Background: The Dorsal transcription factor regulates the dorsal-ventral developmental gene regulatory network by binding to enhancers.
Results: The core promoters of multiple Dorsal target genes are evolutionarily conserved and functionally dependent on specific elements.
Conclusion: The core promoter composition is an important determinant of the transcriptional outcome.
Significance: Transcriptional regulation results from an interplay between enhancers and core promoter composition.
Keywords: Drosophila, Gene Expression, RNA Polymerase II, Transcription Regulation, Transcription/Developmental Factors, DPE (Downstream Core Promoter Element), Dorsal, Core Promoter, Gene Network
Abstract
Developmental processes are highly dependent on transcriptional regulation by RNA polymerase II. The RNA polymerase II core promoter is the ultimate target of a multitude of transcription factors that control transcription initiation. Core promoters consist of core promoter motifs, e.g. the initiator, TATA box, and the downstream core promoter element (DPE), which confer specific properties to the core promoter. Here, we explored the importance of core promoter functions in the dorsal-ventral developmental gene regulatory network. This network includes multiple genes that are activated by different nuclear concentrations of Dorsal, an NFκB homolog transcription factor, along the dorsal-ventral axis. We show that over two-thirds of Dorsal target genes contain DPE sequence motifs, which is significantly higher than the proportion of DPE-containing promoters in Drosophila genes. We demonstrate that multiple Dorsal target genes are evolutionarily conserved and functionally dependent on the DPE. Furthermore, we have analyzed the activation of key Dorsal target genes by Dorsal, as well as by another Rel family transcription factor, Relish, and the dependence of their activation on the DPE motif. Using hybrid enhancer-promoter constructs in Drosophila cells and embryo extracts, we have demonstrated that the core promoter composition is an important determinant of transcriptional activity of Dorsal target genes. Taken together, our results provide evidence for the importance of core promoter composition in the regulation of Dorsal target genes.
Introduction
Transcriptional regulation of gene expression is critical for embryonic development (1–6). Multiple sequence-specific DNA binding transcription factors and co-regulators control gene expression (7–10), but the ultimate target of the transcription machinery is the initiation of transcription at the core promoter, which could hence be referred to as the gateway to transcription (11–16).
Focused core promoters, i.e. promoters in which transcription initiates at a single nucleotide or within a narrow region of several nucleotides, exist in all eukaryotes and are predominant in simple organisms, such as Drosophila (13, 17, 18). Focused core promoters are typically ∼80 nucleotides in length and encompass the RNA start site (13, 17, 18).
Core promoters may contain one or more functional DNA sequence elements, termed core promoter elements or motifs, such as the TATA box, TFIIB recognition elements (BREu and BREd), initiator (Inr),2 TCT motif, motif 10 element (MTE), and downstream core promoter element (DPE), which confer specific properties to the core promoter (13, 19–21). The TATA box, the Inr, the MTE, and the DPE motifs are recognized and bound by subunits of TFIID, the first complex that recognizes and binds the core promoter in the process of RNA polymerase II recruitment to the core promoter.
The TATA box is the first core promoter motif identified and is conserved from archaebacteria to humans (22, 23). The upstream T is typically located at −30 or −31 relative to the transcription start site (24). The TATA box is bound by the TATA box-binding protein (TBP) subunit of TFIID. The Inr encompasses the transcription start site and is the most common core promoter element (11, 19, 25). The Inr is bound by the TAF1 and TAF2 subunits of TFIID (26). The DPE was originally discovered as a TFIID recognition site that is located downstream of the initiator element (precisely from +28 to +33 relative to the A+1 of the Inr) and is conserved from Drosophila to humans (27, 28). It is bound by the TAF6 and TAF9 subunits of TFIID (28). The MTE is located immediately upstream of the DPE at precisely +18 to +27 relative to the A+1 in the Inr and is also conserved from Drosophila to humans (29, 30). Both the DPE and the MTE motifs are dependent on the Inr and function cooperatively with it (28, 30–33). Moreover, we have previously demonstrated that gene expression levels can be modulated via the core promoter (31).
