Background: In cyanobacteria, starvation-induced phycobilisome degradation is caused by NblA.
Results: Synechocystis expresses two NblA proteins that form a heterodimer. The heterodimer binds phycobiliproteins and ClpC and is degraded by a ClpC-ClpP1ClpR protease in vitro.
Conclusion: NblA1/NblA2 is an adaptor protein that mediates degradation of phycobilisomes by the ATP-dependent protease ClpC-ClpP1ClpR.
Significance: These findings improve our understanding of the mechanisms by which cyanobacteria adapt to changing environmental conditions.
Keywords: Cyanobacteria, Fluorescence Resonance Energy Transfer (FRET), Protease, Protein Degradation, Protein Synthesis, NblA, Synechocystis, Nitrogen Starvation, Phycobilisome Degradation
Abstract
When cyanobacteria acclimate to nitrogen deficiency, they degrade their large (3–5-MDa), light-harvesting complexes, the phycobilisomes. This massive, yet specific, intracellular degradation of the pigmented phycobiliproteins causes a color change of cyanobacterial cultures from blue-green to yellow-green, a process referred to as chlorosis or bleaching. Phycobilisome degradation is induced by expression of the nblA gene, which encodes a protein of ∼7 kDa. NblA most likely acts as an adaptor protein that guides a Clp protease to the phycobiliproteins, thereby initiating the degradation process. Most cyanobacteria and red algae possess just one nblA-homologous gene. As an exception, the widely used “model organism” Synechocystis sp. PCC6803 expresses two such genes, nblA16803 and nblA26803, both of whose products are required for phycobilisome degradation. Here, we demonstrate that the two NblA proteins heterodimerize in vitro and in vivo using pull-down assays and a Förster energy-transfer approach, respectively. We further show that the NblA proteins form a ternary complex with ClpC (the HSP100 chaperone partner of Clp proteases) and phycobiliproteins in vitro. This complex is susceptible to ATP-dependent degradation by a Clp protease, a finding that supports a proposed mechanism of the degradation process. Expression of the single nblA gene encoded by the genome of the N2-fixing, filamentous cyanobacterium Nostoc sp. PCC7120 in the nblA1/nblA2 mutant of Synechocystis sp. PCC6803 induced phycobilisome degradation, suggesting that the function of the NblA heterodimer of Synechocystis sp. PCC6803 is combined in the homodimeric protein of Nostoc sp. PCC7120.
Introduction
Cyanobacterial cultures growing in a nutrient-rich medium exhibit a typical blue-green color due to the photosynthetic pigments chlorophyll a and phycobiliproteins. Together with mostly uncolored linker proteins, phycobiliproteins form the phycobilisome (PBS).2 The PBS is a large (3–5 MDa) multiprotein complex associated with the cytoplasmic side of the thylakoid membranes (reviewed in Refs. 1–4). In addition to the non-pigmented linker proteins, the complex is composed of the chromophorylated phycobiliproteins phycocyanin (PC; λmax ≈ 620 nm), allophycocyanin (λmax ≈ 650 nm), and, in some cyanobacteria, phycoerythrin (λmax ≈ 565 nm) or phycoerythrocyanin (λmax ≈ 590 nm). The linker proteins are necessary for the assembly and stabilization of the PBS structure through their interactions with phycobiliprotein subunits. They also play a role in energy transfer from the PBS to photosystem II and, in some cyanobacteria, to photosystem I as well.
PBSs are classified according to their morphology. The most common PBS structure is the hemidiscoidal type, which is made up of a core complex that is anchored in the thylakoid membrane and peripheral rods (1, 3, 5). Size, composition, and position of PBSs on the thylakoid membrane vary according to light quality and quantity so as to optimize light absorption (6–9). Nearly complete degradation of PBSs can be observed in unicellular, non-diazotrophic cyanobacteria upon nitrogen starvation and, in some species, also upon sulfur depletion. Degradation begins with the sequential loss of the peripheral rods and terminates with degradation of the core complex (10, 11). Because phycobiliproteins constitute up to 50% of the total soluble protein of a cyanobacterial cell (2), a considerable amount of cellular protein becomes degraded within the first 24 h of nitrogen deprivation. The loss of phycobiliproteins results in a color change of cyanobacterial cultures from blue-green to yellow-green, a process referred to as bleaching or chlorosis (12). PBS degradation is not essential for acclimation to nutrient deprivation but is rather thought to prevent photo damage under stress and to provide substrates for the de novo synthesis of proteins required for the acclimation process (11, 13).
The small protein NblA (non-bleaching A) plays a key role in the degradation process (14). Cyanobacterial mutants lacking NblA degrade virtually none of their PBS under nitrogen starvation and thus exhibit a non-bleaching phenotype (14–17). The exact function of NblA has been the subject of numerous studies. Binding experiments have shown that NblA interacts with the α-subunits of phycobiliproteins (18, 19). In pull-down experiments, NblA was found to bind to ClpC, an HSP100 chaperone partner of a Clp protease, in an ATP-dependent manner (20). This result led to a proposed model of PBS degradation in which NblA acts as a so-called adaptor protein of a Clp protease (20). Clp degradation complexes consist of two functional elements: a cylinder-like proteolytic core of two heptameric rings and an AAA+ chaperone (21, 22). The hexameric chaperone ring is responsible for substrate recognition, unfolding, and threading of the extended polypeptide chain through a narrow pore into the protease compartment, where the proteolytically active sites are sequestered from the cytoplasm (23, 24). The substrate specificity of a Clp protease is determined by its chaperone partner. Substrate recognition occurs by binding to the chaperone partner of the Clp protease or is mediated by adaptor proteins (25, 26). Adaptor proteins simultaneously bind the substrate and the chaperone, thereby forming a ternary complex that causes degradation of the substrate. In other words, adaptor proteins modulate the substrate specificity of a protease, enabling the degradation of particular proteins at the proper time. This means that substrate degradation is solely regulated by the expression of the adaptor protein. This is precisely what is observed in PBS degradation, which is induced by the expression of the small protein NblA (14). Because the Clp proteases require NblA to recognize phycobiliproteins as substrates, strains lacking NblA degrade the PBS at a low rate, even under conditions of nitrogen starvation (14, 15, 17).
Most phycobiliprotein-containing cyanobacteria and red algae possess nblA genes. The NblA proteins are rather small, consisting of ∼60 amino acids. Sequence alignment of NblA proteins reveals a low homology (about 30% sequence identity on average) (27). Short stretches of highly conserved amino acid residues located near the N and C termini of the proteins are involved in phycobiliprotein and ClpC binding (19, 20). NblA proteins from Nostoc sp. PCC7120 (Nostoc 7120), Thermosynechococcus vulcanus, and Synechococcus elongatus sp. PCC7942 (Synechococcus 7942) have been crystallized (19, 27). All three NblA proteins share a similar structure. They are homodimers that consist of two α-helices: one shorter N-terminal helix and a longer C-terminal helix. The biologically active form of NblA is thus a four-helix bundle.
Cyanobacteria and red algae usually possess one nblA gene. However, two genes have been found in Nostoc 7120 and Synechocystis sp. PCC6803 (Synechocystis 6803). Whereas only one of the two nblA genes is expressed upon nitrogen deprivation in Nostoc 7120 and is sufficient for PBS degradation (17), both genes are expressed in Synechocystis 6803, and both are necessary for induction of the degradation process (15, 16).
In this study, we investigated why the two NblA proteins, NblA16803 and NblA26803, are necessary for PBS degradation in Synechocystis 6803. We present evidence that NblA16803 and NblA26803 act as a heterodimer in which NblA16803 mediates the binding to ClpC6803 and phycobiliproteins. Both NblA proteins are degraded by a Clp protease in vitro, a finding that strongly supports the proposed model of the role of NblA in PBS degradation. We further show that expression of the single nblA gene from Nostoc 7120 complements the non-bleaching phenotype of the nblA1/nblA2 double mutant of Synechocystis 6803.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Strains are listed in supplemental Table S1. Unless stated otherwise, sources of plasmids, antibiotics, medium, restriction enzymes, and chemicals were as described previously (20).
Synechocystis 6803 and its mutants were grown photoautotrophically under constant illumination essentially as described before (17) except that the growth temperature was 28 °C, and the internal diameter of the culture vessels was 3.5 cm. Whole-cell absorbance spectra were recorded from 550 to 750 nm on a Specord® 200 PLUS spectrophotometer (Analytik Jena, Jena, Germany) and were corrected for cell scattering at 750 nm.
Strains of Escherichia coli were grown under standard conditions (28). When appropriate, antibiotics were added to the medium to final concentrations of 50 μg ml−1 ampicillin, 50 μg ml−1 kanamycin, or 25 μg ml−1 chloramphenicol, respectively.
Plasmid and Mutant Constructions
Plasmids and oligonucleotides are listed in supplemental Table S1 and Fig. S1.
