Background: Reinitiation of translation is rare in eukaryotes and depends on specialized cis-acting RNA sequences tethering ribosomes to the reinitiation site.
Results: Ribosomes can reinitiate on calicivirus RNA at different sites.
Conclusion: Sites for translation reinitiation are selected either by ribosome positioning or a scanning-like process.
Significance: Start site selection in prokaryotic-like translation reinitiation in eukaryotes occurs in different modes.
Keywords: Gene Expression, Protein Expression, RNA Structure, RNA Viruses, Translation, Viral Protein, TURBS, Calicivirus, Norovirus, Reinitiation of Translation
Abstract
The calicivirus minor capsid protein VP2 is expressed via termination/reinitiation. This process depends on an upstream sequence element denoted termination upstream ribosomal binding site (TURBS). We have shown for feline calicivirus and rabbit hemorrhagic disease virus that the TURBS contains three sequence motifs essential for reinitiation. Motif 1 is conserved among caliciviruses and is complementary to a sequence in the 18 S rRNA leading to the model that hybridization between motif 1 and 18 S rRNA tethers the post-termination ribosome to the mRNA. Motif 2 and motif 2* are proposed to establish a secondary structure positioning the ribosome relative to the start site of the terminal ORF. Here, we analyzed human norovirus (huNV) sequences for the presence and importance of these motifs. The three motifs were identified by sequence analyses in the region upstream of the VP2 start site, and we showed that these motifs are essential for reinitiation of huNV VP2 translation. More detailed analyses revealed that the site of reinitiation is not fixed to a single codon and does not need to be an AUG, even though this codon is clearly preferred. Interestingly, we were able to show that reinitiation can occur at AUG codons downstream of the canonical start/stop site in huNV and feline calicivirus but not in rabbit hemorrhagic disease virus. Although reinitiation at the original start site is independent of the Kozak context, downstream initiation exhibits requirements for start site sequence context known for linear scanning. These analyses on start codon recognition give a more detailed insight into this fascinating mechanism of gene expression.
Introduction
Translation of mRNAs represents one of the basic processes in living organisms and is a prominent site of gene regulation that occurs at the step of translational initiation. Eukaryotic mRNAs are generally monocistronic, and the translational start site is usually located close to the 5′ end. Initiation of translation is conducted with the help of a set of initiation factors, first accomplishing binding of the 40 S ribosomal subunit to the cap structure, then supporting scanning of the preinitiation complex along the RNA to identify the translational start site that is the first AUG in most cases. After positioning of the AUG in the P-site of the 40 S subunit of the ribosome, the large subunit is bound to the ribosomal complex and translation starts (for scanning model see Refs. 1–3).
In eukaryotes, there is only a limited number of examples of RNAs using alternative initiation mechanisms or coding for more than one protein. However, such principles are quite commonly found in viral RNAs. Among a variety of alternative translation initiation mechanisms employed for expression of these mRNAs (4–18), reinitiation after termination is used and leads to expression of more than one protein from an RNA molecule (5, 19–26). The strategies used for protein translation in viruses ensure translation of their RNAs in competition with cellular RNAs and allow viruses to generate a range of proteins from a limited amount of genetic material. For special purposes, similar alternative initiation mechanisms are also used for cellular RNAs (27–32).
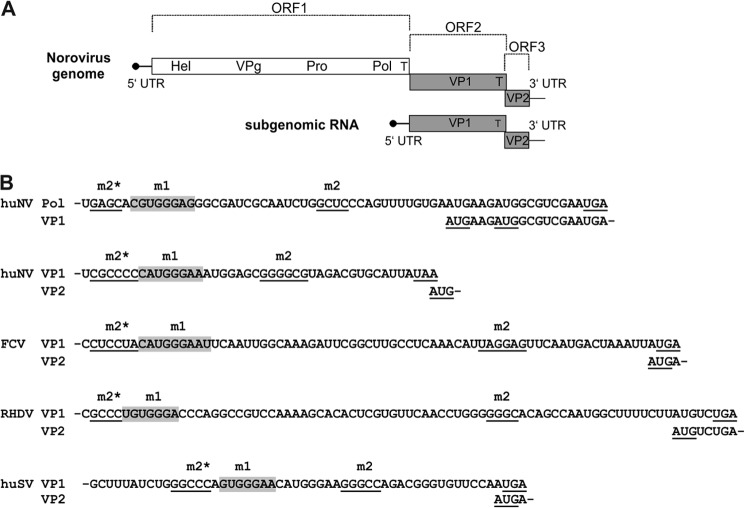
So far, the best studied case of translation reinitiation after translation of a long upstream ORF in higher eukaryotic cells was found in caliciviruses. The single strand positive sense RNA genome of caliciviruses is 7–8 kb long and contains two or three functional ORFs (33). ORF1 codes for the nonstructural proteins, which are translated as a polyprotein that is processed co- and post-translationally by a virus-encoded protease. A 3′-terminal, small ORF codes for the minor capsid protein VP2 with a size that varies among the caliciviruses from 8 to 20 kDa (34). The major capsid protein VP1 is encoded by a separate ORF2 in members of the genera Vesivirus and Norovirus or is part of the ORF1-encoded polyprotein in viruses belonging to the genera Lagovirus, Nebovirus, and Sapovirus. The genomic RNA serves as a template for the synthesis of the nonstructural polyprotein. A 3′-coterminal subgenomic mRNA is used for translation of at least the overwhelming amount of the major capsid protein VP1 and the minor capsid protein VP2. Both the full-length and the subgenomic RNAs do not carry a 5′ cap but are covalently linked to a viral protein (VPg). VPg serves as a primer for genome replication (35) and presumably functions as a cap substitute promoting translation of the 5′-terminal ORFs of the genomic and subgenomic viral RNAs (36–38). Previous studies on FCV3 (genus Vesivirus (23, 26, 39)), RHDV (genus Lagovirus (24, 25)), murine Norovirus (40), and bovine Norovirus (41) showed that the ribosomes gain access to the 3′-terminal frame via a translation termination/reinitiation process, leading to translation of VP2. As a special feature of the calicivirus genome organization, the frames coding for the two capsid proteins overlap by 1–14 nucleotides (nt) (Fig. 1).
FIGURE 1.
A, schematic map of Norovirus genomic and subgenomic RNA. In the scheme, the basic organization of the huNV genomic and subgenomic RNAs is illustrated (not drawn to scale). Gray bars represent ORF2 and ORF3 coding for the capsid. The white bar symbolizes the nonstructural protein coding region (ORF1). VPg (virus protein, genome-linked) present at the 5′ ends of both the genomic and subgenomic RNAs is symbolized by a black circle at the end of a line representing the 5′-nontranslated regions of the RNAs. The encoded proteins are indicated by the following abbreviations: Hel, putative helicase domain; VPg, virus protein, genome-linked; Pro, 3C-like protease; Pol, RNA-dependent RNA polymerase; VP1, major capsid protein; VP2, minor capsid protein; T, TURBS. B, TURBS regions of different caliciviruses. The sequences represent TURBS regions of members of four different genera of caliciviruses. Below the sequence of the upstream ORF containing the TURBS, the overlapping sequence corresponding to the downstream ORF is shown. The names of the proteins encoded in the two frames are given on the left side. The essential motifs identified by sequence analyses are highlighted. Motif 1 (m1) is marked in gray, and motif 2 (m2) and motif 2* (m2*) are underlined. The stop codon of the upstream frame and the start codon of the downstream frame are underlined and shown in boldface. Please note that the huNV genome contains two TURBS, one at the border polymerase (ORF1)/VP1 (ORF2) and one at the border VP1 (ORF2)/VP2 (ORF3). huNV, human Norovirus; FCV, feline calicivirus; RHDV, rabbit hemorrhagic disease virus; huSV, human sapovirus.
Reinitiation of translation in caliciviruses depends on an upstream sequence named TURBS (termination upstream ribosomal binding site; 40–70 nt), which is thought to bind the post-termination ribosome and therefore to increase the chance of reinitiation. The TURBS region contains three essential motifs (Fig. 1). Motif 1 (UGGGA) is found to be conserved among caliciviruses and is located at similar positions in the mRNAs of the different caliciviruses, upstream of the 3′-terminal ORFs. This motif is complementary to the loop region of helix 26 of the 18 S rRNA. Hybridization of motif 1 to 18 S rRNA was demonstrated to be important for reinitiation (39). There is also evidence for an interaction of motif 1 with eukaryotic translation initiation factor 3 (eIF3) (26). In contrast to motif 1, the sequence of motif 2 is not conserved, but this motif is located at similar positions in the mRNA, 12–23 nt upstream of the 3′ ORF start codon. A complementary sequence to motif 2, named motif 2*, is located directly upstream of motif 1. These two motifs were proposed to establish a secondary structure playing a role in positioning the ribosome relative to the start site of the 3′-terminal ORF (23, 25). The proposed secondary structure of the TURBS might also be important for presenting motif 1 for interaction with the 18 S rRNA. The basic mechanism of the reinitiation process is clearly conserved among caliciviruses, but the efficiency of reinitiation varies for the different caliciviruses between 5 and 20% of the VP1 translation rate.
In this study, we describe analysis of the termination-reinitiation mechanism of VP2 translation for human Norovirus (huNV). In contrast to other caliciviral genera, the stop codon of the VP1 coding frame is located upstream of the VP2 start codon (see Fig. 1). Therefore, our analyses were focused on the start site used for the reinitiation event, and we compared the results with data generated for other caliciviruses and found differences in start site recognition but also common features for this interesting mechanism of translation.
EXPERIMENTAL PROCEDURES
Cells and Viruses
BHK-21 cells (kindly provided by T. Rümenapf) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and nonessential amino acids. Vaccinia virus MVA-T7 (42) was kindly provided by B. Moss (National Institutes of Health, Bethesda).
Construction of Recombinant Plasmids
Restriction and subcloning were done according to standard procedures (43). Restriction and modifying enzymes were purchased from New England Biolabs (Schwalbach, Germany) and Fermentas GmbH (Sankt Leon-Rot, Germany). The huNV coding regions for constructs pNVWT and pNPWT were generated by PCR from a full-length cDNA clone (Norovirus Hu/NLV/Dresden174/pUS-NorII/1997/GE, GenBankTM accession number AY741811) kindly provided by J. Rohayem. The huNV sequences were inserted into a two-tag construct with a 5′ Pep6- and a 3′ V5-His6 tag sequence (pSK-cLC3 with a multiple cloning site inserted between the tag coding regions (44)). Point mutations and deletions were introduced by standard PCR-based site-directed mutagenesis methods using thermostable Pfu polymerase (Promega, Heidelberg, Germany) and synthetic primers purchased from Metabion (Munich, Germany). For detection of downstream initiation close to the original stop/start site, pNVΔC, a 3′-terminally truncated version of pNVWT was generated; this construct lacks the codons coding for the C-terminal residues 107–268 of VP2. The huNV expression construct pNVΔCdM lacking all internal methionine codons was established synthetically (Invitrogen). For easier detection, the methionine codons were replaced by cysteine codons, and the codons coding for the C-terminal residues 107–268 of VP2 were deleted as in pNVΔC. Into this construct the desired mutations were introduced via site-directed mutagenesis as described above. The mutant constructs for FCV were established on the basis of plasmid pCH1 (23) and for RHDV on the basis of plasmid pRmRNA (24). The cloned PCR products were all verified by nucleotide sequencing with the BigDye Terminator Cycle Sequencing Kit (PE Applied Biosystems, Weiterstadt, Germany). Details of the cloning procedure and the sequences of the primers are available on request.