The existence of different types of core promoters implies that core promoter elements play regulatory roles beyond the specification of transcription initiation. Transcription of TATA-dependent genes differs from transcription of DPE-dependent genes in many respects. First, the set of basal transcription factors that is necessary to transcribe TATA-dependent promoters in vitro is insufficient to transcribe DPE-dependent promoters (34, 35). Second, enhancers with preference for DPE-containing promoters or TATA-containing promoters have been identified, supporting the existence of enhancer-promoter specificity (16, 36–38). Third, TBP is necessary for TATA-dependent transcription. However, TBP down-regulates DPE-dependent transcription (39). The two transcriptional regulators, NC2 and MOT1, which have been shown to be positive regulators of DPE-dependent transcription, do so by counteracting TBP, thus relieving its inhibition of DPE transcription (39–41).
In this study, we explored the contribution of the DPE motif to the regulation of gene expression in the dorsal-ventral developmental gene network that is governed by the Rel family transcription factor Dorsal. We show that the promoters of many Dorsal target genes contain DPE sequence motifs. We demonstrate that core promoters of multiple Dorsal target genes are evolutionarily conserved and are functionally dependent on the DPE motif. Moreover, we have analyzed the activation of key Dorsal target genes by Dorsal and another Rel family transcription factor, Relish, and the dependence on the DPE motif. Using Drosophila S2R+ cells, we further demonstrate that the TATA box cannot compensate for the loss of DPE in the Dorsal target brinker and can only partially compensate for the loss of DPE in the Dorsal target twist. The addition of a TATA box to the brinker and twist promoter can, however, compensate for the loss of DPE using nuclear extracts derived from Drosophila embryos. We show that Dorsal is able to discriminate between a TATA box-containing promoter and a DPE-containing promoter. Transcription of enhancer-promoter constructs in both Drosophila S2R+ cells and embryo extracts manifests the importance of the core promoter in transcriptional regulation of Dorsal target genes. Collectively, we demonstrate that the DPE motif plays an important role in the expression of Dorsal target genes regulating dorsal-ventral patterning and provides evidence for the key contribution of the core promoter composition to gene regulatory networks.
EXPERIMENTAL PROCEDURES
Sequence Conservation Analysis
Sequence conservation analysis was performed using the UCSC genome browser.
Calculation of Core Promoter Element Frequency
