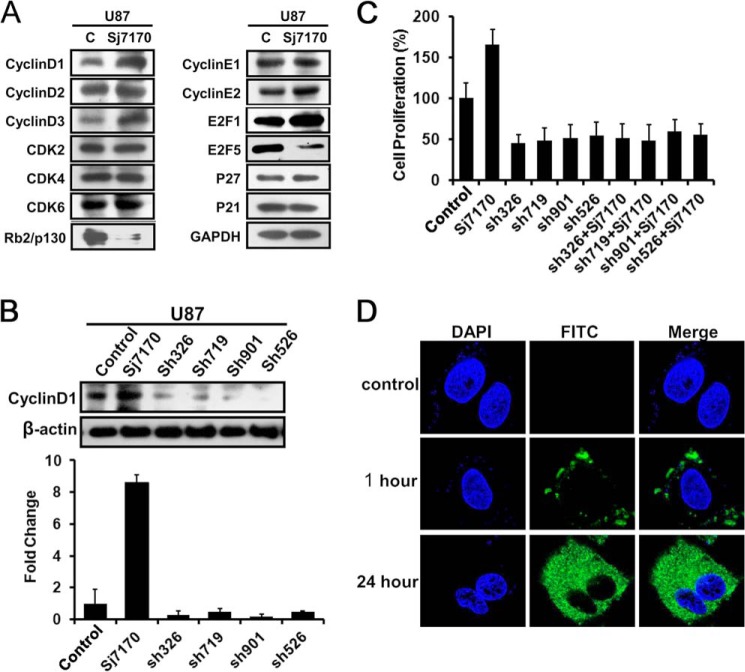
Sj7170 Promotes U87 Cell Proliferation by Up-regulating cyclin D1 through Rb/E2F Signaling
To explore the mechanism of the effect of Sj7170 on the cell cycle, we detected the expression levels of cell cycle-related proteins by Western blot analysis, including cyclin D1, cyclin D2, cyclin E1, cyclin E2, CDK2, CDK4, CDK6, Rb2/p130, E2F1, E2F5, P21, and P27 (Fig. 5A). The results revealed that the expression levels of cyclin D1 and E2F1 were significantly increased in rSj7170-treated U87 cells. Consistently, the expression levels of Rb2/p130 and E2F5 in U87 cells were reduced by rSj7170. As is well known, the activity of cyclin D1 is required for cell cycle G1/S transition. This protein has been shown to interact with tumor suppressor protein Rb, and its expression is regulated positively by Rb. The growth inhibitory activity of Rb2/p130 also correlated with its E2F-binding capacity. E2F is an activator, but E2F5 is an inhibitor (14). The variations of expression levels of cyclin D1, E2F1, Rb2/p130, and E2F5 were consistent with the results of cell cycle analysis. These results indicate that Sj7170 promotes U87 cells proliferation and G1/S phase transition by cyclin D1-Rb-E2F signaling.
FIGURE 5.
Effective mechanism of Sj7170 on the cell proliferation of U87 cells. A, Western blot analysis of cyclin D1, cyclin D2, cyclin D3, CDK2, CDK4, CDK6, cyclin E1, cyclin E2, Rb2/p130, E2F1, E2F5, p21, and p27 in control and Sj7170-treated U87 cells. c, control. B, inhibition of cyclin D1 expression by cyclin D1 shRNAs in U87 cells. Real time PCR and Western blot were used to examine the mRNA and protein expression levels of cyclin D1. C, effects of Sj7170 on cell proliferation in U87 cells by knockdown of cyclin D1. D, subcellular localization of Sj7170 in U87 after 1 and 24 h of incubation. Immunofluorescence results of Sj7170 in U87 cells are shown. Blue indicates DAPI nuclear staining. Green fluorescence was plotted in the cytoplasm as location of Sj7170 in U87 cells.
To determine whether Sj7170 targeted to cyclin D1 protein, four different GenePharma SupersilencingTM cyclin D1 shRNAs (pGPU6/GFP/Neo-cyclin D1–326, -719, -901, and -526) were employed to down-regulate the cyclin D1 expression as a validation. Real time PCR and Western blot analysis showed that all the four shRNAs led to a significant decrease in mRNA and protein levels of cyclin D1 (Fig. 5B). rSj7170 (10 μm) was used to treat U87 cells whose cyclin D1 was silenced, and the cell numbers were counted. As shown in Fig. 5C, rSj7170 did not induce the proliferation of U87 cells after knockdown of cyclin D1. These results suggest that Sj7170 promotes U87 cells proliferation by up-regulating cyclin D1 through Rb/E2F signaling. Cyclin D1 is in fact a predominantly cytoplasmic protein in mammalian cancer cell lines. Cytoplasmic sequestration may additionally serve to regulate cyclin D1 activity. Immunofluorescence results revealed that Sj7170 entered into cells and located in the cytoplasm (Fig. 5D). So the subcellular localizations of both cyclin D1 and Sj7170 were already in the cytoplasm. These data suggested cyclin D1 as a crucial target is a response gene for Sj7170 that drives the G1/S progression of U87 cells.
