Background: The hemolymph of healthy shrimp contains low albeit stable numbers of bacteria.
Results: Knockdown of C-type lectin MjHeCL led to suppressed expression of antimicrobial peptides, bacterial proliferation, and shrimp death.
Conclusion: MjHeCL protects shrimp by inhibiting the proliferation of hemolymph microbiota.
Significance: This study demonstrated a novel role for soluble C-type lectins in antibacterial response.
Keywords: Antimicrobial Peptides, Invertebrates, Lectin, Pattern Recognition Receptor, RNA Interference (RNAi), C-type Lectin, Hemolymph Microbiota, Shrimp
Abstract
Some aquatic invertebrates such as shrimp contain low albeit stable numbers of bacteria in the circulating hemolymph. The proliferation of this hemolymph microbiota in such a nutrient-rich environment is tightly controlled in healthy animals, but the mechanisms responsible had remained elusive. In the present study, we report a C-type lectin (MjHeCL) from the kuruma shrimp (Marsupenaeus japonicus) that participates in restraining the hemolymph microbiota. Although the expression of MjHeCL did not seem to be modulated by bacterial challenge, the down-regulation of its expression by RNA interference led to proliferation of the hemolymph microbiota, ultimately resulting in shrimp death. This phenotype was rescued by the injection of recombinant MjHeCL, which restored the healthy status of the knockdown shrimp. A mechanistic analysis revealed that MjHeCL inhibited bacterial proliferation by modulating the expression of antimicrobial peptides. The key function of MjHeCL in the shrimp immune homeostasis might be related to its broader recognition spectrum of the hemolymph microbiota components than other lectins. Our study demonstrates the role of MjHeCL in maintaining the healthy status of shrimp and provides new insight into the biological significance of C-type lectins, a diversified and abundant lectin family in invertebrate species.
Introduction
Healthy animals, vertebrates or invertebrates, host diverse bacterial communities as commensals or symbionts in the various microenvironments they provide, and significant progress has been achieved in recent years with regard to the mechanisms responsible for the establishment of such consortia (1–3). In vertebrates, the microbiota mostly resides on external surfaces, i.e. skin or cavities directly connected to the external environment such as the gut (2). In some healthy invertebrates such as shrimp, however, bacteria are not only present in the digestive tract but also in the circulating hemolymph (4). This observation has now been extended to other aquatic invertebrate species (5–7). In shrimp, the hemolymph bacterial communities are present at relatively low numbers as compared with the gut microbiota and usually include species that can become opportunistic pathogens under stressful conditions such as those that may occur in the aquaculture context (4, 8). However, the mechanism(s) that regulates homeostasis of the hemolymph microbiota is largely unknown, and the factor(s) that may inhibit its proliferation in such a nutrient-rich environment had remained elusive. Although antimicrobial peptides (AMPs)2 that have been identified in shrimp hemolymph are likely candidates as the inhibitory factors (9, 10), how the bacteria are sensed, how the AMP expression is regulated, and how they restrain the proliferation of the shrimp hemolymph microbiota had remained unknown.
Invertebrates lack the typical antibody- and T/B cell-based adaptive immunity of vertebrates and only rely on physical barriers and innate immunity for defense against infectious agents (11–14). Among the diverse recognition and effector innate immune factors, lectins and antimicrobial peptides play key roles in sensing and controlling or eradicating any potential pathogens not only in invertebrates but also in most vertebrate species (15, 16). By binding to microbial surface glycans, including lipopolysaccharide and peptidoglycan, C-type lectins (CTLs) from invertebrate species effectively participate not only in the initial step of pathogen recognition via the carbohydrate recognition domain but also in various antimicrobial effector functions such as immobilization, phagocytosis, clearance, encapsulation, nodule formation, activation of the prophenoloxidase system/melanization, and others including direct antimicrobial activity (17–24). Thus, CTLs from invertebrates probably carry a heavier burden in pathogen recognition and the activation of pathways leading to antimicrobial effector functions than their vertebrate counterparts. This hypothesis is partially supported by the greater abundance and diversification of CTLs in invertebrates as compared with vertebrates (16, 25, 26). Furthermore, based on our observations and those of others, a larger subset of the CTL proteins is soluble in insects and crustaceans than in mammals (27, 28). The humoral CTL repertoire in shrimp, constituted by at least 49 members as determined by a transcriptomic analysis, is highly expanded and diversified as compared with other invertebrate and vertebrate species.3
In this study, we identified and characterized a CTL that is highly expressed in the hemocytes and present in plasma of the kuruma shrimp (Marsupenaeus japonicus), a species of high commercial value, and that we named MjHeCL for M. japonicus hemocyte C-type lectin. Although MjHeCL expression was not affected by microbial challenge, silencing its expression by RNA interference (RNAi) caused uncontrolled bacterial proliferation in the hemolymph and death of the shrimp. A functional study revealed that the activity of MjHeCL was based on its ability to modulate the expression of AMPs.
EXPERIMENTAL PROCEDURES
Animals
Healthy kuruma shrimp (M. japonicus) with an average weight of 6–7 g each were obtained from an aquaculture farm in Rizhao, Shandong, China; acclimated in aerated artificial seawater (30 parts per thousand) at 20 °C for a week prior to use; and fed daily with a commercial shrimp diet.
Immune Challenge
Vibrio anguillarum (American Type Culture Collection (ATCC) 43305) and Vibrio harveyi (ATCC 33842) were obtained from the Marine College, Shandong University (Weihai, China), cultured overnight in Luria-Bertani medium (3% NaCl), collected by centrifugation at 5,000 × g for 3 min, washed by sterile phosphate-buffered saline (PBS; 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4, pH 7.4) thrice, resuspended in PBS, and plated for colony counting, and the bacterial suspension was adjusted to 2 × 106 CFU/ml. Fifty microliters of the bacterial suspension were injected into each shrimp intramuscularly at the fourth abdominal segment with similar mock PBS injections in the control shrimp. Hemolymph was drawn from the ventral sinus at 2, 6, 12, and 24 h postchallenge (at least six shrimp at each time point) using a sterile syringe fitted with a 26-gauge needle and collected into precooled anticoagulant (450 mm NaCl, 10 mm KCl, 10 mm EDTA, 10 mm HEPES, pH 7.45) at a ratio of 1:1. The hemolymph was centrifuged at 800 × g for 10 min, and total RNA was extracted from the hemocyte pellet using TRIzol (Invitrogen) according to the manufacturer's instructions. The plasma supernatant was subjected to ultracentrifugation at 140,000 × g for 2 h to remove most of the hemocyanin and concentrated in an ultracentrifugal filter (Millipore). The hemocytes and plasma from unchallenged shrimp were collected and processed at the same time points as controls.
Analysis of MjHeCL Expression Profiles
Semiquantitative RT-PCR was used to study the tissue distribution and abundance of MjHeCL transcripts. A pair of specific primers was designed to amplify a 141-bp fragment (Table 1) with β-actin amplified as the reference gene. The PCR amplification protocol consisted of an initial 94 °C for 3 min followed by 30 cycles (22 cycles for β-actin) of 94 °C for 30 s, 54 °C for 30 s, and 72 °C for 30 s and a final 72 °C for 10 min. The PCR products were analyzed by electrophoresis in 1% agarose and stained by ethidium bromide.
TABLE 1.
Primers used in the present study
| Primers | Sequence (5′–3′) |
|---|---|
| (q)RT-PCR | |
| MjHeCLRTF | ACGCTGGTGTGATGCCCG |
| MjHeCLRTR | ACCGAGTCTGAGCCGCCTAA |
| ActinRTF | CAGCCTTCCTTCCTGGGTATGG |
| ActinRTR | GAGGGAGCGAGGGCAGTGATT |
| Alf1RTF | CGGTGGTGGCCCTGGTGGCACTCTTCG |
| Alf1RTR | GACTGGCTGCGTGTGCTGGCTTCCCCTC |
| Alf4RTF | CGCTTCAAGGGTCGGATGTG |
| Alf4RTR | CGAGCCTCTTCCTCCGTGATG |
| Alf5RTF | CTGGTCGGTTTCCTGGTGGC |
| Alf5RTR | CCAACCTGGGCACCACATACTG |
| Alf6RTF | TCCTGGTGGTGGCAGTGGCT |
| Alf6RTR | TGCGGGTCTCGGCTTCTCCT |
| Alf8RTF | CGCAGGCTTATGGAGGAC |
