Background: IFITM3 restricts the fusion of viruses within endolysosomes.
Results: Phosphorylation of IFITM3 on Tyr20 blocks IFITM3 endocytosis and ubiquitination.
Conclusion: Tyr20 of IFITM3 is part of a YXXΦ endocytosis signal and has a dual role in regulating IFITM3 ubiquitination.
Significance: Identifying mechanisms regulating IFITM3 trafficking and activity is crucial for understanding and manipulating IFITM3 biology for combating virus infections.
Keywords: Endocytosis, Influenza Virus, Innate Immunity, Interferon, Phosphorylation, Phosphotyrosine, Ubiquitination, IFITM, IFITM3, Posttranslational Modification
Abstract
Interferon-inducible transmembrane protein 3 (IFITM3) is essential for innate defense against influenza virus in mice and humans. IFITM3 localizes to endolysosomes where it prevents virus fusion, although mechanisms controlling its trafficking to this cellular compartment are not fully understood. We determined that both mouse and human IFITM3 are phosphorylated by the protein-tyrosine kinase FYN on tyrosine 20 (Tyr20) and that mouse IFITM3 is also phosphorylated on the non-conserved Tyr27. Phosphorylation led to a cellular redistribution of IFITM3, including plasma membrane accumulation. Mutation of Tyr20 caused a similar redistribution of IFITM3 and resulted in decreased antiviral activity against influenza virus, whereas Tyr27 mutation of mouse IFITM3 showed minimal effects on localization or activity. Using FYN knockout cells, we also found that IFITM3 phosphorylation is not a requirement for its antiviral activity. Together, these results indicate that Tyr20 is part of an endocytosis signal that can be blocked by phosphorylation or by mutation of this residue. Further mutagenesis narrowed this endocytosis-controlling region to four residues conforming to a YXXΦ (where X is any amino acid and Φ is Val, Leu, or Ile) endocytic motif that, when transferred to CD4, resulted in its internalization from the cell surface. Additionally, we found that phosphorylation of IFITM3 by FYN and mutagenesis of Tyr20 both resulted in decreased IFITM3 ubiquitination. Overall, these results suggest that modification of Tyr20 may serve in a cellular checkpoint controlling IFITM3 trafficking and degradation and demonstrate the complexity of posttranslational regulation of IFITM3.
Introduction
The interferon-inducible transmembrane proteins (IFITMs)3 inhibit cellular infection by a wide range of significant viral pathogens (1–9). IFITM3 is particularly important for the restriction of influenza virus. IFITM3 knockout mice succumb to sublethal doses of virus (10, 11), and a deleterious polymorphism in the human IFITM3 gene has been associated with increased severity of infection in at least three independent studies (10, 12, 13). Despite the clear importance of IFITM3 in restricting influenza virus, many questions remain regarding its mechanism of action, cellular trafficking patterns, and regulation by cellular enzymes.
IFITM3 localizes to acidic compartments, staining positively for endosomal and lysosomal markers (2, 4, 14), where it prevents viral fusion through an unknown mechanism (3, 14, 15). Experiments with pseudotyped viruses demonstrated that inhibition of viruses by IFITM3 is dependent upon the viral fusion glycoprotein used for cellular entry (1). Likewise, nearly all of the viruses shown to be inhibited by IFITM3 enter cells via endocytosis (16, 17). Conversely, Sendai virus, which fuses at the cell surface, is largely unaffected by IFITM3 (18). Similarly, exogenous protease treatment of severe acute respiratory syndrome-associated coronavirus allows it to fuse at the cell surface, thereby bypassing its pH-dependent activation in lysosomes and restriction by IFITM3 (2). Thus, endolysosomes appear to be the site of antiviral action by IFITM3, and the targeting signals that control IFITM3 trafficking to and from this compartment warrant further study.
IFITM3 is a highly regulated protein with at least four posttranslational modifications occurring on multiple residues reported to date. We first reported palmitoylation of IFITM3 occurring on three cysteines that are essential for proper membrane anchoring and antiviral activity (4, 19–21). We also reported ubiquitination of IFITM3 occurring on four lysines (4). This modification negatively regulates IFITM3 by targeting the protein away from endolysosomes for degradation (4). Set7-dependent methylation on a single lysine has also been described to negatively regulate IFITM3 antiviral activity through an unknown mechanism (22). Finally, IFITM3 phosphorylation by the protein-tyrosine kinase FYN on tyrosine 20 (Tyr20) has been reported (23). Mutation of Tyr20 resulted in decreased antiviral activity against vesicular stomatitis virus and an accumulation of IFITM3 at the plasma membrane (23). These intriguing findings prompted us to further investigate the role of phosphorylation in controlling IFITM3 trafficking and in restricting influenza virus.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfections, and Western Blotting
Mouse embryonic fibroblasts deficient in SRC, YES, and FYN kinases (SYF cells, ATCC), WT mouse embryonic fibroblasts, and HEK293T cells were cultured in DMEM supplemented with 4.5 g/liter d-glucose, l-glutamine, 110 mg/liter sodium pyruvate, and 10% fetal bovine serum (Thermo Scientific) at 37 °C and 5% CO2 in a humidified incubator. For Western blotting, cells were plated for 90% confluency in 6-well plates 24 h prior to transfection with 2 μg/well of the indicated plasmids using Lipofectamine 2000 (Invitrogen). Cells for microscopy were plated for 50% confluency on glass coverslips in 12-well plates and transfected with 1 μg/well of the indicated plasmids. IFITM3 constructs were expressed from the pCMV-HA or pCMV-myc vectors as described previously (19). Mutants were made using the QuikChange Multi site-directed mutagenesis kit (Stratagene). The plasmid expressing human FYN was provided by Dr. Marilyn Resh (Memorial Sloan Kettering Cancer Center) (24), and the plasmid expressing FLAG-ubiquitin was provided by Drs. Rebecca Delker and F. Nina Papavasiliou (Rockefeller University). Control siRNA or IFITM3 siRNA (Ambion, catalog no. 4390816) was transfected into SYF cells using Lipofectamine RNAiMax transfection reagent (Invitrogen).