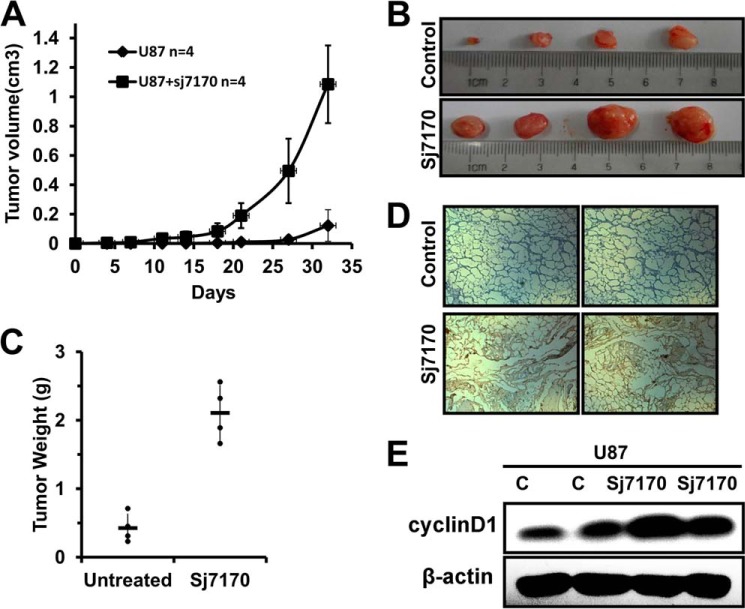
Sj7170 Promotes Tumor Growth in Vivo Tumor Xenograft Model
To further investigate the effect of Sj7170 on the progression of tumors, we detected the proliferated activity of rSj7170 against U87 xenografted tumor in vivo. Control U87 cells (n = 4) and rSj7170-treated U87 cells (n = 4) were used to inject mice, respectively. The growth of tumors was monitored every 3 days, and tumors were excised and weighed 32 days after injection. As shown in Fig. 6A, the tumor volumes significantly increased during the 5-week follow-up period, suggesting that rSj7170 effectively promotes the growth of U87 tumor engraftments. After 32 days, tumor growth was completely enhanced in each mouse (100%) inoculated with rSj7170-treated U87 cells (Fig. 6B). The average tumor weights of the untreated and rSj7170-treated mice were 0.4 ± 0.21 and 2.1 ± 0.4 g, respectively (Fig. 6C). After acquired specimens of U87 xenografts, IHC analysis of samples, including both the control group and the rSj7170-treated group, revealed a higher expression level of cyclin D1 in the rSj7170-treated group, compared with the untreated group (Fig. 6D). Consistently with the results of cell proliferation in vitro, cyclin D1 also plays an important role in tumorigenesis in vivo. Furthermore, we obtained tumor tissue to detect the cyclin D1 expression by Western blot analysis. The result is shown in Fig. 6E. Consistently with the IHC analysis, the rSj7170-treated group revealed a higher expression level of cyclin D1, compared with the untreated group. All these results demonstrate the tumor-activated function of Sj7170 in U87 cells.
FIGURE 6.
Effect of Sj7170 on tumor growth in vivo. A, tumor volume curve of nude mice treated by Sj7170. After tumor cells were injected subcutaneously into male athymic nude mice, the short and long diameters of the tumors were measured every 3 days, and tumor volumes (cm3) were calculated. Tumor-bearing mice were then sacrificed after 32 days. B, photograph of representative tumors derived from control and Sj7170-treated cells in nude mice. C, comparison of tumor weights of nude mice treated and not treated by Sj7170. D, representative immunohistochemistry image of cyclin D1 expression in two xenograft tumor tissues. E, cyclin D1 expression of tumor tissue in both Sj7170-treated and -untreated groups. Western blot was used to detect the expression of cyclin D1. Lanes c and c represent the control group.