Expression, Detection, and Quantification of Proteins
Transient expression of plasmids in BHK-21 cells using vaccinia virus MVA-T7, metabolic labeling with [35S]cysteine and -methionine (MP Biomedicals, Heidelberg, Germany, or Hartmann Analytics, Braunschweig, Germany), preparation of cell extracts, and recovery of immunoprecipitates with double precipitation were done as described previously (24). Briefly, protein expression efficiency was quantified after SDS-PAGE separation of the precipitated proteins. huNV proteins were precipitated with the Pep6 antisera (44) and V5 antisera (Invitrogen). Precipitation for FCV and RHDV capsid proteins was done as described in Refs. 23, 24, respectively. The 4–5-fold volume of cell lysate was used for precipitation of the protein resulting from translation of the second frame compared with that for precipitation of the first frame. AU1 tag-dependent precipitation for the pNV56 constructs was done with an AU-1 antiserum from BIOZOL Diagnostic (Munich, Germany). Double precipitation was used to ensure quantitative recovery of the proteins as tested before. The precipitates were combined, and aliquots thereof were separated by 10–16% PAGE using a gel system published previously (45). The gels were analyzed with a Fujifilm BAS-1500 phosphorimager, and the intensities of the signals were determined with TINA 2.0 software (Raytest, Straubenhardt, Germany). The molar ratio of VP1 and VP2 was calculated based on the number of labeled residues within the proteins and the measured radioactivity. For comparison of expression efficiencies of different constructs, the VP2 expression level of the wild type (WT) construct was defined as 100%. The amounts of VP2 expression of the other constructs were normalized to the values determined for VP1 as an internal standard. The corrected value for VP2 was then used for calculation of the expression efficiency, given as a percentage of the WT value. An analogous quantification was done for analysis of the polymerase and VP1 proteins of the constructs established for analysis of the second NV TURBS located in the polymerase coding region (construct pNPWT and mutants thereof). The presented quantitative data represent the averages of at least three independent experiments. In Figs. 3–9, different exposure times for VP1 and VP2 are shown as follows: 1–3 days for VP1 and 7–28 days for VP2.
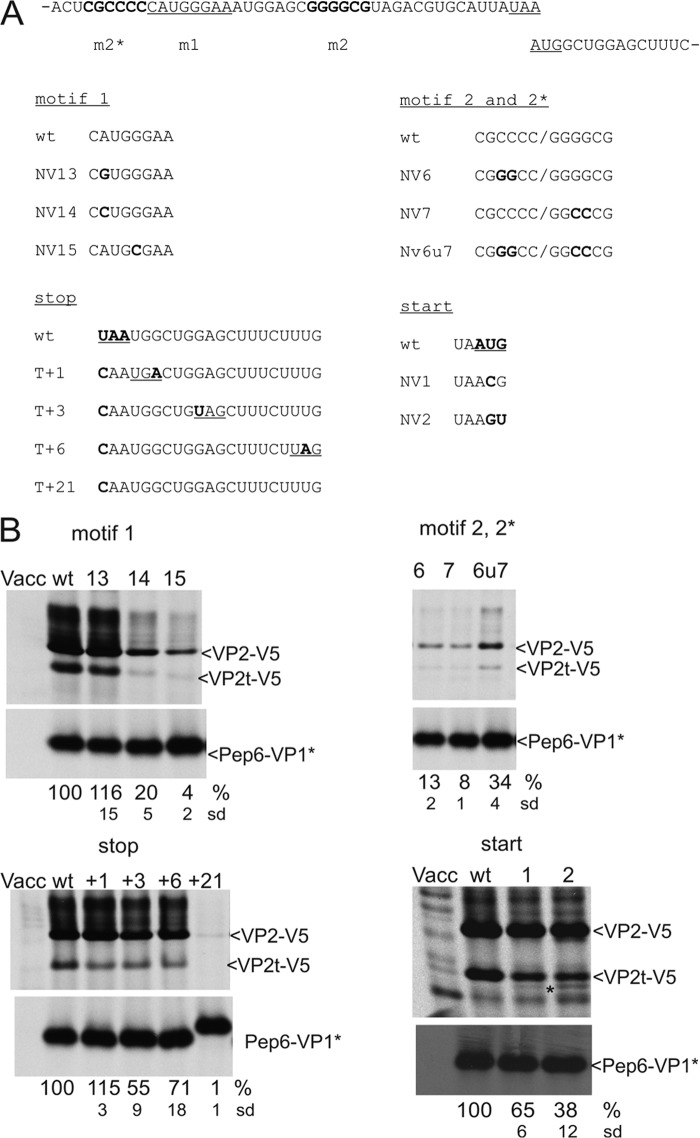
FIGURE 3.
Importance of the motifs of the huNV TURBS. A, sequence on top represents the TURBS of huNV as determined by sequence analysis. The two reading frames overlapping by one A residue are given in separate lines with essential regions of the TURBS highlighted: motif 1 (m1), the stop codon of the VP1 coding ORF and the start codon of the VP2 coding sequence are underlined, the complementary motifs 2* (m2*) and motif 2 (m2) are written in boldface. The sequences below show the mutations introduced into the different important regions of the TURBS. The affected motifs are indicated above the sequences. The substitutions are highlighted in boldface. The termination or start codon is shown in boldface and underlined within the wild type sequence, respectively. B, gels with the proteins precipitated with antiserum anti-V5 (upper part) or anti-pep6 (lower part) after transient expression of the indicated constructs. The calculated VP2 expression efficiencies in percent of the WT level are given below the autoradiographs (pNVWT VP2 expression rate set to 100%, normalized to the amount of Pep6-VP1*). The standard deviations (sd) are indicated. Cells infected with vaccinia virus (Vacc) MVA-T7 served as a negative control. For VP2t-V5, see legend to Fig. 2. A protein even smaller than VP2t-V5, detected by V5 precipitation in cells expressing the pNV2 construct, is marked by an asterisk (lane 2, right bottom panel).
FIGURE 4.
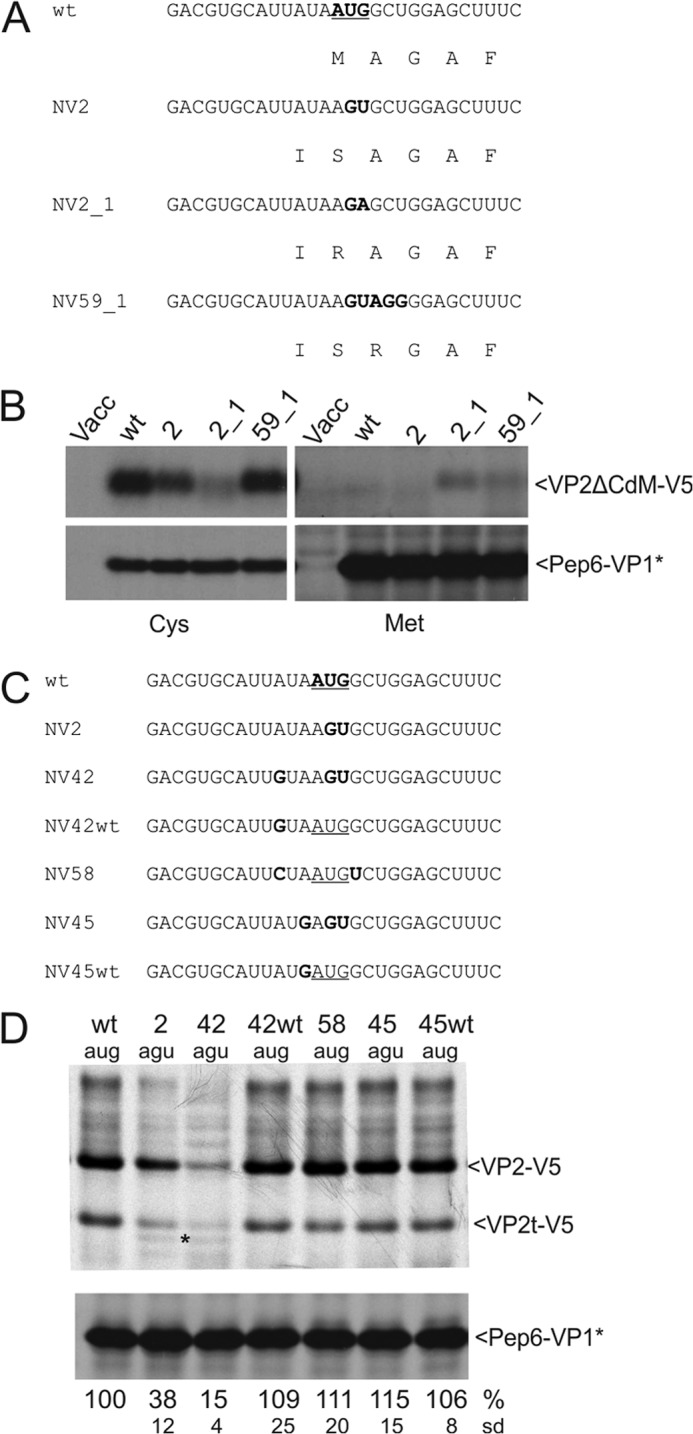
Importance of start site sequence and location for initiation position and efficiency. A, sequences represent part of the TURBS. Top, wild type sequence is shown with the VP2 start codon given in boldface. Below the nucleotide sequence, the deduced VP2 amino acid sequence starting with the Met encoded by the original start site is given. Below the WT sequences, the equivalent information is given for the tested mutants. For constructs with mutated VP2 start codon, the amino acid encoded by the altered codon plus the residue upstream thereof is given. The AUG in WT as well as the residues altered in the mutants are highlighted in boldface. B, gels with the proteins precipitated with antiserum anti-V5 (upper part) or anti-pep6 (lower part) after transient expression of the indicated constructs. All constructs are shown twice, either after labeling with [35S]Cys (left part) or [35S]Met (right part). Please note that in the constructs expressed here, all internal Met codons were replaced by Cys codons and that the VP2 coding ORF contains a deletion of the C-terminal codons 107–268. The designation VP2ΔCdM reflects the truncation (ΔC) and absence of internal Met codons (dM). Vacc, vaccinia virus. C, sequences represent part of the TURBS. Top, wild type sequence is shown with the VP2 start codon given in boldface. Below, mutants with substitutions affecting the start codon or the codon upstream thereof are shown with the changes highlighted in boldface. Please note that in constructs NV45 and NV45WT, the VP1 stop codon was changed from UAA to UGA. D, gel with the proteins precipitated after transient expression (antisera as described for Fig. 3). Numbers indicating the transfected constructs and the sequences present at the position of the original start site are given above the gel. As described in legend to Fig. 3, the calculated VP2 expression efficiencies in percent and the standard deviations (sd) are given below the autoradiographs (relative to pNVWT, normalized to the expression level of Pep6-VP1*). For VP2t-V5, see legend Fig. 2.
FIGURE 5.