Construction of Plasmids for Protein Overexpression in E. coli
Vectors for expression of GST-tagged NblA16803 and NblA26803 in E. coli were constructed as follows. The plasmid pGEX-2TK/NdeI (20) was digested with NdeI and SmaI in order to obtain the fragment encoding GST, the fragment was ligated into the expression vector pACYCDuet-1, which was digested with NdeI and EcoRV prior to ligation. The resultant plasmid pACYC/GST was digested with NdeI and, after dephosphorylation, was ligated to a fragment harboring nblA16803 (ssl0452) or nblA26803 (ssl0453), respectively, yielding plasmids pACYC/A1_GST and pACYC/A2_GST. Fragments encoding nblA16803 and nblA26803 were generated by PCR from genomic Synechocystis 6803 DNA using primers nbl7 and nbl28 (amplification of nblA16803) or nbl24 and nbl31 (amplification of nblA26803). Each primer sequence contained an NdeI restriction site for cloning. The primers nbl28 and nbl31 additionally contained a sequence encoding the PreScissionTM protease recognition sequence, enabling cleavage of the NblA16803/NblA26803-GST fusion protein. For coexpression of His-tagged NblA16803 and GST-tagged NblA26803, the nblA16803 gene was amplified by PCR using primers nbl32 (EcoRI site incorporated) and nblA-4 (HindIII site incorporated) and was ligated into plasmid pACYC/A2_GST to yield plasmid pACYC/His_A1 and A2_GST. For simultaneous expression of His-tagged NblA26803 and GST-tagged NblA16803, nblA26803 was amplified by PCR using primers nbl33 (HindIII site inserted) and nbl34 (EcoRI site inserted). The PCR fragment was cloned into the plasmid pACYC/A1_GST, generating plasmid pACYC/His_A2 and A1_GST. For coexpression of NblA26803 and GST-tagged NblA16803, the nblA26803 gene was amplified by PCR using the primer nbl14 (NcoI site incorporated) and nbl21 (EcoRI site incorporated). The resulting PCR fragment was ligated into plasmid pACYC/A1_GST, generating plasmid pACYC/A2 and A1_GST. For binding experiments, the ClpC6803 protein was expressed without any affinity tag. For construction of the expression plasmid, clpC6803 (sll0020) was amplified by PCR from total Synechocystis 6803 DNA using primers clpC_3 (NdeI site inserted) and clpC_5 (BamHI site inserted). The PCR fragment was digested with NdeI (partial digestion) and BamHI and cloned into plasmid pET11a, yielding plasmid pET11/ClpC. For degradation experiments, the ClpC6803 protein was expressed with an N-terminal His tag. ClpC6803 was amplified by PCR with clpC_3 (NdeI site inserted) and clpC_5 (BamHI site inserted) and ligated into plasmid pET22b, resulting in plasmid pET22b/HisClpC6803. For expression of the phycocyanin α-subunit N-terminally fused to GST, the chromosomal gene cpcA (sll1578) from Synechocystis 6803 was amplified by PCR using primers cpcA_1 (BamHI site inserted) and cpcA_2 (EcoRI site inserted) and cloned into plasmid pGEX-2TK via BamHI and EcoRI to yield plasmid pGEX/cpcA. For expression of HisClpP1 and ClpR, the clpP1 (slr0542) gene was first amplified with primers clpP1_1 (NcoI site incorporated) and clpP1_2 (NcoI site incorporated) from total Synechocystis 6803 DNA. The PCR product was digested with NcoI cloned into plasmid pACYCDuet-1, yielding the plasmid pACYC/HisClpP1. The clpR (slr0164) gene was then PCR-amplified with primers clpR_1 (NdeI site inserted) and clpR_2 (XhoI site inserted). The PCR fragment was digested with NdeI and XhoI and cloned into the plasmid pACYC/HisClpP1, resulting in the plasmid pACYC/HisClpP1, ClpR.
All DNA constructs were checked by DNA sequencing. All plasmids and oligonucleotides are listed in supplemental Table S1.
PBS Isolation from Synechocystis 6803 Crude Extract
The PBS preparation was performed as described before (29).
Coexpression and Purification of HisClpP1 and ClpR and Expression of His-tagged ClpC6803
After induction with isopropyl β-d-1-thiogalactopyranoside (IPTG) for 3 h at 30 °C, cells were pelleted and resuspended in buffer A (10 mm Tris-HCl buffer (pH 7.5), 75 mm KCl, 1 mm DTT). Cells were disrupted by sonication with a homogenizer (Sonopuls) for 2 × 2 min (settings: KS76, pulsation 2, 55% power), and the extract was clarified by centrifugation and incubated with Protino® Ni-IDA resins (Macherey-Nagel). Bound proteins were purified according to the manufacturer's protocol and eluted with buffer A containing 250 mm imidazole, followed by buffer change and concentration with a centrifugal concentrator (Vivaspin® 500, molecular weight cut-off 10,000) in buffer A.
Coexpression and Purification of NblA26803 and GST-tagged NblA16803
After induction with IPTG overnight at 18 °C, cells were pelleted, resuspended in buffer B (10 mm Tris-HCl buffer (pH 7,5), 200 mm KCl, 20 mm NaCl, 20 mm MgCl2, 10% glycerol), cell-free extracts were prepared as described above. The soluble crude extract was incubated with glutathione-agarose (Macherey-Nagel). After washing with buffer B, the bound proteins were eluted using Pre-ScissionTM protease according to the manufacturer's protocol (GE Healthcare). The NblA16803 and NblA26803 proteins were further purified by size exclusion chromatography on a Superdex 75TM column (GE Healthcare).
Coexpression and Purification of the NblA16803 and NblA26803 Heterodimers
GST-NblA16803 and His-NblA26803 or His-NblA16803 and GST-NblA26803 were coexpressed after induction with IPTG (overnight, 18 °C). Crude extracts were divided; one half was incubated with glutathione-agarose (Macherey-Nagel) for binding of GST-tagged NblA, and the other half was incubated with Protino® Ni-IDA resins (Macherey-Nagel) for binding of His-tagged NblA. Purification was performed as described above. Eluted proteins were immediately analyzed by Tricine-SDS-PAGE with 6 m urea (30).
Protein Expression, Purification, and in Vitro Binding Assays Using GST-tagged PC or GST-tagged NblA16803/NblA26803
The N-terminally GST-tagged α-subunit of PC (GST-PC) was expressed after induction with IPTG for 3 h at 30 °C and purified according to the manufacturer's protocol (GE Healthcare). Expression of the C-terminally GST-tagged NblA16803 (NblA16803-GST) was induced with IPTG for 80 h at 18 °C. NblA26803-GST was expressed with IPTG overnight at 18 °C. GST fusion proteins were purified as described above. GST-free NblA16803 and NblA26803 were obtained by on-column cleavage with PreScissionTM protease (GE Healthcare). The NblA16803-GST and His-tagged NblA26803 were purified as described above. The expression of ClpC6803 was induced by IPTG for 3 h at 30 °C. The soluble fraction of the crude extract containing overexpressed ClpC6803 without affinity tag was used for the in vitro binding assays.
In vitro binding assays were started by adding glutathione-agarose to the soluble cell extract containing the overexpressed GST-PC fusion protein. After incubation for 30 min at room temperature with gentle agitation, unbound proteins were removed by three washes with five column volumes of buffer B. The PC-loaded agarose was divided into four equal aliquots and incubated with crude extract containing overexpressed ClpC6803. Additionally ATP, NblA16803, NblA26803, or NblA16803 and His-NblA26803 were added in surplus. After incubation for 30 min at room temperature with gentle agitation, unbound proteins were removed by washing with buffer B containing 1 mm ATP as described above. Bound proteins were eluted by the addition of 40 mm glutathione in 50 mm Tris-HCl (pH 8.0). Eluted proteins were directly analyzed by Tricine-SDS-PAGE with 6 m urea.
In Vitro Degradation of α-Casein, PBS, NblA16803, and NblA26803 by ClpC6803-ClpP1-ClpR
Each purified protein was used in a final concentration of 2 μm in buffer C (50 mm Tris-HCl (pH 7.5), 75 mm KCl, 1 mm DTT, 5 mm MgCl2). All reactions were performed at 37 °C with an ATP-regenerating system consisting of 20 ng of pyruvate kinase (Roche Applied Science), 4 mm phosphoenolpyruvate, and 2 mm ATP. Degradation of α-casein (Sigma-Aldrich), the NblA16803/NblA26803 heterodimers, and the PBSs was analyzed by Tricine-SDS-PAGE in the presence of 6 m urea.
Cloning and Expression of Fluorescent Proteins in Synechocystis 6803
For FRET measurements, the donor cerulean (Cer) and the acceptor YFP were used. Due to the spectral overlap between donor (Cer) emission (excitation/emission maxima, 433/475 nm) and acceptor (YFP) absorbance (excitation/emission maxima, 514/527 nm), excitation of the donor molecule leads to emission from the acceptor molecule when the proteins are in close proximity (1–10 nm). For labeling, we used the monomeric forms of YFP and Cer. These forms were generated by a single amino acid exchange (A206K) in order to eliminate self-association of the proteins and thus avoid false positive FRET signals (31).
For expression of fluorescent NblA proteins in Synechocystis 6803, a ∼1480-bp fragment containing nblA16803 and nblA26803, flanked by putative promoter and terminator regions, was amplified using primers nbl11 (XhoI incorporated) and nbl27 (XbaI incorporated). The PCR product was digested by the restriction enzymes XbaI and XhoI and ligated into the XbaI-XhoI-digested plasmid pET22b, yielding plasmid pET/A1 and A2, which served to introduce an NcoI site at the 3′- or 5′-end of the nblA16803 coding region and an NdeI site at the 3′- or 5′-end of the nblA26803 coding region. The restriction sites were successively generated by mutagenesis (QuikChange® site-directed mutagenesis kit; Stratagene) using primers nblA1QCM1 and nblA1QCM2 (3′-end) or nblA1QCM3 and nblA1QCM4 (5′-end) as well as nblA2QCM1 and nblA2QCM2 (3′-end) or nblA2QCM3 and nblA2QCM4 (5′-end). Mutagenesis at the 3′-end of the nblA16803 gene resulted in the change of the base triplet for Leu61 to a methionine residue and the concomitant loss of the base triplet for one amino acid (Gly62). The amino acid sequence of the NblA26803 protein was altered by site-directed mutagenesis from Leu59 to a histidine residue and from Pro60 to a methionine residue, whereby the mutagenesis of both 5′-ends did not result in any changes. The resultant plasmids were restricted with NcoI or NdeI and, after dephosphorylation, ligated to a PCR fragment harboring the coding region for yfp or mCerulean, yielding the plasmids pET22/A1_Y and A2, pET22/A1 and A2_C, pET22/Y_A1 and A2, and pET22/A1 and A2_C. The yfp gene was amplified by PCR using primers YFP1 (NcoI site inserted) and YFP2 (NcoI site inserted) and plasmid pEmYFP as a template. The fragment coding for mCerulean was PCR-amplified using primers CFP1 (NdeI site inserted) and CFP2 (NdeI site inserted); here the plasmid pEmCer served as a template. Using the described cloning strategy, we also constructed the plasmids pET22/A1_Y and A2_C and pET22/A1_Y and A2_C for coexpression of NblA16803 and NblA26803 C-terminally or N-terminally fused to different fluorophores in Synechocystis 6803 cells.
For the FRET controls, each of the two fluorophores used in this study, mCerulean and YFP, was C-terminally linked to an NblA26803 molecule. To this end, the pET22/A1_Y and A2_C constructs described above were used to generate a second NcoI restriction site at the 5′-end of the nblA16803 coding region by QuikChange® mutagenesis using primers nblA1QCM3 and nblA1QCM4. The resultant plasmid pVZ/Y and A2_C was partially restricted with NcoI, dephosphorylated, and ligated to a PCR fragment harboring the coding region of nblA26803, yielding plasmid pET22/A2_Y and A2_C. The nblA26803 fragment was amplified with the primers nbl14 and nbl37, whereby the inserted NcoI site at the 3′-end led to a change from Leu59 to a proline residue and from Pro60 to a tryptophan residue. In addition to the A2_Y and A2_C control, we also used the yielded construct pVZ/Y and A2_C as a control.