| Alf8RTR | AGGTGACAGTGCCGAGGA |
| Alf9RTF | TGCCGTGTTCTCCTGCTTAT |
| Alf9RTR | TTGGTGGGATTCGTGTGGT |
| Pen2RTF | AGGCGAGGAGAAAATCAA |
| Pen2RTR | AGAGAAGAAGCAACTACCAATC |
| cLysRTF | TTGCGGAGTTGCTGGAGA |
| cLysRTR | TTCGGTATCACGGCGGA |
| iLys1RTF | TACTGGATGGACGGCGG |
| iLys1RTR | ATTCGGCATTAGGTGGTGG |
| Cru4RTF | CAAGCCCTCCACCACTCTCG |
| Cru4RTR | TTCCTGGGTTGCGGTCACA |
| Cru11RTF | GCGTTTTCGTCTTCGTCCTG |
| Cru11RTR | AATGATTGGTGGTTTCACGGTAG |
| RNAi | |
| MjHeCL RNAiF | GCGTAATACGACTCACTATAGGGTCCTGACGACTTCTTCCT |
| MjHeCL RNAiR | GCGTAATACGACTCACTATAGGACTCGCACTGACGCAAT |
| GFPRNAiF | GCGTAATACGACTCACTATAGGTGGTCCCAATTCTCGTGGAAC |
| GFPRNAiR | GCGTAATACGACTCACTATAGGCTTGAAGTTGACCTTGATGCC |
| Alf4RNAiF | GCGTAATACGACTCACTATAGGATCTCGCTCTACAGCAACG |
| Alf4RNAiR | GCGTAATACGACTCACTATAGGCATCTGATACCACGACCTTT |
| Alf5RNAiF | GCGTAATACGACTCACTATAGGCAGTCAGCGGCGGAGAA |
| Alf5RNAiR | GCGTAATACGACTCACTATAGGCTGCGTAAAACAAACACCC |
| Alf6RNAiF | GCGTAATACGACTCACTATAGGAGTGGCTGCGTCCTTC |
| Alf6RNAiR | GCGTAATACGACTCACTATAGGATTATTTCATTGAGTTGGTC |
| Pen2RNAiF | GCGTAATACGACTCACTATAGGTCATCTGGCTCCTACGG |
| Pen2RNAiR | GCGTAATACGACTCACTATAGGTCTTTTTTTCACAAGCATTT |
| Recombinant expression | |
| MjHeCLYEF | TACTCAGAATTCAAGGTATCCTGTCCTGAC |
| MjHeCLYER | TACTCAGCGGCCGCTTAGTGGTGGTGGTGGTGGTGATTAAATGCCTCCACTATG |
| MjHeCLEF | CCGGAATTCAAGGTATCCTGTCCTGAC |
| MjHeCLER | CCCAAGCTTTTAATTAAATGCCTCCAC |
| MjCL1EF | CCGGAATTCAACCACACGCCCACAGAA |
| MjCL1ER | CCCAAGCTTTTATGAGGGTTCCACTTG |
| MjCL2EF | CCGGAATTCAGCGTTCGTTCTAACGAG |
| MjCL2ER | CCCAAGCTTTTAATAATTCTTGAGGCA |
| MjCL3EF | CCGGAATTCTGTCCAGATGGCTTCTTT |
| MjCL3ER | CCCAAGCTTTCAGCTGGACTTAGCCCT |
| MjCL4EF | CCGGAATTCTGCGAGATCGGATGGGTA |
| MjCL4ER | CCCAAGCTTTTAGAGCATCATGCAGAG |
| MjCL5EF | CCGGAATTCAAGTGCACGGCGCCCTTC |
| MjCL5ER | CCCAAGCTTGAAGGTGTCCGGGCTGAA |
| MjCL6EF | CCGGAATTCTCCCACATGACGGCCGAG |
| MjCL6ER | CCCAAGCTTGTCTCCGCCATGTTATTT |
| MjCL7EF | CCGGAATTCTCGGAGGCCTCCAGAGGC |
| MjCL7ER | CCCAAGCTTTGATCATCCATGATAAAC |
| MjCL8EF | CCGGAATTCCAACAACCTGATAACGCA |
| MjCL8ER | CCCAAGCTTTTACGTCACCTGCGCCCACGT |
| MjCL9EF | CCGGAATTCAAACAAGACTGGGCGTCA |
| MjCL9ER | CCCAAGCTTGCTTTATTTCTGGCAAAG |
| 16 S rRNA amplification | |
| 27F | AGAGTTTGATCCTGGCTCAG |
| 1492R | GGTTACCTTGTTACGACTT |
Quantitative changes of MjHeCL expression were assessed by real time PCR (qRT-PCR) in a CFX96 Real-Time System (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad) and the same primers as for semiquantitative RT-PCR. PCR was performed with an initial 94 °C for 3 min, 35 cycles of 94 °C for 10 s and 60 °C for 1 min, and then a melt period from 65 to 95 °C. Results were processed by using a 2−ΔΔCt method by which the data were normalized in two steps. First, the expression of the gene of interest (MjHeCL) was normalized to that of the reference gene (β-actin) in the same sample. Second, the relative expression of the gene in the experimental shrimp sample was normalized to that in the control shrimp sample. Results were expressed as the mean ± S.D. from three independent repeats. The expression of the antimicrobial peptides was evaluated with the primers listed in Table 1 using a similar strategy as for MjHeCL.
MjHeCL levels in shrimp plasma were assessed by Western blot in both challenged and control animals. Plasma proteins were separated by SDS-PAGE in 12.5% gels and transferred onto nitrocellulose membranes. After blocking in nonfat milk (5% in PBS) for 1 h, the membrane was immersed in rabbit anti-MjHeCL antiserum (1:200 in blocking milk) overnight. The membrane was washed thrice with PBST (0.02% Tween 20) and incubated with HRP-conjugated goat anti-rabbit antibodies (1:10,000 in blocking milk; Zhongshan, Beijing, China) for 3 h. After three washes with PBST, the protein bands were visualized by oxidation of 4-chloro-1-naphthol.
RNAi in Vivo
A 597-bp MjHeCL fragment was amplified by PCR with the specific primers linked to the T7 promoter (Table 1). The resulting DNA was used as the template to synthesize dsRNA using an in vitro transcription T7 kit (Takara) following the manufacturer's instructions. GFP dsRNA was synthesized as the control. Each shrimp was injected with 10 μg of dsRNA (MjHeCL or GFP), and hemolymph was collected at 24, 48, and 72 h postchallenge (at least three shrimp per time point for experimental samples and controls). RNAi efficiency was assessed by RT-PCR on total RNAs from hemocytes and by Western blot on plasma. RNAi for the AMPs was performed in the same way as for MjHeCL using the primers listed in Table 1.
RNAi in Vitro
Primary cultures of shrimp hemocytes were established by separating the cells from plasma by centrifugation at 800 × g for 10 min and resuspending the cells in Leibovitz L-15 medium (Sangon, Shanghai, China) supplemented with 15% fetal bovine serum (Sigma), 5% shrimp plasma (collected without anticoagulant and centrifuged at 20,000 × g for 30 min to obtain the supernatant), 1 g/liter glucose, 2 g/liter NaCl, 0.3 g/liter glutamine, 0.1 mg/liter vitamin C, 100 IU/ml penicillin, and 100 μg/ml streptomycin sulfate. The hemocyte suspensions (5 × 105 cells/ml) were distributed in 6-well plates at 2 ml/well and incubated at 28 °C for 12 h. Two micrograms of dsRNA (for MjHeCL and for GFP as control) and 5 μl of Lipofectamine 2000 (Invitrogen) were mixed with 250 μl of serum-free medium, respectively, incubated separately for 5 min, mixed together, incubated for an additional 20 min, and added to serum-free medium (1.5 ml). The final mixture was added to each well after removing the initial culture medium, incubated for 10 h, and replaced by serum-containing medium (2 ml). Total RNA was extracted 24 h later to evaluate expression of the target genes in both experimental and control (GFP dsRNA) wells.
Assessment of Survival Rates
Shrimp were randomly divided into three groups with 30 animals in each group. In the first group, each shrimp was injected with 10 μg of MjHeCL dsRNA, 10 μg of GFP dsRNA was injected in the second group, and PBS was injected in the third group. The numbers of dead shrimp were recorded for each group daily for 3 days, and survival rates were calculated at 72 h postinjection.
Bacterial Counts in Hemolymph
Hemolymph from experimental and control shrimp was collected as described above and immediately mixed with an equal volume of sterile anticoagulant buffer. One hundred microliters of each mixture were plated onto a modified 2216E agar plate (0.5% tryptone, 0.1% yeast extract, 0.01% FeCl3, 2.4% sea salt, and 1.5% agar). The plates were placed at 28 °C for 36 h, and the colony counts were recorded for each plate.
Antibiotic Treatments
To ensure that any proliferating bacteria in shrimp hemolymph originated in the endogenous microbiota, ampicillin and kanamycin were added to the culture water to a final concentration of 25 mg/liter to generate an axenic environment. To exclude the potential interference of the endogenous microbiota on the expression of AMPs, each shrimp was injected with ampicillin and kanamycin (25 μg each/animal) to generate axenic animals.
Recombinant Protein Expression and Purification
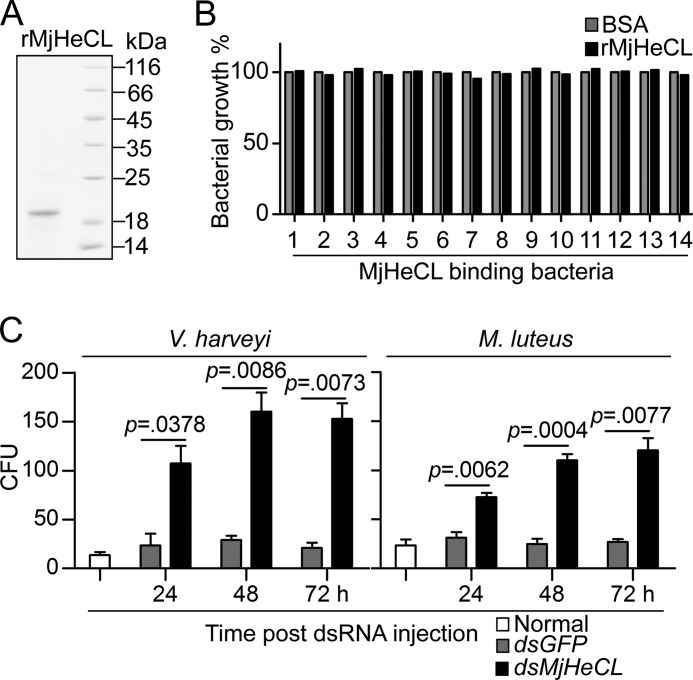
For expression of the recombinant MjHeCL in yeast, the fragment corresponding to the mature MjHeCL was amplified with the primers shown in Table 1 and cloned into the pPIC9k vector (Invitrogen). The recombinant plasmid was linearized with SacI (New England Biolabs) and transformed into the Pichia pastoris GS115 cells by electroporation. The transformants were screened with the histidine-deficient minimal dextrose medium. Positive transformants were inoculated into BMGY medium (1% yeast extract, 2% tryptone, 1.34% yeast nitrogen base, 4 × 10−5% biotin, 1% glycerol, and 100 mm potassium phosphate, pH 6.0) and cultured at 30 °C for 18 h and transferred into BMMY medium (1% yeast extract, 2% tryptone, 1.34% yeast nitrogen base, 4 × 10−5% biotin, 0.5% methanol, and 100 mm potassium phosphate, pH 6.0) for protein induction. Methanol was supplemented into the medium to a final concentration of 0.5% (w/v) every 24 h for 3 days. The culture supernatant (600 ml) was collected by centrifugation and applied to a nickel-nitrilotriacetic acid His-Bind resin column (Novagen). The recombinant protein was eluted by 250 mm imidazole in 50 mm Tris-HCl, pH 7.5 and 300 mm NaCl. The eluted protein was dialyzed in PBS thrice at 4 °C to remove the imidazole. Protein concentrations were determined by the standard Bradford method with BSA as the control and adjusted to 200 μg/ml. After being filtered through a sterile 0.45-μm filter, the protein was stored at −80 °C with 5% glycerol (v/v).