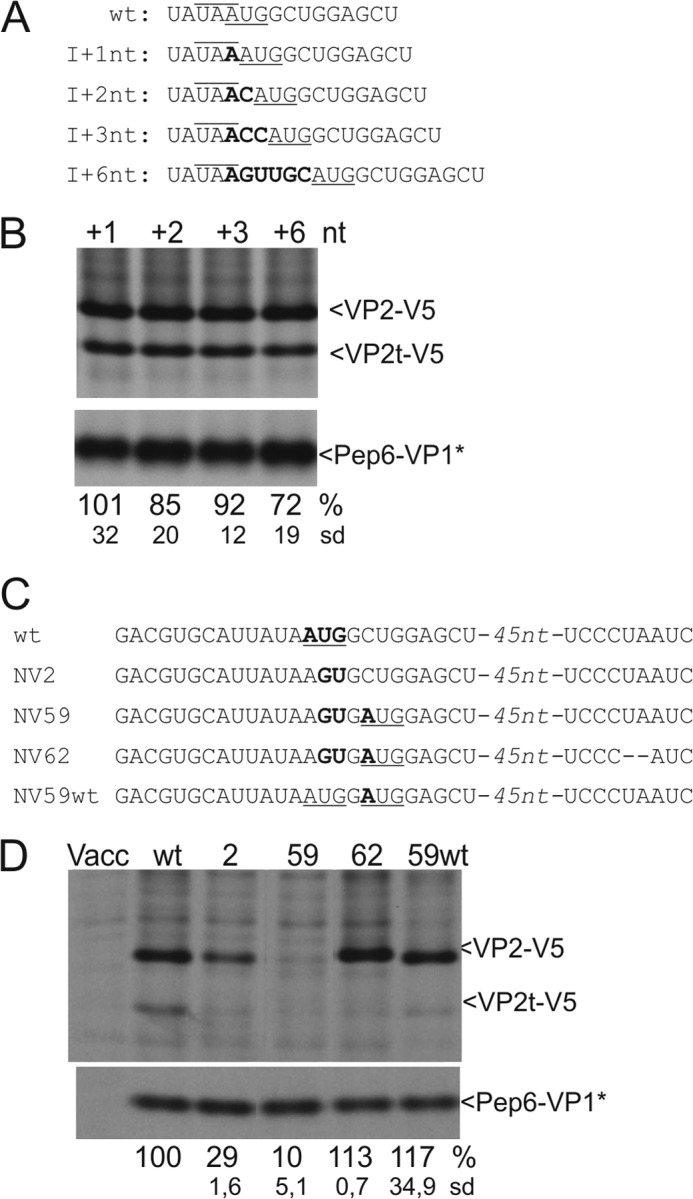
Insertions between stop and start. A, sequences represent part of the TURBS. Top, wild type sequence is shown with the VP2 start codon underlined and the VP1 stop codon indicated by an overline. Below, sequences of the mutants are given, with insertions of 1–6 nucleotides between the stop codon of VP1 and the start codon of VP2. The inserted nucleotides are shown in boldface, and the number of inserted nucleotides is specified in the mutant's name. B, gels with the proteins precipitated after transient expression of the indicated constructs, C, as in A. Please note that the 5′ part of the VP2 coding sequence is not completely shown here; the gap is indicated by −45nt-. In NV62, two residues were deleted that represent the first to nucleotides of the UAA stop codon terminating in the other constructs the ORF started with the codon at positions 5–7 with respect to the original VP2 start site. D, gels with the proteins precipitated after transient expression of the indicated constructs (antisera as described for Fig. 3). The calculated VP2 expression efficiencies in percent and the standard deviations (sd) are given below the autoradiographs (relative to pNVWT, normalized to the expression of Pep6-VP1*). VP2t-V5, truncated VP2-V5 protein. Vacc, vaccinia virus.
FIGURE 6.
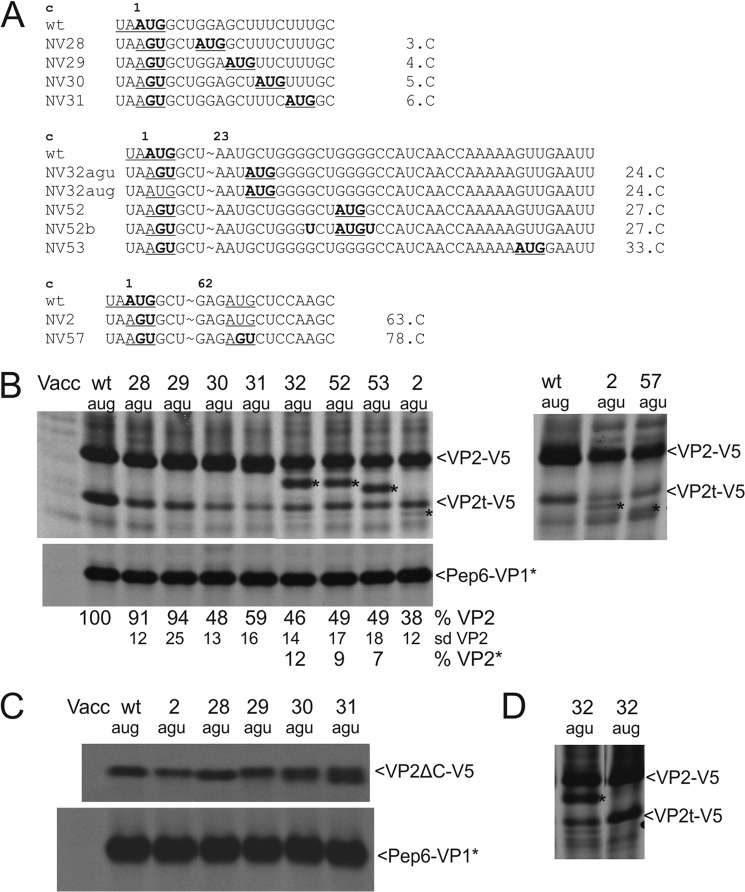
Reinitiation of translation at downstream AUGs. A, sequences represent the 5′ region of the VP2 coding frame. Top, wild type (wt) sequence is given with the stop/start site underlined and the VP2 start codon shown in boldface. Above the sequence, codon numbers are given with codon 1 representing the VP2 start codon in the WT sequence. Below, mutants with a 2-nt substitution of the start codon and/or introduction of a downstream AUG, which is highlighted in boldface and underlined, are shown. The position of the codon of the VP2 frame that is mutated to AUG is given on the right side of the mutant sequences (number.C, expressed as codon number with the VP2 AUG representing codon 1). B–D, gels with the proteins precipitated after transient expression of the indicated constructs (antisera as described for Fig. 3). Proteins precipitated by V5 antibody that is smaller than VP2-V5, detected in addition to the long protein and VP2t-V5, are marked by an asterisk. B, left panel, calculated VP2 expression efficiencies in percent and the standard deviations (sd) are given below the autoradiographs. The upper lane presents the values for the full-length VP2-V5 protein (VP2); the middle lane shows the standard deviations for the full-length protein, and the lower lane shows the results for the smaller protein VP2* marked by an asterisk within the gel. Each protein was quantified relative to the full-length VP2-V5 protein of the pNVWT construct and normalized to the expression of Pep6-VP1*. The right panel stems from a different experiment and shows the shortened products more clearly. C, start site mutations of the expressed constructs are indicated on top and are equivalent to those in B. However, to shorten the respective expression product, the constructs expressed here differ from those in B by a deletion of the VP2 coding codons 107–268 and in analogy to Fig. 4 are designated VP2ΔC-V5. D, two constructs shown here differ in the absence (agu) or presence (aug) of the AUG in the stop/start site. For VP2t-V5 see legend to Fig. 2. Vacc, vaccinia virus.
FIGURE 7.
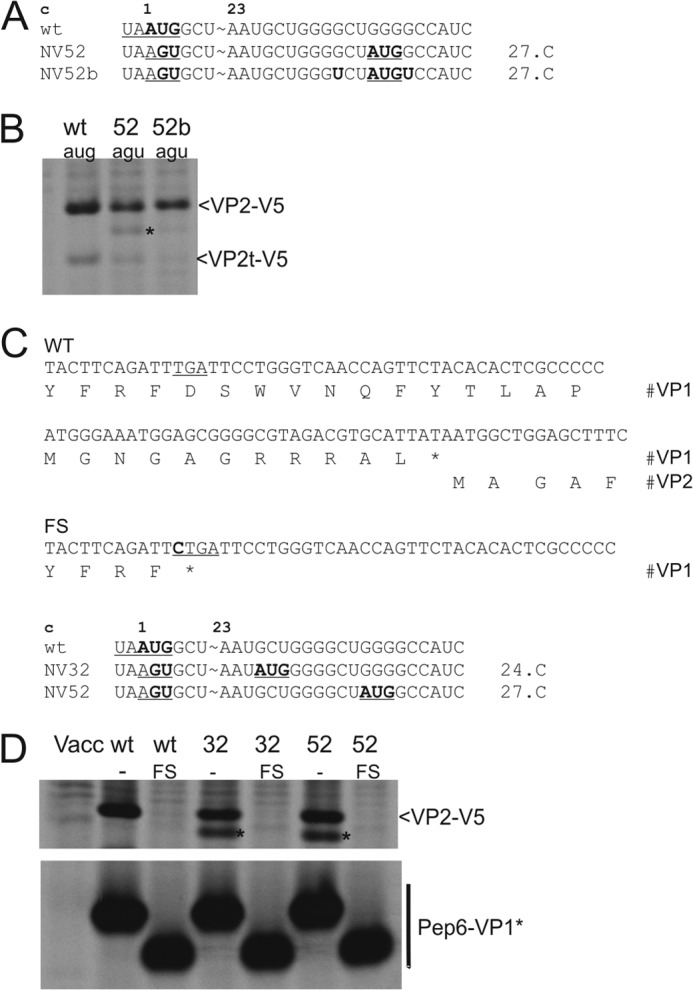
Reinitiation at downstream AUGs depends on the Kozak context and translation of the upstream ORF down to the UAA in the stop/start site. A, sequences represent the 5′ region of the VP2 coding frame. Top, the wild type (wt) sequence is given with the stop/start site underlined, and the VP2 start codon is shown in boldface. Above the sequence, codon numbers are given with codon 1 representing the VP2 start codon in the WT sequence. Below, mutants with a 2-nt substitution of the start codon and introduction of a downstream AUG, which is highlighted in boldface and underlined, are shown. The position of the codon of the VP2 frame that is mutated to AUG is given on the right side of the mutant sequences (expressed as codon number with the VP2 AUG representing codon 1). In construct pNV52b, two further mutations were introduced to create a bad Kozak context of the downstream AUG. B, gels with the proteins precipitated after transient expression of the indicated constructs (antisera as described for Fig. 3). A protein precipitated by V5 antibody that is smaller than VP2-V5, detected in addition to the long protein and VP2t-V5, is marked by an asterisk. C, nucleotide and encoded amino acid sequence of the stop/start region of the WT construct (top) and of the region equivalent to the upper row of the WT sequence of the frameshift (FS) construct containing an inserted C residue leading to premature termination of Pep6-VP1 translation (inserted C in boldface and stop codon underlined). Below these two sequences, the sequence context around the stop/start region of the tested start site mutants is given (see A). Please note that for each start site mutant, two versions were tested, one with the WT Pep6-VP1 coding sequence and one with the frameshift insertion. D, gels with the proteins precipitated after transient expression of the indicated constructs (antisera as described for Fig. 3). A protein precipitated by V5 antibody that is smaller than VP2-V5, detected in addition to the long protein and VP2t-V5, is marked by an asterisk. Lanes FS show products expressed from the frameshift versions of the indicated constructs. Vacc, vaccinia virus.
FIGURE 8.