All DNA constructs were confirmed by DNA sequence analysis. Finally, the plasmids were restricted with XbaI and XhoI, obtaining DNA fragments for expression of fluorophor-tagged nblA16803 and nblA26803 together with the promoter and terminator region, which were then ligated into the conjugative, self-replicating plasmid pVZ321. The plasmids pVZ/A1_Y and A2, pVZ/A1 and A2_C, pVZ/A1_Y and A2_C, pVZ/Y_A1 and A2, pVZ/A1 and C_A2, pVZ/Y_A1 and Cer_A2, and pVZ/A2_Y and A2_C were transferred to the nblA1/nblA2 double mutant of Synechocystis 6803, and the plasmid pVZ/YFP and A2_Cer was transferred to Δnbl2 mutant cells of Synechocystis 6803 by conjugation (32). Exconjugants were selected on BG11 agar containing 50 μg ml−1 kanamycin and 14 μg ml−1 chloramphenicol.
FRET Measurements
The FRET signal was detected with a fluorescence spectrometer (FluoroMax-4-Horiba) in a suspension of living Synechocystis 6803 cells with an A750 of ∼0.3. Cer expressed in the cells was excited by monochromatic 458-nm light, and fluorescence emission spectra were recorded in the wavelength range from 500 to 600 nm.
Fluorescence Lifetime Measurements
Fluorescence lifetime measurements were performed using a FluoTime 200 instrument (PicoQuant) in suspension of living Synechocystis 6803 cells with an A750 of ∼0.3 as described before (33).
Complementation Experiments with NblA of Nostoc 7120
The plasmid for expression of nblA from Nostoc 7120 was constructed as follows. A 669-bp fragment, bearing upstream sequences of nblA16803 (ssl0452) and nblA26803 (ssl0453) and a 417-bp fragment containing downstream sequences of nblA16803 and nblA26803, was amplified by PCR from total Synechocystis 6803 DNA using primers nbl11 (XhoI site inserted) and nbl13 (NcoI site inserted) and primers nbl26 (NdeI site inserted) and nbl27 (XbaI site inserted), respectively. The fragments were cloned into the plasmid pET22b, yielding plasmid pET22/P6803T6803. Subsequently, the plasmid pET22/P6803T6803 was digested with NcoI and NdeI and ligated to a fragment harboring the nblA gene from Nostoc 7120, which was amplified by PCR from total Nostoc 7120 DNA using primers nblana2.23 (NcoI site inserted) and nblana2.24 (NdeI site inserted), yielding plasmid pET22/P6803NblA7120T6803. The fragment harboring the nblA gene from Nostoc 7120, flanked by the putative promoter and terminator regions of nblA16803 and nblA26803 from Synechocystis 6803, was excised by digestion with XbaI and XhoI and inserted into the conjugative, self-replicating plasmid pVZ321 (32), replacing the plasmid's Km resistance cassette, yielding plasmid pVZ/P6803NblA7120T6803. The plasmid pVZ/P6803NblA7120 was transferred to nblA1/nblA2 double mutant cells by conjugation, and exconjugants were selected on BG11 agar containing 50 μg ml−1 kanamycin and 14 μg ml−1 chloramphenicol.
Zinc-induced Fluorescence
For zinc-induced fluorescence, the Tricine-SDS-PAGE was run with 1 mm zinc acetate in cathode buffer (30). The fluorescence was visualized by a UV transilluminator (Bio-Rad).
RESULTS
Homodimeric NblA of Nostoc 7120 Complements the Non-bleaching Phenotype of the nblA1/nblA2 Double Mutant of Synechocystis 6803
Genomes of most cyanobacteria, such as Synechococcus 7942 and Nostoc 7120, encode just one nblA gene, whose expression suffices to initiate the proteolytic degradation of the PBS. The intensively studied unicellular strain Synechocystis 6803, however, simultaneously expresses two sequence-related nblA genes, nblA16803 and nblA26803, to induce PBS degradation (15, 16).
Complementation experiments were performed to determine whether the two NblA proteins from Synechocystis 6803 act in the same manner as NblA from Nostoc 7120 (20). To this end, the coding region of Nostoc 7120 nblA (ORF asr4517) and a DNA fragment encoding Synechocystis 6803 nblA16803 and nblA26803 were amplified by PCR. Both fragments were ligated into the self-replicating plasmid pVZ321 containing the predicted promoter and terminator regions of Synechocystis 6803 nblA16803 and nblA26803, which are cotranscribed from a common promoter (15). The generated plasmids were subsequently transferred to the Synechocystis 6803 nblA1/nblA2 double mutant by triparental mating (34).
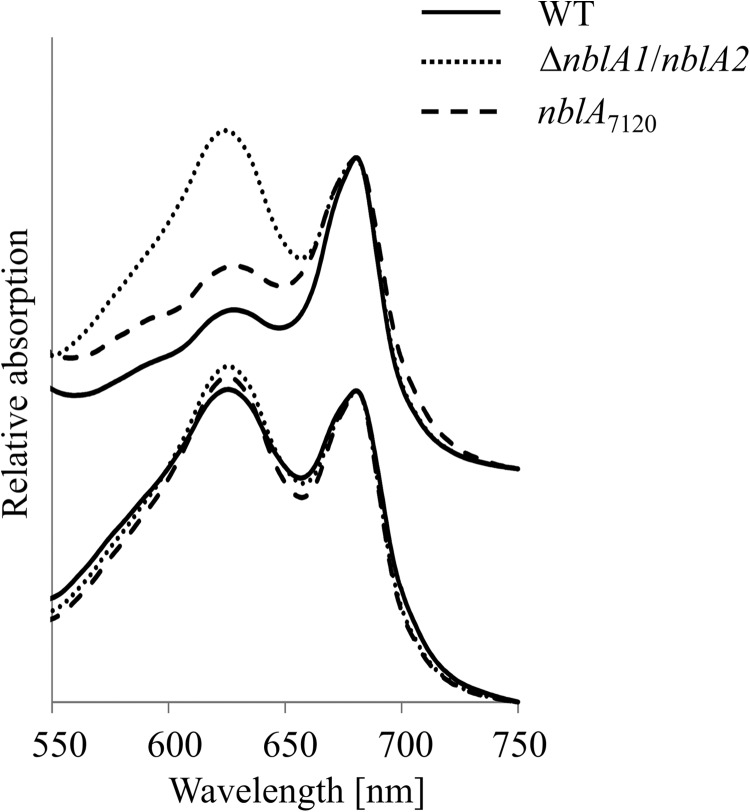
When grown in nitrate-containing medium, the pigmentation of the resulting exconjugants was indistinguishable from that of the wild type, as judged from whole-cell absorbance spectra (Fig. 1). After transfer to nitrogen-free medium, nblA16803 and nblA26803, expressed in trans in the nblA1/nblA2 double mutant, complemented the mutant phenotype, visible in the absorbance spectra shown in Fig. 1 as a decrease in the PC absorbance peak at ∼625 nm. A similar complementation effect was obtained with nblA from Nostoc 7120. This clearly demonstrates that the homodimeric NblA from Nostoc 7120 is able to replace the function of NblA16803 and NblA26803 in Synechocystis 6803.
FIGURE 1.
Complementation experiment. Absorbance spectra of the Synechocystis 6803 wild type, the nblA1/nblA2 double mutant of Synechocystis 6803, and the nblA1/nblA2 mutant of Synechocystis 6803 complemented in trans with nblA from Nostoc 7120, either grown in nitrogen-replete medium (+N) or starved of combined nitrogen for 24 h (−N), are shown.
Detecting the Interaction between NblA16803 and NblA26803 in Vivo by FRET
The biologically active form of NblA from Nostoc 7120 is a homodimer (19). Dimer formation seems to be a characteristic feature of NblA proteins (27) and is assumed to be essential for their function. To monitor the dimerization of the two NblA proteins during the PBS degradation process, we used a method based on FRET. As a donor-acceptor pair, we chose Cer (35), a variant of enhanced cyan fluorescent protein (CFP), and YFP.
Plasmids containing the monomeric forms of YFP and Cer (31), designated Y and C, respectively, were used for labeling. The NblA-fluorophore fusion proteins were expressed in the nblA1/nblA2 double mutant under the control of the NblA promoter and terminator region in the self-replicating vector pVZ321. The following controls were used for evaluation of FRET measurements: (i) nblA1/nblA2 double mutant transformed with plasmids expressing only a single fluorophore fused to NblA16803 or NblA26803; (ii) nblA1/nblA2 double mutant transformed with plasmid pVZ/A2_Y and A2_C expressing two NblA26803 proteins, one tagged with YFP and one tagged with Cer; (iii) nblA2 single mutant (15) transformed with plasmid pVZ/Y and A2_C, which allows expression of the YFP protein and C-terminal Cer-tagged NblA26803; and (iv) nblA2 single mutant transformed with plasmid pVZ/A2_Y and A2_C (see above).
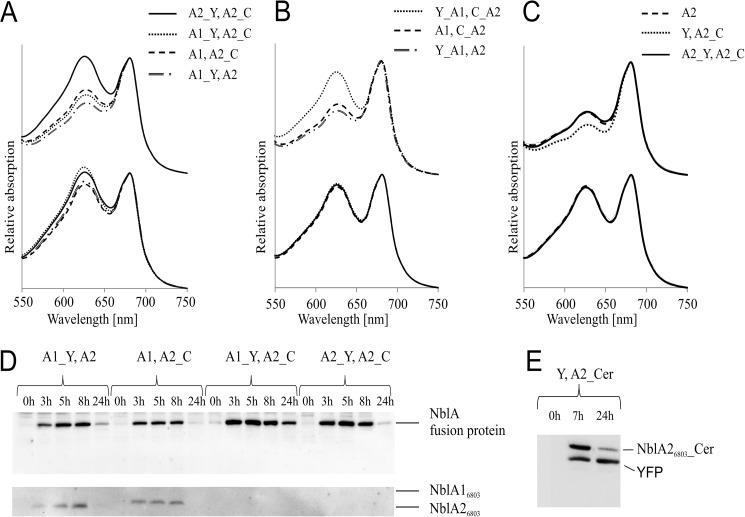
In preliminary experiments, we determined whether NblA-fluorophore fusion proteins were functional in vivo. Mutants grown in nitrogen-containing medium (BG11) were transferred to medium without combined nitrogen (BG110), and whole-cell absorbance spectra were recorded after 24 h (Fig. 2, A–C). PBS degradation after nitrogen deprivation took place in all strains expressing only a single fluorophore, regardless of whether the fluorophore was fused to the C or N terminus of the respective NblA protein (Fig. 2, A and B). Complementation was also observed in mutants expressing C-terminally tagged NblA16803 and NblA26803 (A1_Y and A2_C) (Fig. 2A). However, expression of N-terminally tagged NblA proteins (Y_A1 and C_A2) did not induce PBS degradation (Fig. 2B). This observation can be explained by our previous finding that NblA triggers PBS degradation by binding to ClpC via its N-terminal helix (20). Accordingly, fluorophores fused to the N terminus of NblA presumably sterically hinder the interaction with ClpC, thereby interfering with the in vivo function of NblA. In this context, it is remarkable that complementation occurred when only one of the two NblA proteins was N-terminally fused to a fluorophore, whereas it did not matter which NblA protein was labeled or what fluorophore it was labeled with (Fig. 2B). Furthermore, as shown in Fig. 2C, expression of YFP and C-terminally tagged NblA26803 (Y and A2_C) and simultaneous expression of two NblA26803 proteins with C-terminal YFP or Cer (A2_Y and A2_C) complemented the non-bleaching phenotype of the nblA2-deficient mutant.