The bacterial expression of recombinant MjHeCL and other shrimp CTLs (MjCL1–9) was performed according to the standard method using the pET32a(+) (Novagen) vector and Escherichia coli Rosetta (DE3) cells (primers are listed in Table 1). The recombinant proteins were purified on a nickel-nitrilotriacetic acid His-Bind column. Potential endotoxin contaminants were removed by adding an additional wash before final elution with 0.1% Triton X-114 (Sigma) in the wash buffer (50× column volume) at 4 °C (29). The recombinant proteins were processed and stored following the protocol described above.
Antiserum Preparation
The recombinant MjHeCL (rMjHeCL-tag) was expressed in bacteria, purified, and concentrated to 3 mg/ml with an ultracentrifugal filter. Equal volumes of rMjHeCL-tag solution (1 ml) and complete Freund's adjuvant (Sigma) were mixed thoroughly and injected into a New Zealand White rabbit. The injection was repeated 25 days later with a similar rMjHeCL-tag inoculum but mixed with incomplete Freund's adjuvant. Periodic bleeds were tested for anti-MjHeCL titer and specificity, and after the kill bleed, the anti-MjHeCL antiserum was stored at −80 °C for further use.
Binding of MjHeCL to Components of the Hemolymph Microbiota
The bacteria present in the shrimp hemolymph were isolated and characterized by 16 S rRNA. The bacteria growing in plates corresponding to the experimental and control shrimp from the experiment described above were selected based on colony morphology, isolated, and grown as individual cultures in 2216E medium at 30 °C overnight. From each culture the 16 S rRNA was amplified using the standard primers (Table 1) and sequenced for species identification. The remaining bacterial cultures were used for MjHeCL binding assays and stored at −80 °C.
To test whether MjHeCL could bind to bacteria, a preliminary study was carried out with a Vibrio spp. isolated from shrimp hemolymph. An overnight bacterial culture was harvested as described above and resuspended in PBS. The binding was performed by incubating the rMjHeCL-tag (expressed in bacteria; 4 μg) with the selected bacteria (108 CFU) in 1 ml of PBS. The mixture was incubated at 25 °C for 1 h with gentle rotation. The bacteria were collected by centrifugation at 5,000 × g for 3 min and washed thrice with PBST. The resultant bacterial pellet was resuspended in PBS, processed for SDS-PAGE by adding regular sample buffer and heating at 100 °C for 5 min, and subjected to Western blot. rMjHeCL-tag that bound to the bacteria was detected by an antibody specific for the pET32a(+) tag. Negative controls consisted in replacing the rMjHeCL-tag by the pET32a(+) tag. To determine whether calcium was necessary for the binding, a similar experiment was carried out with either CaCl2 (5 mm), EDTA (5 mm), or both added to the mixture of bacteria and rMjHeCL-tag.
After the preliminary experiments revealed that rMjHeCL could bind to Vibrio spp., all other bacterial isolates from shrimp hemolymph were tested similarly to determine the binding spectrum of rMjHeCL and other recombinant shrimp CTLs (MjCL1–9). The authentic MjHeCL was also tested to examine whether it exhibited binding behavior similar to that of the rMjHeCL. For this, ∼107 hemocytes were resuspended in 1.5 ml of PBS and sonicated at 300 Hz for 5 min. The hemocyte lysate was centrifuged at 20,000 × g for 20 min. The supernatant (1 ml) was used as the source of native MjHeCL. Bound proteins were detected with an antibody specific for the tag (for recombinant proteins) or the anti-MjHeCL antibody (for the native MjHeCL).
Antibacterial Activity of MjHeCL
The bacteria isolated from the shrimp hemolymph and recognized by MjHeCL were used to determine whether MjHeCL possesses antibacterial activity. The bacterial strain suspensions were prepared as described above, adjusting their concentration to 106 CFU/ml. To assess the antibacterial activity of MjHeCL, 10 μl of the bacterial suspension were incubated with 2 μg of rMjHeCL for 2 h at 25 °C. The mixture was then introduced to 200 μl of fresh LB medium (3% NaCl) and cultured at 25 °C for 24 h. The A600 of each sample was recorded as a measure of bacterial growth.
The antimicrobial activity of plasma was evaluated after the initial centrifugation of hemolymph at 800 × g for 10 min to separate it from the hemocytes followed by centrifugation at 20,000 × g for 20 min to obtain a clear supernatant. Equal volumes (10 μl) of the supernatant and bacterial suspension were mixed and placed at 25 °C for 1 h. The mixtures were plated onto LB (3% NaCl for V. harveyi) agar plates and cultured at 30 (V. harveyi) or 37 °C (Micrococcus luteus) for 30 h, and the colony counts were recorded. Results were expressed as the mean ± S.D. derived from three independent repeats.
Phenotype Rescue Experiments
Phenotypes (AMP expression, bacterial proliferation, and shrimp survival rates) resulting from the down-regulated MjHeCL expression by RNAi were rescued by co-injection of the recombinant MjHeCL together with the dsRNA. For the AMP expression analysis, MjHeCL dsRNA (10 μg) was mixed with 0.1, 0.5, 2.5, or 8 μg of recombinant protein, respectively, in a total volume of 50 μl that was injected in each shrimp (six shrimp per experiment). GFP dsRNA or MjHeCL dsRNA together with BSA (2 μg) served as controls. Total RNA was extracted from hemocytes at 24 h postinjection, and the expression of AMPs was analyzed by qRT-PCR.
For the bacterial number determination, each shrimp was co-injected with MjHeCL dsRNA (10 μg) together with either rMjHeCL or BSA (2.5 μg each). Thirty shrimp were used for the MjHeCL group, whereas 70 animals were used for the BSA group. Bacterial numbers in hemolymph were determined at 24, 48, and 72 h postinjection. For each time point, eight shrimp were analyzed.
For the assessment of survival rates, each shrimp was injected with MjHeCL dsRNA (10 μg) together with 2.5 μg of either rMjHeCL or BSA. The survival rates were evaluated at 72 h postinjection. Twenty shrimp were used for MjHeCL group, whereas 30 were used for the BSA group.
To assess whether MjHeCL alone could induce the expression of AMPs in the absence of hemolymph microbiota in vitro, hemocytes were cultured in 6-well plates. Either rMjHeCL or BSA (3 μg) was applied to each well, and the expression of AMPs was evaluated 6 h later. Results were expressed as the mean ± S.D. derived from three independent repeats. For the in vivo studies, rMjHeCL (5 μg) was injected into axenic shrimp that had been pretreated by antibiotics. An equal amount of BSA was used as a control. The expression of AMPs was determined 24 h later. The experiment was repeated three times with at least six shrimp in each sample.
Binding of MjHeCL to Hemocytes
Hemocytes were cultured in 6-well plates. Either rMjHeCL or pET32a tag (3 μg) was added into the well and incubated for 3 h. The medium was removed, and the cells were washed five times with PBS. Cells were mixed with SDS-PAGE sample buffer and subjected to Western blot. The recombinant proteins bound to hemocytes were detected with an anti-His tag antibody (Zhongshan).
RESULTS
MjHeCL Expression and Release to Plasma Are Not Affected by Infectious Agents
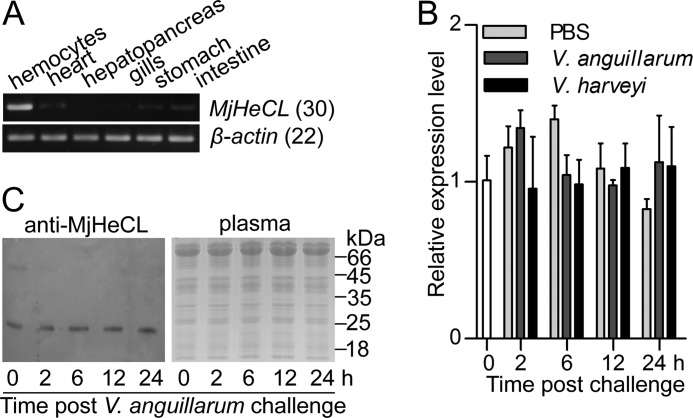
The open reading frame of MjHeCL contains 474 base pairs encoding a protein of 157 residues with a 17-amino acid signal peptide and a 125-amino acid carbohydrate recognition domain (GenBankTM accession number KJ175168). As shown in Fig. 1A, MjHeCL transcripts were mostly localized to shrimp hemocytes with lower levels detected in heart, gills, stomach, and intestine. Surprisingly, no expression was observed in the hepatopancreas, an organ in which most other shrimp lectins are synthesized. qRT-PCR results showed that MjHeCL expression was high and relatively stable, showing negligible changes even upon challenge by the pathogenic V. anguillarum and V. harveyi (Fig. 1B). Consistent results were observed for the MjHeCL protein for which the bacterial challenge had little effect on its release into the circulating plasma (Fig. 1C).
FIGURE 1.
Expression of MjHeCL is not significantly affected by bacterial challenge. A, MjHeCL was expressed mainly in hemocytes as analyzed by semiquantitative RT-PCR with β-actin as the reference. B, MjHeCL did not respond to external bacterial challenge. Shrimp were challenged by V. anguillarum or V. harveyi, and the expression of MjHeCL in hemocytes was assessed postchallenge by qRT-PCR. Data represent mean ± SD from three independent repeats. Error bars represent S.D. C, MjHeCL was constitutively secreted into plasma. Shrimp plasma was collected after bacterial challenge, subjected to ultracentrifugation to remove the majority of hemocyanin, concentrated, and subjected to Western blot analyzed by MjHeCL antiserum.