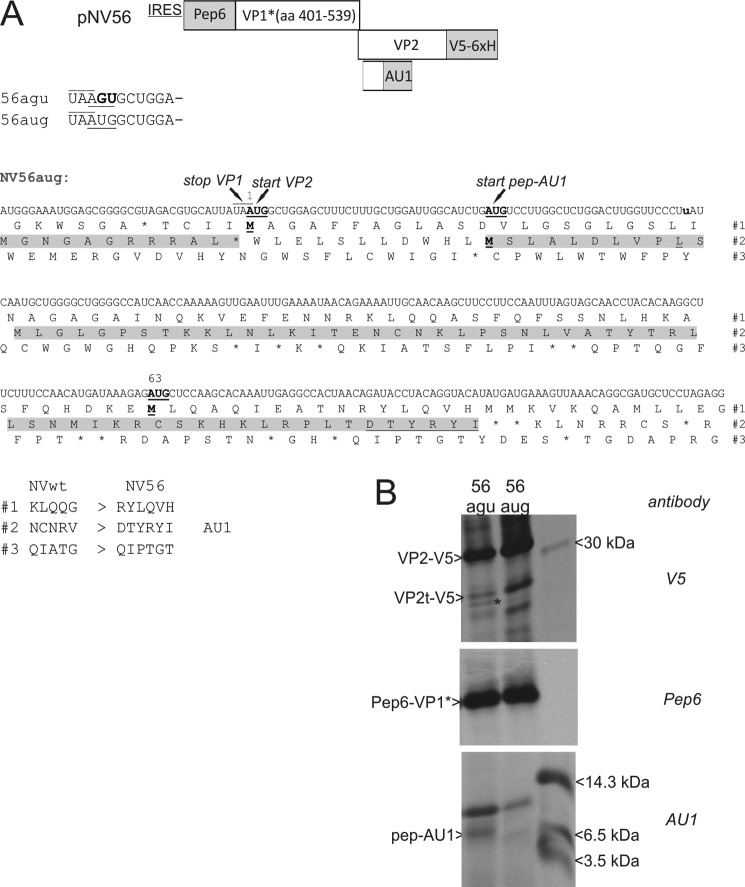
Reinitiation at downstream AUGs in different frames. A, on top, a schematic presentation of the NV56 construct is given (not drawn to scale). The basic structure is as in the pNVWT construct described in legend to Fig. 2. In addition to the sequences coding for pep6-VP1* and VP2-V5-His6 that are equivalent to the pNVWT construct, a newly engineered expression cassette with an AU1 tag, which is located within the VP2 coding sequence, is shown. Below, 5′ nucleotide sequence of the VP2 coding region is shown for the constructs NV56agu and NV56aug with a line above the VP1 stop codon and the (mutated) VP2 start codon underlined. The sequence presentation below represents the 3′ region of the VP1 coding frame and the 5′ region of the VP2 coding frame of the NV56aug construct. The nucleotide sequence and the amino acid sequences of all three frames are given. The amino acids (aa) being part of VP1 and the engineered AU1 tag protein are marked in gray. The AUGs of the VP2 frame, codon 1 and codon 63, and the start AUG of the AU1 tag-containing protein are marked in boldface and underlined. The mutation of a stop codon in frame 2 is highlighted in boldface (UAA > UuA). The amino acid sequence of the AU1 tag is underlined. For easier orientation, the VP1 stop, VP2 start, and pep-AU1 start sites are marked with arrows above the sequence. Below the sequence presentation, amino acid sequences of the frames 1 and 2 of the mutant construct NV56 (right) are compared with the wild type sequence (left). B, gels with the proteins precipitated after transient expression of the constructs NV56agu and NV56aug with the antisera given on the right side of the gels. A size marker with the indicated molecular mass bands is shown in the right lane of the gel. VP2t-V5, truncated VP2-V5 protein; pep-AU1, protein resulting from translation of frame 2. A smaller protein precipitated by V5 antibody is marked by an asterisk.
FIGURE 9.
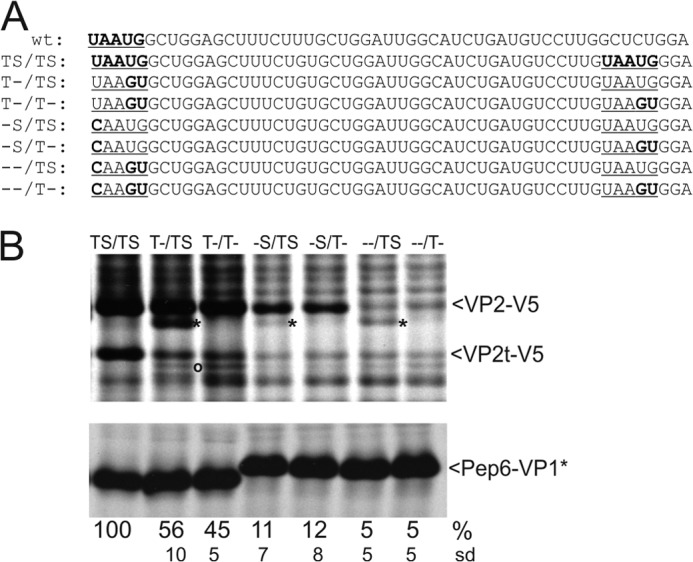
Reinitiation in constructs with a complete downstream stop/start site. A, sequences represent the 5′ region of the VP2 coding frame of huNV. Top, wild type sequence is shown with the stop-start site in bold face and underlined. The 2nd line displays the sequence of the basic construct TS/TS with the original stop/start site and the introduced second stop-start site highlighted in bold face and underlined. Below, sequences of the stop and/or start mutants are given with the stop/start sites underlined and the mutations highlighted in bold face. B, gels with the proteins precipitated after transient expression of the indicated constructs (antisera as described for Fig. 3). A second smaller protein detected by precipitation with V5 antiserum is marked by an asterisk. A protein resulting from initiation at codon 63 is marked with an o. VP2t-V5, truncated VP2-V5 protein. The calculated expression of the VP2-V5 full-length protein efficiencies is given in percent below the autoradiographs (relative to the TS/TS construct, normalized to the expression of Pep6-VP1*) together with the standard deviations (sd).
RESULTS
The caliciviral termination-reinitiation mechanism depends on motif 1 containing the conserved pentamer “UGGGA.” Further essential sequences are represented by the complementary motifs 2 and 2* (Fig. 1).
For the human Norovirus, important elements for reinitiation leading to VP2 translation are the ORF2 and ORF3 overlap region UAAUG containing the stop of the upstream ORF and the start of the downstream ORF. As expected, motifs 1 “CAUGGGAA“, 2 “GGGGCG,” and 2* “CGCCCC” were also found (see Figs. 1 and 3). To analyze VP2 translation of huNV, a plasmid was generated that contains the 3′ part of the VP1-coding sequence with a 5′ fused tag-coding sequence (Pep6 tag) and the VP2 coding region with a sequence coding for a V5-His6 tag fused to the 3′ end (Fig. 2) under control of a T7 promotor. Translation of VP1 and VP2 was analyzed in T7-MVA vaccinia virus-infected BHK cells. The vaccinia virus infection prior to transfection of the different cDNA constructs provides T7-RNA polymerase that ensures transcription of the desired RNAs. Translation of the upstream frame of our mRNAs is mediated by the encephalomyocarditis virus IRES contained in the constructs (Fig. 2). As analyzed before, the mechanism used for initiating translation of the upstream frame has no influence on the principles of reinitiation. IRES-driven translation has just been chosen here and in earlier analyses to ensure efficient translation of the upstream ORF that is important for obtaining a reasonable amount of the downstream ORF product (23–25, 39).4 Precipitation of VP1 was done with a Pep6 serum, and the VP2-containing fusion protein was precipitated with a V5 antibody. In addition to the expected bands, a second band precipitated with the V5 antibody was detected that is shorter than the full-length VP2 fusion protein. The amount of this product correlates with that of VP2-V5 and it is not present in the vaccinia control. This truncated VP2-V5 protein (VP2t-V5) is generated in addition to the full-length fusion protein (VP2-V5) and is probably a result of cleavage of VP2-V5 within the cells (Figs. 2–8). There are also bands above of the VP2-V5 band that are not seen in the control. These can also be visible as a smear of different intensity (see Figs. 2–8). We have no idea of the nature of these proteins and have not included them in the quantification. The calculated reinitiation frequency for the huNV system was ∼3% of the VP1 translation rate. To show that the motifs determined by sequence analyses are indeed essential for reinitiation of huNV VP2 translation, substitution mutants were generated and analyzed for each of the essential regions (Fig. 3).
FIGURE 2.
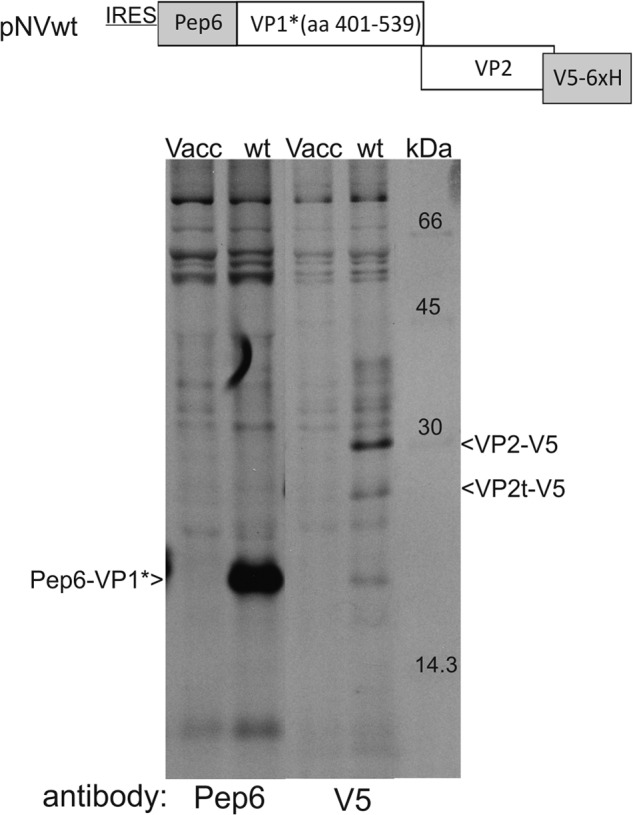
Expression of huNV VP1 and VP2 proteins. Top, schematic representation of the basic structure of the pNVWT construct is given (not drawn to scale). The pep6 and V5-His6 tag sequences are given as gray bars, and VP1- and VP2-coding sequences are shown as white bars. The amino acid (aa) numbers of the VP1 fragment contained in VP1* are indicated. Below the scheme, a gel with the proteins precipitated after transient expression of the wild type construct pNVWT is shown. Lanes Vacc contain the products precipitated from control cells (vaccinia virus-infected cells but no plasmid transfection). Lanes wt show the proteins precipitated from cells transiently transfected with pNVWT. The antibodies used for precipitation are given below the lanes, and the precipitated proteins are specified on the left and right side of the gel. IRES, encephalomyocarditis virus IRES; Pep6, peptide tag derived from pestivirus sequence; Pep6 is fused to the N terminus of the C-terminal part of the VP1 protein of huNV (Pep6-VP1*, 18 kDa); a V5 and His6 tag is fused to the C terminus of the VP2 protein of huNV (VP2-V5, 31 kDa). A second smaller protein detected after V5 precipitation presumably representing a truncated VP2-V5 (named VP2t-V5) is indicated.
Single substitutions within motif 1 interfering with the proposed hybridization with 18 S rRNA led to a prominent reduction of VP2 translation (NV14 and NV15, Fig. 3). In contrast, a mutation at the same position as in NV14 but still allowing hybridization with the 18 S rRNA via a G-U pairing showed a WT level of VP2 translation (NV13, Fig. 3). Two nucleotide substitutions within motif 2 or motif 2* also led to a reduced reinitiation rate (NV6 and NV7, Fig. 3). Reciprocal substitutions within these complementary regions restored reinitiation efficiency to some extent but did not restore wild type levels of reinitiation (NV6 and NV7, Fig. 3). Thus, the motif 1, motif 2, and motif 2* regions are indeed important for reinitiation of translation. Motif 2 and/or motif 2* could have functions beyond establishment of the proposed stem structure, as mutations preserving this structure reduced the VP2 translation level. However, it has to be stressed that we have not demonstrated that the reciprocal mutants fold appropriately so that inappropriate secondary structure can still account for the observed lower reinitiation efficiency.
Relocation of the VP1 stop codon to positions downstream of the original termination site led to reduced VP2 translation (Fig. 3). A shift of the termination signal one codon downstream had no negative effect on VP2 translation (T+1, Fig. 3), but termination of VP1 translation 3 or 6 codons downstream of the original site reduced the level of VP2 translation (T+3 and T+6, Fig. 3). When the original stop codon is mutated in the wild type sequence, the next stop codon in-frame is located 21 codons downstream. For this mutant, VP2 translation was nearly abolished (T+21, Fig. 3). Thus, a VP1 termination signal close to the VP2 start site is important for efficient VP2 translation.
Mutation of the VP2 start codon led to some reduction of the reinitiation level, but VP2 translation still occurred at a significant frequency (Fig. 3). Mutation of one nucleotide resulted in the reduction of VP2 translation to 65% of the wild type level (NV1, Fig. 3), whereas substitution of two nucleotides of the AUG codon induced a more pronounced drop in VP2 translation (38% of WT level) (NV2, Fig. 3). Thus, an AUG as start codon is not essential, but reinitiation occurs at a lower level on non-AUG codons.