Sj7170 Enhances Cell Migration and Invasion of U87 by EMT
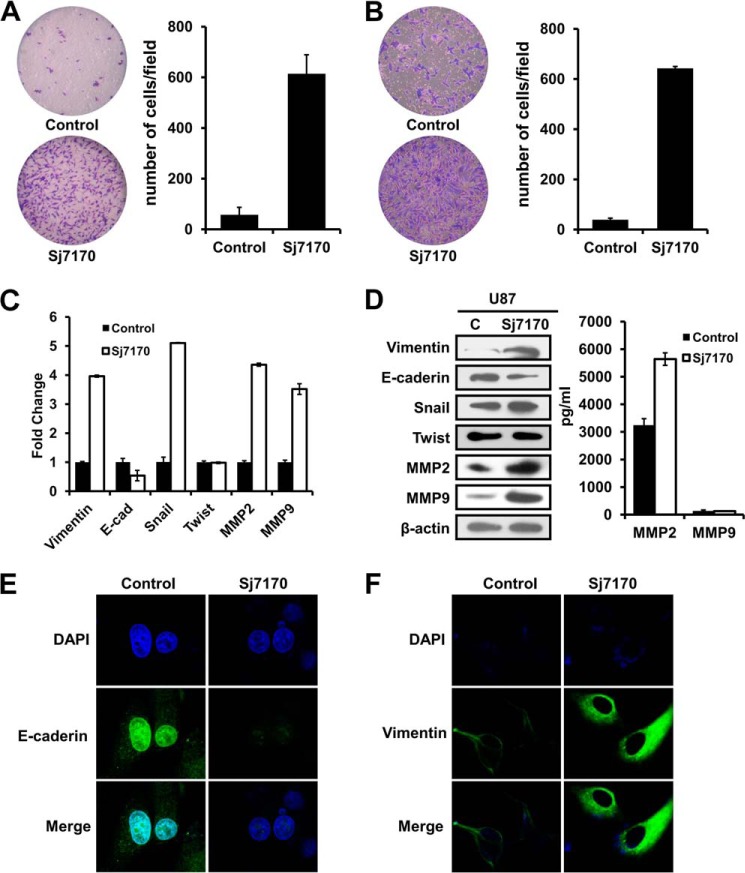
To determine the role of Sj7170 in metastasis, we examined cell migration using a transwell assay chamber with PET track-etched membranes. Cells that migrated toward the bottom side of the membrane were fixed and stained with crystal violet. The results showed that rSj7170 resulted in markedly increased cellular motility, as the numbers of migrating rSj7170-treated cells were significantly higher than those of the untreated cells (Fig. 7A). In addition, we examined the cellular invasive activity of rSj7170 using invasion chamber assays with a Matrigel basement membrane matrix. Consistent with the migration results, rSj7170 significantly enhanced cell invasion capacity by ∼20-fold compared with the control cells (p < 0.01; Fig. 7B). Taken together, these results indicate that Sj7170 promotes cell metastasis.
FIGURE 7.
Effects of Sj7170 on migration and invasion of U87 cells. A and B, transwell migration and invasion assay. Left panel, representative fields of migration and invasion cells on the membrane (magnification ×40). Right panel, average migration and invasion cell number per field. Migration and invasiveness were determined by counting cells in six randomly selected microscopic fields per well. C, real time-PCR analysis of the EMT markers E-cadherin, vimentin, Snail, Twist, MMP2, and MMP9. D, Western blot analysis of the EMT markers E-cadherin, vimentin, Snail, Twist, MMP2 and MMP9. C, control. E and F, immunofluorescence analysis of E-cadherin and vimentin. The expression level/patterns of the epithelial and mesenchymal markers in the control and Sj7170-treated U87 cells were determined by immunofluorescence staining.
EMT is an important mechanism associated with cancer invasiveness and metastasis (15). To examine whether the Sj7170-induced cell migration and invasion were associated with EMT, we first assessed the status of epithelial and mesenchymal cell markers by real time PCR, Western blot, and immunofluorescence analyses. As shown in Fig. 7, C and D, mRNA and protein levels of the epithelial cell markers E-cadherin were down-regulated in the rSj7170-treated U87 cells. Correspondingly, the control U87 cells expressed low levels of the mesenchymal cell markers vimentin and Snail. Transcription factor Twist expression had no change. Cell metastasis-related proteins MMP2 and MMP9 also were up-regulated by Sj7170. Consistent with the results of real time PCR and Western blot, the down-regulation of E-cadherin and up-regulation of vimentin were also observed by immunofluorescence analysis in the rSj7170-treated U87 cells (Fig. 7, E and F). In particular, the vimentin intermediate filaments localized in a concentrated and polarized pattern in rSj7170-treated U87 cells.