MjHeCL Inhibits the Proliferation of Bacteria Present in Shrimp Hemolymph
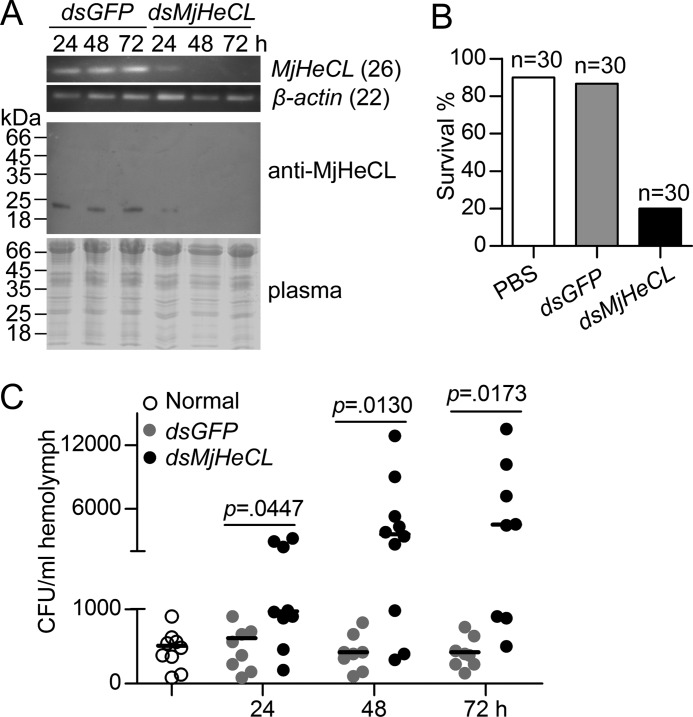
To reveal the function of MjHeCL, RNAi was performed on healthy shrimp to suppress its expression. The expression of MjHeCL could be significantly knocked down by dsRNA injection for at least 3 days (Fig. 2A) with a survival rate of about 20% compared with 90% in the control group (Fig. 2B). This suggested that MjHeCL expression is critical to maintain the healthy status of shrimp. To elucidate the potential causes of shrimp death, the hemolymph of dsRNA-injected shrimp was examined for bacterial proliferation. The results (Fig. 2C) revealed that the number of bacteria in the circulating hemolymph significantly increased from 24 to 72 h post-MjHeCL dsRNA injection, whereas the bacterial counts in the hemolymph of the control group did not change.
FIGURE 2.
MjHeCL inhibits the proliferation of endogenous bacteria. A, RNAi efficiency of MjHeCL. Shrimp were injected with 10 μg of dsRNA, and the expression of MjHeCL was analyzed at 24, 48, and 72 h postinjection by RT-PCR (upper panel) and Western blot (lower panel). GFP dsRNA (dsGFP) was used as a control. B, knockdown of MjHeCL led to shrimp death. Thirty shrimp were injected with 10 μg of MjHeCL dsRNA (dsMjHeCL) each, and shrimp death was recorded 72 h postinjection. PBS and GFP dsRNA were used as controls. C, the bacterial counts in MjHeCL knockdown shrimp were higher than in the control animals. Shrimp were injected with 10 μg of MjHeCL dsRNA, and the hemolymph was drawn out at 24, 48, and 72 h postinjection and plated onto agar plates, and the bacterial counts were determined. The horizontal bars represent the median. The p value was calculated by the t test for paired samples, and significant differences was accepted when p was <0.05.
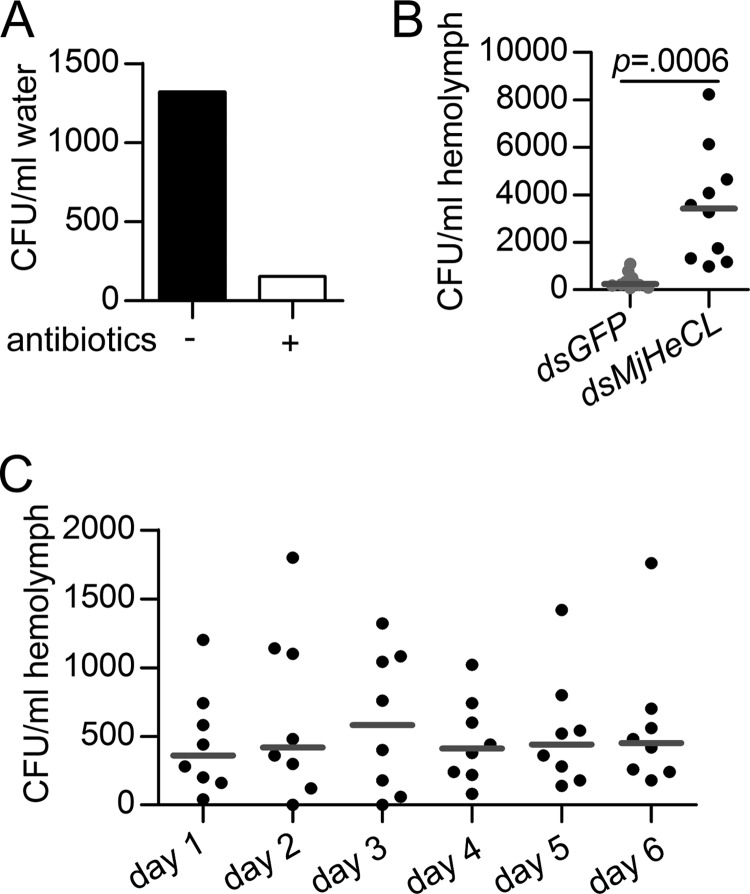
To identify the source of the proliferating bacteria in the knockdown shrimp, that is whether they were originating from endogenous or environmental bacterial populations, we treated the water with antibiotics to generate an axenic (i.e. bacterium-free) environment (Fig. 3A). The bacterial counts in the knockdown shrimp that had been maintained in axenic water increased similarly as those in the untreated water (Fig. 3B). These results suggest that the proliferating bacteria originate from the endogenous shrimp microbiota, which in the healthy shrimp was inhibited by MjHeCL expression.
FIGURE 3.
The source of proliferating bacteria was the endogenous microbiota. A, antibiotics were used to eliminate the bacteria in the aquarium water. Ampicillin and kanamycin were added to the water to a final concentration of 25 mg/liter. B, endogenous bacteria still proliferated when MjHeCL knockdown shrimp were cultured in the antibiotic-treated water. Shrimp were injected with 10 μg of MjHeCL dsRNA (dsMjHeCL), and the bacterial counts in hemolymph at 24 h postinjection were determined. GFP dsRNA (dsGFP) was used as the control. The horizontal bars represent the median. The p value was calculated by the t test for paired samples. C, healthy shrimp host bacteria in the hemolymph. The bacterial counts in the hemolymph from eight shrimp maintained under standard conditions were determined every day for 6 continuous days. The horizontal bars represent the median.
Although bacterial communities colonize most surfaces and cavities of shrimp with by far the most abundant bacterial populations present in the gut microbiota, bacteria have been detected in shrimp hemolymph of healthy animals (4). Thus, considering the possibility that the source of the proliferating bacteria in the knockdown shrimp could actually be the hemolymph, we assessed the presence and abundance of bacteria in the hemolymph of healthy shrimp. Bacteria were detected in the hemolymph of all healthy animals examined, and the bacterial counts remained unchanged during the 6-day monitoring period, while the shrimp remained healthy and with no signs/symptoms of infectious disease (Fig. 3C). This observation was consistent with a former study that showed that the relatively low and constant numbers of bacteria detected in the hemolymph of healthy shrimp rapidly increased under stressful conditions (4).
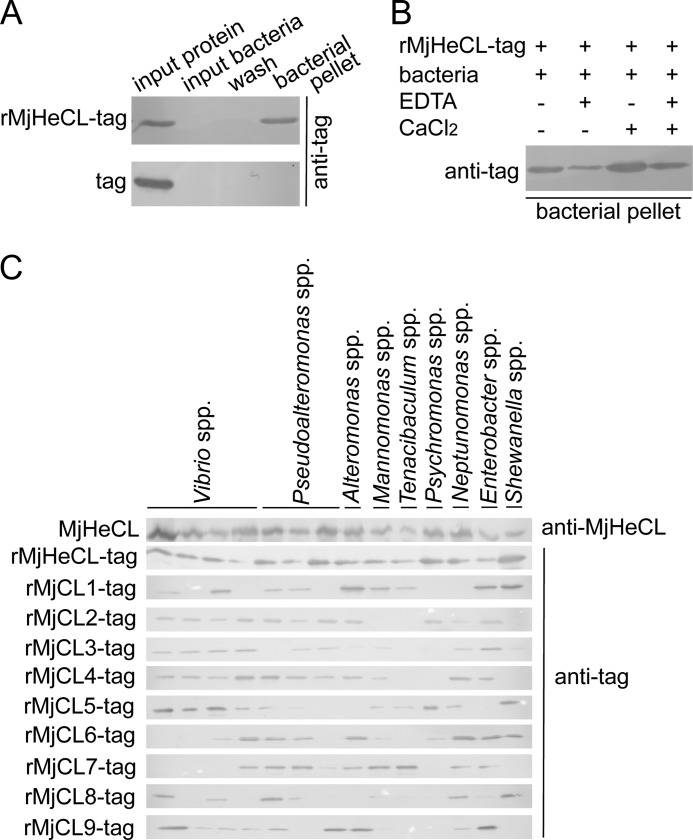
MjHeCL Recognizes and Binds to the Strains of the Hemolymph Microbiota but Lacks Antimicrobial Activity
As some lectins can recognize and kill microbial pathogens, we explored the possibility that the proliferation of hemolymph bacteria in the MjHeCL RNAi shrimp was due to the loss of the MjHeCL-mediated recognition and control of the bacterial populations. For this, we first examined the recognition spectrum of MjHeCL for selected components of the hemolymph microbiota. Based on 16 S rRNA sequencing, from the 14 colony types we most frequently isolated from shrimp hemolymph, four were identified as Vibrio spp.; three were identified as Pseudoalteromonas spp.; and other isolates were identified as Alteromonas spp., Marinomonas spp., Tenacibaculum spp., Psychromonas spp., Neptunomonas spp., Enterobacter spp., and Shewanella spp. A preliminary experiment revealed that rMjHeCL-tag could bind to a Vibrio spp. isolate from the hemolymph microbiota (Fig. 4A), and the calcium requirement of this interaction was examined. The removal of calcium by EDTA chelation partially reduced binding of rMjHeCL to Vibrio spp., whereas calcium addition enhanced the binding, and the addition of both had no effect (Fig. 4B). Based on this preliminary information, the binding spectra of MjHeCL and other shrimp CTLs for the hemolymph microbiota were determined. Both the native and recombinant MjHeCL recognized and bound all the bacterial strains isolated. However, the binding spectrum of each of the nine additional recombinant CTLs (MjCLs) we had identified was restricted to a limited subset of isolates (Fig. 4C). The broad recognition activity of MjHeCL for hemolymph bacteria supports a critical role, direct or indirect, in the inhibition of proliferation of the hemolymph microbiota.
FIGURE 4.