Drosophila transcripts that initiate at different chromosomal positions (based on the RefSeq database) were used to calculate the frequency of the core promoter elements (see Tables 1 and 2). Inr elements were identified in the region −10 to +10 relative to the transcription start site if there was an A at +1 and at least three of five additional matches to the consensus (TCAKTY, where the A is the +1 of the transcript). DPE motifs that were precisely located at +28 relative to the A + 1 of the Inr, were identified based on at least five of six matches to the DPE functional range set (DSWYVY). Putative TATA box elements were identified based on a match to a TATA sequence located from −45 to −19 relative to the RefSeq transcription start site. The significance of differences in core promoter composition between Drosophila transcripts and Dorsal target genes was calculated using the chi-square test.
TABLE 1.
Core promoter composition of Dorsal target genes
The abbreviations used are: Acp1, adult cuticle protein; Act57B, Actin57B; Ance, Angiotensin converting enzyme; Argk, Arginine kinase; Asph, Aspartyl beta-hydroxylase; brk, brinker; BRWD3, bromodomain and WD repeat domain containing 3; bun, bunched; cv-2, crossveinless 2; Cyp310a1, Probable cytochrome P450 310a1; DNaseII, Deoxyribonuclease II; doc3, Dorsocross3; dpp, decapentaplegic; Dr, Drop; Dscam, Down syndrome cell adhesion molecule; Ect1, Dorsal target gene in the Dorsal ectoderm 1, eiger; Ect2, Dorsal target gene in the Dorsal ectoderm 2, Dorsocross 1; Ect3, Dorsal target gene in the Dorsal ectoderm 3; Ect4, Dorsal target gene in the Dorsal ectoderm 4; Ect5, Dorsal target gene in the Dorsal ectoderm 5, C15; ed, echinoid; E(spl), Enhancer of split; fog, folded gastrulation; gcm, Glial cells missing; hbr, Heartbroken, Stumps, Dof; hoip, hoi-polloi; htl, heartless; ind, intermediate neuroblasts defective; lea, leak; Lcp65Ag2, larval cuticle protein 65Ag2; loco, locomotion defects; Mbs, Myosin binding subunit; Mdr49, Multi drug resistance 49, Mes5; Mef2, Myocyte enhancer factor 2; Mes 2, Dorsal target gene in the mesoderm 2; Mes 3, Insulin-like peptide 4; Mes 4, Dorsal target gene in the mesoderm 4; Neu2, Dorsal target gene in the neuroectoderm 2; Neu3, Dorsal target gene in the neuroectoderm 3; neur, neuralized; PGRP-SC2, Peptidoglycan-recognition protein-SC2; Phk-3, Pherokine 3; phm, phantom; Pif1A, PFTAIRE-interacting factor 1A; pnr, pannier; ptc, patched; pyr, pyramus; rho, rhomboid; RhoL, rho-like, Mes1; ry, rosy; sca, scabrous; sim, single-minded; sna, snail; Socs36E, Suppressor of cytokine signaling at 36E; sog, short gastrulation; SoxN, SoxNeuro; Sulf1, Sulfated; T3, lethal of scute, l(1) sc; ths, thisbe, Neu4; tin, tinman; tld, tolloid; trbl, tribbles; trh, trachealess; Trim9, tripartite motif-containing 9; tup, tailup; TwdlM, Tweedle M; TwdlN, TweedleN; twi, twist; ush, u-shaped; vn, vein; vnd, ventral nervous system defective; wntD, wnt inhibitor of Dorsal; wts, warts; zen, zerknult; zfh 1, Zn finger homeodomain 1. Promoters containing Inr and DPE with ++ or +++ score are framed in red.
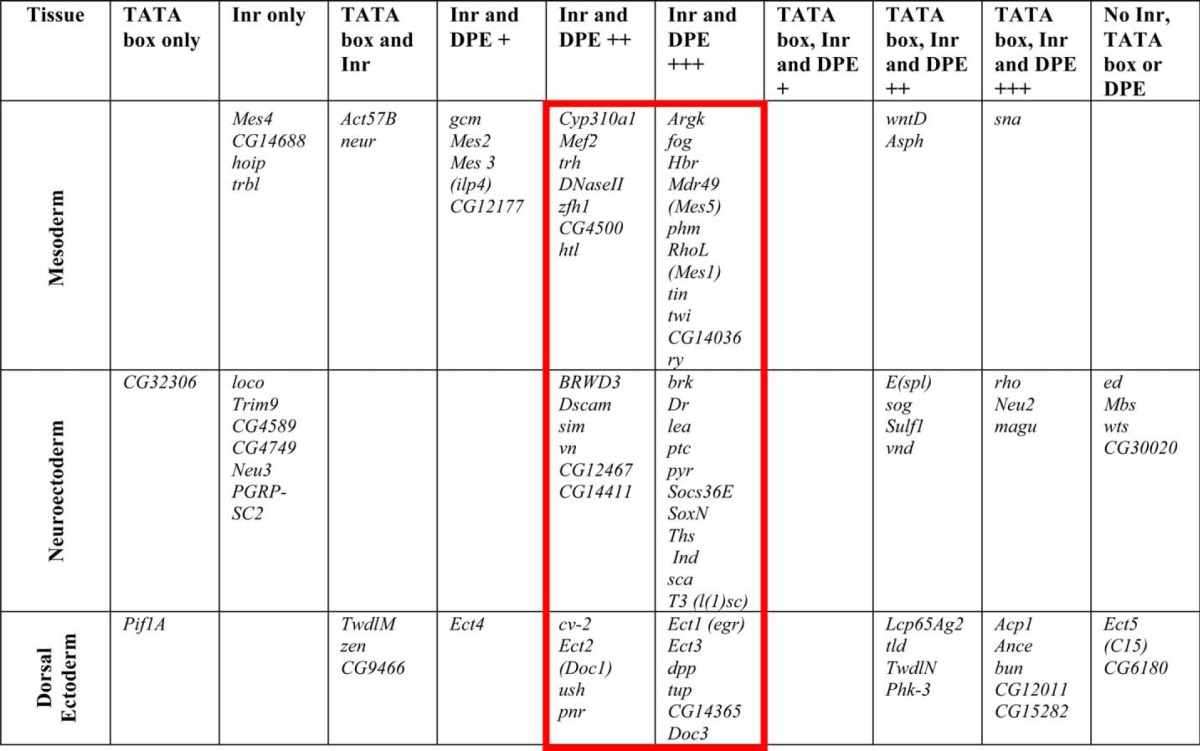
TABLE 2.