Sj7170 Increases Cell Motility by Up-regulating Snail
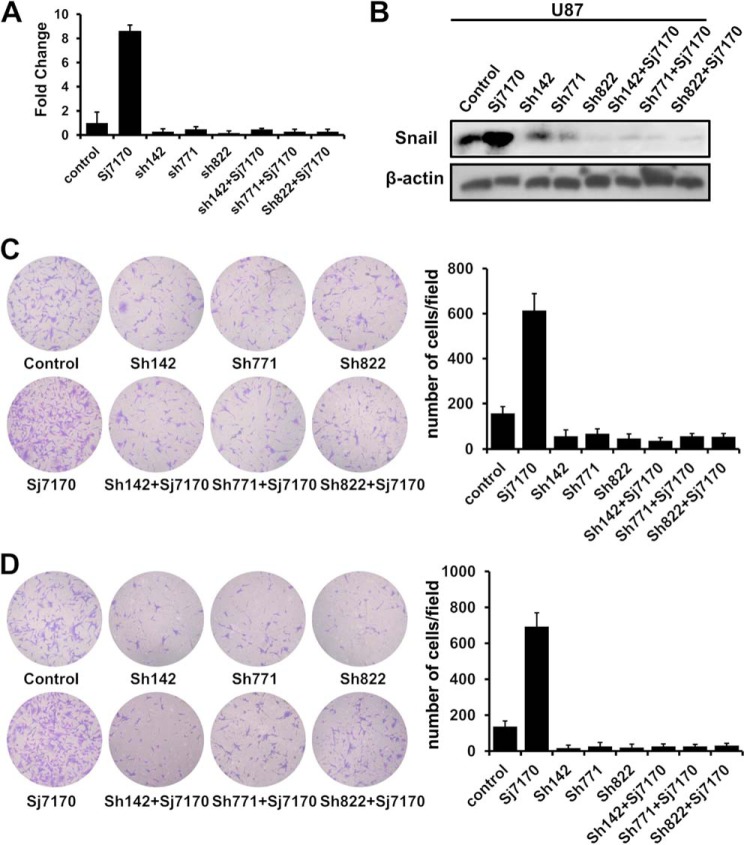
To further confirm the relationship between cell motility and EMT, three different GenePharma SupersilencingTM Snail shRNAs (pGPU6/GFP/Neo-Snail-142, -771, and -822) were employed to down-regulate the Snail expression as a validation. Real time PCR and Western blot analysis showed that all the four shRNAs led to a significant decrease in mRNA and protein levels of Snail (Fig. 8, A and B). rSj7170 (10 μm) was used to treat U87 cells whose Snail was silenced, and the cell migration and invasion were assayed. As shown in Fig. 8, C and D, rSj7170 did not induce the motility of U87 cells after knockdown of Snail. In other words, when EMT progress was inhibited, Sj7170 lost the ability to enhance cell motility. Therefore, we conclude that Sj7170 promotes cell migration and invasion by inducing EMT through increasing the expression of Snail.
FIGURE 8.
Effective mechanism of Sj7170 on the metastasis of U87 cells. A, inhibition of Snail mRNA expression by Snail shRNAs in U87 cells. B, inhibition of Snail protein expression by Snail shRNAs in U87 cells. C and D, effects of Sj7170 on cell motility of U87 cells with Snail knockdown. Transwell migration and invasion assay was used to detect whether Sj7170 increases cell motility after down-regulation of Snail in U87 cells.
Sj7170 Leads to Morphological Changes and Actin Cytoskeleton Rearrangement by Perturbing Rho GTPase Signaling
Our initial observations suggested that Sj7170 induced morphological changes in U87 cells. Untreated U87 cells were relatively small, morphologically heterogeneous, and loosely attached, with many cells eventually rounding up. In contrast, rSj7170-treated U87 cells were significantly larger, more homogeneous, firmly attached, and well spread (Fig. 9A). In addition, rSj7170 decreased the tumor sphere number of U87 cells in a dose-dependent manner (Fig. 9B). These results suggested that Sj7170 induced a profound cytoskeletal rearrangement. Therefore, we evaluated the organization of the actin cytoskeleton of rSj7170-treated and -untreated U87 cells. F-actin staining indicated that polymerized actin was mostly located within the cell periphery in rSj7170-treated U87 cells, which were largely devoid of actin stress fibers (Fig. 9C). Small GTPases of the Rho-GTPase family (RhoA, Rac1, and Cdc42) are known to act directly on the cytoskeleton and are responsible for the development of membrane ruffles, stress fibers, lamellipodia, and filopodia (16). In this study, the protein levels of RhoA, Rac1, and Cdc42 were significantly increased in rSj7170-treated U87 cells, compared with the control U87 cells (Fig. 9D). These results indicate that Sj7170 promotes the rearrangement of actin cytoskeleton through the modulation of Rho GTPase signaling.
FIGURE 9.
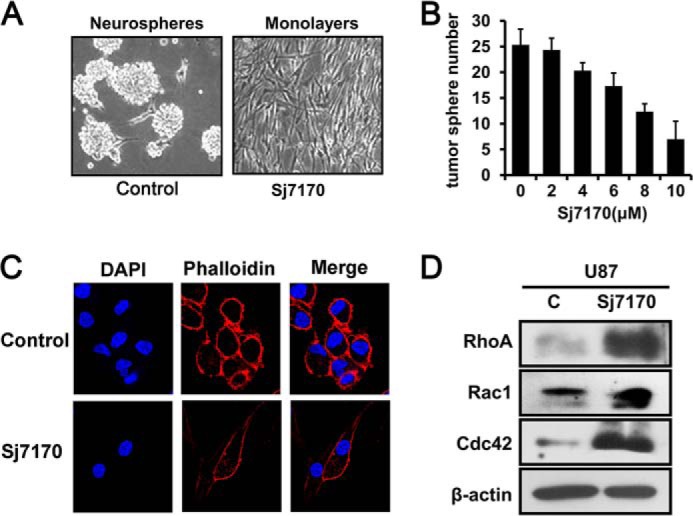
Effects of Sj7170 on morphology and actin cytoskeleton reorganization of U87 cells. A, representative phase-contrast micrographs of U87 cells left untreated (control) or treated for 72 h with 10 μm Sj7170. B, dose-dependent effect of Sj7170 on tumor sphere number in U87 cells. C, distribution of F-actin in U87 cells. U87 cells grown on coverslips were upside untreated (control) or were treated for 24 h with 10 μm Sj7170, fixed, and stained with TRITC-labeled phalloidin. Scale bars, 10 μm. D, Western blot analysis of the Rho GTPase family RhoA, Rac1, and Cdc42 in 10 μm Sj7170-treated U87 cells compared with control (c) U87 cells.