FIGURE 2.
FRET constructs and their functionalities. Analyses of different NblA proteins, C- or N-terminally fused to the FRET donor Cer or FRET acceptor YFP, in Synechocystis 6803. A, NblA proteins C-terminally fused to the FRET donor Cer or FRET acceptor YFP complement the nblA1/nblA2 double mutant of Synechocystis 6803. Absorbance spectra of mutants either grown in nitrogen-replete medium (+N) or starved of combined nitrogen for 24 h (−N) are shown. B, NblA proteins N-terminally fused to the FRET donor Cer and FRET acceptor YFP cannot complement the nblA1/nblA2 double mutant of Synechocystis 6803. Mutants were either grown in nitrogen-replete medium (+N) or starved of combined nitrogen for 24 h (−N). A decrease in absorbance at 620 nm indicates degradation of PBS. C, expression of NblA26803 proteins in the nblA2-deficient mutant of Synechocystis 6803 complements the non-bleaching phenotype. Mutants were either grown in nitrogen-replete medium (+N) or starved of combined nitrogen for 24 h (−N). A decrease in absorbance at 620 nm indicates degradation of PBS. D, immunoblot analysis of the NblA fusion proteins in crude extracts of Synechocystis 6803 immediately (0 h) and at various times after nitrogen step-down. Blots were probed with a mixture of NblA16803 and NblA26803 antisera. E, immunoblot analysis of YFP and A2_C proteins in crude extracts of Synechocystis 6803 mutants immediately (0 h) and at 7 and 24 h after nitrogen step-down. The blots were probed with a GFP antiserum.
On the basis of the results of complementation experiments, we used the mutant expressing functional C-terminally tagged NblA16803 and NblA26803 for our initial FRET measurements. To monitor possible interactions between NblA16803-YFP and NblA26803-Cer, we used fluorescence spectroscopy. We excited the donor molecule (Cer) by actinic light with a wavelength of 458 nm and detected the emitted light in the range of 525–575 nm. YFP-specific emission was detected in the range of 500 and 600 nm, indicating the occurrence of FRET due to the interaction of fluorophore-tagged NblA16803 and NblA26803.
Expression of NblA proteins was induced by nitrogen depletion and monitored by immunoblot analysis (Fig. 2, D and E). PBS degradation was verified by recording emission spectra of whole cells immediately as well as 3, 5, 8, and 24 h after nitrogen deprivation. There were no significant differences in emission spectra among analyzed mutants grown on nitrate-containing medium (Fig. 3, A and B; 0 h); the emission peak at ∼540 nm corresponds to Synechocystis 6803 autofluorescence. Mutants were then switched to nitrogen starvation conditions, and fluorescence emission was monitored for 24 h. The mutant expressing C-terminally tagged NblA16803 and NblA26803 (A1_Y and A2_C) showed an increase in the amplitude of acceptor (YFP) emission at ∼530 nm over the first 8 h. After 24 h of nitrogen starvation, acceptor emission was still detected, albeit with a lower amplitude (Fig. 3A, 24 h).
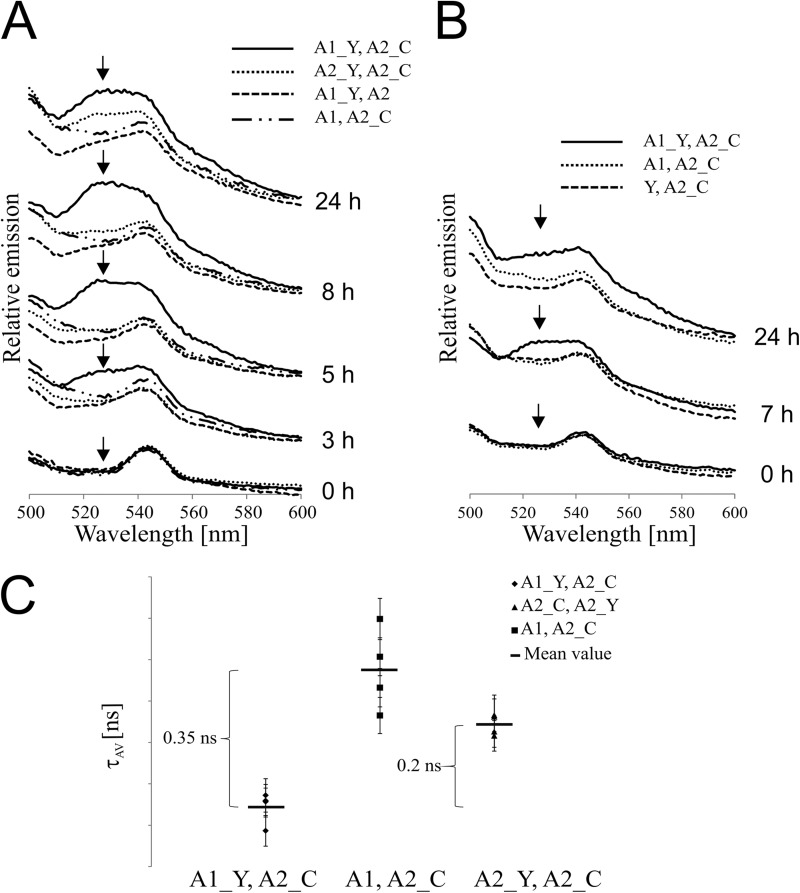
FIGURE 3.
Formation of NblA16803–NblA26803 heterodimers in vivo. The nblA1/nblA2 double mutant of Synechocystis 6803 was complemented in trans, under the control of the nblA promoter and terminator, with one NblA protein C-terminally fused to the FRET donor Cerulean (C) and the other fused to the FRET acceptor YFP (Y). For expression of NblA fusion proteins, the Synechocystis 6803 mutants were starved of combined nitrogen. A, fluorescence emission spectra of Synechocystis 6803 cells. Emission spectra of A1_Y, A2_C, A1_Y, A2, A1, A2_C, and A2_Y, A2_C mutants were recorded on cultures adjusted to an A750 of 0.3. Cerulean was excited at 458 nm, and emission of the FRET partner YFP was recorded between 500 and 600 nm after 0, 3, 5, 8, and 24 h of nitrogen depletion. B, fluorescence emission spectra of Synechocystis 6803 cells. Emission spectra of A1_Y and A2_C, A1 and A2_C, and Y and A2_C mutants were recorded at cell densities corresponding to an A750 of 0.3. Cerulean expressed in the cells was excited at 458 nm, and emission of the FRET partner YFP was recorded between 500 and 600 nm after 0, 7, and 24 h of nitrogen depletion. C, fluorescence lifetime spectroscopy of the A1_Y and A2_Cer, A2_C and A2_Y, and A1 and A2_C mutants. Cells at a density corresponding to an A750 of 0.3 were analyzed after 8 h of nitrogen starvation. Fluorescence intensity measurements were carried out in 1-cm cuvettes with excitation at 440 nm and emission of cerulean-based fluorescence decay recorded at 475 nm. Only a single photon every 100 laser pulses was registered (1 count); therefore, measurements were repeated for a total of 10,000 counts, and the measured time differences were sorted into a histogram. Each measurement was repeated seven times. The resulting decay curves were analyzed by fitting to the sum of two exponential terms using a non-linear least squares iterative procedure (FluoFit, PicoQuant). For each measurement, the amplitude-weighted lifetime (τAV) of cerulean was calculated. The mean values of τAV for each biological replicate are shown in the diagram, and the S.D. for each value is indicated. Furthermore the mean value of τAV for cerulean-based fluorescence of each mutant is shown.
Whole-cell emission spectra of control samples of mutants containing only Cer donor (A1 and A2_C) or YFP acceptor (A1_Y and A2) fluorophores showed no YFP emission at 530 nm following excitation at 458 nm (Fig. 3A), indicating the absence of FRET. Expression of the fluorophores was verified by immunoblot analysis (Fig. 2D). The emission spectra of control samples indicated that YFP is not directly excited by the donor (Cer) excitation wavelength and that the detected emission signal is YFP-specific and hence results from FRET. Our attempts to measure Cer emission in cells expressing only Cer fused to NblA26803 failed due to the strong autofluorescence of the photosynthetic pigments, which spectrally overlaps with the emission of Cer (475–525 nm; not shown).
As noted above, we also analyzed mutants expressing both fluorophores (YFP and Cer) together in one Synechocystis 6803 cell but not fused to interacting NblA proteins. Protein expression was once again monitored by immunoblot analysis with anti-NblA (Fig. 2D) and anti-GFP antibodies (Fig. 2E). Both fluorophores were presumed to be expressed, and FRET was measured as described above. The emission spectrum of the mutant expressing YFP and Cer coupled to NblA26803 showed no FRET signal (Fig. 3B, Y and A2_C), and its spectrum did not differ significantly from that of mutants containing only a single fluorophore (Fig. 3, A and B). The A2_Y and A2_C mutant also showed no FRET signal during the first 8 h after nitrogen depletion (Fig. 3A). After 24 h, however, an increase in acceptor fluorescence indicative of FRET was detected, albeit to a lesser extent than in the A1_Y and A2_C mutant. Interestingly, immunoblot analyses revealed an induction of protein expression within the first hours after nitrogen step-down and a decrease in protein levels after 24 h, a pattern inconsistent with the temporal changes in the FRET signal (Fig. 2D).