Frequency of core promoter elements among Drosophila transcripts that initiate at different chromosomal positions and Dorsal target genes
| Combination of core promoter elements |
Drosophila transcripts |
Dorsal target genes |
p value | ||
|---|---|---|---|---|---|
| No. of Genes | Frequency | No. of Genes | Frequency | ||
| % | % | ||||
| Inr | 16,352 | 82 | 83 | 91 | <0.02 |
| TATA | 3,494 | 18 | 26 | 29 | <0.01 |
| TATA and no DPE | 2,568 | 13 | 7 | 8 | <0.15 |
| TATA and no Inr | 461 | 2 | 2 | 2 | 1 |
| Inr and DPE | 4,628 | 23 | 63 | 69 | <0.0001 |
| Inr, DPE and TATA | 836 | 4 | 19 | 21 | <0.0001 |
| Total | 19,865 | (100) | 91 | (100) | |
In Vitro Transcription Assays
Double-stranded oligonucleotides comprising core promoter sequences from −10 to +40 (see Fig. 1 and supplemental Fig. S1) of the tested Dorsal target genes core promoters were inserted into the PstI and XbaI sites of pUC119. Mutation of the DPE in the core promoters was identical to that used previously (30), where the mutant DPE contains CATA at +30 to +33 relative to A + 1. Natural tinman (tin), brinker (brk), twist (twi), and leak (lea) enhancer-promoter constructs driving the luciferase reporter gene were used in Figs. 3–5, and tin-twi hybrid enhancer-promoter constructs driving the luciferase reporter gene were used in Fig. 6. In vitro transcription reactions were carried out as described previously (42) using 250 ng of supercoiled DNA templates with Drosophila high salt nuclear extracts (43). The resulting transcripts were subjected to primer extension analysis with an M13 reverse sequencing primer (AGCGGATAACAATTTCACACAGGA; see Fig. 2) or with a reverse luciferase primer (TCTTCCAGCGGATAGAATGGCGCC; see Figs. 5 and 6). Quantitation of reverse transcription products was carried out using ImageQuant, ImageJ, and GelQuantNET. All experiments were carried out a minimum of three independent times to ensure reproducibility of the data.
FIGURE 1.
The core promoters of multiple Drosophila Dorsal target genes contain conserved DPE motifs. The core promoter sequences (−40 to +40 relative to the A + 1 of the Inr) of the Drosophila Dorsal target genes brinker (A) and twist (B) are shown. Nucleotides conforming to the Inr consensus sequence that are identical to D. melanogaster Inr sequence are highlighted in yellow. Nucleotides that are identical to the D. melanogaster DPE functional range set sequence are highlighted in light blue. Nucleotides that are not conserved to D. melanogaster but still conform to the DPE functional range set are highlighted in gray. Sequence conservation of a nucleotide in all 12 species is marked on the bottom by a colon (:), whereas sequence conservation of a nucleotide in 10 or 11 species is marked on the bottom by a single dot.
FIGURE 3.
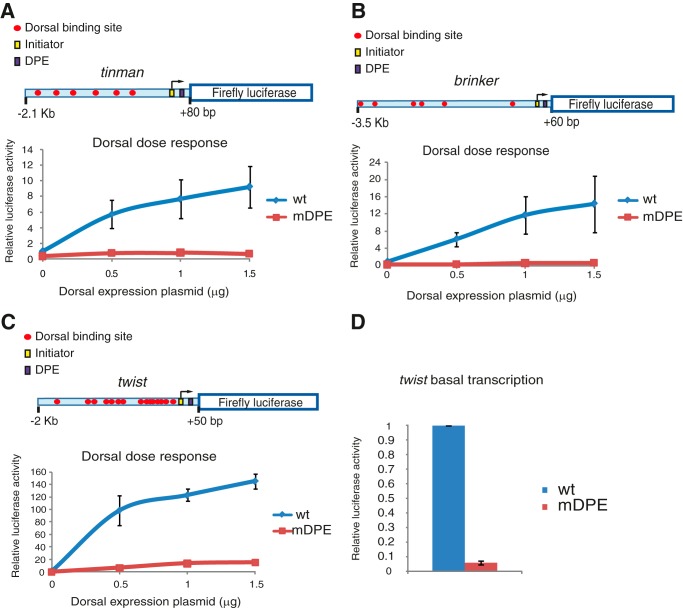
The activation of transcription of the natural tinman, brinker, and twist promoters in Drosophila S2R+ cells by Dorsal and the dependence of their activation on the DPE motif. Drosophila S2R+ cells were co-transfected with firefly luciferase reporter constructs containing wt or mDPE promoter, as well as varying amounts of a Dorsal expression plasmid, as indicated. To normalize for transfection efficiency, cells were co-transfected with a Pol III-Renilla luciferase control plasmid and assayed for dual luciferase activity. The activities are reported relative to the wild-type promoter in the absence of co-transfected Dorsal expression plasmid, which was defined to be 1. A schematic diagram of the genomic fragments is shown on top of panels A–C. A, transcriptional activation of the natural tinman promoter by Dorsal (n = 3). B, transcription activation of the natural brinker promoter by Dorsal (n = 3). C, transcription activation of the natural twist promoter by Dorsal (n = 4). D, basal transcription levels of twi. To enable the visualization of the basal transcription levels of wt and mDPE twist reporters, these data are presented in a separate panel. In all panels, error bars represent S.E.