DISCUSSION
Despite the progress in understanding carcinogenesis, cancer is still a major cause of morbidity and mortality worldwide. The development of novel therapies based on biologically active peptides and protein has emerged as a new strategy to defeat cancer. Venom-producing animals are usually known solely for the negative effects they cause after accidental contact with humans. They carry a variety of toxins with different physiological activities that cause mild symptoms. Even though the effects of the envenomation might lead to a negative reputation, these animals are also seen as a rich source of pharmacologically active principles, and many of their toxins have been the subject of research projects aiming at the development of new molecules for the diagnosis, treatment, and cure of some types of disease (17). Among the animals that produce pharmacologically active molecules capable of interfering in human cellular physiology, the highlights are venomous animals, such as scorpions, bees, wasps, spiders, snakes, and conus. The substances found in the venom of these animals present great potential as anti-tumor or pro-tumor agents (18–22).
Venom from these animals may hold the promise for curing many types of malignancies, especially after analyzing the results from studies that show a complete remission of tumor cells after treatment with molecules derived from animal venom. However, studies focusing on the mechanism by which these venoms act are still very recent, and much has yet to be found out about these molecules. The first clinical trials against cancer using synthetic peptides derived from animal venom (called Capoten, copied from a pit viper venom peptide) are beginning to show results. As more positive results are obtained, researchers and patients find reasons to believe that these small substances found in nature may have extraordinary applications.
Developing venoms to kill or paralyze prey provides an important means for venomous animals to interact with their environment. Under the natural selection pressure, venomous animals strive to construct more efficient toxins so as to be evolutionarily successful. It was hypothesized that the evolutionary process of the animal venom system includes a series of important events, including recruitment of existing ancestor genes, gene duplications, and mutations, eventually forming diverse types of toxins. Peptide protease inhibitors are considered to be the earliest protein toxins recruited from fluid protein (23). Several protease inhibitors have been isolated from venomous animals, mainly sea anemones, snakes, and anurans. Nevertheless, the characterization of protease inhibitors from scorpions, spiders, and Hymenopterans has recently attracted increasing interest from the scientific community. Protease inhibitors can be primarily grouped as serine, cysteine, aspartic, and metalloprotease inhibitors because of targeting the four major protease classes.
Many known protease inhibitors from venomous animals are characterized as the group of SPIs, the largest and most widely distributed superfamily of protease inhibitors. Based on their possession of conserved functional motifs, SPIs can be subdivided in many classes, in which the Kunitz-type inhibitor class was the best characterized. Most of SPIs discovered thus far from animal venom belong to the Kunitz-type class, such as AEAPI and AXAPI from sea anemone, dendrotoxin E from snake, BSTI from anuran, HWTX-XI from spider, As-fr-19 from Hymenopteran, and Hg1 from scorpion.
In this study, a new venom peptide, termed Sj7170, was characterized from the venom of the scorpion S. jendeki. Sj7170 shares high homology with the known Ascaris-type peptides from other phyla and possesses the same cysteine framework stabilized by five disulfide bridges as them. Based on a protease inhibition assay in vitro and in vivo, Sj7170 was confirmed to be a potent α-chymotrypsin inhibitor. These results suggest the idea that Sj7170 from the venom of the scorpion S. jendeki is a typical Ascaris-type serine protease inhibitor. Thus, this work enriches the member and type of SPIs from animal venoms.
Ascaris-type peptides constitute an important class of SPIs. But compared with other types of serine protease inhibitors, only 20 Ascaris-type peptides have been characterized from animals and fewer Ascaris-type peptide inhibitors have been reported from venomous animals. To date, the first identified Ascaris-type scorpion toxin is SjAPI with α-chymotrypsin and elastase inhibitory activities from S. jendeki (8). Although both Sj7170 and SjAPI are characterized from the venom of the scorpion S. jendeki, Sj7170 has a specific inhibitory activity against α-chymotrypsin, different from SjAPI with α-chymotrypsin and elastase inhibitory activities.