For Western blotting, cells were lysed with 1% Brij buffer (0.1 mm triethanolamine, 150 mm NaCl, 1% BrijO10 (pH 7.4)) containing EDTA-free protease inhibitor mixture. For phospho-tyrosine Western blot analyses, cells were treated with sodium orthovanadate (Sigma) for 1 h prior to harvesting, and PhosSTOP phosphatase inhibitor mixture was added to the lysis buffer (Roche). Immunoprecipitations were performed using EZview Red anti-c-myc or anti-HA affinity gel (Sigma) as indicated. Western blotting was performed with anti-phospho-Tyr-100 (Cell Signaling Technology, catalog no. 9411), anti-myc (Clontech, catalog no. 51826), anti-FYN (Cell Signaling Technology, catalog no. 4023), anti-actin (Abcam, catalog no. ab3280), anti-FLAG (Sigma, catalog no. F7425), anti-HA (Clontech, catalog no. 631207), anti-IFITM3 (Abcam, catalog no. ab65183), and anti-GAPDH (Invitrogen, catalog no. 398600). All primary antibodies were used at a 1:1000 dilution.
Fluorescence Microscopy
Cells were fixed for 10 min with 3.7% paraformaldehyde, permeabilized with 0.1% Triton X-100 in PBS for 10 min, and blocked for 10 min with 2% FBS in PBS. Primary antibodies, anti-myc (1:500), anti-FYN (1:500), and anti-CD4 (directly conjugated to Alexa Fluor 488, 1:100, BD Biosciences, catalog no. 557695), and Alexa Fluor-labeled secondary antibodies (Invitrogen, 1:1000) were diluted in 0.1% Triton X-100 in PBS for staining of permeabilized cells. Cells were treated with antibodies sequentially for 20 min at room temperature and washed five times with 0.1% Triton X-100 in PBS after each antibody treatment. For non-permeabilized cells, anti-CD4 was added to live cells in ice-cold PBS for 15 min, followed by five washes with ice-cold PBS and paraformaldehyde fixation. Glass slides were mounted in ProLong Gold antifade reagent containing DAPI (Invitrogen). Images were captured using a Fluoview FV10i confocal microscope (Olympus).
Infections and Flow Cytometry
Influenza virus A/PR/8/34 (H1N1) was propagated in 10-day embryonated chicken eggs for 48 h at 37 °C and titrated using Madin-Darby canine kidney cells as described previously (25). HEK293T were infected at a multiplicity of infection of 2.5 for 6 h. SYF cells were infected at a multiplicity of infection of 5 for 24 h. Infected cells were washed with PBS and harvested in 0.25% trypsin EDTA. Cells were fixed in 3.7% paraformaldehyde for 10 min and permeabilized with 0.1% Triton X-100 for 10 min. Cells were stained with anti-myc and anti-influenza nucleoprotein (Abcam, catalog no. ab20343, 1:333) directly conjugated to Alexa Fluor 488 and Alexa Fluor 647, respectively, using a 100-μg antibody labeling kit (Invitrogen). All antibodies were diluted in 0.1% Triton X-100 in PBS, and cells were stained for 20 min. Cells were washed twice with 0.1% Triton X-100 in PBS after each antibody treatment. PBS was used for final resuspension of cells for cytometric analysis using a FACSCanto II flow cytometer (BD Biosciences).