Location of the Start Site Is Flexible within a Few Codons
For noroviruses, the stop codon of the VP1 coding region is located upstream of the start codon of the VP2 gene (see Figs. 1 and 2). This arrangement differs from what is found at the equivalent site in other caliciviruses (see Fig. 1). We therefore wanted to have a closer look at the AUG recognition during reinitiation at the huNV VP1-VP2 stop/start site.
First we analyzed the importance of AUG as initiation codon. An AUG start codon is not essential for reinitiation. Most substitutions within the AUG led to a rather mild decrease of VP2 translation (Fig. 3) (23, 24). The codon directly upstream from the NV-VP2 start codon is AUA, and thus it may serve as an alternative start codon (23, 24, 46). We tried to get support for this hypothesis by mass spectrometric analysis of the VP2 expressed from construct pNV2. Unfortunately, we were not able to obtain sequence information of the N-terminal fragment in different approaches. To provide experimental support for reinitiation at the AUA, we established the expression construct pNVΔCdM, in which all Met codons in the VP2 frame, except for the one in the stop/start site, were replaced by Cys codons. Based on pNVΔCdM, a variety of mutants was generated (Fig. 4A). These constructs were tested in transient expression studies with [35S]Met. According to the Sherman rules, the N-terminal methionine is lost when small amino acids are present at position 2. Thus, a labeled product can only be detected when translation starts with methionine followed by a large residue that prevents removal of the initiator Met. For demonstration of correct expression of all mutants, a second set of expression studies was conducted with [35S]Cys. As expected, Met-labeled VP2-V5 was not detected for pNVΔCdM, the construct without internal methionine codons and WT stop/start site. The protein encoded by this construct starts with Met-Ala so that the initiator Met is lost (Fig. 4B). This result demonstrates that reinitiation at the WT stop/start site starts at the AUG overlapping the VP1 gene stop codon. In the WT construct, the preceding AUA or the AUU at codon position −2 do not serve as start sites, because the respective translation products would both preserve the initiator Met. pNV2ΔCdM, the construct corresponding to pNV2 in Fig. 3, is expected to yield products without Met, when translation starts at the original start site (mutated to AGU in the pNV2ΔCdM RNA) or at the preceding AUA. A labeled product would result from initiation at the AUU at position −2. Similar to the parental pNVΔCdM, pNV2ΔCdM also did not give rise to a detectable amount of Met-labeled VP2-V5. Thus, translation of the reinitiation product does not start at the AUU. As a next step, the original start site was mutated to AGA (construct pNV2_1ΔCdM). Restart of translation at the AUA at position −1 would result in a labeled protein now, because the initiator Met is followed by Arg. In contrast, reinitiation at the original start site would again not lead to a Met-labeled product due to the Ala at position +1. In fact, construct pNV2_1ΔCdM yielded a clearly visible product upon Met labeling proving that translation can start here at the AUA just upstream of the original start site (Fig. 4B). This conclusion is supported by the notion that the signal observed for pNV2_1ΔCdM after [35S]Cys was rather weak compared with the other three tested RNAs. Accordingly, the overall reinitiation rate is only moderate for this construct (Fig. 4B). To demonstrate that reinitiation also takes place at a mutated original start site, construct pNV59_1ΔCdM was established. In this construct, the original start site is changed to AGU, and the next codon is mutated to AGG so that a Met introduced during reinitiation at the original start site would be followed by Arg and would therefore be preserved. In contrast, a restart at the preceding AUA would result in loss of the initiation Met. Indeed, expression of this construct resulted in a Met-labeled product proving our hypothesis that reinitiation occurs at the mutated original site.
Taken together, the experiments described above showed that translation is reinitiated at the AUG of the WT stop/start site. When this AUG is mutated, reinitiation occurs both at the original start site and the AUA, which is located upstream. More insight into the reinitiation start site selection was gained by analyzing the influence of a variety of further mutations on reinitiation. These mutants were established in the background of the WT construct pNVWT containing the internal Met codons. Mutation of the AUA codon to GUA and mutation of the original start codon to AGU (NV42) led to a further reduction of the VP2 level (15%) compared with the start codon single mutant NV2 (38%) (Fig. 4, C and D). Thus, the reduction from 38 to 15% is most likely founded in the loss of reinitiation at the AUA. Mutation of the upstream AUA to GUA in context of the wild type start codon (NV42WT) had no significant influence on the reinitiation level (109%, Fig. 4, C and D). This result demonstrates again that initiation at the AUA codon occurs only when the original start codon is mutated. The residual reinitiation rate of 15% detected for the double mutant NV42 is due to translation starting at the AGU replacing the WT AUG as demonstrated above. When the AUA is mutated to an AUG codon combined with the mutation of the original start codon to AGU (NV45), the reinitiation rate is at the wild type level (115%, Fig. 4D). Thus, an AUG located just upstream of the original start codon functionally replaces the WT AUG. Interestingly, the reinitiation rate is not raised when an upstream AUG is present in addition to the original start codon (45 WT, 106%, Fig. 4D).
Taken together, the data show that the location of the reinitiation start site is flexible for at least one codon. Translation starts preferentially at an AUG codon located within this region, but in the absence of an AUG codon alternative codons are used.
The sequence context of a start site is important for initiation when the initiation site is selected via the scanning mechanism (47). The optimal context is ACCAUGG, in which the underlined positions −3 and +4 are especially important for initiation. To analyze if the context of the start codon is important for the reinitiation event in noroviruses, the sequence surrounding the AUG was changed from a good (−3 = A, +4 = G) to a poor Kozak consensus sequence (−3 = C, +4 = U, NV58, Fig. 4C). The reinitiation rate observed for this mutant is like wild type (111%) (Fig. 4D). Thus, the Kozak sequence context seems to play no role in this type of translation initiation.
For further information on the importance of the start codon position, we inserted 1–3 or 6 nucleotides upstream of the AUG, thereby constantly increasing the distance between the stop codon and the AUG codon. Compared with the WT, we observed for these constructs just a mild reduction of reinitiation efficiency to levels between 70 and 90% that might not even be statistically significant (Fig. 5). We have shown above that VP2 translation can start at both the upstream AUA and at the mutated original start codon (see Fig. 4). Accordingly, for the mutant I+6 nt, a reinitiation could occur at the AUA or AGU as was shown in Fig. 4 for mutant NV2. However, the downstream AUG present in construct I+6 nt is obviously also used for reinitiation as VP2 translation is much more efficient in the I+6-nt RNA than in NV2 (72 and 38%, respectively, see Fig. 4). Thus, reinitiation can obviously occur a few codons downstream from the stop in this system. Because we were not able to differentiate between initiation at the upstream AUA, the original start site, and the downstream AUG, the exact level of reinitiation at each of these sites cannot be determined. However, the significant increase of the reinitiation observed upon the introduction of the downstream AUG codon strongly argues in favor of initiation at this downstream site.
The insertion of 1 or 2 nucleotides changed the arrangement of ORFs 2 and 3. Nevertheless, reinitiation rates comparable with those of in-frame insertions of 3 or 6 nt were found (Fig. 5, A and B). This finding is further proof of the fact that reinitiation represents true de novo initiation of translation.
To provide additional support for reinitiation independent of the reading frame of the downstream ORF, we analyzed constructs pNV59WT, pNV59, and pNV62. In the RNA derived from the latter two, the AUG of the stop/start site is changed to AGU, and an AUG is presented at nucleotide positions 2–4 with respect to the original start site. In pNV59, initiation at this codon would result in a short out of the VP2 frame product. To obtain pNV62, a two-nucleotide deletion was introduced into pNV59 just at the stop codon terminating translation of this short product from the NV59 RNA. Therefore, the frame is shifted back again into the VP2-coding frame (Fig. 5C). Expression of these constructs led to a WT level of the VP2-V5 product for construct pNV62, although almost no product was detected with V5 antibody for pNV59 (Fig. 5D). To show that the latter finding is not a general problem of the pNV59 construction, pNV59WT was established. It encodes an RNA that contains the original start codon together with the downstream out of frame AUG. pNV59WT yielded WT VP2-V5 levels (Fig. 5D). These results show that reinitiation within the NV stop/start region is not dependent on a specific reading frame. Moreover, the data demonstrate that an AUG close to the original start site is preferred over a mutated codon at the original start position because pNV59 does not yield a significant level of reinitiation product in the VP2 frame.
Initiation at a Downstream AUG
The results shown above indicated that an AUG located downstream of the original start site can be used for reinitiation when the original AUG is mutated. To further support this point and to find out how far downstream of the termination site reinitiation can occur, we established a series of expression constructs with newly created AUG codons. These AUGs were located at different downstream positions ranging from codon position 3–33 of the VP2 frame. In addition, this approach was supposed to allow differentiation of initiation at the original start site from the translational start at a downstream AUG. These mutations were combined with a change of the original AUG start codon to AGU (Fig. 6A). For the mutants with AUG at codon positions 3–5 (encoded by constructs pNV28–30), the products resulting from initiation at the two possible start sites cannot be distinguished. For construct pNV31, with the second initiation site located six codons downstream of the original start site, a broadening of the VP2-V5 band is visible on the gels, but a double band cannot be definitely identified (Fig. 6B). Shortening of the VP2-V5 coding sequences of the constructs resulted in better separation of the two bands in NV31 and a broad band in NV30 (Fig. 6C). A more obvious separation was obtained for those constructs containing the second start site at codon +24 or further downstream (Fig. 6B, NV32, NV52, and NV53). Thus, it is rather likely also that the introduced AUGs in the RNAs derived from constructs pNV28 and pNV29 can be used for initiation when the original start codon is mutated. Most of the initiation processes still occurred within the original stop/start region (about 80% of the total initiation events in those cases where the two bands can be separated and quantified), but also initiation at the downstream AUG occurred at significant frequency.
The total reinitiation rate is around 90% of the WT level for the constructs with a mutated original start site and the new AUG at codons 3 or 4 (NV28 and NV29) (Fig. 6B). It cannot be distinguished whether reinitiation occurs here almost exclusively at the newly created downstream AUG or at the original start site and the downstream AUG. The former possibility was suggested by the results for the frameshift constructs pNV59 and pNV62 (Fig. 5). Importantly, the overall reinitiation rates decreased for the codon 5 AUG mutant (NV30) and the constructs with the new AUG introduced further downstream (Fig. 6B). Thus, it can be concluded that reinitiation efficiency at an AUG inserted very close to the original start site is considerably higher than at AUGs located further downstream. For the NV2 RNA with the start codon mutated to AGU, a weak second protein band was detected after precipitation with the V5 antiserum in addition to the VP2-V5 protein. This weak band migrates in the gel between the truncated VP2t-V5 and a faster migrating unspecified band (specific band marked with an asterisk in Fig. 3B, right bottom panel, and Figs. 4D and 6B for NV2). The respective protein is somewhat difficult to detect, but it can be seen for all mutants that have a mutated original start site and no AUG nearby. It could result from initiation at an AUG found in the wild type sequence at codon 63 of the VP2 frame. Mutation of codon 63 together with mutation of the original start codon led to loss of this band (NV57, Fig. 6B). In agreement with our hypothesis of reinitiation at downstream sites, a new band was detected after expression of NV57 that could start at the next AUG located at position 78 (marked with an asterisk for NV57 in Fig. 6B, right panel).
Substitution of a downstream codon by AUG in the wild type context (data shown for position 24, NV32aug, Fig. 6D) did not result in detection of the shortened VP2-V5 protein and had no influence on the reinitiation rate as can be concluded from comparison of the results obtained for NV32agu and NV32aug. Both of the transcripts derived from these constructs have a new engineered AUG at the VP2 codon position +24. In case of NV32agu, the original start codon is mutated to AGU, whereas NV32aug contains the original AUG start codon in addition to the new one (Fig. 6D). Thus, initiation at the downstream AUG occurs only when the original AUG is mutated.