The Ascaris-type peptide was almost isolated from the parasitic worm A. suum and has been implicated in the survival of the parasite within the host by inhibiting the activities of exogenous host proteases (13). In a different way, Ascaris-type SPIs from animal venoms were deduced to inhibit the proteases of their preys or predators, thereby protecting other toxic peptides to avoid degradation and inactivation. Almost all Ascaris-type SPIs have a common structural characteristic with four short β-strands arranged in two approximately perpendicular β-sheets and stabilized by five disulfide bridges: Cy- I–Cys-VII, Cys-II–Cys-VI, Cys-III–Cys-V, Cys-IV–Cys-X, and Cys-VIII–Cys-IX (24). Consistent with the previous results, Sj7170 shares the similar structural characteristic with Ascaris-type SPIs.5
Tumor progression is generally associated with extensive tissue remodeling to provide a proper environment for tumor growth, angiogenesis, invasion, and metastasis of cancer cells. An impressive amount of data reveals that, among many factors, proteases expressed by cancer cells are key players in this process. Indeed, because of their abilities to activate and release cytokines and growth factors and to degrade components of the extracellular matrix, proteases are necessary to provide optimal conditions for growth and invasion of cancer and endothelial cells.
Because protease inhibitors can inhibit these proteases, they are therefore considered to counteract tumor progression and metastasis. Moreover, there is growing evidence that serine protease inhibitors may even inhibit the malignancy of cancer cells.
Conversely, SPIs are often overexpressed in different tumor types, suggesting that overexpression of these inhibitors might favor tumor progression. Indeed, a number of SPIs from Serpin and Kunitz families have been reported to result in enhancement of cancer cell malignancy. For example, serpinE2, a serine protease inhibitor, is a novel target of ERK signaling involved in human colorectal tumorigenesis. Other oncogenic pathways have been associated with induction of serpinE2 expression. MET and PTEN deletion can up-regulate the expression of serpinE2 (25). SPINK1, a Kazal type 1 serine protease inhibitor, acts as a growth factor involved in cancer progression and local invasion through EGF receptor signaling and its downstream signaling pathway MAPK/ERK (26). After the discovery of the Sj7170 peptide, our pursuit of the potential underlying molecular mechanisms of Sj7170 in tumor progression revealed a significant divergence from other SPIs. In in vitro and in vivo proliferations, cyclin D1 is co-localized in the cytoplasm of U87 cells with Sj7170 and acts as a crucial target for Sj7170 that promotes the proliferation of U87 cells and tumor growth in nude mice. Previously, none of the Ascaris-type and endogenous SPIs have yet been shown to promote malignancy of cancer cells; the discovery of Sj7170 expends the diversity of tumorigenesis/metastasis activators among SPIs, which possibly helps to point out the functional evolution of SPIs with tumorigenesis or metastasis activation.
The majority of cancer-related deaths result from tumor metastasis, rather than from effects of the primary tumors. Tumor metastasis includes a series of biological processes that move tumor cells from the primary neoplasm to the distal metastatic nodules and acquires the ability to proteolytically remodel the extracellular matrix to coordinate cell adhesion and contractility. EMT was originally identified as a crucial differentiation and morphogenetic process during embryogenesis; it has now been found to be important in cancer progression and metastasis (27). Many molecular mechanisms contribute to EMT progress. These mechanisms usually involve growth factors/cytokines, their cognate receptors, and downstream signaling molecules. Protease activities have been implicated in EMT by activating growth factors and their receptors and cleaving cellular adhesion molecules (28). Recently, in addition to MMPs, the membrane-bound serine protease TMPRSS4 and the membrane-associated serine protease inhibitor HAI-1/SPINT1 were reported to initiate EMT (29, 30).
In our study, we show for the first time that exogenous serine protease inhibitor Sj7170 increases the capacity of migration and invasion of cancer cells by inducing EMT and up-regulating Snail expression. During the EMT, cell-cell junctions are disrupted, the actin cytoskeleton is extensively reorganized, and the cells acquire increased migratory characteristics. Interestingly, we also observed that Sj7170 alters the morphology and motility of glioma cell U87 by activating the Rho GTPase pathway. Altogether, cyclin D1 and Snail expression and Rho GTPase activity both have a significant regulatory role within the context of tumorigenesis and metastasis of tumor cells by inducing EMT. Mutants and other chymotrypsin inhibitor assays also demonstrate that tumorigenic activity of Sj7170 was not induced by protease inhibitory activity, and the two had no necessary associations (data not shown). All these results illustrate that Sj7170, as a serine protease inhibitor, is not just a growth factor but has more active roles in cancer development.
In summary, a new Ascaris-type serine protease inhibitor Sj7170 was identified from the venom of the scorpion S. jendeki. The recombinant Sj7170 peptide was expressed in a prokaryotic expression system and purified by HPLC and affinity chromatography. We found that rSj7170 played an important role in the tumorigenesis and metastasis of glioma U87 cells by accelerating cellular proliferation and colony formation in vitro and in vivo, significantly enhancing cell invasiveness by inducing EMT and changing cell morphology and cytoskeleton through the cyclin D1 and Rho GTPase pathway. Furthermore, Sj7170 has functions in promoting cell proliferation and enhancing cell migration and invasion independent of anti-protease activity. There are no relations between the two functions. Thus, Sj7170, as a unique dual-function peptide, may be a pro-cancer agent or a tumor activator of glioblastoma. This study provides a new understanding of the natural peptide library of scorpion venom, supports new findings of tumorigenesis/metastasis modulators from serine protease inhibitors, and strengthens the functional link between serine protease inhibitors and tumorigenesis/metastasis activators, which help to further reveal the cancer development mechanism and provide a possible breakthrough of cancer treatment.