RESULTS
IFITM3 Phosphorylation by FYN Is Not Required for Anti-influenza Virus Activity
The previous finding that mutagenesis of the phosphorylated residue Tyr20 results in decreased antiviral activity of IFITM3 might suggest that phosphorylation is necessary for the function of IFITM3 (23). In addressing this hypothesis, we first sought to confirm that human (h) IFITM3 is phosphorylated and also examined whether this modification is conserved on murine (m) IFITM3, which possesses Tyr20 and also a non-conserved tyrosine nearby at amino acid position 27 (Fig. 1A). For these experiments, we utilized a murine fibroblast cell line deficient in SRC, YES, and FYN kinases (SYF cells) in which we observed nearly undetectable tyrosine phosphorylation of myc-tagged IFITM3 (Fig. 1, B and C). Upon FYN coexpression, tyrosine phosphorylation of both mIFITM3 and hIFITM3 could be detected, in agreement with the conclusion published previously showing that FYN can indeed phosphorylate hIFITM3 (23) (Fig. 1, B and C). Of note, sodium orthovanadate pretreatment of cells to inhibit phosphatases was required, as reported previously (23), to observe IFITM3 phosphorylation, indicating that IFITM3 phosphorylation is a dynamic and reversible process. Tyrosine mutagenesis indicated that hIFITM3 is phosphorylated solely on Tyr20 (Fig. 1B), whereas mIFITM3 phosphorylation can occur on both Tyr20 and Tyr27 (Fig. 1C). In these experiments, in which we utilized broad-spectrum phosphatase inhibitors in our lysis buffer, we reproducibly observed an upper band in anti-myc blots that likely represents phosphorylated IFITM3 (Fig. 1, B and C). However, this band does not correlate directly with anti-phospho-tyrosine blots, suggesting that additional phosphorylation sites on serine or threonine residues may exist on IFITM3. This should be a fruitful area for future investigation. Nonetheless, tyrosine-specific phosphorylation clearly occurs primarily on Tyr20/27.
FIGURE 1.
IFITM3 phosphorylation by FYN. A, alignment of hIFITM3 and mIFITM3 amino acids 15–30. Tyrosines are highlighted with gray shading. B and C, SYF cells were cotransfected overnight with the indicated myc-tagged hIFITM3 (B) or mIFITM3 (C) constructs and either a vector control or a plasmid expressing FYN. Cells were then treated for 1 h with sodium orthovanadate. Cell lysates were subjected to anti-FYN and anti-actin Western blotting or anti-myc immunoprecipitation (IP) followed by blotting for anti-phospho-tyrosine (p-Tyr) and anti-myc. Blots are representative of at least three experiments.
Having confirmed tyrosine phosphorylation on both mIFITM3 and hIFITM3 by FYN, we sought to test whether IFITM3 is active in SYF cells, i.e. in cells in which IFITM3 phosphorylation is lacking. We and others (1, 4) have shown that murine fibroblast lines express a basal amount of IFITM3 that limits infection of these cells. Thus, we performed siRNA knockdown of IFITM3 in SYF cells (Fig. 2A) and, subsequently, examined infection by influenza virus. The percentage of infected SYF cells increased significantly upon IFITM3 knockdown, indicating that endogenous IFITM3 is active in these cells and that IFITM3 tyrosine phosphorylation by FYN is not a requirement for its antiviral function (Fig. 2B).
FIGURE 2.
IFITM3 is active in the absence of FYN. A and B, SYF cells were transfected for 18 h with control siRNA (siCont) or siRNA targeting IFITM3 (siIFITM3). A, cells were collected just prior to infection for confirmation of IFITM3 knockdown by anti-IFITM3 Western blotting. Anti-GAPDH blotting served as a loading control. B, following siRNA treatment, cells were infected with influenza virus strain PR8 at a multiplicity of infection of 5 for 24 h. Cells were then fixed and stained with anti-influenza nucleoprotein to measure the percentage of cells that were infected using flow cytometry. The results shown are an average of six samples from two independent experiments. The average percentage of infection of siCont cells was set to 1 for the calculation of relative fold infection. Error bars represent mean ± S.D. Student's t test was used to calculate the indicated p value.
IFITM3 Tyr20 Is Required for Complete Antiviral Activity
We next examined several of our mIFITM3 and hIFITM3 tyrosine mutants for the ability to inhibit influenza virus compared with WT IFITM3. HEK293T cells are a commonly used cell line for the analysis of IFITM3 mutants because they express no detectable amount of endogenous IFITM3, are readily transfectable, and retain the ability to be highly infected with influenza virus even after transfection, allowing a large dynamic range for observations of differences in antiviral protective effects between various IFITM3 mutants (3, 4, 6, 15, 18, 19, 26). Tyr20 mutants of both mIFITM3 and hIFITM3 are expressed at levels similar to their respective WT proteins (Fig. 3A) but lose anti-influenza virus activity, as determined by an established flow cytometry assay that measures the percentage of cells infected in each condition (Fig. 3B) (4, 18, 19). Interestingly, mutation of Tyr27, although phosphorylatable (Fig. 1C), has no apparent effect on antiviral activity of mIFITM3, and Tyr27 does not significantly compensate for mutation of Tyr20 (Fig. 3). These results demonstrate that Tyr20 is a critical amino acid for the anti-influenza virus activity of both mIFITM3 and hIFITM3.
FIGURE 3.