The analyses of the substitution mutants showed that reinitiation of translation occurs at downstream sites when the original start codon is mutated. This is even true for constructs in which the next AUG in the VP2 frame is located 78 codons downstream of the original start site, but with a low efficiency. In all analyzed cases, reinitiation at the downstream site is observed in addition to a prominent restart at the mutated original start site.
Downstream initiation obviously occurs only at AUG codons. We therefore wanted to know if the downstream initiation is affected by the Kozak sequence context. Therefore, we changed the favorable context of the engineered downstream AUG of the NV52 RNA to an unfavorable sequence (NV52b, Fig. 7A). The initiation rate at the mutated original start site in the stop/start region was nearly the same for the NV52 and NV52b RNAs (about 50% compared with the wild type). However, the amount of the protein initiated at the downstream AUG decreased from ∼10 to ∼2% for a good or bad Kozak context, respectively (Fig. 7B). Thus, in contrast to initiation at the original reinitiation start site, initiation at a downstream site occurs in agreement with the Kozak rules.
The above described results show that initiation at downstream sites relies at least in part on a different mechanism than initiation at the stop/start site. It was therefore important to investigate whether translational start at downstream sites is still a reinitiation event. Reinitiation is characterized by the absolute requirement of translation of the upstream ORF down to the stop/start site. To provoke premature termination of upstream frame translation, we engineered a stop codon in the VP1 frame by insertion of a C residue at position −70 with regard to the stop/start site. This mutation was not only introduced in the WT construct but also in pNV32 and pNV52, two constructs showing initiation at engineered downstream sites (Fig. 7C). Translation of the VP1 frame stops 24 codons upstream of the WT termination site in these constructs. Transient expression revealed that upstream frame translation resulted in the expected shortened VP1 proteins, whereas VP2 expression was neither observed from the original nor from the downstream start site (Fig. 7D). These results demonstrate that initiation at downstream sites represents true reinitiation. However, in contrast to restart at the original start site, reinitiation at downstream sites is dependent on the Kozak context.
To understand this fascinating mechanism in detail, we next wanted to analyze whether the downstream initiation occurs exclusively at the first AUG in-frame or whether initiation is possible at multiple AUGs in different frames. V5 precipitation alone can only detect the proteins encoded in the VP2 frame but not products resulting from initiation in other frames. Therefore, we wanted to insert a tag for protein precipitation. Frame 1 is the VP2 coding frame. Frame 3 has no AUG codon in the vicinity of the original VP2 start site and contains many stop codons (see Fig. 8A) so that this frame can hardly be translated into a detectable protein. In contrast, ORF2, which is in-frame with the VP1 ORF but separated from the VP1 coding region by the VP1 stop codon, contains an AUG 34 nt downstream of the VP2 start codon. ORF2 has only one stop codon close to this possible start site (nine codons downstream of the AUG in ORF2). We substituted this stop codon of frame 2 (UAA to UUA) and inserted an AU1 tag (48) directly upstream of the next stop codon in this frame (see Fig. 8A). This arrangement should allow identification of a protein with a calculated molecular mass of 7.6 kDa, with an AU1 tag fused to the C terminus and three methionine residues for 35S labeling. Two versions of this construct were established. Construct pNV56aug contained the AU1 tag in a WT background, whereas pNV56agu contained the AU1 tag and a mutated original VP2 start codon. The latter plasmid revealed a reduced VP2 expression level and some initiation at the second AUG in-frame, which is marked with an asterisk in the gel (Fig. 8B, upper panel). This result is similar to what was obtained for the different constructs with mutation of the start codon, e.g. pNV2. Importantly, we were also able to precipitate an AU1-tagged protein of ∼7 kDa from cells transfected with the plasmid pNV56agu but not pNV56aug (Fig. 8B, lower panel). The translation of this small protein obviously starts at the downstream AUG in ORF2. Thus, for the NV56agu construct, reinitiation occurs at three sites. The dominant band results from initiation at the mutated original start site. The two bands of lower intensity are initiated downstream at the next AUG in the VP2 coding frame or at the AUG located 34 nt downstream from the original start site in another frame. For construct 56aug where the original AUG start codon is still present, no downstream initiation was detectable. So initiation occurs independent of the frame at different AUGs located further downstream of the stop/start site, if the original AUG start codon is mutated.
Start and Stop Site of the TURBS Are Not Linked
The data presented above show flexibility with regard to the site of reinitiation of translation in the huNV system. We wanted to know in which way reinitiation is linked to the site of termination. Termination close to the reinitiation start site is essential for reinitiation, as already shown for other caliciviruses (23, 24, 40) and here for huNV VP2 translation (Fig. 3). Some flexibility with regard to the exact location of the termination site was found in this work and in earlier experiments. The range, in which translation termination has to occur, varies for different caliciviruses. In general, the reinitiation frequency is reduced with increasing distance between the start and stop site.
To analyze whether termination and reinitiation could be transferred to a downstream position, we introduced a complete second stop/start site with a termination codon in the VP1 frame and an AUG in the VP2 frame just as in the original context 16 codons downstream of the original site (construct TS/TS in Fig. 9). When the AUG is mutated to AGU in this construct, downstream initiation results in a separate protein band (Fig. 9B, construct T-/TS, band marked with asterisk). We used this construct to analyze the linkage between reinitiation and termination in detail. To understand the requirements for downstream reinitiation, we analyzed the reinitiation event for different substitution mutants affecting the original stop and/or start site and/or the second start site of the basic construct TS/TS (Fig. 9A).
As already shown above, mutation of the original start codon leads to reduction of the total reinitiation efficiency (see Figs. 3–7). This is also true for the analyzed mutants with a second stop/start site (Fig. 9, construct T-/TS). The mutant T-/TS lacking the original start codon shows reinitiation not only at the mutated original start site but also at the introduced downstream AUG of the second stop/start site, located at codon position +16 (marked with an asterisk in the gel). In addition, a faint band can be detected for this construct. This finding reveals reinitiation even further downstream, most likely at the AUG at position 63 (Fig. 9B, construct T-/TS, marked with an o). Reinitiation at codon +16 in the construct T-/TS was proven by the fact that the respective band disappeared when the AUG at position +16 was changed to AGU (construct T-/T-).
Mutation of the original stop codon (-S/TS) leads to a prominent reduction of the reinitiation rate (Fig. 9B), as the next termination signal in the VP1 frame is located 15 codons downstream. For this configuration, the AUG-dependent initiation at the second start site can also be detected but less frequently than for the construct with a mutation of the original start codon (compare -S/TS with T-/TS). Accordingly, most reinitiation events still occur at the original site even when the genuine stop codon is mutated and a new potential AUG start site is introduced in connection with the new stop codon. Thus, the site, at which reinitiation occurs, is obviously not linked to the position of the termination signal. Again, initiation at the second start site is no longer detectable when this AUG is substituted to AGU (-S/T-, Fig. 9).
Mutation of both the original start and stop codon (–/TS) leads to a further reduction of reinitiation. Remarkably, reinitiation in the original stop/start region is still observed, despite the fact that termination takes place 15 codons further downstream and the original start codon is mutated.
For this construct (–/TS) also some VP2 translation is initiated at the downstream site. The respective band (asterisk in Fig. 9B) is specific because mutation of the corresponding AUG codon to AGU in plasmid –/T- leads to absence of this band.
Taken together, the results described above clearly show that the site, at which reinitiation occurs, is neither linked to the termination site nor relies on the presence of an AUG codon. Accordingly, the location of the TURBS defines the reinitiation position that most likely depends on a positioning effect of the TURBS-bound ribosome in a certain distance to the motif 2*/2 stem structure.
Downstream Initiation in Different Caliciviruses
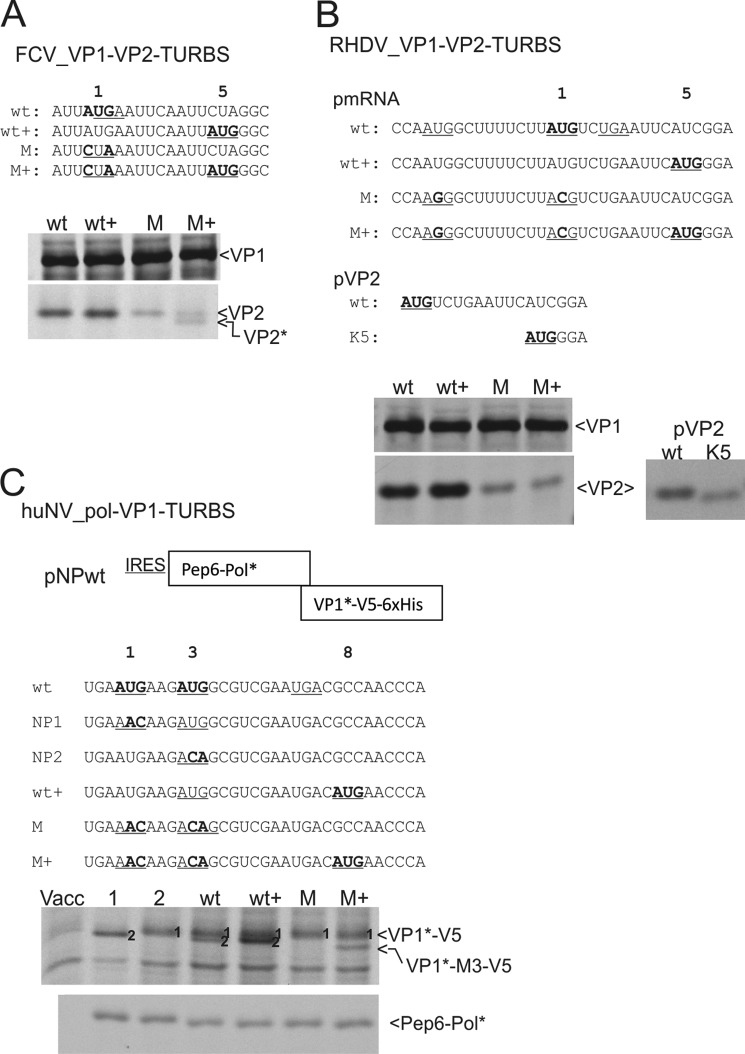
The data presented above clearly prove that in NV the reinitiating ribosome is able to start at sites downstream of the original start site. We wanted to know if this is a unique feature of the reinitiation mechanism in NV subgenomic viral mRNA, due to the fact that here the stop of the first frame (VP1 gene) is located upstream of the start codon of the second frame (VP2 gene). The TURBS in the subgenomic viral mRNAs of all other caliciviruses and also the reinitiation site in the genomic RNA of the noroviruses at the polymerase/VP1 border display an arrangement with the stop of the first frame located downstream of the start codon of the second frame (see Fig. 1). Therefore, we analyzed FCV and RHDV sequences for the ability to reinitiate at a downstream AUG (Fig. 10, A and B). In addition, we conducted similar analyses for the TURBS at the polymerase-VP1 border for huNV (Fig. 10C). Two sets of constructs were generated with the WT+ constructs containing the original start codon together with an AUG codon a few codons further downstream. The M+ constructs display a mutation of the former triplet together with the downstream AUG (Fig. 10). The WT and M constructs display the same sequences as WT+ and M+, respectively, but without the downstream AUG.
FIGURE 10.
Initiation at downstream AUGs in different caliciviral TURBS. A–C, top, sequences of the TURBS at the borders of the genes coding for the given proteins are shown. The top sequence represents the wild type construct with the start codon of the downstream frame highlighted in boldface and the stop codon of the upstream frame underlined. Below, sequences of the start codon and AUG substitution mutants are given with the substitutions within the start codon and the engineered downstream AUG codon given in boldface and underlined. Important codons are numbered above the sequences with 1 for the known reinitiation site The gels show the proteins precipitated after transient expression of the indicated constructs. A, analysis of the reinitiation at the VP1-VP2 TURBS in FCV. VP2*, VP2 resulting from initiation at the 5 codon, B, RHDV, analysis of the reinitiation for VP2 after translation of VP1. pVP2 represents a construct designed for expression of VP2 (WT) or a version lacking the N-terminal four residues (K5) established for demonstration of the size difference between the two expression products. C, huNV, analysis of the reinitiation for VP1 after translation of the polymerase. The construct generated for these analyses is given as a scheme on top. Initiation for VP1 translation at the first or second AUG of the wild type sequence is indicated within the gel by 1 or 2, respectively. VP1*-M3-V5, protein resulting from initiation at codon 8 of the VP1 coding frame.