This work was supported by National Key Basic Research Program Grants 2010CB529800 and 2010CB530100, China Specific Project for Developing New Drugs Grant 2011ZX09401-302, Fundamental Research Funds for the Central Universities in China and the National Nature Science Foundation of China Grant 81273540.
Y. Song, K. Gong, H. Yan, W. Hong, L. Wang, Y. Wu, W. Li, W. Li and Z. Cao, unpublished data.
- SPI
- serine protease inhibitor
- IPTG
- isopropyl β-d-thiogalactoside
- EMT
- epithelial-mesenchymal transition
- IHC
- immunohistochemistry
- rSj7170
- recombinant Sj7170 peptide
- TRITC
- tetramethylrhodamine isothiocyanate
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. DeAngelis L. M. (2001) Brain tumors. N. Engl. J. Med. 344, 114–123 [DOI] [PubMed] [Google Scholar]
- 2. Lee H. C., Park I. C., Park M. J., An S., Woo S. H., Jin H. O., Chung H. Y., Lee S. J., Gwak H. S., Hong Y. J., Yoo D. H., Rhee C. H., Hong S. I. (2005) Sulindac and its metabolites inhibit invasion of glioblastoma cells via down-regulation of Akt/PKB and MMP-2. J. Cell. Biochem. 94, 597–610 [DOI] [PubMed] [Google Scholar]
- 3. Silverman G. A., Bird P. I., Carrell R. W., Church F. C., Coughlin P. B., Gettins P. G., Irving J. A., Lomas D. A., Luke C. J., Moyer R. W., Pemberton P. A., Remold-O'Donnell E., Salvesen G. S., Travis J., Whisstock J. C. (2001) The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 276, 33293–33296 [DOI] [PubMed] [Google Scholar]
- 4. Gessler F., Voss V., Seifert V., Gerlach R., Kögel D. (2011) Knockdown of TFPI-2 promotes migration and invasion of glioma cells. Neurosci. Lett. 497, 49–54 [DOI] [PubMed] [Google Scholar]
- 5. Kataoka H., Itoh H., Koono M. (2002) Emerging multifunctional aspects of cellular serine proteinase inhibitors in tumor progression and tissue regeneration. Pathol. Int. 52, 89–102 [DOI] [PubMed] [Google Scholar]
- 6. Ma Y., He Y., Zhao R., Wu Y., Li W., Cao Z. (2012) Extreme diversity of scorpion venom peptides and proteins revealed by transcriptomic analysis: implication for proteome evolution of scorpion venom arsenal. J. Proteomics 75, 1563–1576 [DOI] [PubMed] [Google Scholar]
- 7. Zeng X. C., Li W. X., Zhu S. Y., Peng F., Jiang D. H., Yang F. H., Wu K. L. (2000) Cloning and characterization of the cDNA sequences of two venom peptides from Chinese scorpion Buthus martensii Karsch (BmK). Toxicon 38, 893–899 [DOI] [PubMed] [Google Scholar]
- 8. Chen Z., Wang B., Hu J., Yang W., Cao Z., Zhuo R., Li W., Wu Y. (2013) SjAPI, the first functionally characterized Ascaris-type protease inhibitor from animal venoms. PLoS One 8, e57529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cierpicki T., Bania J., Otlewski J. (2000) NMR solution structure of Apis mellifera chymotrypsin/cathepsin G inhibitor-1 (AMCI-1): structural similarity with Ascaris protease inhibitors. Protein Sci. 9, 976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang K., Strynadka N. C., Bernard V. D., Peanasky R. J., James M. N. (1994) The molecular structure of the complex of Ascaris chymotrypsin/elastase inhibitor with porcine elastase. Structure 2, 679–689 [DOI] [PubMed] [Google Scholar]
- 11. Grasberger B. L., Clore G. M., Gronenborn A. M. (1994) High-resolution structure of Ascaris trypsin inhibitor in solution: direct evidence for a pH-induced conformational transition in the reactive site. Structure 2, 669–678 [DOI] [PubMed] [Google Scholar]
- 12. Mignogna G., Pascarella S., Wechselberger C., Hinterleitner C., Mollay C., Amiconi G., Barra D., Kreil G. (1996) BSTI, a trypsin inhibitor from skin secretions of Bombina bombina related to protease inhibitors of nematodes. Protein Sci. 5, 357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Babin D. R., Peanasky R. J., Goos S. M. (1984) The isoinhibitors of chymotrypsin/elastase from Ascaris lumbricoides: the primary structure. Arch. Biochem. Biophys. 232, 143–161 [DOI] [PubMed] [Google Scholar]
- 14. Dowdy S. F., Hinds P. W., Louie K., Reed S. I., Arnold A., Weinberg R. A. (1993) Physical interaction of the retinoblastoma protein with human D cyclins. Cell 73, 499–511 [DOI] [PubMed] [Google Scholar]