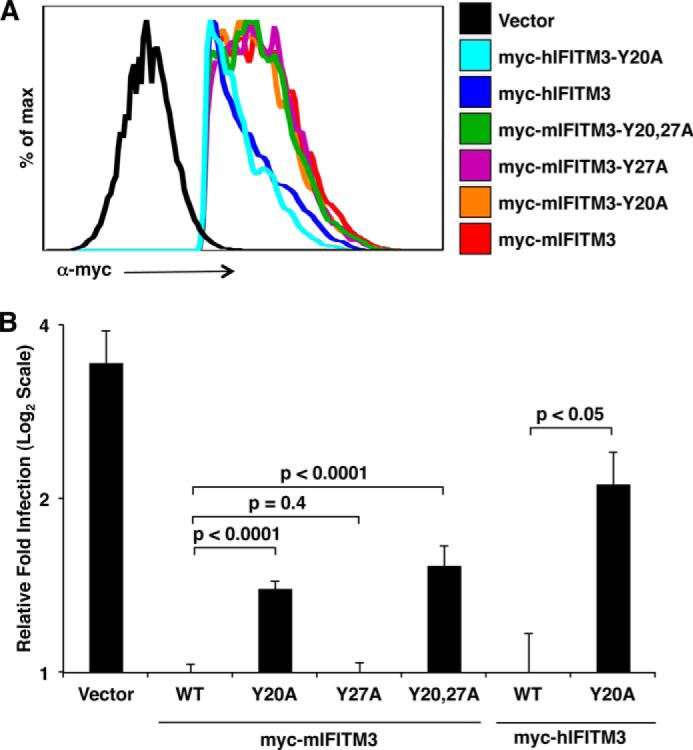
Tyr-20 of IFITM3 is necessary for complete antiviral activity against influenza virus. A and B, HEK293T cells were transfected overnight with the indicated myc-tagged IFITM3 constructs or a vector control infected for 6 h with influenza virus strain PR8 at a multiplicity of infection of 2.5. Cells were then fixed and stained with anti-myc and anti-influenza nucleoprotein to confirm the expression of IFITM3 and to measure the percentage of cells that were infected, respectively, using flow cytometry. A, myc-positive cells were gated on the basis of lack of staining in the vector control and analyzed in histogram form to confirm the comparable expression of IFITM3 and tyrosine mutants. B, myc-positive cells were gated as in A and analyzed for percentage of infection on the basis of anti-influenza nucleoprotein staining using non-infected samples as a baseline for gating. The results shown are an average of at least nine samples from a minimum of three independent experiments. The average percentage of infection of cells expressing WT mIFITM3 was set to 1 for the calculation of relative fold infection. Error bars represent mean ± S.D. Results are presented on a log2 scale for ease of visualizing differences between WT IFITM3 and tyrosine mutants. Student's t test was used to calculate the indicated p values.
Phosphorylation of IFITM3 on Tyr20 Leads to Plasma Membrane Accumulation
Previous imaging of hIFITM3 Tyr20 mutants demonstrated that mutation of this residue results in retention of IFITM3 at the plasma membrane (5, 23). However, these experiments did not address whether phosphorylation of Tyr20 or the Tyr20 residue itself is necessary for IFITM3 internalization. We reasoned that imaging IFITM3 in SYF cells lacking FYN or coexpressing FYN would allow effects of phosphorylation on IFITM3 localization to be observed. Although IFITM3 in the absence of FYN was localized in punctate clusters intracellularly, as is normally observed for IFITM3 staining in most cell types (1, 4, 5, 14), IFITM3 in the presence of overexpressed FYN was redistributed, including accumulation at the plasma membrane (Fig. 4, A and B). This effect was seen for both mIFITM3 and hIFITM3 and mimics the effect we also observed for Tyr20 mutants (Fig. 4, A and B). Interestingly, Tyr27 mutation of mIFITM3 had no apparent effect on its cellular distribution, agreeing with the lack of effect observed on antiviral activity for this mutant compared with WT mIFITM3 (Figs. 3 and 4B). Retention of IFITM3 at the plasma membrane through a direct interaction with FYN is a possible interpretation of these results. However, it has been observed previously that the interaction between FYN and IFITM3 depends on the presence of Tyr20 (23), and Tyr20 mutants also show plasma membrane accumulation, arguing against a direct role for interaction with FYN in sequestering IFITM3 at the plasma membrane. Thus, we conclude that both Tyr20 phosphorylation and Tyr20 mutation result in a similar change in the cellular distribution of IFITM3, including plasma membrane accumulation.
FIGURE 4.
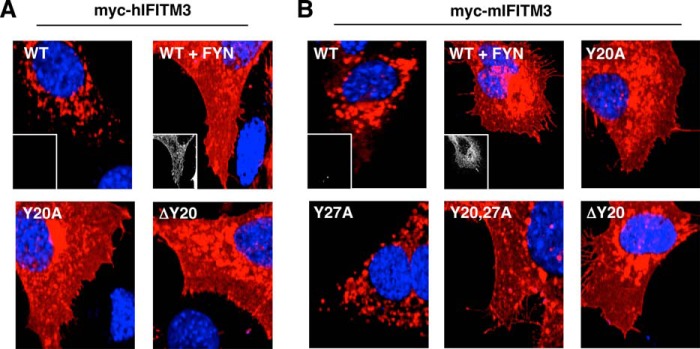
IFITM3 Tyr-20 phosphorylation or mutation affects IFITM3 localization, including plasma membrane accumulation. A and B, SYF cells were cotransfected with the indicated myc-tagged hIFITM3 (A) or mIFITM3 (B) constructs and either a vector control or a plasmid expressing FYN. Representative merged fluorescent confocal microscopy images for nuclear staining (DAPI, blue) and anti-myc staining (red) are shown. Insets depicting anti-FYN staining (grayscale) are shown for some conditions to confirm FYN expression or the lack thereof.