MjHeCL binds to all strains isolated from the hemolymph microbiota. A, recombinant MjHeCL binds to a Vibrio spp. isolated from hemolymph. Overnight cultured bacteria were collected, resuspended at about 108 CFU/ml, and incubated with 4 μg of recombinant proteins (from bacteria) for 1 h at 25 °C. The bacteria were then washed and analyzed by Western blot using antibodies against the tag of pET32a(+). B, calcium requirement of MjHeCL for binding to bacteria (Vibrio spp.). Either EDTA (5 mm), CaCl2 (5 mm), or both were added to the bacterium-rMjHeCL mixture, and the binding was evaluated by Western blot. C, MjHeCL shows a broader binding spectrum than other shrimp CTLs. Binding of shrimp recombinant CTLs and native MjHeCL was determined as described above. Hemocyte lysate supernatant (1 ml) was used as the source of native MjHeCL, and binding was detected with antibody against MjHeCL.
To find out why the endogenous bacteria proliferated in the MjHeCL RNAi shrimp, we examined whether, in addition to its broad microbial recognition properties, MjHeCL itself displays antibacterial activity. The purified recombinant MjHeCL expressed in yeast (Fig. 5A) was used to treat the bacteria, and it did not show antimicrobial activity against any of the bacterial species or strains tested (Fig. 5B).
FIGURE 5.
The antimicrobial activity of plasma was suppressed in the MjHeCL knockdown shrimp. A, expression and purification of recombinant MjHeCL. The recombinant MjHeCL was expressed in P. pastoris GS115 and purified by affinity chromatography. B, the recombinant MjHeCL did not show antimicrobial activity. Recombinant protein (2 μg) was incubated with the bacterial suspension (∼104 CFU) for 2 h, the mixture was added to 200 μl of fresh medium, and the bacterial growth was recorded 24 h later. The strains used here are Vibrio spp. (1–4), Pseudoalteromonas spp. (5–7), Alteromonas spp. (8), Marinomonas spp. (9), Tenacibaculum spp. (10), Psychromonas spp. (11), Neptunomonas spp. (12), Enterobacter spp. (13), and Shewanella spp. (14). C, the plasma antimicrobial activity was suppressed in the MjHeCL knockdown shrimp. Equal volumes (10 μl) of plasma and bacterial suspension were incubated at 25 °C for 1 h. The mixture was then plated onto agar plates to determine the surviving bacteria. Data show the mean ± S.D. from three independent repeats. Error bars represent S.D. The p value was calculated by t test for paired samples, and significant differences were accepted when p was <0.05. dsMjHeCL, MjHeCL dsRNA; dsGFP, GFP dsRNA.
MjHeCL Regulates the Expression of Several Antimicrobial Peptides
Based on the observation that MjHeCL itself did not kill or inhibit the growth of bacteria, we investigated the potential antimicrobial activity of the shrimp plasma to find out whether the endogenous bacterial proliferation was due to a change in plasma properties. As shown in Fig. 5C, after incubation with the plasma of the MjHeCL RNAi shrimp, the survival of both the Gram-negative pathogenic bacterium V. harveyi and the Gram-positive non-pathogenic bacterium M. luteus was higher than that following incubation with the control plasma. This indicated that the antimicrobial ability of plasma was suppressed after MjHeCL expression was knocked down by RNAi.
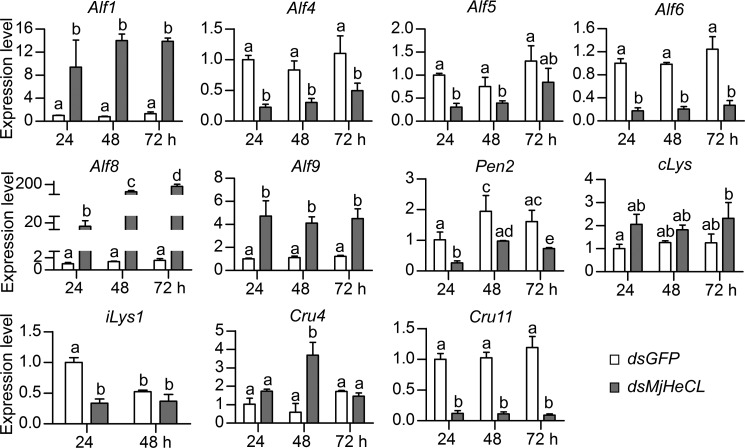
Because as indicated above our results revealed that MjHeCL lacked antimicrobial activity, we explored the possibility that the decrease of antimicrobial activity in the MjHeCL RNAi shrimp was due to the suppression of AMP expression. Those AMPs that are expressed in hemocytes (our observation3) were selected for this study, and the results showed that the expression of some AMPs including antilipopolysaccharide factors 4, 5, and 6 (Alf4, Alf5, and Alf6), penaeidin 2 (Pen2), i-type lysozyme 1 (iLys1), and crustin 11 (Cru11) was down-regulated by MjHeCL dsRNA injection (Fig. 6). However, the expression of other AMPs was significantly up-regulated. This observation prompted us to discern whether these opposite changes in expression of AMPs were due to the MjHeCL RNAi itself or the bacterial proliferation resulting from MjHeCL silencing.
FIGURE 6.
Knockdown of MjHeCL induced changes in expression of AMPs in hemocytes. Shrimp were injected with 10 μg of MjHeCL dsRNA (dsMjHeCL), and the expression of AMPs was analyzed by qRT-PCR at 24, 48, and 72 h postinjection. GFP dsRNA (dsGFP) was injected as a control. Data represent mean ± SD from three independent repeats. Error bars represent S.D. Data were subjected to one-way analysis of variance from triplicate assays. Significant differences (p < 0.05) are represented by different letters.
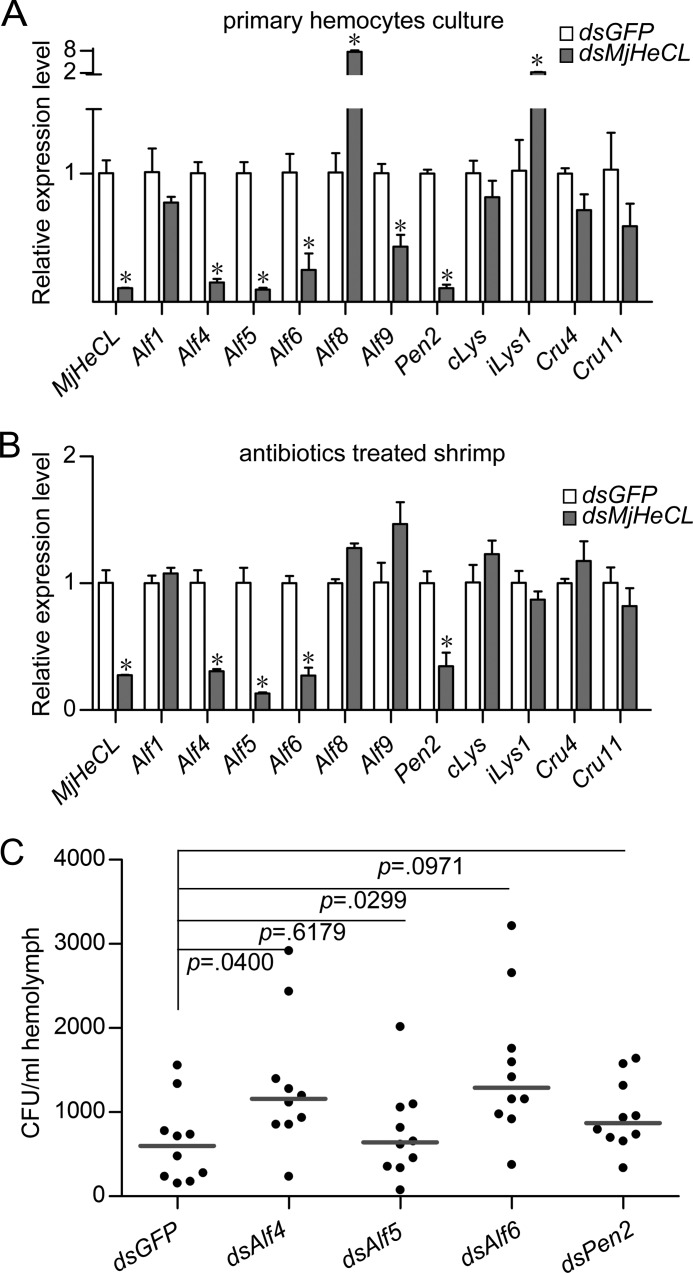
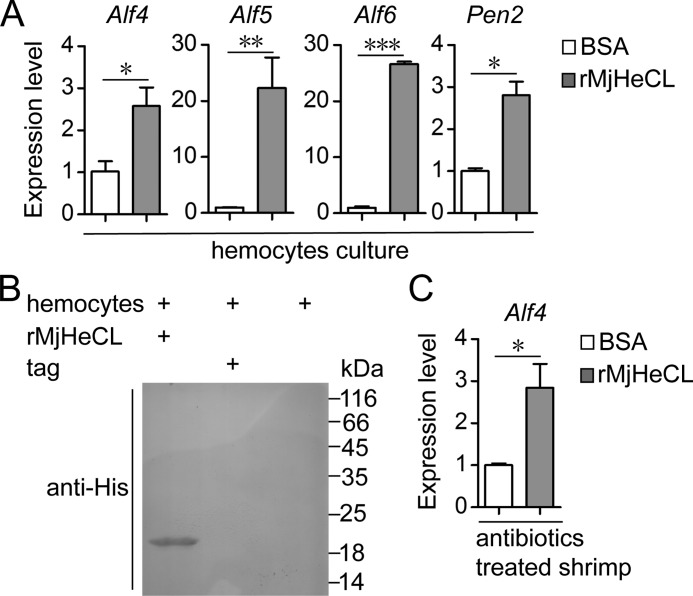
To examine the first alternative, that is the possibility that instead of the endogenous bacteria actually MjHeCL regulates the hemolymph levels of AMPs, we carried out an in vitro study in which an axenic hemocyte primary culture was treated with MjHeCL dsRNA. The results revealed that the expression of the AMPs Alf4, Alf5, Alf6, and Pen2 was suppressed upon MjHeCL knockdown (Fig. 7A). To test in vivo the potential role(s) of the endogenous microbiota on the regulation of expression of AMPs in the MjHeCL RNAi and control shrimp, we treated the shrimp with antibiotics to eliminate the endogenous bacteria, subsequently injected them with dsRNA, and measured the expression of selected AMPs. The expression of Alf4, Alf5, Alf6, and Pen2 was down-regulated upon MjHeCL RNAi, revealing that MjHeCL, not the endogenous bacterial populations, regulates the levels of the AMPs indicated above (Fig. 7B).