In order to attribute the observable changes in steady-state fluorescence characteristics to changes in the donor fluorescence lifetime, we performed time-resolved measurements of the donor (Cer) lifetime in addition to measuring steady-state FRET. Donor fluorescence lifetime might decrease, for example, when the acceptor molecule is in close proximity to the donor molecule and absorbs energy through FRET. These measurements were performed 8 h after nitrogen step-down, the time at which the strongest FRET signal occurred (Fig. 3A). The average fluorescence lifetime of Cer fused to NblA26803 was 2.29 ns (Fig. 3C). In the presence of YFP fused to NblA16803, the fluorescence lifetime of Cer decreased to 1.94 ns, a difference (ΔτAV) of 0.35 ns. This faster decay of Cer fluorescence is attributable to the energy transfer (FRET) from Cer to YFP that occurred when the two fluorophores were brought close together by interactions between NblA16803 and NblA26803. When YFP was linked to NblA26803 instead of NblA16803, the fluorescence lifetime of Cer fused to another NblA26803 molecule also decreased, albeit to a lesser extent (ΔτAV = 0.15 ns; Fig. 3C). The possibility that the reduced fluorescence lifetime is due to nonspecific energy transfer from the donor to the acceptor cannot be excluded. However, the resulting difference in fluorescence lifetimes between Cer and Cer expressed together with YFP (ΔτAV = = 0.2 ns) still indicates a clear FRET signal.
Taken together, the results from time-resolved fluorescence measurements confirm our steady-state measurements, indicating that FRET occurred when Cer and YFP were connected to NblA16803 and NblA26803, respectively. The much weaker FRET signal that occurred when both fluorophores were coupled to NblA26803 suggests a weak tendency of NblA2 to form homodimers.
Detecting the Interaction between NblA16803 and NblA26803 in Vitro by Pull-down Experiments
To validate our in vivo results, we performed in vitro binding studies. We first cloned nblA16803 and nblA26803 into the expression vector pACYCDuet-1, which is designed for coexpression of two target proteins. This vector was chosen to imitate expression in Synechocystis 6803, where nblA16803 and nblA26803 are adjacent cotranscribed genes (15).
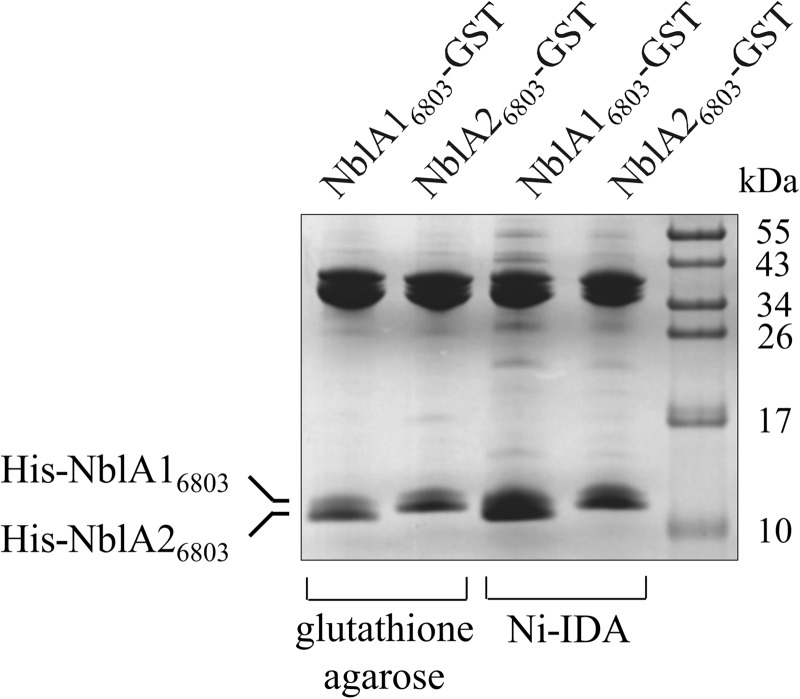
Affinity purification of NblA16803-GST invariably resulted in coprecipitation of HisNblA26803 and vice versa (Fig. 4), clearly demonstrating an interaction between NblA16803 and NblA26803 in vitro. These findings corroborate the results obtained with in vivo binding studies, described above. Taken together, in vivo and in vitro experiments provide evidence for an interaction between NblA16803 and NblA26803, indicating that the biologically active form is a heterodimer.
FIGURE 4.
Coprecipitation of affinity-tagged NblA16803 and NblA26803. Crude extracts from E. coli coexpressing either GST-tagged NblA16803 and His-tagged NblA26803 or His-tagged NblA16803 and GST-tagged NblA26803 were incubated with glutathione-agarose and Protino Ni-IDA resins, respectively. Protein complexes bound to the agarose were eluted with glutathione- or imidazole-containing buffer, respectively, separated by Tricine-SDS-PAGE, and stained with Coomassie Blue R-250.
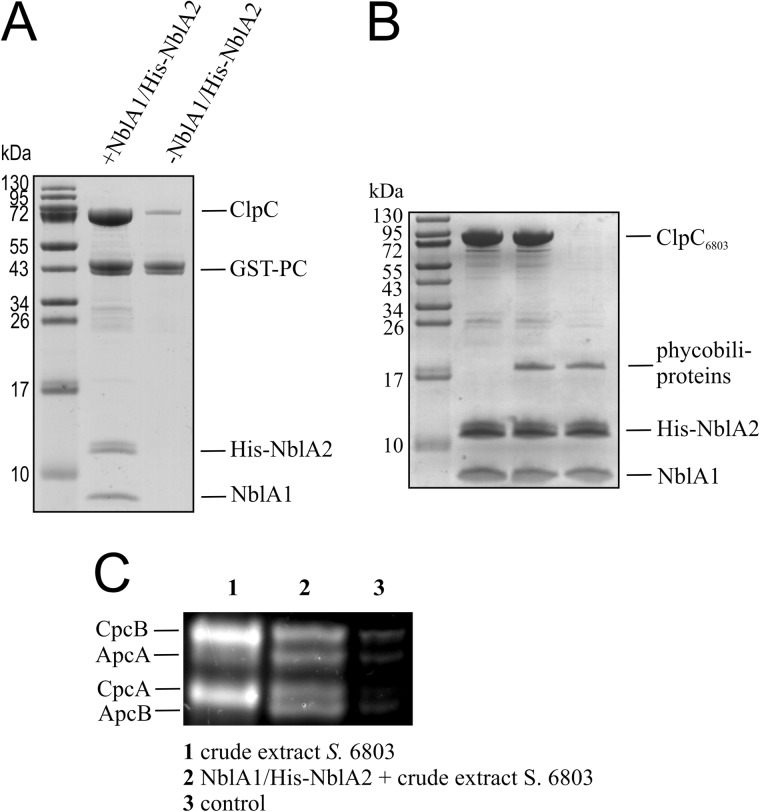
NblA16803 and NblA26803 Heterodimers Interact with ClpC6803 and the α-Subunit of PC in Vitro
Previous investigations of NblA function in Nostoc 7120 revealed that formation of a ternary complex among NblA, phycobiliproteins, and ClpC6803, the chaperone partner of Clp proteases, is indispensable for PBS degradation (20). Here, we performed binding experiments with NblA16803, NblA26803, ClpC6803, and the apoprotein of the α-subunit of PC from Synechocystis 6803. The N-terminally GST-tagged apoprotein of the PC α-subunit was immobilized on glutathione-agarose. The loaded matrix was then divided into two equal aliquots and incubated with crude extracts of ClpC6803-overexpressing E. coli in the presence or absence of NblA16803 and NblA26803. As expected, copurification of ClpC6803 was observed only when both NblA16803 and NblA26803 were present (Fig. 5A). Similar results were obtained using another experimental approach. In this case, heterodimers consisting of NblA16803 and His-tagged NblA26803 were immobilized on nickel-nitrilotriacetic acid, incubated first with a crude extract of E. coli expressing ClpC6803 and then with a crude extract of Synechocystis 6803. As shown in Fig. 5 (B and C), ClpC6803 and phycobiliproteins (PC and apophycocyanin) copurified with NblA16803 and His-tagged NblA26803.
FIGURE 5.
The NblA16803-NblA26803 heterodimer interacts simultaneously with phycobiliproteins and ClpC6803. A, the α-subunit of phycocyanin was expressed with an N-terminal GST tag, immobilized on glutathione-agarose, and incubated with soluble crude extract from ClpC6803-expressing E. coli cells in the presence or absence of NblA16803 and His-tagged NblA26803. After washing, bound protein complexes were eluted with glutathione, resolved by Tricine-SDS-PAGE, and stained with Coomassie Blue R-250. Experiments were performed in buffer B containing 1 mm Mg-ATP. B, the NblA16803/His-NblA26803 protein complex was immobilized on Protino Ni-IDA resin and incubated with crude extract from E. coli cells expressing ClpC6803. After washing, the matrix was incubated with Synechocystis 6803 crude extract. The bound protein complexes were eluted with imidazole-containing buffer, resolved by Tricine-SDS-PAGE, and stained with Coomassie Blue R-250. Experiments were performed in buffer B containing 1 mm Mg-ATP. C, NblA1 and His-NblA2 were immobilized on Protino Ni-IDA resin and incubated with Synechocystis 6803 crude extract (lane 2). As a control, HisClpC-loaded Protino Ni-IDA resin was incubated with Synechocystis 6803 crude extract (lane 3). The bound proteins were eluted as described above. Additionally, Synechocystis 6803 crude extract was loaded to visualize the phycobili subunits (lane 1) and resolved by Tricine-SDS-PAGE. The phycobiliproteins were detected by zinc-induced fluorescence after SDS-PAGE.