Identification of a Putative IFITM3 Endocytosis Motif
Our imaging results suggest that Tyr20 may be part of an endocytosis signal that can be blocked by phosphorylation or perturbed by mutating Tyr20 (Fig. 4). Previous IFITM3 mutagenesis has shown that the 17PPN19 residues immediately prior to Tyr20 are dispensable for antiviral activity (Fig. 5A and Ref. 23). Likewise, we observed that mutation of Lys24 does not affect IFITM3 localization or activity (Fig. 5A and Ref. 4). This narrows the hIFITM3 endocytic motif to the region 20YEML23 (Fig. 5A). Interestingly, this sequence conforms to the pattern of a YXXΦ motif (where X is any amino acid and Φ is Val, Leu, or Ile) that is involved in the adaptor protein complex-mediated endocytosis of a multitude of membrane proteins (27). Our experiments with mIFITM3 also support that this motif is an endocytosis signal because the 20YERI23 sequence of mIFITM3 also conforms to this pattern, whereas the 27YEVA30 motif involving Tyr27 does not (Fig. 5A), agreeing with our data that Tyr27, although phosphorylatable, does not play a major role in IFITM3 cellular distribution or antiviral activity (Figs. 3 and 4B). To test the hypothesis that the 20YEML23 sequence in IFITM3 is an endocytosis signal, we made a conservative mutation of the Φ residue in hIFITM3 to valine (L23V) and a non-conservative mutation to the polar residue glutamine (L23Q). Imaging of these mutants revealed a similar localization of the L23V mutant, which preserves the canonical YXXΦ pattern, whereas the L23Q mutant was redistributed, including visible plasma membrane localization similar to what we saw previously with Tyr20 mutants (Figs. 4, A and B, and 5B).
FIGURE 5.
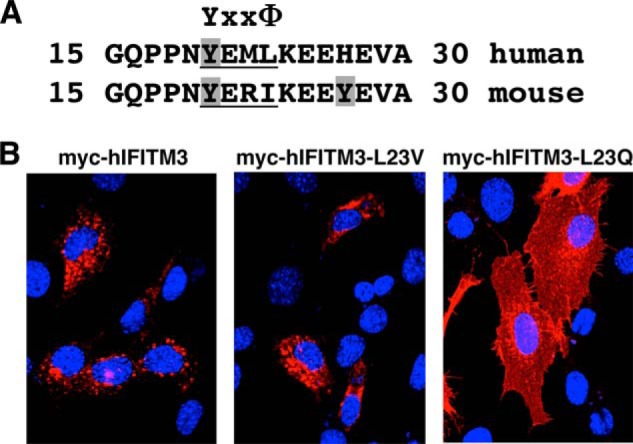
Identification of a YXXΦ endocytosis motif in IFITM3. A, alignment of amino acids 15–30 of hIFITM3 and mIFITM3. Tyrosines are highlighted with gray shading, and the conserved YXXΦ motif is labeled and underlined. B, SYF cells were transfected overnight with the indicated myc-tagged hIFITM3 constructs. Representative merged fluorescent confocal microscopy images for nuclear staining (DAPI, blue) and anti-myc staining (red) are shown.
To further confirm that the YEML sequence of hIFITM3 functions as an endocytosis signal, we transferred this tetrapeptide to the cytoplasmic C-terminal region of CD4, which normally localizes in part to the plasma membrane. Anti-CD4 staining of the cell surface of non-permeabilized cells showed outlining of cells transfected with a plasmid encoding WT myc-tagged CD4 (Fig. 6A). Under the same conditions, minimal staining of a CD4 construct containing the YEML peptide was observed (Fig. 6A), although this construct was expressed strongly and localized in internal compartments, as indicated by staining of permeabilized cells with anti-CD4 (Fig. 6A) or anti-myc (Fig. 6B). Overall, these results indicate that the YEML motif from hIFITM3 causes a robust internalization of CD4 (Fig. 6). Taken together, our results provide evidence that the hIFITM3 YEML motif functions as an endocytosis signal that can be regulated by FYN-mediated phosphorylation.
FIGURE 6.

The YEML motif from hIFITM3 causes internalization of CD4. A and B, mouse embryonic fibroblasts were transfected overnight with myc-tagged CD4 or CD4-YEML constructs. A, non-permeabilized or permeabilized cells were stained with anti-CD4 antibody for fluorescent confocal microscopy. Representative merged images for nuclear staining (DAPI, blue) and anti-CD4 staining (green) are shown. B, permeabilized cells were stained with anti-myc antibody for confocal fluorescent microscopy. Representative merged images for nuclear staining (DAPI, blue) and anti-myc staining (red) are shown.