FIGURE 7.
MjHeCL maintains the expression of Alf4, Alf5, Alf6, and Pen2, which are responsible to inhibit proliferation of endogenous bacteria. A, in vitro knockdown of MjHeCL in hemocytes suppressed the expression of Alf4, Alf5, Alf6, and Pen2. Shrimp hemocytes were precultured in 6-well plates. Two micrograms of dsRNA were transfected into hemocytes with 5 μl of Lipofectamine 2000 according to the manufacturer's instructions. The medium was replaced 10 h later. The expression of AMPs was analyzed after 24 h. GFP dsRNA (dsGFP) was used as the control. B, knockdown of MjHeCL in antibiotic-treated shrimp suppressed the expression of Alf4, Alf5, Alf6, and Pen2. Shrimp were preinjected with ampicillin and kanamycin (25 μg each), and 24 h later, 10 μg of dsRNA were injected into each shrimp. The expression of AMPs was analyzed by qRT-PCR 24 h later. For A and B, data represent mean ± SD from three independent repeats. Error bars represent S.D. Asterisks represent significant differences (calculated by the t test for paired samples from three repeats, and significant differences were accepted when p was <0.05). C, knockdown of Alf4, Alf5, Alf6, and Pen2 triggered proliferation of endogenous bacteria. Shrimp were injected with 10 μg of Alf4 (dsAlf4), Alf5 (dsAlf5), Alf6 (dsAlf6), and Pen2 (dsPen2) dsRNA, respectively. The bacterial counts in the hemolymph were determined 24 h later. The horizontal bars represent the median. GFP dsRNA was used as a control. The p value was calculated by the t test for paired samples against controls, and significant differences were accepted when p was <0.05.
MjHeCL-regulated Antimicrobial Peptides Maintain the Homeostasis of the Hemolymph Microbiota
To further test whether the bacterial proliferation was due to the down-regulation of expression of AMPs caused by MjHeCL RNAi, expression of the four AMPs of interest, Alf4, Alf5, Alf6, and Pen2, was silenced by RNAi, and the bacterial counts were determined in the hemolymph of the knockdown animals. Results showed that in the Alf4- and Alf6-deficient animals the hemolymph microbiota proliferated significantly, whereas the effects of Alf5 and Pen2 silencing were not significantly dramatic (Fig. 7C). Taken together, these results demonstrated that MjHeCL maintains the homeostasis of the hemolymph microbiota and thus the health of the animals by maintaining the expression of two AMPs, which are responsible for the inhibition of bacterial proliferation in hemolymph.
Recombinant MjHeCL Can Rescue the MjHeCL RNAi Phenotype
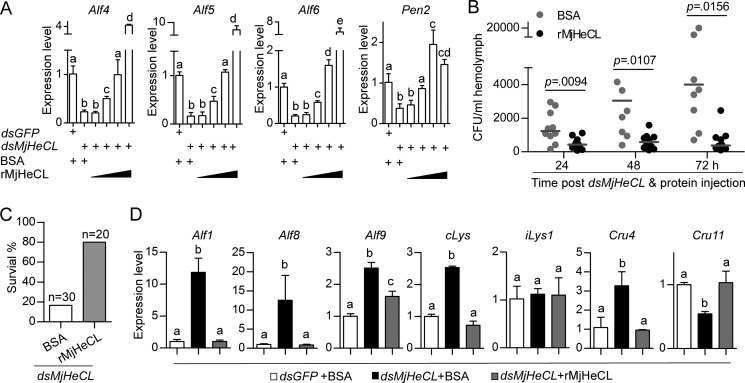
To confirm the role of MjHeCL in the regulatory functions proposed above, we injected the recombinant protein in the MjHeCL RNAi shrimp to inquire whether it could restore their healthy status. When the shrimp were injected with MjHeCL dsRNA together with recombinant MjHeCL, both the decreases of AMP expression and bacterial proliferation were restored to the control levels. As shown in Fig. 8A, the rescue by recombinant MjHeCL took place in a dose-dependent manner with 2.5 μg of protein fully counteracting the effects of dsRNA. It is noteworthy that injection of excess recombinant MjHeCL up-regulated the expression of AMPs. Furthermore, the bacterial counts did not increase in the MjHeCL RNAi shrimp that had been co-injected with recombinant MjHeCL, whereas co-injection of BSA was unable to rescue the phenotype (Fig. 8B). Most importantly, the survival rate of dsRNA-treated shrimp that had been co-injected with recombinant MjHeCL was about 80% compared with <20% in the BSA group (Fig. 8C). In addition, the changes in expression of other AMPs that were caused by the proliferating bacteria in the MjHeCL RNAi shrimp were also restored to the control levels by the co-injection of recombinant MjHeCL (Fig. 8D). These results further confirmed the protective regulatory role of MjHeCL in the AMP-controlled homeostasis of the hemolymph microbiota.
FIGURE 8.
Injection of recombinant MjHeCL rescued the phenotype observed in the MjHeCL knockdown shrimp. A, injection of recombinant MjHeCL restored the expression of Alf4, Alf5, Alf6, and Pen2. Shrimp were injected with MjHeCL dsRNA (dsMjHeCL) (10 μg) together with increasing amounts of rMjHeCL (0.1, 0.5. 2.5, and 8 μg). The expression of Alf4, Alf5, Alf6, and Pen2 was analyzed by qRT-PCR 24 h later. GFP dsRNA (dsGFP) and BSA were used to control MjHeCL dsRNA and rMjHeCL. Data represent mean ± SD from three independent repeats with error bars representing S.D. The data were subjected to one-way analysis of variance analysis. Different letters represent significant differences (p < 0.05). B, injection of recombinant MjHeCL suppressed the bacterial proliferation caused by the MjHeCL knockdown. Shrimp were injected with a mixture of MjHeCL dsRNA (10 μg) and rMjHeCL (2.5 μg). The bacterial counts in hemolymph were determined 24, 48, and 72 h later. BSA was used as a control. The horizontal bars represent the median. C, injection of rMjHeCL restored shrimp survival. Shrimp were injected with a mixture of MjHeCL dsRNA (10 μg) and rMjHeCL (2.5 μg). The survival rate was recorded at 72 h postinjection. BSA was used as a control. p values were calculated by the t test for paired samples, and significant differences were accepted when p was <0.05. D, injection of recombinant MjHeCL restored the expression of AMPs down-regulated in the MjHeCL knockdown shrimp. Shrimp were injected with a mixture of MjHeCL dsRNA (10 μg) and rMjHeCL (2.5 μg). The expression of AMPs was analyzed by qRT-PCR 24 h later. GFP dsRNA and BSA were used to control MjHeCL dsRNA and rMjHeCL. Data represents mean ± SD from three independent repeats with error bars representing S.D. The data were subjected to one-way analysis of variance analysis. Different letters represent significant differences (p < 0.05).
MjHeCL Binds to Hemocytes to Modulate AMP Expression
To investigate whether the MjHeCL alone or the MjHeCL-bacterium complex modulates the AMP expression, hemocytes were cultured in vitro, and rMjHeCL was added. As shown in Fig. 9A, rMjHeCL alone (in the absence of bacteria) could stimulate the expression of AMPs. A further study showed that the exogenous rMjHeCL could bind to the surface of hemocytes (Fig. 9B), suggesting that interaction of the MjHeCL with a hemocyte receptor is responsible for the up-regulation of AMP expression. Similarly, an in vivo study showed that the exogenous rMjHeCL alone could stimulate AMP expression (Fig. 9C).
FIGURE 9.
MjHeCL binds to hemocytes to modulate AMP expression. A, recombinant MjHeCL alone stimulates AMPs expression in vitro. Hemocytes were cultured in 6-well plates, and recombinant MjHeCL or BSA (3 μg) was applied to the wells. Expression of AMPs was detected 6 h later. Data represents mean ± SD from three independent repeats with error bars representing S.D. Asterisks represent significant differences (calculated by the t test for paired samples from three repeats: *, p < 0.05; **, p < 0.01; **, p < 0.001). B, recombinant MjHeCL binds to hemocytes. Recombinant MjHeCL or control tag (3 μg) was added to the wells to incubate with the hemocytes for 3 h. Medium was removed, and cells were washed by PBS five times, lysed, and subjected to SDS-PAGE. Bound exogenous proteins were detected with the anti-His tag antibody. C, recombinant MjHeCL alone (in the absence of bacteria) stimulates AMP expression in vivo. Shrimp were pretreated with antibiotics to remove the hemolymph microbiota. Recombinant MjHeCL or BSA (5 μg) was injected into shrimp, and AMP expression was checked 24 h later. Data represent mean ± S.D. from three independent repeats with error bars representing S.D., and the asterisk represents a significant difference calculated by the t test for paired samples.
DISCUSSION
It is currently well established through our study and those of others that the hemolymph of some aquatic invertebrates is not sterile but populated by relatively low, fairly stable numbers of multiple bacterial species and strains (4). Until now, however, the mechanistic basis of this homeostatic balance between host and the hemolymph microbiota had remained elusive. As all bacterial and eukaryotic cells possess a rich glycocalyx that decorates the surface and encodes complex information, carbohydrate-binding proteins such as lectins play critical roles in the establishment and homeostatic balance of the microbiome-host consortium (30). In this study, we identified and functionally characterized in shrimp a C-type lectin, MjHeCL, as playing a central role in the homeostatic regulation of the hemolymph microbiota.