For FCV, the basic construct is pCH1 (WT) (23), which contains the VP1 and VP2 coding frames. VP2 translation initiates at the original start site even when a second AUG is offered in the wild type sequence at position 5 of the VP2 frame (wt+, Fig. 10A). Mutation of the start codon to CUA led to a strong reduction of VP2 translation compared with the wild type (M, Fig. 10A) (23). Despite the mutation, initiation obviously took place at the altered original start site. When in this context a second AUG was offered downstream at codon 5 (M+), initiation at both the mutated original start codon and the downstream AUG occurred as demonstrated by the presence of two distinct bands (Fig. 10A).
For analysis of the RHDV TURBS, the construct pRmRNA was used. This plasmid contains both frames coding for the capsid proteins (24). Reinitiation for VP2 translation of RHDV is reduced when both possible AUG start codons of the TURBS are mutated (M, Fig. 10B) (24). No initiation is observed at an engineered downstream AUG (codon 5) even when both possible start AUGs in the TURBS region are mutated (M+, Fig. 10B). The expected size difference between the products initiated at the standard or the downstream sites was similar to that of the clearly separated FCV products. To prove that the downstream initiation product would have been detected, we engineered constructs for separate expression of the WT VP2 frame product and the polypeptide that would be generated by downstream initiation. A clearly detectable difference in migration rate was observed showing that the protein translated from the downstream site would be detected (Fig. 10B, right panel). To exclude that the position chosen for the introduced downstream AUG is not suitable for reinitiation, we also tested AUG codons engineered at two other positions, codon +4 or +7 of the VP2 coding frame. But also for these mutants the initiation of translation at the engineered downstream AUG was not detected (data not shown).
In a further step, we tested whether also the reinitiation of VP1 translation from the huNV genomic RNA can be started at a downstream AUG. We generated a construct analogous to the one used for analysis of the huNV VP2 translation, with the 3′ part of the polymerase coding sequence fused with a 5′ Pep6 tag and the 5′ part of the VP1 coding sequence combined with a 3′ V5 and His6 tag coding sequence (Fig. 10C). The TURBS upstream of the huNV VP1 start site (located within the polymerase coding sequence) has two AUGs that could be used for reinitiation. We mutated each of them individually (NP1 and NP2) or both of them (M, Fig. 10C) to learn which AUG is used for VP1 translation. Analysis of the wild type construct showed reinitiation at both AUGs (Fig. 10C). Mutation of one of the AUGs by the two-nucleotide substitution led to a reduced reinitiation frequency at the remaining AUG codon (NP1 and NP2, Fig. 10C). Engineering of a third AUG at codon 8 did not lead to a third translation product in the wild type sequence (wt+, Fig. 10C). Mutation of both original start AUGs led to reduced VP1 expression, and interestingly, the product starting at the second AUG codon (here ACA) was obviously missing for this start codon mutant (M, Fig. 10C). Introduction of a downstream AUG at codon 8 of the VP1 frame in context with the two mutated start codons led to initiation at this engineered start site (M+, band VP1*-M3-V5, Fig. 10C). Thus, the NV polymerase-VP1 TURBS can also drive reinitiation at a downstream start site.
DISCUSSION
Translation started by reinitiation after termination is one mechanism that allows expression of two or more proteins from one mRNA. In eukaryotes, cases of reinitiation after termination of translation of long ORFs have first been found in viral RNAs. The prototype of this mechanism has been described for caliciviruses (23, 24). Recently, a first case of coupled translation termination/reinitiation relying on a novel mechanism was found in cellular mRNAs (49).
In the cases of reinitiation on viral RNAs analyzed so far, just a small portion of the ribosomes restart translation of the second frame after termination of translation of the upstream ORF. This mechanism depends on the TURBS located in the 3′ coding region of the upstream ORF. TURBS regions are found in all caliciviruses upstream of the VP2 coding frame. For noroviruses, a second TURBS was identified upstream of the VP1 coding frame in the polyprotein gene. The importance of the three essential TURBS motifs was first shown for the FCV and RHDV VP1-VP2 TURBS and later on for bovine and murine Norovirus (40, 41) and human Norovirus (this work). In influenza B virus, translation of the proteins M1 and BM2 from the mRNA transcribed from segment 7 RNA is also driven by a TURBS (21, 50, 51). In all viral TURBS, the two coding frames overlap in a small region. Although we know the motifs essential for reinitiation, we do not know how this mechanism works in detail. Three open questions are as follows. What happens to the ribosome after termination of upstream frame translation? How is the start site for reinitiation determined? Why do only a few ribosomes reinitiate translation after termination of VP1 translation? Similarly, it is an open question whether the mechanism of AUG recognition and the variation in the level of reinitiation between the different caliciviruses from less than 1% (for huNV VP1 translation) to about 20% (for RHDV VP2 translation) are correlated.
We have shown that an AUG is not essential and that alternative start codons can be used for reinitiation (RHDV (24), FCV (23), and huNV, Fig. 3). For huNV, FCV, and influenza B, substitution of 1 nt of the start codon leads to reduction of reinitiation to about 50–60% of the wild type level (Fig. 3) (23, 51). For RHDV, the reduction is more prominent with a translation level of 30% for the mutant compared with the WT (24). A stronger effect on reinitiation frequency was observed when 2 nt of the AUG were substituted. This change leads to a reinitiation level of 30–40% for huNV (Fig. 3), FCV, and influenza B (23, 51). For complete substitution of the start codon, the reinitiation rate is still 12% in FCV but nearly 0 in RHDV. Thus, mutations affecting the start codon have in general a stronger effect in RHDV than in FCV, huNV, and influenza B. But despite sequence changes affecting the start codon, there is still a significant rate of initiation in the different viruses.
We have shown for FCV that at least most VP2 molecules translated from constructs with an altered start codon begin with a methionine. Thus, reinitiation depends on a Met-tRNA and therefore represents a de novo initiation (23). Interestingly, the Kozak context has no influence on the reinitiation rate at the original start site (NV58, Fig. 4) (51).
In summary, our data show that reinitiation of translation driven by the calicivirus TURBS neither depends on codon-anticodon pairing nor on the Kozak context. This finding raises the question after the principle that determines reinitiation start site selection. For huNV, we were able to show here that an AUA codon just upstream of the original start site can be used for reinitiation even though there is clearly a preference for AUG as the start site in this region (Fig. 4). Efficient initiation can occur at a downstream AUG, located up to four codons downstream of the original start site (Figs. 4 and 5). Thus efficient reinitiation in huNV is somewhat flexible within a few codons upstream or downstream of the original start site, but it shows at least a strong preference for the region very close to the original site even in the absence of a classical start codon. Similar results were also obtained for other caliciviruses. Accordingly, the preferred position for reinitiation must be defined by intrinsic features of the TURBS that are independent of the start site sequence. A reasonable explanation supported by all available data is that the distance from an essential element of the TURBS defines the start site.
The termination signal is essential for reinitiation but can be moved downstream leading to a less efficient but still prominent level of reinitiation (Fig. 3) (23, 51). Importantly, reinitiation in the termination site-shifted mutants still begins at the original start site. This is even true for mutants, where the total stop/start site is moved (huNV, Fig. 9). Thus, the site of initiation is not determined by location of the termination signal but probably by the distance to motif 2, e.g. the secondary structure is built by the interaction between motif 2* and motif 2. Because of the interaction of motif 1 with the 18 S rRNA of the 40 S ribosomal subunit, the bound post-termination ribosome is positioned for reinitiation within a certain distance from the TURBS structure, which ranges from 12 to 24 in different caliciviruses (23, 25).
For noroviruses, translation of VP1 from the genomic RNA could start at two AUGs located 12 or 18 nt downstream of motif 2, respectively. Indeed, both are used for reinitiation in the wild type sequence (Fig. 10C). However, only the first position is used for reinitiation in cases where both AUG codons are mutated. This finding indicates that a 12-nt distance from motif 2 is preferred when the start site alone is determined by the positioning of the ribosome on the genomic viral RNA TURBS of huNV. Also for RHDV, two AUG codons are found within the TURBS. One AUG is 8–10 nt downstream of motif 2, and the second is 20–22 nt. Only the second one is used for initiation (24), indicating that the first one is probably too close to the TURBS motif 2/2* stem and therefore is out of the start site window. The range of the possible start codon localization is probably determined by the size of the ribosome (40 S) bound to the TURBS (motif 1) and more precisely by the localization of the P-site of the ribosome on the RNA.
Recently, several cases of translation initiation have been analyzed that exhibit in part similar characteristics as those of the TURBS-driven reinitiation in caliciviruses. Examples are hepatitis C virus (HCV)-like IRES elements and the 5′-nontranslated region of the 26 S mRNA of Sindbis virus. These RNA elements were shown to bind and position small ribosomal subunits at the translational start site. Similar to reinitiation at a TURBS, initiation of translation occurs here without scanning and is not dependent on the presence of an AUG codon. eIF2 is not necessary for translation initiation driven by HCV-like IRESs or the 5′-nontranslated regions of Sindbis 26 S mRNA but can be replaced by a protein called ligatin or a combination of two truncated ligatin homologues (MCT-1 and DENR) (52). Alternatively, eIF2 can also be replaced by eIF5B, which is reminiscent of bacterial translation initiation where the eIF5B homologue IF2 accelerates binding of the initiator fMet-tRNAfiMet to the initiation complex (53, 54). It is not yet known whether the factors described above are or can also be active in TURBS-driven reinitiation. However, regardless of this point, there is a clear difference between HCV IRES-dependent initiation and the TURBS-driven reinitiation because ribosomes recruited by the HCV IRES cannot reach downstream start sites (55) as shown here for the huNV and FCV TURBS.
As mentioned above, reinitiation for huNV VP2 translation can occur at a downstream AUG when the original start codon is mutated. Also in this case, the main proportion of the VP2 protein is generated by initiation at the original start site, in accordance with the above described positioning effect, but a significant number of ribosomes obviously reinitiate at the engineered downstream site (Fig. 6). This finding is of interest because we can learn more about the parameters of AUG recognition during reinitiation. Initiation occurs at the next downstream AUG independent of the reading frame, indicating that the ribosome has to move along the RNA in a 3′ direction and has to recognize the AUG. This is reminiscent of the scanning process in standard cap-dependent translation initiation. In agreement with the scanning mechanism, the Kozak context is important for reinitiation efficiency at downstream sites (Fig. 7, NV52b).
Thus, start site selection during reinitiation after termination in caliciviruses can be achieved in two ways. The usual way depends on positioning of the TURBS-bound ribosome at the start site, which is obviously guided by the TURBS structure itself. In FCV and huNV codon-anticodon interaction between the AUG and the initiator-tRNA seems to contribute significantly to stabilizing the ribosome positioned at the start site. If this stabilizing effect is missing, a certain percentage of the ribosomes start moving in a 3′ direction and select a new start site. This occurs in a process dependent on an AUG in good Kozak sequence context, which is reminiscent of classical scanning. This scanning-like mode indicates that the 40 S subunit bound to the TURBS seems to get into a fully initiation-competent state loaded with the initiation factors and initiator t-RNA basically as in a standard scanning-based initiation. Recognition of an AUG start codon with its Kozak context follows this standard initiation process. This hypothesis is supported by the finding of Pöyry et al. (26) for FCV and Powell et al. (50) for influenza B virus that eIF3, an initiation factor that is involved in ribosome recycling and translation initiation, plays a role in the TURBS-dependent reinitiation of translation.