Tyrosine Phosphorylation Regulates IFITM3 Ubiquitination
Having previously discovered IFITM3 ubiquitination (4) and noting the often-observed cross-talk between phosphorylation and ubiquitination (28), we sought to investigate a potential link between these two modifications on IFITM3. In our previous studies of IFITM3, we found that, upon overexposure of IFITM3 Western blot analyses, bands above the expected IFITM3 molecular weight could be observed. We identified this banding pattern as ubiquitination through the use of mass spectrometry, anti-ubiquitin blotting, and lysine mutagenesis (4). Here we employed a straightforward assay whereby we transfected mIFITM3 and hIFITM3 into 293T cells with or without overexpression of FYN and examined the banding pattern of IFITM3 by Western blotting. Interestingly, bands that we identified previously as mono- and diubiquitinated IFITM3 (4) were diminished upon FYN overexpression for both mIFITM3 and hIFITM3, whereas the bands at the expected molecular weight were largely unaffected by FYN (Fig. 7A). We then compared the banding patterns for WT IFITM3 and tyrosine mutants. Tyr20 mutants of both mIFITM3 and hIFITM3 showed a decreased intensity of ubiquitinated bands, whereas a Tyr27 mutant of mIFITM3 was ubiquitinated similarly to WT mIFITM3 (Fig. 7B). A ubiquitination-deficient lysine-less mutant of mIFITM3 (termed UbΔ, note that the myc tag contains one lysine) we generated previously (4) was included as a control in this experiment to confirm that the higher molecular weight bands we observed indeed represent ubiquitination (Fig. 7B).
FIGURE 7.
Unmodified Tyr-20 is necessary for proper IFITM3 ubiquitination. A, 293T cells were cotransfected overnight with the indicated myc-tagged IFITM3 constructs and a vector control or a plasmid expressing FYN. Anti-myc and anti-fyn blotting were performed. B, 293T cells were transfected overnight with the indicated myc-tagged IFITM3 constructs, and anti-myc blotting was performed. A and B, anti-myc blots were overexposed to allow visualization of ubiquitinated IFITM3. Single arrows and doubleheaded arrows indicate monoubiquitinated and diubiquitinated IFITM3, respectively. IFITM3 at the expected molecular mass of 15 kDa serves as a loading control. C, 293T cells were cotransfected overnight with the indicated HA-tagged IFITM3 constructs and a plasmid expressing FLAG-tagged ubiquitin (Ub). Anti-HA immunoprecipitation was performed, followed by anti-HA and anti-FLAG blotting. The results in A–C are representative of at least three experiments.
To further visualize IFITM3 ubiquitination, including polyubiquitinated IFITM3, and to be sure that the lysine residue present within the myc epitope tag was not altering our results, we cotransfected HA-tagged mIFITM3 and tyrosine mutants with FLAG-ubiquitin and performed anti-HA immunoprecipitation followed by blotting for HA and FLAG. mIFITM3 constructs were chosen for this experiment because of the availability of the mIFITM3-UbΔ construct, which served as a negative control, and because we also previously generated a palmitoylation-deficient mIFITM3 construct (PalmΔ) that does not have a defect in ubiquitination that served as an additional control (4). Ubiquitination patterns observed in this experiment agreed with our previous results in that Tyr20 mutants showed a partial defect in ubiquitination, whereas Tyr27 mutation had no effect compared with WT mIFITM3 (Fig. 7C). Overall, these data suggest that the phosphorylatable residue Tyr20 is involved in promoting IFITM3 ubiquitination in addition to its role in promoting endocytosis.
DISCUSSION
The posttranslational regulation of IFITM3 is rich with complexity. At least eight distinct amino acids within this small, 15-kDa protein are modified with at least four different posttranslational modifications, including phosphorylation, palmitoylation, ubiquitination, and methylation (4, 19, 22, 23). We demonstrate here, for the first time, that there is cross-regulation of posttranslational modifications on IFITM3, namely that phosphorylation of IFITM3 by FYN at Tyr20 decreases IFITM3 ubiquitination (Fig. 7). This is in contrast to the previously observed independent nature of IFITM3 ubiquitination with respect to its palmitoylation (4).
Cross-talk between phosphorylation and ubiquitination has been documented extensively for multiple other proteins (28). Particularly, phosphorylation often serves as a signal for modification by E3 ubiquitin ligases. For example, CBL family E3 ubiquitin ligases recognize phosphorylated tyrosines on their substrates (29), which, coincidentally, include FYN (30). Thus, it may have been expected that IFITM3 phosphorylation would promote ubiquitination. However, the opposite was observed in that IFITM3 phosphorylation by FYN led to a defect in ubiquitination, and mutating the Tyr20 phosphorylation site resulted in the same effect (Fig. 7).