Most of the MjHeCL transcripts were localized to shrimp hemocytes. The relatively weaker signals detected in heart, gills, stomach, and intestine probably originate in hemocytes infiltrating those tissues. In most invertebrates such as shrimp, hemocytes are key participants in both the cellular and humoral immune responses against infectious agents (31). Although hemocyte subsets are responsible for phagocytosis, encapsulation, and nodulation responses (32), they can also function as the source of soluble recognition and effector factors such as lectins and antimicrobial peptides. In any given invertebrate species, both lectin and antimicrobial peptide repertoires are both structurally and functionally diverse. The immune functions of lectins have been mostly characterized by their functions in agglutination, immobilization, opsonization, and activation of enzymatic pathways leading to melanization, whereas the AMPs carry out effector functions by directly binding to and killing the target pathogens. The unique site of MjHeCL expression in the hemocytes, the cells that must rapidly respond to humoral infectious agents, and its secretion into plasma are suggestive of a key role for MjHeCL in immune defense. However, in contrast with lectins such as C-reactive protein and members of the CTL family such as the mannose-binding lectin that can function as acute phase reactants by increasing their plasma levels in minutes or hours as a response to infectious agents (33, 34), expression of MjHeCL was not affected by injection of bacteria. The constitutive expression of MjHeCL in hemocytes and its relatively high and constant levels in plasma even in the presence of infectious agents, however, supports an important and unique regulatory role specifically directed to maintaining homeostasis of the host endogenous microbiota.
As the shrimp soluble CTL repertoire is highly diverse, we examined the recognition spectrum of MjHeCL and the other nine CTLs (MjCL1–9) for the isolated components of the hemolymph microbiota. Surprisingly and in contrast will all other shrimp CTLs, MjHeCL recognized all bacterial species and strains tested. Binding of MjHeCL to bacteria appears to be only partially dependent on the presence of soluble calcium. The broad recognition activity of MjHeCL for hemolymph bacteria supports a central role, direct or indirect, in the inhibition of proliferation of the hemolymph microbiota. Because some lectins can kill microbial pathogens directly (35) or indirectly (36), we tested the potential bacteriocidal activity of the pure recombinant MjHeCL. MjHeCL did not show any antimicrobial activity against any of the Gram-negative or the Gram-positive bacterial species tested, clearly revealing that the regulatory mechanism of MjHeCL over the hemolymph microbiota was most likely indirect.
The lack of direct antimicrobial activity of MjHeCL prompted us to consider the possibility that this effect could be exerted by other hemolymph factors, and our results revealed a significant decrease in the antibacterial activity in the plasma of the MjHeCL RNAi animals as compared with the controls. Among the potential candidates, the decreased expression of those AMPs that were expressed in the hemocytes, namely Alf4, Alf5, Alf6, and Pen2, was found to be related with the effects observed. A gene silencing study on the four AMPs indicated that those mostly responsible for the bacterial inhibition were Alf4 and Alf6, whereas Alf5 and Pen2 had a more moderate inhibitory effect on the bacteria tested. ALFs are cationic peptides widely distributed in crustacean species such as shrimps, crabs, and crayfish (37–39). This family is characterized by an LPS-binding region with a cluster of positive charges lying within the two conserved cysteine residues. The ability of ALFs to neutralize LPS confers them broad antimicrobial function to inhibit bacteria (mainly Gram-negative bacteria) in vivo and in vitro (40). The ALF best characterized to date, ALFPm3 from the black tiger shrimp Penaeus monodon, actively inhibits bacterial infection. Gene silencing of ALFPm3 increased bacterial counts in the hepatopancreas and hemolymph (10), suggesting that it plays a role similar to that of the three ALFs found in this study. Our results showed that Alf4 and Alf6 have a more important role in protection than Alf5. Alignments of the sequences of these three ALFs showed that Alf4 and Alf6 shared a higher identity (78%) than either did with Alf5 (Alf4:Alf5, 43%; Alf6:Alf5, 47%), suggesting a potential relationship between the primary structures of ALFs and their protective roles. Conversely, penaeidins are AMPs unique to shrimp and are characterized by proline-rich and cysteine-rich N- and C-terminal regions, respectively (41, 42). Based on their sequence diversity, penaeidins are classified into four subgroups and preferentially display antimicrobial activities against Gram-positive bacteria and fungus as compared with Gram-negative bacteria (43). The moderate protective activity of Pen2 as compared with Alf4 and Alf6 may be related to the relative proportions and pathogenicity of the Gram-positive and Gram-negative bacteria present in the shrimp hemolymph microbiota.
Previous studies have shown that the soluble CTLs participate in antibacterial responses in several ways. First, most multivalent members of this family have the capacity to cross-link ligands on the microbial surface and thus agglutinate and immobilize potential pathogens (16). Second, soluble CTLs such as the mannose-binding lectin, shrimp FcLec4, and insect immulectin-2 can also function as opsonins by promoting the phagocytosis and removal of the invading bacteria (19, 21, 23, 36). Third, some C-type lectins are endowed with microbicidal activity. For example, the mouse CTL RegIIIγ expressed in the gut epithelium binds to and displays bacteriocidal activity for intestinal Gram-positive bacteria by interacting with peptidoglycans (44), another RegIII family member exerts its bactericidal activity through binding to lipid A (45), and similar activity against Staphylococcus aureus was revealed for a CTL from amphioxus (46). Finally, some CTLs can activate enzymatic pathways leading to complement activation in vertebrates such as mannose-binding lectin and ficolins (36) or melanization by activation of the prophenoloxidase pathway in invertebrates by the insect immulectin-1 and the crayfish Pacifastacus leniusculus mannose-binding lectin (47, 48). Thus, our finding that MjHeCL expression directly regulates the AMP plasma levels, which in turn maintain the homeostasis of the hemolymph microbiota, constitutes a yet undescribed role for any CTL characterized to date.
Although the mechanistic aspects of regulation of AMP expression in shrimp are still unclear, it is widely accepted that recognition of nonself molecular structures by pattern recognition receptors such as peptidoglycan recognition proteins and Gram-negative binding proteins in Drosophila constitutes the first step in the activation of Toll and immune deficiency pathways (49). Thus, it is tempting to speculate that MjHeCL could function as a pattern recognition receptor that recognizes the hemolymph microbiota components and activates the AMP expression pathways by targeting hemocyte surface receptors. This would either promote phagocytosis in a typical opsonic effect or facilitate recognition of the bacterial products by the hemocyte surface Toll-like receptors. The interaction of the lectin-bacterium complex by these mechanisms would maintain a constitutive pathway activation and continued expression of AMPs, mainly Alf4 and Alf6, to maintain the homeostatic balance of the hemolymph microbiota. The constitutive expression of a limited subset of key AMPs would represent an effective and energy-economic strategy for healthy shrimp to thrive in a suitable environment, although the animals are clearly endowed with highly diversified lectin and AMP repertoires that would allow them to rapidly respond to very diverse and potentially stressful conditions.
This work was supported by the National Natural Science Foundation of China (Grants 31130056 and 31302217), the Provincial Natural Science Foundation of Shandong, China (Grant ZR2011CM014), the Ph.D. Program Foundation of the Ministry of Education of China (Grant 20110131130003), and the China Postdoctoral Science Foundation (Grant 2013M540553) (to J.-X. W. and X.-W. W.) and by National Science Foundation Grants IOS-1050518, IOB-0618409, and IOS-0822257 (to G. R. V.). This work was supported, in whole or in part, by National Institutes of Health Grant 5R01GM070589-06 (to G. R. V.).
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) KJ175168.
X.-W. Wang, J.-D. Xu, X.-F. Zhao, G. R. Vasta, and J.-X. Wang, unpublished data.
- AMP
- antimicrobial peptide
- ALF
- antipolysaccharide factor
- CTL
- C-type lectin
- Lys
- lysozyme
- Cru
- crustin
- MjCL
- M. japonicus C-type lectin
- MjHeCL
- M. japonicus hemocyte C-type lectin
- Pen
- penaeidin
- qRT-PCR
- quantitative RT-PCR
- dsRNA
- double-stranded RNA
- rMjHeCL
- recombinant MjHeCL.