NblA16803 Mediates the Interaction of ClpC6803 with Phycobiliproteins
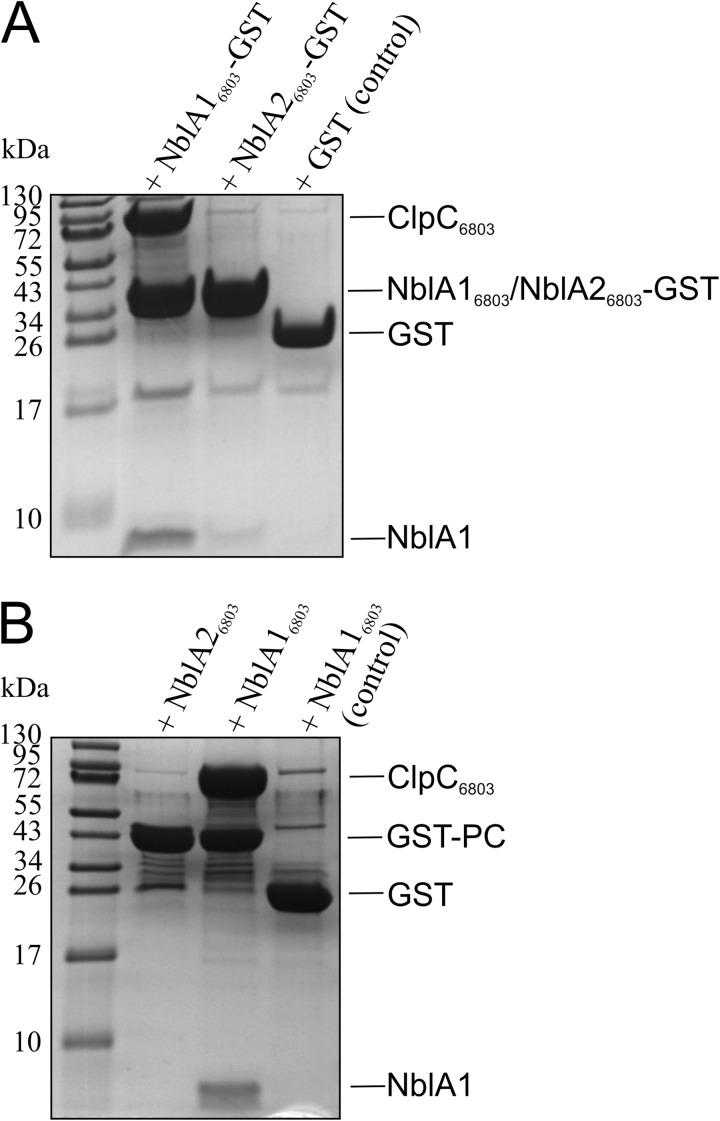
As shown previously, a highly conserved stretch of amino acids near the N terminus of NblA from Nostoc 7120 is involved in ClpC binding (20), whereas residues near the C terminus are involved in phycobiliprotein binding (19). Assuming that the biologically active form of NblA in Synechocystis 6803 is a heterodimer, we would expect that both NblA16803 and NblA26803 would be able to bind ClpC6803 and phycobiliproteins and that both NblA proteins would be essential for formation of a complex with ClpC6803 and phycobiliproteins. To verify this hypothesis, we first analyzed whether NblA16803 and NblA26803 interact with ClpC6803 in vitro. To this end, we immobilized C-terminally GST-tagged NblA16803 and NblA26803 on glutathione-agarose and incubated each with crude extracts of E. coli expressing ClpC6803. After washing, proteins were eluted and separated by Tricine-SDS-PAGE (Fig. 6A). ClpC6803 copurified only with GST-tagged NblA16803. There was no interaction between ClpC6803 and NblA26803 (lane 2). We conclude that the ClpC6803 binding site on the heterodimer is located on NblA16803.
FIGURE 6.
NblA16803 mediates binding to ClpC6803 and phycobiliproteins. A, binding of ClpC6803 to GST-tagged NblA16803 in vitro. NblA16803 and NblA26803 were expressed with a C-terminal GST tag, immobilized on glutathione-agarose, and incubated (in the presence of 5 mm Mg-ATP) with soluble crude extract from E. coli cells expressing ClpC6803. After washing, NblA-GST complexes were eluted with glutathione, resolved by Tricine-SDS-PAGE, and stained with Coomassie Blue R-250. GST protein served as control. B, NblA16803 binds simultaneously to ClpC6803 and the GST-tagged apoprotein of phycocyanin. The α-subunit of phycocyanin was expressed with an N-terminal GST tag and preincubated with NblA16803 or NblA26803. After the addition of glutathione-agarose, proteins were incubated with soluble crude extract from ClpC6803-expressing E. coli cells in the presence of 5 mm Mg-ATP. After washing, GST-PC complexes were eluted with glutathione, separated by Tricine-SDS-PAGE, and stained with Coomassie Blue R-250. GST protein served as a control.
To demonstrate that NblA16803 is sufficient to induce formation of the ternary complex with ClpC6803 and phycobiliproteins in vitro, we first immobilized the GST-tagged apoprotein of α-PC on glutathione-agarose. We then incubated the protein-agarose conjugate with a crude extract of ClpC6803-expressing E. coli in the presence of NblA16803 or NblA26803. As expected, given the inability of NblA26803 to interact with ClpC6803 (Fig. 6A), ClpC6803 did not copurify with NblA26803 (Fig. 6B). Notably, NblA26803 itself also did not copurify with GST-tagged PC, whereas NblA16803 bound simultaneously to the PC apoprotein and ClpC6803 (Fig. 6B). These results indicate that the presence of NblA16803 is sufficient for in vitro formation of the ternary complex required for PBS degradation.
Heterologously Coexpressed ClpP1 and ClpR Form a Proteolytic Complex
Clp proteases of cyanobacteria were first identified in Synechococcus sp. PCC7942 (36). In this organism, the Clp machinery consists of two chaperones (ClpC and ClpX), three different proteolytic subunits (ClpP1, ClpP2, and ClpP3), and the ClpP variant ClpR, which lacks the typical catalytic triad of serine-type proteases. In Synechococcus 7942, two Clp proteases could be identified by gel filtration and immunoblot analyses. Each protease is a complex composed of a unique proteolytic core, containing ClpP1 and ClpP2 or ClpP3 and ClpR, and a particular chaperone partner: ClpC in the case of ClpP3ClpR and ClpX in the case of ClpP1ClpR. In addition to these two soluble Clp proteases, a third, membrane-associated proteolytic complex consisting of ClpP1 and ClpR is assumed.
Homologs of the entire Clp machinery, including the ClpP1-ClpR complex, can be found in Synechocystis 6803. This protease has been suggested to interact with ClpC and, as shown by cross-linking studies, probably binds to the PBS (36, 37), making it a logical candidate for subsequent in vitro studies in Synechocystis 6803. Here, we expressed the proteolytic core complex composed of ClpP1 and ClpR using the pACYCDuet-1 vector. Expecting that ClpP1 and ClpR would spontaneously associate with each other, we used only one affinity tag, a His tag fused to the C terminus of ClpP1, for purification of the core complex. At 25.2 and 24.8 kDa, respectively, HisClpP1 and ClpR have nearly the same molecular mass; thus, they cannot be separated from each other by Tricine-SDS-PAGE (Fig. 7A). To determine whether ClpR copurifies with HisClpP1, we performed size-exclusion chromatography3 and native PAGE.3 The apparent molecular mass of the protein complex was 200 kDa, indicating that ∼3–4 copies of ClpR and HisClpP1 assemble into an oligomeric structure.
FIGURE 7.
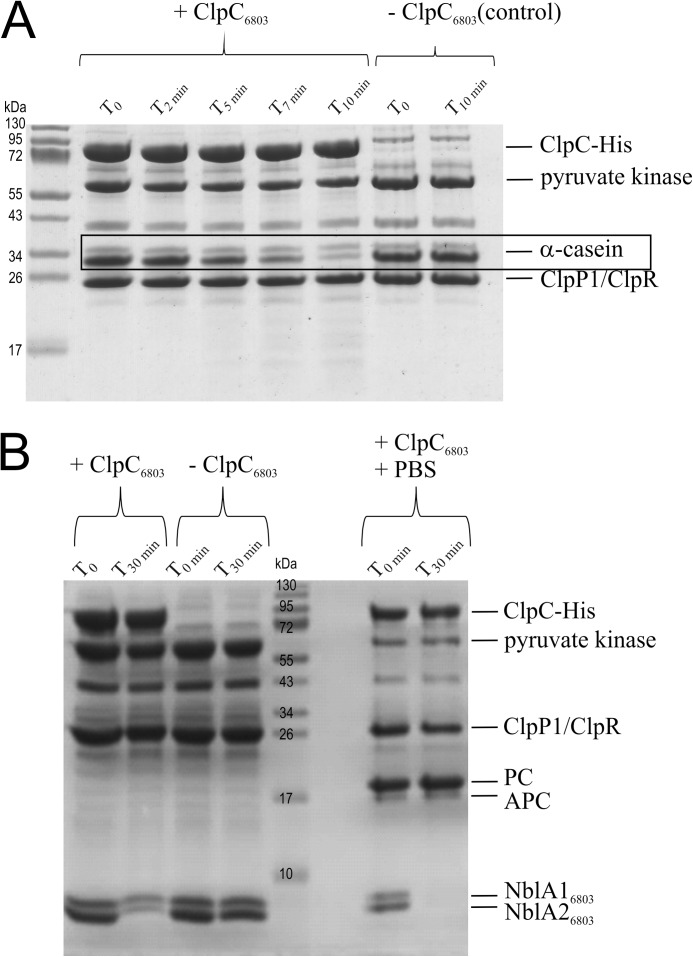
ClpC6803 forms an active proteolytic complex with ClpP1-ClpR and degrades the adaptor proteins NblA16803 and NblA26803. A, in vitro degradation of α-casein. α-Casein was incubated in the presence of pyruvate kinase (20 ng/μl), 4 mm phosphoenolpyruvate, 2 mm ATP, and purified ClpC6803 and ClpP1/R (2 μm each) at 37 °C. Samples were taken at the indicated time points, separated by SDS-PAGE, and visualized by Coomassie Blue R-250 staining. B, in vitro degradation of purified NblA16803 and NblA26803. NblA16803/NblA26803 (5 μm each) were incubated in the presence of pyruvate kinase (20 ng/μl), 4 mm phosphoenolpyruvate, 2 mm ATP, purified ClpC6803 and ClpP1-ClpR (2 μm each), and purified PBS from Synechocystis 6803 (29) at 37 °C. Samples were taken at the indicated time points, separated by SDS-PAGE, and visualized by Coomassie Blue R-250 staining.