The observed decrease in ubiquitination of IFITM3 upon phosphorylation may have at least two possible explanations. First, IFITM3 endocytosis may be required for ubiquitination to occur. We found that phosphorylation blocks an endocytosis motif that is necessary for proper localization and full antiviral activity of IFITM3 (Figs. 1–6). Our imaging data indicate that IFITM3 traffics to the plasma membrane and is either retained there upon phosphorylation or is internalized (Figs. 4 and 5). During the writing of this manuscript, a second group also reported identification of the 20YEML23 motif of hIFITM3 as a critical determinant for its endocytosis (31), thus validating and complementing our observations regarding phosphorylation of this motif. Thus, if ubiquitination takes place at the endolysosome or another cellular compartment, it may be inhibited by retention of IFITM3 at the plasma membrane upon Tyr20 mutation or phosphorylation. Second, Tyr20 may be part of an additional motif recognized by ubiquitin ligases, again explaining the similar results we obtained when either blocking Tyr20 by phosphorylation or by mutating Tyr20 (Fig. 7). The sequence 17PPNY20 in both mIFITM3 and hIFITM3 presents a potential E3 ligase interaction motif because it conforms to the PPXY pattern recognized by HECT E3 ubiquitin ligases (Fig. 5A) (32). Both of these aforementioned possibilities are currently under active investigation in our laboratory, and our findings should aid in the future identification of the IFITM3 ubiquitin ligase(s) responsible for modifying IFITM3.
It remains to be determined what effect relocalization of IFITM3 upon Tyr20 mutation or upon increasing FYN activity would have on its range of viral restrictions. For instance, IFITM1, which lacks a YXXΦ motif and localizes in part to the plasma membrane, has been described to have an overlapping but somewhat distinct specificity for different viruses compared with IFITM3 (2, 8, 17). Likewise, it will be interesting to examine what effect drugs that inhibit SRC family tyrosine kinases such as FYN would have on IFITM3 activity because phosphorylation of IFITM3 by FYN has both potentially negative and positive effects through modulating endocytosis and ubiquitination, respectively. Overall, the continued study of IFITM3 posttranslational modifications, their cross-talk, and their mechanisms of regulating antiviral activity will be important for understanding and controlling IFITM3 biology for combating influenza and other viruses.
Acknowledgments
We thank Dr. Marilyn Resh (Memorial Sloan Kettering Cancer Center) for FYN constructs and Drs. Rebecca Delker and F. Nina Papavasiliou (Rockefeller University) for FLAG-ubiquitin. We also thank Dr. Howard Hang (Rockefeller University) for helpful discussions.
This work was supported, in whole or in part, by NIAID, National Institutes of Health Grant R00AI095348 (to J. S. Y.). This work was also supported by the Ohio State University Public Health Preparedness for Infectious Disease program.
- IFITM
- interferon-inducible transmembrane protein
- h
- human
- m
- murine.
REFERENCES
- 1. Brass A. L., Huang I. C., Benita Y., John S. P., Krishnan M. N., Feeley E. M., Ryan B. J., Weyer J. L., van der Weyden L., Fikrig E., Adams D. J., Xavier R. J., Farzan M., Elledge S. J. (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang I. C., Bailey C. C., Weyer J. L., Radoshitzky S. R., Becker M. M., Chiang J. J., Brass A. L., Ahmed A. A., Chi X., Dong L., Longobardi L. E., Boltz D., Kuhn J. H., Elledge S. J., Bavari S., Denison M. R., Choe H., Farzan M. (2011) Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 7, e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lu J., Pan Q., Rong L., He W., Liu S. L., Liang C. (2011) The IFITM proteins inhibit HIV-1 infection. J. Virol. 85, 2126–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yount J. S., Karssemeijer R. A., Hang H. C. (2012) S-palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. J. Biol. Chem. 287, 19631–19641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. John S. P., Chin C. R., Perreira J. M., Feeley E. M., Aker A. M., Savidis G., Smith S. E., Elia A. E., Everitt A. R., Vora M., Pertel T., Elledge S. J., Kellam P., Brass A. L. (2013) The CD225 domain of IFITM3 is required for both IFITM protein association and inhibition of influenza A virus and dengue virus replication. J. Virol. 87, 7837–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weidner J. M., Jiang D., Pan X. B., Chang J., Block T. M., Guo J. T. (2010) Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J. Virol. 84, 12646–12657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anafu A. A., Bowen C. H., Chin C. R., Brass A. L., Holm G. H. (2013) Interferon-inducible transmembrane protein 3 (IFITM3) restricts reovirus cell entry. J. Biol. Chem. 288, 17261–17271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mudhasani R., Tran J. P., Retterer C., Radoshitzky S. R., Kota K. P., Altamura L. A., Smith J. M., Packard B. Z., Kuhn J. H., Costantino J., Garrison A. R., Schmaljohn C. S., Huang I. C., Farzan M., Bavari S. (2013) IFITM-2 and IFITM-3 but not IFITM-1 restrict Rift Valley fever virus. J. Virol. 87, 8451–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jiang D., Weidner J. M., Qing M., Pan X. B., Guo H., Xu C., Zhang X., Birk A., Chang J., Shi P. Y., Block T. M., Guo J. T. (2010) Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J. Virol. 