Interestingly, the scanning-like start site selection cannot be found in RHDV, where reinitiation occurs at the highest frequency. One possible explanation for this difference could be that the state of the post-termination ribosome bound to the TURBS differs between RHDV on the one hand and FCV and huNV on the other hand, which could be accomplished by differences with regard to the loaded initiation factors.
In contradiction to the scanning-like mode found for the FCV and huNV downstream initiation, Powell et al. (50) have shown for the TURBS-dependent reinitiation mechanism of influenza B virus that downstream initiation is not dependent on the scaffold initiation factor eIF4G. This finding suggests that the reinitiation site can be located in a scanning-independent manner in influenza B virus.
The recent work on start codon recognition for reinitiation in different caliciviruses discovers similarities and differences in the detailed requirements of the TURBS-dependent reinitiation of translation and so added significantly to our understanding of this special mechanism of translation initiation. It will be interesting to figure out in future experiments whether reinitiation works differently in caliciviruses.
Acknowledgments
We thank Maren Ziegler, Petra Wulle, and Gaby Stooss for excellent technical assistance. We are very grateful to Jacques Rohayem (Dresden, Germany) for providing the full-length huNV cDNA clone and to Axel Karger (Friedrich-Loeffler-Institut) and Dirk Albrecht (University of Greifswald) for intensive but finally unsuccessful attempts to get mass spectroscopic data on the expressed proteins.
This work was supported by Grant Me 1367/3 from the Deutsche Forschungsgemeinschaft.
C. Luttermann and G. Meyers, unpublished data.
- FCV
- feline calicivirus
- huNV
- human norovirus
- IRES
- internal ribosome entry site
- RHDV
- rabbit hemorrhagic disease virus
- TURBS
- termination upstream ribosomal binding site
- nt
- nucleotide
- HCV
- hepatitis C virus.
REFERENCES
- 1. Hinnebusch A. G. (2011) Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 75, 434–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kozak M. (1987) An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15, 8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kozak M. (1989) The scanning model for translation: An update. J. Cell Biol. 108, 229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kozak M. (1991) An analysis of vertebrate mRNA sequences: Intimations of translational control. J. Cell Biol. 115, 887–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ryabova L. A., Pooggin M. M., Hohn T. (2006) Translation reinitiation and leaky scanning in plant viruses. Virus Res. 119, 52–62 [DOI] [PubMed] [Google Scholar]
- 6. Cuesta R., Xi Q., Schneider R. J. (2001) Preferential translation of adenovirus mRNAs in infected cells. Cold Spring Harbor Symp. Quant. Biol. 66, 259–267 [DOI] [PubMed] [Google Scholar]
- 7. Fütterer J., Kiss-László Z., Hohn T. (1993) Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell 73, 789–802 [DOI] [PubMed] [Google Scholar]
- 8. Latorre P., Kolakofsky D., Curran J. (1998) Sendai virus Y proteins are initiated by a ribosomal shunt. Mol. Cell. Biol. 18, 5021–5031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park H. S., Himmelbach A., Browning K. S., Hohn T., Ryabova L. A. (2001) A plant viral “reinitiation” factor interacts with the host translational machinery. Cell 106, 723–733 [DOI] [PubMed] [Google Scholar]
- 10. Ryabova L. A., Pooggin M. M., Hohn T. (2002) Viral strategies of translation initiation: Ribosomal shunt and reinitiation. Prog. Nucleic Acids Res. Mol. Biol. 72, 1–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xi Q., Cuesta R., Schneider R. J. (2005) Regulation of translation by ribosome shunting through phosphotyrosine-dependent coupling of adenovirus protein 100k to viral mRNAs. J. Virol. 79, 5676–5683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hellen C. U., Sarnow P. (2001) Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15, 1593–1612 [DOI] [PubMed] [Google Scholar]
- 13. Pestova T. V., Kolupaeva V. G., Lomakin I. B., Pilipenko E. V., Shatsky I. N., Agol V. I., Hellen C. U. (2001) Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 98, 7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sarnow P., Cevallos R. C., Jan E. (2005) Takeover of host ribosomes by divergent IRES elements. Biochem. Soc. Trans. 33, 1479–1482 [DOI] [PubMed] [Google Scholar]
- 15. Bertram G., Innes S., Minella O., Richardson J., Stansfield I. (2001) Endless possibilities: Translation termination and stop codon recognition. Microbiology 147, 255–269 [DOI] [PubMed] [Google Scholar]
- 16. Dreher T. W., Miller W. A. (2006) Translational control in positive strand RNA plant viruses. Virology 344, 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brierley I., Dos Ramos F. J. (2006) Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res. 119, 29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dinman J. D., Icho T., Wickner R. B. (1991) A −1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc. Natl. Acad. Sci. U.S.A. 88, 174–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gould P. S., Easton A. J. (2005) Coupled translation of the respiratory syncytial virus M2 open reading frames requires upstream sequences. J. Biol. Chem. 280, 21972–21980 [DOI] [PubMed] [Google Scholar]
- 20. Gould P. S., Easton A. J. (2007) Coupled translation of the second open reading frame of M2 mRNA is sequence dependent and differs significantly within the subfamily Pneumovirinae. J. Virol. 81, 8488–8496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Horvath C. M., Williams M. A., Lamb R. A. (1990) Eukaryotic coupled translation of tandem cistrons: identification of the influenza B virus BM2 polypeptide. EMBO J. 9, 2639–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kozak M. (1987) Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol. 7, 3438–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luttermann C., Meyers G. (2007) A bipartite sequence motif induces translation reinitiation in feline calicivirus RNA. J. Biol. Chem. 282, 7056–7065 [DOI] [PubMed] [Google Scholar]
- 24. Meyers G. (2003) Translation of the minor capsid protein of a calicivirus is initiated by a novel termination-dependent reinitiation mechanism. J. Biol. Chem. 278, 34051–34060 [DOI] [PubMed] [Google Scholar]
- 25. Meyers G. (2007) Characterization of the sequence element directing translation reinitiation in RNA of the calicivirus rabbit hemorrhagic disease virus. J. Virol. 81, 9623–9632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pöyry T. A., Kaminski A., Connell E. J., Fraser C. S., Jackson R. J. (2007) The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes Dev. 21, 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hinnebusch A. G. (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 [DOI] [PubMed] [Google Scholar]
- 28. Jackson R. J. (2005) Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 33, 1231–1241 [DOI] [PubMed] [Google Scholar]
- 29. Chappell S. A., Dresios J., Edelman G. M., Mauro V. P. (2006) Ribosomal shunting mediated by a translational enhancer element that base pairs to 18 S rRNA. Proc. Natl. Acad. Sci. U.S.A. 103, 9488–9493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith E., Meyerrose T. E., Kohler T., Namdar-Attar M., Bab N., Lahat O., Noh T., Li J., Karaman M. W., Hacia J. G., Chen T. T., Nolta J. A., Müller R., Bab I., Frenkel B. (2005) Leaky ribosomal scanning in mammalian genomes: significance of histone H4 alternative translation in vivo. Nucleic Acids Res. 33, 1298–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vagner S., Galy B., Pyronnet S. (2001) Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2, 893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stoneley M., Willis A. E. (2004) Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene 23, 3200–3207 [DOI] [PubMed] [Google Scholar]
- 33. Green K. Y., Ando T., Balayan M. S., Berke T., Clarke I. N., Estes M. K., Matson D. O., Nakata S., Neill J. D., Studdert M. J., Thiel H. J. (2000) in Virus Taxonomy (Regenmortel M. H., Fauquet C. M., Bishop D. H., eds) pp. 725–734, Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- 34. Clarke I. N., Lambden P. R. (2000) Organization and expression of calicivirus genes. J. Infect. Dis. 181, S309–S316 [DOI] [PubMed] [Google Scholar]
- 35. Rohayem J., Robel I., Jäger K., Scheffler U., Rudolph W. (2006) Protein-primed and de novo initiation of RNA synthesis by norovirus 3Dpol. J. Virol. 80, 7060–7069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chaudhry Y., Nayak A., Bordeleau M. E., Tanaka J., Pelletier J., Belsham G. J., Roberts L. O., Goodfellow I. G. (2006) Caliciviruses differ in their functional requirements for eIF4F components. J. Biol. Chem. 281, 25315–25325 [DOI] [PubMed] [Google Scholar]
- 37. Daughenbaugh K. F., Fraser C. S., Hershey J. W., Hardy M. E. (2003) The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. EMBO J. 22, 2852–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goodfellow I., Chaudhry Y., Gioldasi I., Gerondopoulos A., Natoni A., Labrie L., Laliberté J. F., Roberts L. (2005) Calicivirus translation initiation requires an interaction between VPg and eIF 4 E. EMBO Rep. 6, 968–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Luttermann C., Meyers G. (2009) The importance of inter- and intramolecular base pairing for translation reinitiation on a eukaryotic bicistronic mRNA. Genes Dev. 23, 331–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Napthine S., Lever R. A., Powell M. L., Jackson R. J., Brown T. D., Brierley I. (2009) Expression of the VP2 protein of murine norovirus by a translation termination-reinitiation strategy. PLoS One 4, e8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McCormick C. J., Salim O., Lambden P. R., Clarke I. N. (2008) Translation termination reinitiation between open reading frame 1 (ORF1) and ORF2 enables capsid expression in a bovine norovirus without the need for production of viral subgenomic RNA. J. Virol. 82, 8917–8921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wyatt L. S., Moss B., Rozenblatt S. (1995) Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology 210, 202–205 [DOI] [PubMed] [Google Scholar]
- 43. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 44. Fricke J., Voss C., Thumm M., Meyers G. (2004) Processing of a pestivirus protein by a cellular protease specific for light chain 3 of microtubule-associated proteins. J. Virol. 78, 5900–5912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schägger H., von Jagow G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 46. Peabody D. S. (1989) Translation initiation at non-AUG triplets in mammalian cells. J. Biol. Chem. 264, 5031–5035 [PubMed] [Google Scholar]
- 47. Kozak M. (1986) Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44, 283–292 [DOI] [PubMed] [Google Scholar]
- 48. Lim P. S., Jenson A. B., Cowsert L., Nakai Y., Lim L. Y., Jin X. W., Sundberg J. P. (1990) Distribution and specific identification of papillomavirus major capsid protein epitopes by immunocytochemistry and epitope scanning of synthetic peptides. J. Infect. Dis. 162, 1263–1269 [DOI] [PubMed] [Google Scholar]
- 49. Gould P. S., Dyer N. P., Croft W., Ott S., Easton A. J. (2014) Cellular mRNAs access second ORFs using a novel amino acid sequence-dependent coupled translation termination-reinitiation mechanism. RNA 20, 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Powell M. L., Leigh K. E., Pöyry T. A., Jackson R. J., Brown T. D., Brierley I. (2011) Further characterisation of the translational termination-reinitiation signal of the influenza B virus segment 7 RNA. PLoS One 6, e16822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Powell M. L., Napthine S., Jackson R. J., Brierley I., Brown T. D. (2008) Characterization of the termination-reinitiation strategy employed in the expression of influenza B virus BM2 protein. RNA 14, 2394–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Skabkin M. A., Skabkina O. V., Dhote V., Komar A. A., Hellen C. U., Pestova T. V. (2010) Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 24, 1787–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Terenin I. M., Dmitriev S. E., Andreev D. E., Shatsky I. N. (2008) Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol. 15, 836–841 [DOI] [PubMed] [Google Scholar]
- 54. Pestova T. V., de Breyne S., Pisarev A. V., Abaeva I. S., Hellen C. U. (2008) eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J. 27, 1060–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rijnbrand R. C., Abbink T. E., Haasnoot P. C., Spaan W. J., Bredenbeek P. J. (1996) The influence of AUG codons in the hepatitis C virus 5′ nontranslated region on translation and mapping of the translation initiation window. Virology 226, 47–56 [DOI] [PubMed] [Google Scholar]