Hydrolysis of α-Casein by the ClpP1-ClpR Core Complex Depends on ClpC6803 and ATP
To analyze the proteolytic activity of the recombinant ClpP1-HisClpR complex, we tested its capacity to degrade α-casein. Accordingly, we incubated the complex with α-casein, HisClpC6803, and ATP in the presence of an ATP-regenerating system (38). With numerous exposed hydrophobic amino acids on its surface (39), α-casein from cow's milk is a nonspecific substrate for Clp proteases (40, 41). ClpC6803 is the putative chaperone partner of the ClpP1-ClpR proteolytic core complex and is required for substrate recognition, ATP-dependent unfolding, and translocation into the proteolytic chamber. At intervals after combining reaction components, aliquots of the assay mixture were withdrawn, proteins were separated by Tricine-SDS-PAGE, and α-casein concentration was estimated by Coomassie Blue staining (Fig. 7A). The ClpC6803-ClpP1-ClpR complex completely degraded α-casein within 10 min (Fig. 7A). As expected, given the functional roles of ClpC6803, no degradation occurred in the absence of this putative chaperone (Fig. 7A, lanes 7 and 8). Moreover, no degradation of α-casein was observed upon incubation with ClpC6803 alone (Fig. 7A, lanes 9 and 10), demonstrating that all three proteins (ClpP1, ClpR, and ClpC6803) are required to form a proteolytically active Clp complex.
A Clp Protease Composed of ClpC6803, ClpP1, and ClpR Degrades NblA16803 and NblA26803 in Vitro
Previous investigations of the interaction between NblA and ClpC from Nostoc 7120 showed that exchanging amino acids in the ClpC binding site of NblA results in accumulation of NblA in vivo.3 From this we inferred that NblA acts as an adaptor protein that is codegraded with its substrate as part of the overall regulatory mechanism. We therefore asked whether NblA proteins are degraded in vitro by recombinant Synechocystis 6803 Clp protease.
To test the hypothesis that NblA itself may be codegraded by the Clp protease, we performed degradation experiments with NblA16803 and NblA26803 essentially as described above. Degradation of NblA16803 and NblA26803 was monitored by Coomassie Blue staining of samples resolved on Tricine-SDS-polyacrylamide gels. Fig. 7B illustrates the simultaneous degradation of NblA proteins within 30 min. No degradation was observed in the absence of ClpC6803 or ATP.3 These results demonstrate that the putative Clp protease ClpC6803-ClpP1-ClpR degrades NblA16803 and NblA26803 in vitro at similar rates. Attempts to degrade the GST-tagged PC apoprotein or PBS isolated from Synechocystis 6803 in vitro have not yet proved successful (see below).
DISCUSSION
Complementation of the Non-bleaching Phenotype of the nblA1/nblA2 Double Mutant of Synechocystis 6803 with the NblA Protein from Nostoc 7120
In this study, we solved part of the mystery of why Synechocystis 6803 possesses two nblA genes, which are both essential for PBS degradation (15), when other cyanobacteria accomplish this by expressing only one nblA gene (14, 17, 18).
The complementation experiment shown in Fig. 1 demonstrates that the NblA protein from Nostoc 7120 is capable of restoring the wild-type phenotype to the non-bleaching nblA1/nblA2 double mutant of Synechocystis 6803, suggesting that the functions of NblA16803 and NblA26803 are combined in one protein in Nostoc 7120. A similar result was obtained by Dines et al. (27), who reported successful complementation of the non-bleaching phenotype of the nblA mutant of Synechococcus 7942 by expression of the Nostoc 7120 nblA gene. These results support the conclusion that NblA proteins are functionally interchangeable among different cyanobacteria. Consistent with this, the various NblA proteins contain highly conserved regions near the N and C termini that mediate binding to phycobiliproteins and ClpC6803 (19, 20), and the three existing crystal structures are very similar (19, 27).
Formation of NblA16803/NblA26803 Heterodimers
FRET measurements, fluorescence lifetime measurements, and in vitro pull-down experiments demonstrated that NblA16803 and NblA26803 form a heterodimer. In contrast, the tendency toward homodimer formation was observed only under conditions that do not reflect those that exist in the cell, where nblA16803 and nblA26803 are cotranscribed (15, 16). On the basis of our results, we postulate that the biologically active form is a heterodimer, and we assume that the structure of the heterodimer is similar to that of other NblA proteins that have been crystallized to date.
Heterologous Expression of the Putative Protease ClpC6803-ClpP1-ClpR and Degradation of NblA16803 and NblA26803
To provide evidence that PBSs are degraded by a Clp protease, we performed in vitro degradation assays. Investigations of Clp proteases in Synechococcus 7942 have demonstrated that cyanobacteria possess two distinct Clp proteases in the soluble fraction of the cell and probably a third that is associated with the thylakoids. This membrane-associated protease appears to be involved in PBS degradation because (i) the protease has been reported to interact with PBS (37), and (ii) ClpC, the putative HSP100 chaperone partner of this protease, interacts with NblA (20).
The membrane-associated protease consists of a unique proteolytic core complex, composed of the Clp subunits, ClpP1 and ClpR, and the HSP100 chaperone partner ClpC, as noted above. To heterologously express the protease, we first identified orthologs of ClpP1, ClpR, and ClpC in Synechocystis 6803.3 We then successfully cloned, expressed, and purified the corresponding proteins. As expected, coexpression of the two proteolytic subunits, ClpR and ClpP1, enabled the purification of a multiprotein complex. The complex had a molecular mass of about 200 kDa,3 which is similar to that of the ClpR-ClpP3 proteolytic core complex of Synechococcus 7942 (41), and appeared to correspond to a single mixed heptameric ring of ClpR and ClpP1, as expected. The protease activity of the complex, determined by analyzing the degradation of α-casein, was also similar to that of the proteolytic core ClpP3-ClpR from Synechococcus 7942 (41). Both cyanobacterial proteases are thus much slower than the model protease ClpA-ClpP from E. coli (41). The lower activity of cyanobacterial Clp proteases compared with that of E. coli may be due, at least in part, to differences in the structures of the core complexes (41, 42). We further demonstrated that NblA16803 and NblA26803 are degraded by the recombinant Clp protease ClpC-ClpP1ClpR.
The Clp protease-mediated degradation of NblA16803 and NblA26803 represents an elegant NblA regulatory mechanism. During the first hours after nitrogen starvation, the heterodimer is highly expressed, forming a ternary complex with ClpC6803 and the PBS that leads to ClpC6803 and PBS degradation. When all PBSs are degraded, the remaining NblA proteins are also degraded by the protease, eliminating the need for further regulation. This finding that the protease degrades NblA reinforces previous reports by our group that NblA accumulates when the ClpC binding site on NblA is inactivated by mutagenesis.3 Collectively, these observations indicate that the NblA protein acts like other identified adaptor proteins that undergo protease-mediated degradation, even when they are not delivering a substrate (43, 44).
Despite the fact that all conditions necessary for PBS degradation were fulfilled, including demonstrable protease activity and formation of a ternary complex between NblA16803/NblA26803, phycobiliproteins, and ClpC, we were unable to demonstrate in vitro degradation of PBS isolated from Synechocystis 6803 or the GST-tagged PC apoprotein.3 In further attempts to measure PBS degradation, we used a crude extract of wild-type cells grown under nitrogen or harvested immediately after nitrogen deprivation and also tried adding recombinant ClpC, NblA16803, NblA26803, ClpP1-ClpR, ATP, and an ATP regeneration system. Both strategies failed to induce degradation. One possible explanation for these unexpected results might be that dephosphorylation of PBS linker proteins is necessary for the degradation process. In this context, Piven et al. (45) reported that nitrogen starvation and long term exposure to higher light intensities resulted in an increase in dephosphorylated linker proteins and partial disassembly of the PBS. Even studies in Nostoc 7120, which exhibit a bottom-to-top disassembly of the PBS in early heterocysts, suggest that dephosphorylation of linker proteins may act as a signal for degradation (46). Accordingly, one possible scenario could be that, when the V-shaped NblA-dimers penetrate the gap formed by four back-to-back-assembled PBS β-subunits (27), the PBS structure disassembles (27), enabling dephosphorylation of the linker proteins by a phosphatase and opening the door for subsequent degradation. However, this explanation cannot account for the observation that the PC apoprotein is not degraded in vitro3 despite binding to NblA16803/NblA26803 (Fig. 6B). It is also possible that a bilin lyase could be involved in the degradation process. Bilin lyases catalyze the covalent ligation of chromophores to specific binding sites of phycobiliproteins and also control subsequent detachment (47). In Synechococcus 7942, the bilin lyase NblB participates in the degradation of PBS (48); in contrast, the two homologs of NblB in Synechocystis 6803 are not essential for PBS degradation in this strain (16). Thus, it may be that another unidentified lyase remains be found. A further possibility could be that NblA16803 and NblA26803 must be activated by phosphorylation or dephosphorylation. Examples in which adaptor proteins must be activated are known from other organisms, including Bacillus subtilis, where the tyrosine kinase MecB is activated by phosphorylation (49). In this context, it is conceivable that light could induce NblA1/NblA2 activation, perhaps through a light-dependent phosphatase like NblS (50, 51). Clearly, further biochemical analyses will be required to establish the relevance of the mechanisms discussed here and close out the topic of PBS degradation.
Supplementary Material
Acknowledgments
We thank Sabine Nicklisch and Katja Zado for skillful technical assistance.

This article contains supplemental Table S1 and Fig. S1.