REFERENCES
- 1. Duron O., Hurst G. D. (2013) Arthropods and inherited bacteria: from counting the symbionts to understanding how symbionts count. BMC Biol. 11, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kamada N., Seo S. U., Chen G. Y., Núñez G. (2013) Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 13, 321–335 [DOI] [PubMed] [Google Scholar]
- 3. Maynard C. L., Elson C. O., Hatton R. D., Weaver C. T. (2012) Reciprocal interactions of the intestinal microbiota and immune system. Nature 489, 231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fagutao F. F., Koyama T., Kaizu A., Saito-Taki T., Kondo H., Aoki T., Hirono I. (2009) Increased bacterial load in shrimp hemolymph in the absence of prophenoloxidase. FEBS J. 276, 5298–5306 [DOI] [PubMed] [Google Scholar]
- 5. Olafsen J. A., Mikkelsen H. V., Giaever H. M., Høvik Hansen G. (1993) Indigenous bacteria in hemolymph and tissues of marine bivalves at low temperatures. Appl. Environ. Microbiol. 59, 1848–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sizemore R. K., Colwell R. R., Tubiash H. S., Lovelace T. E. (1975) Bacterial flora of the hemolymph of the blue crab, Callinectes sapidus: numerical taxonomy. Appl. Microbiol. 29, 393–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tubiash H. S., Sizemore R. K., Colwell R. R. (1975) Bacterial flora of the hemolymph of the blue crab, Callinectes sapidus: most probable numbers. Appl. Microbiol. 29, 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gomez-Gil B., Tron-Mayen L., Roque A., Turnbull J. F., Inglis V., Guerra-Flores A. L. (1998) Species of Vibrio isolated from hepatopancreas, haemolymph and digestive tract of a population of healthy juvenile Penaeus vannamei. Aquaculture 163, 1–9 [Google Scholar]
- 9. Kaizu A., Fagutao F. F., Kondo H., Aoki T., Hirono I. (2011) Functional analysis of C-type lysozyme in penaeid shrimp. J. Biol. Chem. 286, 44344–44349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ponprateep S., Tharntada S., Somboonwiwat K., Tassanakajon A. (2012) Gene silencing reveals a crucial role for anti-lipopolysaccharide factors from Penaeus monodon in the protection against microbial infections. Fish Shellfish Immunol. 32, 26–34 [DOI] [PubMed] [Google Scholar]
- 11. Hanington P. C., Forys M. A., Dragoo J. W., Zhang S. M., Adema C. M., Loker E. S. (2010) Role for a somatically diversified lectin in resistance of an invertebrate to parasite infection. Proc. Natl. Acad. Sci. U.S.A. 107, 21087–21092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Loker E. S., Adema C. M., Zhang S. M., Kepler T. B. (2004) Invertebrate immune systems—not homogeneous, not simple, not well understood. Immunol. Rev. 198, 10–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vasta G. R., Ahmed H., Odom E. W. (2004) Structural and functional diversity of lectin repertoires in invertebrates, protochordates and ectothermic vertebrates. Curr. Opin. Struct. Biol. 14, 617–630 [DOI] [PubMed] [Google Scholar]
- 14. Amparyup P., Sutthangkul J., Charoensapsri W., Tassanakajon A. (2012) Pattern recognition protein binds to lipopolysaccharide and β-1,3-glucan and activates shrimp prophenoloxidase system. J. Biol. Chem. 287, 10060–10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Christophides G. K., Vlachou D., Kafatos F. C. (2004) Comparative and functional genomics of the innate immune system in the malaria vector Anopheles gambiae. Immunol. Rev. 198, 127–148 [DOI] [PubMed] [Google Scholar]
- 16. Zelensky A. N., Gready J. E. (2005) The C-type lectin-like domain superfamily. FEBS J. 272, 6179–6217 [DOI] [PubMed] [Google Scholar]
- 17. Rabinovich G. A., van Kooyk Y., Cobb B. A. (2012) Glycobiology of immune responses. Ann. N.Y. Acad. Sci. 1253, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun Y. D., Fu L. D., Jia Y. P., Du X. J., Wang Q., Wang Y. H., Zhao X. F., Yu X. Q., Wang J. X. (2008) A hepatopancreas-specific C-type lectin from the Chinese shrimp Fenneropenaeus chinensis exhibits antimicrobial activity. Mol. Immunol. 45, 348–361 [DOI] [PubMed] [Google Scholar]
- 19. Wang X. W., Zhang X. W., Xu W. T., Zhao X. F., Wang J. X. (2009) A novel C-type lectin (FcLec4) facilitates the clearance of Vibrio anguillarum in vivo in Chinese white shrimp. Dev. Comp. Immunol. 33, 1039–1047 [DOI] [PubMed] [Google Scholar]
- 20. Watanabe A., Miyazawa S., Kitami M., Tabunoki H., Ueda K., Sato R. (2006) Characterization of a novel C-type lectin, Bombyx mori multibinding protein, from the B. mori hemolymph: mechanism of wide-range microorganism recognition and role in immunity. J. Immunol. 177, 4594–4604 [DOI] [PubMed] [Google Scholar]
- 21. Yu X. Q., Kanost M. R. (2000) Immulectin-2, a lipopolysaccharide-specific lectin from an insect, Manduca sexta, is induced in response to gram-negative bacteria. J. Biol. Chem. 275, 37373–37381 [DOI] [PubMed] [Google Scholar]
- 22. Zhao Z. Y., Yin Z. X., Xu X. P., Weng S. P., Rao X. Y., Dai Z. X., Luo Y. W., Yang G., Li Z. S., Guan H. J., Li S. D., Chan S. M., Yu X. Q., He J. G. (2009) A novel C-type lectin from the shrimp Litopenaeus vannamei possesses anti-white spot syndrome virus activity. J. Virol. 83, 347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang X. W., Zhao X. F., Wang J. X. (2014) C-type lectin binds to β-integrin to promote hemocytic phagocytosis in an invertebrate. J. Biol. Chem. 289, 2405–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schnitger A. K., Yassine H., Kafatos F. C., Osta M. A. (2009) Two C-type lectins cooperate to defend Anopheles gambiae against Gram-negative bacteria. J. Biol. Chem. 284, 17616–17624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dodd R. B., Drickamer K. (2001) Lectin-like proteins in model organisms: implications for evolution of carbohydrate-binding activity. Glycobiology 11, 71R–79R [DOI] [PubMed] [Google Scholar]
- 26. Drickamer K., Dodd R. B. (1999) C-type lectin-like domains in Caenorhabditis elegans: predictions from the complete genome sequence. Glycobiology 9, 1357–1369 [DOI] [PubMed] [Google Scholar]
- 27. Christophides G. K., Zdobnov E., Barillas-Mury C., Birney E., Blandin S., Blass C., Brey P. T., Collins F. H., Danielli A., Dimopoulos G., Hetru C., Hoa N. T., Hoffmann J. A., Kanzok S. M., Letunic I., Levashina E. A., Loukeris T. G., Lycett G., Meister S., Michel K., Moita L. F., Müller H. M., Osta M. A., Paskewitz S. M., Reichhart J. M., Rzhetsky A., Troxler L., Vernick K. D., Vlachou D., Volz J., von Mering C., Xu J., Zheng L., Bork P., Kafatos F. C. (2002) Immunity-related genes and gene families in Anopheles gambiae. Science 298, 159–165 [DOI] [PubMed] [Google Scholar]
- 28. Wang X. W., Wang J. X. (2013) Diversity and multiple functions of lectins in shrimp immunity. Dev. Comp. Immunol. 39, 27–38 [DOI] [PubMed] [Google Scholar]
- 29. Reichelt P., Schwarz C., Donzeau M. (2006) Single step protocol to purify recombinant proteins with low endotoxin contents. Protein Expr. Purif. 46, 483–488 [DOI] [PubMed] [Google Scholar]
- 30. Ohtsubo K., Marth J. D. (2006) Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867 [DOI] [PubMed] [Google Scholar]
- 31. Gross P. S., Bartlett T. C., Browdy C. L., Chapman R. W., Warr G. W. (2001) Immune gene discovery by expressed sequence tag analysis of hemocytes and hepatopancreas in the Pacific white shrimp, Litopenaeus vannamei, and the Atlantic white shrimp, L. setiferus. Dev. Comp. Immunol. 25, 565–577 [DOI] [PubMed] [Google Scholar]
- 32. Lin X., Söderhäll I. (2011) Crustacean hematopoiesis and the astakine cytokines. Blood 117, 6417–6424 [DOI] [PubMed] [Google Scholar]
- 33. Dean M. M., Minchinton R. M., Heatley S., Eisen D. P. (2005) Mannose binding lectin acute phase activity in patients with severe infection. J. Clin. Immunol. 25, 346–352 [DOI] [PubMed] [Google Scholar]
- 34. Gewurz H., Mold C., Siegel J., Fiedel B. (1982) C-reactive protein and the acute phase response. Adv. Intern. Med. 27, 345–372 [PubMed] [Google Scholar]
- 35. Stowell S. R., Arthur C. M., Dias-Baruffi M., Rodrigues L. C., Gourdine J. P., Heimburg-Molinaro J., Ju T., Molinaro R. J., Rivera-Marrero C., Xia B., Smith D. F., Cummings R. D. (2010) Innate immune lectins kill bacteria expressing blood group antigen. Nat. Med. 16, 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Epstein J., Eichbaum Q., Sheriff S., Ezekowitz R. A. (1996) The collectins in innate immunity. Curr. Opin. Immunol. 8, 29–35 [DOI] [PubMed] [Google Scholar]
- 37. Imjongjirak C., Amparyup P., Tassanakajon A., Sittipraneed S. (2007) Antilipopolysaccharide factor (ALF) of mud crab Scylla paramamosain: molecular cloning, genomic organization and the antimicrobial activity of its synthetic LPS binding domain. Mol. Immunol. 44, 3195–3203 [DOI] [PubMed] [Google Scholar]
- 38. Liu H., Jiravanichpaisal P., Söderhäll I., Cerenius L., Söderhäll K. (2006) Antilipopolysaccharide factor interferes with white spot syndrome virus replication in vitro and in vivo in the crayfish Pacifastacus leniusculus. J. Virol. 80, 10365–10371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Somboonwiwat K., Marcos M., Tassanakajon A., Klinbunga S., Aumelas A., Romestand B., Gueguen Y., Boze H., Moulin G., Bachère E. (2005) Recombinant expression and anti-microbial activity of anti-lipopolysaccharide factor (ALF) from the black tiger shrimp Penaeus monodon. Dev. Comp. Immunol. 29, 841–851 [DOI] [PubMed] [Google Scholar]
- 40. Rosa R. D., Vergnes A., de Lorgeril J., Goncalves P., Perazzolo L. M., Sauné L., Romestand B., Fievet J., Gueguen Y., Bachère E., Destoumieux-Garzón D. (2013) Functional divergence in shrimp anti-lipopolysaccharide factors (ALFs): from recognition of cell wall components to antimicrobial activity. PLoS One 8, e67937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Destoumieux D., Munoz M., Bulet P., Bachère E. (2000) Penaeidins, a family of antimicrobial peptides from penaeid shrimp (Crustacea, Decapoda). Cell Mol. Life Sci. 57, 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cuthbertson B. J., Yang Y., Bachère E., Büllesbach E. E., Gross P. S., Aumelas A. (2005) Solution structure of synthetic penaeidin-4 with structural and functional comparisons with penaeidin-3. J. Biol. Chem. 280, 16009–16018 [DOI] [PubMed] [Google Scholar]
- 43. Cuthbertson B. J., Deterding L. J., Williams J. G., Tomer K. B., Etienne K., Blackshear P. J., Büllesbach E. E., Gross P. S. (2008) Diversity in penaeidin antimicrobial peptide form and function. Dev. Comp. Immunol. 32, 167–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cash H. L., Whitham C. V., Behrendt C. L., Hooper L. V. (2006) Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313, 1126–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miki T., Holst O., Hardt W. D. (2012) The bactericidal activity of the C-type lectin RegIIIβ against Gram-negative bacteria involves binding to lipid A. J. Biol. Chem. 287, 34844–34855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu Y., Yu Y., Huang H., Feng K., Pan M., Yuan S., Huang S., Wu T., Guo L., Dong M., Chen S., Xu A. (2007) A short-form C-type lectin from amphioxus acts as a direct microbial killing protein via interaction with peptidoglycan and glucan. J. Immunol. 179, 8425–8434 [DOI] [PubMed] [Google Scholar]
- 47. Wu C., Charoensapsri W., Nakamura S., Tassanakajon A., Söderhäll I., Söderhäll K. (2013) An MBL-like protein may interfere with the activation of the proPO-system, an important innate immune reaction in invertebrates. Immunobiology 218, 159–168 [DOI] [PubMed] [Google Scholar]
- 48. Yu X. Q., Gan H., Kanost M. R. (1999) Immulectin, an inducible C-type lectin from an insect, Manduca sexta, stimulates activation of plasma prophenol oxidase. Insect Biochem. Mol. Biol. 29, 585–597 [DOI] [PubMed] [Google Scholar]
- 49. Lemaitre B., Hoffmann J. (2007) The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 [DOI] [PubMed] [Google Scholar]