84, 8332–8341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Everitt A. R., Clare S., Pertel T., John S. P., Wash R. S., Smith S. E., Chin C. R., Feeley E. M., Sims J. S., Adams D. J., Wise H. M., Kane L., Goulding D., Digard P., Anttila V., Baillie J. K., Walsh T. S., Hume D. A., Palotie A., Xue Y., Colonna V., Tyler-Smith C., Dunning J., Gordon S. B., GenISIS Investigators, MOSAIC Investigators, Smyth R. L., Openshaw P. J., Dougan G., Brass A. L., Kellam P. (2012) IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484, 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bailey C. C., Huang I. C., Kam C., Farzan M. (2012) IFITM3 limits the severity of acute influenza in mice. PLoS Pathog. 8, e1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Z., Zhang A., Wan Y., Liu X., Qiu C., Xi X., Ren Y., Wang J., Dong Y., Bao M., Li L., Zhou M., Yuan S., Sun J., Zhu Z., Chen L., Li Q., Zhang Z., Zhang X., Lu S., Doherty P. C., Kedzierska K., Xu J. (2014) Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc. Natl. Acad. Sci. U.S.A. 111, 769–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Y. H., Zhao Y., Li N., Peng Y. C., Giannoulatou E., Jin R. H., Yan H. P., Wu H., Liu J. H., Liu N., Wang D. Y., Shu Y. L., Ho L. P., Kellam P., McMichael A., Dong T. (2013) Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat. Commun. 4, 1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feeley E. M., Sims J. S., John S. P., Chin C. R., Pertel T., Chen L. M., Gaiha G. D., Ryan B. J., Donis R. O., Elledge S. J., Brass A. L. (2011) IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 7, e1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li K., Markosyan R. M., Zheng Y. M., Golfetto O., Bungart B., Li M., Ding S., He Y., Liang C., Lee J. C., Gratton E., Cohen F. S., Liu S. L. (2013) IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 9, e1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perreira J. M., Chin C. R., Feeley E. M., Brass A. L. (2013) IFITMs restrict the replication of multiple pathogenic viruses. J. Mol. Biol. 425, 4937–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Diamond M. S., Farzan M. (2013) The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 13, 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hach J. C., McMichael T., Chesarino N. M., Yount J. S. (2013) Palmitoylation on conserved and non-conserved cysteines of murine IFITM1 regulates its stability and anti-influenza A virus activity. J. Virol. 87, 9923–9927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yount J. S., Moltedo B., Yang Y. Y., Charron G., Moran T. M., López C. B., Hang H. C. (2010) Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat. Chem. Biol. 6, 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yount J. S., Zhang M. M., Hang H. C. (2013) Emerging roles for protein S-palmitoylation in immunity from chemical proteomics. Curr. Opin. Chem. Biol. 17, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yount J. S., Charron G., Hang H. C. (2012) Bioorthogonal proteomics of 15-hexadecynyloxyacetic acid chemical reporter reveals preferential targeting of fatty acid modified proteins and biosynthetic enzymes. Bioorg. Med. Chem. 20, 650–654 [DOI] [PubMed] [Google Scholar]
- 22. Shan Z., Han Q., Nie J., Cao X., Chen Z., Yin S., Gao Y., Lin F., Zhou X., Xu K., Fan H., Qian Z., Sun B., Zhong J., Li B., Tsun A. (2013) Negative regulation of interferon-induced transmembrane protein 3 by SET7-mediated lysine monomethylation. J. Biol. Chem. 288, 35093–35103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jia R., Pan Q., Ding S., Rong L., Liu S. L., Geng Y., Qiao W., Liang C. (2012) The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J. Virol. 86, 13697–13707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alland L., Peseckis S. M., Atherton R. E., Berthiaume L., Resh M. D. (1994) Dual myristylation and palmitylation of Src family member p59fyn affects subcellular localization. J. Biol. Chem. 269, 16701–16705 [PubMed] [Google Scholar]
- 25. Moltedo B., Li W., Yount J. S., Moran T. M. (2011) Unique type I interferon responses determine the functional fate of migratory lung dendritic cells during influenza virus infection. PLoS Pathog. 7, e1002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bailey C. C., Kondur H. R., Huang I. C., Farzan M. (2013) Interferon-induced transmembrane protein 3 is a type II transmembrane protein. J. Biol. Chem. 288, 32184–32193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pandey K. N. (2009) Functional roles of short sequence motifs in the endocytosis of membrane receptors. Front. Biosci. 14, 5339–5360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hunter T. (2007) The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 [DOI] [PubMed] [Google Scholar]
- 29. Huang F., Gu H. (2008) Negative regulation of lymphocyte development and function by the Cbl family of proteins. Immunol. Rev. 224, 229–238 [DOI] [PubMed] [Google Scholar]
- 30. Ghosh A. K., Reddi A. L., Rao N. L., Duan L., Band V., Band H. (2004) Biochemical basis for the requirement of kinase activity for Cbl-dependent ubiquitinylation and degradation of a target tyrosine kinase. J. Biol. Chem. 279, 36132–36141 [DOI] [PubMed] [Google Scholar]
- 31. Jia R., Xu F., Qian J., Yao Y., Miao C., Zheng Y. M., Liu S. L., Guo F., Geng Y., Qiao W., Liang C. (2014) Identification of an endocytic signal essential for the antiviral action of IFITM3. Cell. Microbiol. 10.1111/cmi.12262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rotin D., Kumar S. (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]