A. Baier, W. Winkler, T. Korte, W. Lockau, and A. Karradt, unpublished data.
- PBS
- phycobilisome
- Nostoc 7120
- Nostoc sp. PCC7120
- Synechocystis 6803
- Synechocystis sp. PCC6803
- Synechococcus 7942
- Synechococcus elongatus PCC7942
- PCC
- Pasteur Culture Collection
- PC
- phycocyanin
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- Cer
- cerulean
- CFP
- cyan fluorescent protein
- Ni-IDA
- nickel-iminodiacetic acid
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Glazer A. N. (1982) Phycobilisomes: structure and dynamics. Annu. Rev. Microbiol. 36, 173–198 [DOI] [PubMed] [Google Scholar]
- 2. Grossman A. R., Schaefer M. R., Chiang G. G., Collier J. L. (1993) The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol. Rev. 57, 725–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adir N. (2005) Elucidation of the molecular structures of components of the phycobilisome: reconstructing a giant. Photosynth. Res. 85, 15–32 [DOI] [PubMed] [Google Scholar]
- 4. MacColl R. (1998) Cyanobacterial phycobilisomes. J. Struct. Biol. 124, 311–334 [DOI] [PubMed] [Google Scholar]
- 5. MacColl R., Malak H., Gryczynski I., Eisele L. E., Mizejewski G. J., Franklin E., Sheikh H., Montellese D., Hopkins S., MacColl L. C. (1998) Phycoerythrin 545: monomers, energy migration, bilin topography, and monomer/dimer equilibrium. Biochemistry 37, 417–423 [DOI] [PubMed] [Google Scholar]
- 6. Tandeau de Marsac N. (1977) Occurrence and nature of chromatic adaptation in cyanobacteria. J. Bacteriol. 130, 82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kalla R., Bhalerao R. P., Gustafsson P. (1993) Regulation of phycobilisome rod proteins and mRNA at different light intensities in the cyanobacterium. Synechococcus 6301. Gene 126, 77–83 [DOI] [PubMed] [Google Scholar]
- 8. de Lorimier R. M., Smith R. L., Stevens S. E. (1992) Regulation of phycobilisome structure and gene expression by light intensity. Plant Physiol. 98, 1003–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Raps S., Kycia J. H., Ledbetter M. C., Siegelman H. W. (1985) Light Intensity Adaptation and Phycobilisome Composition of Microcystis aeruginosa. Plant Physiol. 79, 983–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yamanaka G., Glazer A. N., Williams R. C. (1980) Molecular architecture of a light-harvesting antenna. Comparison of wild type and mutant Synechococcus 6301 phycobilisomes. J. Biol. Chem. 255, 11104–11110 [PubMed] [Google Scholar]
- 11. Collier J. L., Grossman A. R. (1992) Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: not all bleaching is the same. J. Bacteriol. 174, 4718–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Allen M. M., Smith A. J. (1969) Nitrogen chlorosis in blue-green algae. Arch. Mikrobiol. 69, 114–120 [DOI] [PubMed] [Google Scholar]
- 13. Adir N., Zer H., Shochat S., Ohad I. (2003) Photoinhibition: a historical perspective. Photosynth. Res. 76, 343–370 [DOI] [PubMed] [Google Scholar]
- 14. Collier J. L., Grossman A. R. (1994) A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 13, 1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baier K., Nicklisch S., Grundner C., Reinecke J., Lockau W. (2001) Expression of two nblA-homologous genes is required for phycobilisome degradation in nitrogen-starved Synechocystis sp. PCC6803. FEMS Microbiol. Lett. 195, 35–39 [DOI] [PubMed] [Google Scholar]
- 16. Li H., Sherman L. A. (2002) Characterization of Synechocystis sp. strain PCC 6803 and Δnbl mutants under nitrogen-deficient conditions. Arch. Microbiol. 178, 256–266 [DOI] [PubMed] [Google Scholar]
- 17. Baier K., Lehmann H., Stephan D. P., Lockau W. (2004) NblA is essential for phycobilisome degradation in Anabaena sp. strain PCC 7120 but not for development of functional heterocysts. Microbiology 150, 2739–2749 [DOI] [PubMed] [Google Scholar]
- 18. Luque I., Ochoa De Alda J. A., Richaud C., Zabulon G., Thomas J. C., Houmard J. (2003) The NblAI protein from the filamentous cyanobacterium Tolypothrix PCC 7601: regulation of its expression and interactions with phycobilisome components. Mol. Microbiol. 50, 1043–1054 [DOI] [PubMed] [Google Scholar]
- 19. Bienert R., Baier K., Volkmer R., Lockau W., Heinemann U. (2006) Crystal structure of NblA from Anabaena sp. PCC 7120, a small protein playing a key role in phycobilisome degradation. J. Biol. Chem. 281, 5216–5223 [DOI] [PubMed] [Google Scholar]
- 20. Karradt A., Sobanski J., Mattow J., Lockau W., Baier K. (2008) NblA, a key protein of phycobilisome degradation, interacts with ClpC, a HSP100 chaperone partner of a cyanobacterial Clp protease. J. Biol. Chem. 283, 32394–32403 [DOI] [PubMed] [Google Scholar]
- 21. Grimaud R., Kessel M., Beuron F., Steven A. C., Maurizi M. R. (1998) Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J. Biol. Chem. 273, 12476–12481 [DOI] [PubMed] [Google Scholar]
- 22. Wang J., Hartling J. A., Flanagan J. M. (1997) The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell 91, 447–456 [DOI] [PubMed] [Google Scholar]
- 23. Kim Y. I., Levchenko I., Fraczkowska K., Woodruff R. V., Sauer R. T., Baker T. A. (2001) Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat. Struct. Biol. 8, 230–233 [DOI] [PubMed] [Google Scholar]
- 24. Ortega J., Lee H. S., Maurizi M. R., Steven A. C. (2002) Alternating translocation of protein substrates from both ends of ClpXP protease. EMBO J. 21, 4938–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dougan D. A., Mogk A., Zeth K., Turgay K., Bukau B. (2002) AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett. 529, 6–10 [DOI] [PubMed] [Google Scholar]
- 26. Levchenko I., Seidel M., Sauer R. T., Baker T. A. (2000) A specificity-enhancing factor for the ClpXP degradation machine. Science 289, 2354–2356 [DOI] [PubMed] [Google Scholar]
- 27. Dines M., Sendersky E., David L., Schwarz R., Adir N. (2008) Structural, functional, and mutational analysis of the NblA protein provides insight into possible modes of interaction with the phycobilisome. J. Biol. Chem. 283, 30330–30340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29. Ducret A., Sidler W., Wehrli E., Frank G., Zuber H. (1996) Isolation, characterization and electron microscopy analysis of a hemidiscoidal phycobilisome type from the cyanobacterium Anabaena sp. PCC 7120. Eur. J. Biochem. 236, 1010–1024 [DOI] [PubMed] [Google Scholar]
- 30. Schägger H., von Jagow G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 31. Zacharias D. A. (2002) Sticky caveats in an otherwise glowing report: oligomerizing fluorescent proteins and their use in cell biology. Sci. STKE 2002, pe23. [DOI] [PubMed] [Google Scholar]
- 32. Zinchenko V. V., Piven I. V., Melnik V. A., Shestakov S. V. (1999) Vectors for the complementation analysis of cyanobacterial mutants. Russ. J. Genet. 35, 228–232 [Google Scholar]
- 33. Finkenwirth F., Neubauer O., Gunzenhäuser J., Schoknecht J., Scolari S., Stöckl M., Korte T., Herrmann A., Eitinger T. (2010) Subunit composition of an energy-coupling-factor-type biotin transporter analysed in living bacteria. Biochem. J. 431, 373–380 [DOI] [PubMed] [Google Scholar]
- 34. Wolk C. P., Vonshak A., Kehoe P., Elhai J. (1984) Construction of shuttle vectors capable of conjugative transfer from Escherichia coli to nitrogen-fixing filamentous cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 81, 1561–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rizzo M. A., Springer G. H., Granada B., Piston D. W. (2004) An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 22, 445–449 [DOI] [PubMed] [Google Scholar]
- 36. Schelin J., Lindmark F., Clarke A. K. (2002) The clpP multigene family for the ATP-dependent Clp protease in the cyanobacterium Synechococcus. Microbiology 148, 2255–2265 [DOI] [PubMed] [Google Scholar]
- 37. Stanne T. M., Pojidaeva E., Andersson F. I., Clarke A. K. (2007) Distinctive types of ATP-dependent Clp proteases in cyanobacteria. J. Biol. Chem. 282, 14394–14402 [DOI] [PubMed] [Google Scholar]
- 38. Dougan D. A., Reid B. G., Horwich A. L., Bukau B. (2002) ClpS, a substrate modulator of the ClpAP machine. Mol. Cell 9, 673–683 [DOI] [PubMed] [Google Scholar]
- 39. Herskovits T. T. (1966) Biochemistry, American Chemical Society, pp. 1018–1026, Columbus, OH [Google Scholar]
- 40. Turgay K., Hamoen L. W., Venema G., Dubnau D. (1997) Biochemical characterization of a molecular switch involving the heat shock protein ClpC, which controls the activity of ComK, the competence transcription factor of Bacillus subtilis. Genes Dev. 11, 119–128 [DOI] [PubMed] [Google Scholar]
- 41. Andersson F. I., Tryggvesson A., Sharon M., Diemand A. V., Classen M., Best C., Schmidt R., Schelin J., Stanne T. M., Bukau B., Robinson C. V., Witt S., Mogk A., Clarke A. K. (2009) Structure and function of a novel type of ATP-dependent Clp protease. J. Biol. Chem. 284, 13519–13532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tryggvesson A., Ståhlberg F. M., Mogk A., Zeth K., Clarke A. K. (2012) Interaction specificity between the chaperone and proteolytic components of the cyanobacterial Clp protease. Biochem. J. 446, 311–320 [DOI] [PubMed] [Google Scholar]
- 43. Persuh M., Mandic-Mulec I., Dubnau D. (2002) A MecA paralog, YpbH, binds ClpC, affecting both competence and sporulation. J. Bacteriol. 184, 2310–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schlothauer T., Mogk A., Dougan D. A., Bukau B., Turgay K. (2003) MecA, an adaptor protein necessary for ClpC chaperone activity. Proc. Natl. Acad. Sci. U.S.A. 100, 2306–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Piven I., Ajlani G., Sokolenko A. (2005) Phycobilisome linker proteins are phosphorylated in Synechocystis sp. PCC 6803. J. Biol. Chem. 280, 21667–21672 [DOI] [PubMed] [Google Scholar]
- 46. Ke S., Haselkorn R. (2013) Fluorescence spectroscopy study of heterocyst differentiation in Anabaena PCC 7120 filaments. Microbiology 159, 253–258 [DOI] [PubMed] [Google Scholar]
- 47. Scheer H., Zhao K. H. (2008) Biliprotein maturation: the chromophore attachment. Mol. Microbiol. 68, 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dolganov N., Grossman A. R. (1999) A polypeptide with similarity to phycocyanin α-subunit phycocyanobilin lyase involved in degradation of phycobilisomes. J. Bacteriol. 181, 610–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kirstein J., Dougan D. A., Gerth U., Hecker M., Turgay K. (2007) The tyrosine kinase McsB is a regulated adaptor protein for ClpCP. EMBO J. 26, 2061–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van Waasbergen L. G., Dolganov N., Grossman A. R. (2002) nblS, a gene involved in controlling photosynthesis-related gene expression during high light and nutrient stress in Synechococcus elongatus PCC 7942. J. Bacteriol. 184, 2481–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kappell A. D., Bhaya D., van Waasbergen L. G. (2006) Negative control of the high light-inducible hliA gene and implications for the activities of the NblS sensor kinase in the cyanobacterium Synechococcus elongatus strain PCC 7942. Arch. Microbiol. 186, 403–413 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.