- 15. Lamouille S., Connolly E., Smyth J. W., Akhurst R. J., Derynck R. (2012) TGF-β-induced activation of mTOR complex 2 drives epithelial-mesenchymal transition and cell invasion. J. Cell Sci. 125, 1259–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goel A., Chhabra R., Ahmad S., Prasad A. K., Parmar V. S., Ghosh B., Saini N. (2012) DAMTC regulates cytoskeletal reorganization and cell motility in human lung adenocarcinoma cell line: an integrated proteomics and transcriptomics approach. Cell Death Dis. 3, e402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heinen T. E., da Veiga A. B. (2011) Arthropod venoms and cancer. Toxicon 57, 497–511 [DOI] [PubMed] [Google Scholar]
- 18. Jäger H., Dreker T., Buck A., Giehl K., Gress T., Grissmer S. (2004) Blockage of intermediate-conductance Ca2+-activated K+ channels inhibit human pancreatic cancer cell growth in vitro. Mol. Pharmacol. 65, 630–638 [DOI] [PubMed] [Google Scholar]
- 19. Deshane J., Garner C. C., Sontheimer H. (2003) Chlorotoxin inhibits glioma cell invasion via matrix metalloproteinase-2. J. Biol. Chem. 278, 4135–4144 [DOI] [PubMed] [Google Scholar]
- 20. Mamelak A. N., Jacoby D. B. (2007) Targeted delivery of antitumoral therapy to glioma and other malignancies with synthetic chlorotoxin (TM-601). Expert Opin. Drug Deliv. 4, 175–186 [DOI] [PubMed] [Google Scholar]
- 21. Arbiser J. L., Kau T., Konar M., Narra K., Ramchandran R., Summers S. A., Vlahos C. J., Ye K., Perry B. N., Matter W., Fischl A., Cook J., Silver P. A., Bain J., Cohen P., Whitmire D., Furness S., Govindarajan B., Bowen J. P. (2007) Solenopsin, the alkaloidal component of the fire ant (Solenopsis invicta), is a naturally occurring inhibitor of phosphatidylinositol-3-kinase signaling and angiogenesis. Blood 109, 560–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saini S. S., Chopra A. K., Peterson J. W. (1999) Melittin activates endogenous phospholipase D during cytolysis of human monocytic leukemia cells. Toxicon 37, 1605–1619 [DOI] [PubMed] [Google Scholar]
- 23. Yuan C. H., He Q. Y., Peng K., Diao J. B., Jiang L. P., Tang X., Liang S. P. (2008) Discovery of a distinct superfamily of Kunitz-type toxin (KTT) from tarantulas. PLoS One 3, e3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gronenborn A. M., Nilges M., Peanasky R. J., Clore G. M. (1990) Sequential resonance assignment and secondary structure determination of the Ascaris trypsin inhibitor, a member of a novel class of proteinase inhibitors. Biochemistry 29, 183–189 [DOI] [PubMed] [Google Scholar]
- 25. Bergeron S., Lemieux E., Durand V., Cagnol S., Carrier J. C., Lussier J. G., Boucher M. J., Rivard N. (2010) The serine protease inhibitor serpinE2 is a novel target of ERK signaling involved in human colorectal tumorigenesis. Mol. Cancer 9, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ohmuraya M., Yamamura K. (2011) Roles of serine protease inhibitor Kazal type 1 (SPINK1) in pancreatic diseases. Exp. Anim. 60, 433–444 [DOI] [PubMed] [Google Scholar]
- 27. Cristofanilli M., Hayes D. F., Budd G. T., Ellis M. J., Stopeck A., Reuben J. M., Doyle G. V., Matera J., Allard W. J., Miller M. C., Fritsche H. A., Hortobagyi G. N., Terstappen L. W. (2005) Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J. Clin. Oncol. 23, 1420–1430 [DOI] [PubMed] [Google Scholar]
- 28. Wang Z., Li Y., Sarkar F. H. (2010) Signaling mechanism(s) of reactive oxygen species in epithelial-mesenchymal transition reminiscent of cancer stem cells in tumor progression. Curr. Stem. Cell Res. Ther. 5, 74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jung H., Lee K. P., Park S. J., Park J. H., Jang Y. S., Choi S. Y., Jung J. G., Jo K., Park D. Y., Yoon J. H., Park J. H., Lim D. S., Hong G. R., Choi C., Park Y. K., Lee J. W., Hong H. J., Kim S., Park Y. W. (2008) TMPRSS4 promotes invasion, migration and metastasis of human tumor cells by facilitating an epithelial-mesenchymal transition. Oncogene 27, 2635–2647 [DOI] [PubMed] [Google Scholar]
- 30. Cheng H., Fukushima T., Takahashi N., Tanaka H., Kataoka H. (2009) Hepatocyte growth factor activator inhibitor type 1 regulates epithelial to mesenchymal transition through membrane-bound serine proteinases. Cancer Res. 69, 1828–1835 [DOI] [PubMed] [Google Scholar]