FIGURE 4.
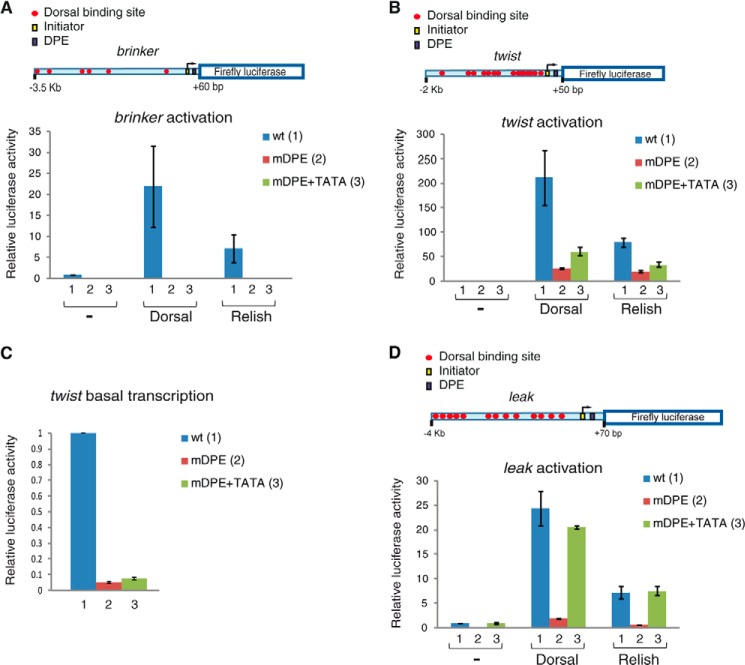
The TATA box motif cannot substitute for the loss of a DPE motif in brk transcription and can only partially substitute for the loss of a DPE motif in twi in Drosophila S2R+ cells, whereas transcriptional activation of the natural leak promoter by Dorsal is dependent on the DPE motif and could fully be restored via a TATA box. Drosophila S2R+ cells were transfected with firefly luciferase reporter constructs containing wt (columns 1), mDPE (columns 2), or mDPE + TATA box promoter (columns 3), as well as a Dorsal expression vector, a Relish expression vector, or an empty vector, as indicated. To normalize for transfection efficiency, cells were co-transfected with a Pol III-Renilla luciferase control plasmid and assayed for dual luciferase activity. The activities are reported relative to the wild-type promoter in the absence of co-transfected Dorsal or Relish expression plasmid, which was defined to be 1. A, transcriptional activation of the natural brinker promoter by Dorsal is dependent on the DPE motif and cannot be compensated via a TATA box (n = 3). B, transcriptional activation of the natural twist promoter by Dorsal is dependent on the DPE and could partially be restored via a TATA box (n = 3). C, basal transcription levels of twi. To enable the visualization of the basal transcription levels of wt (columns 1), mDPE (columns 2), and mDPE + TATA box (columns 3) twist reporters (in the absence of transfected Dorsal), these data are presented in a separate panel. D, transcriptional activation of the natural leak promoter by Dorsal is dependent on the DPE and could fully be restored via a TATA box (n = 3). In all panels, error bars represent S.E.
FIGURE 5.
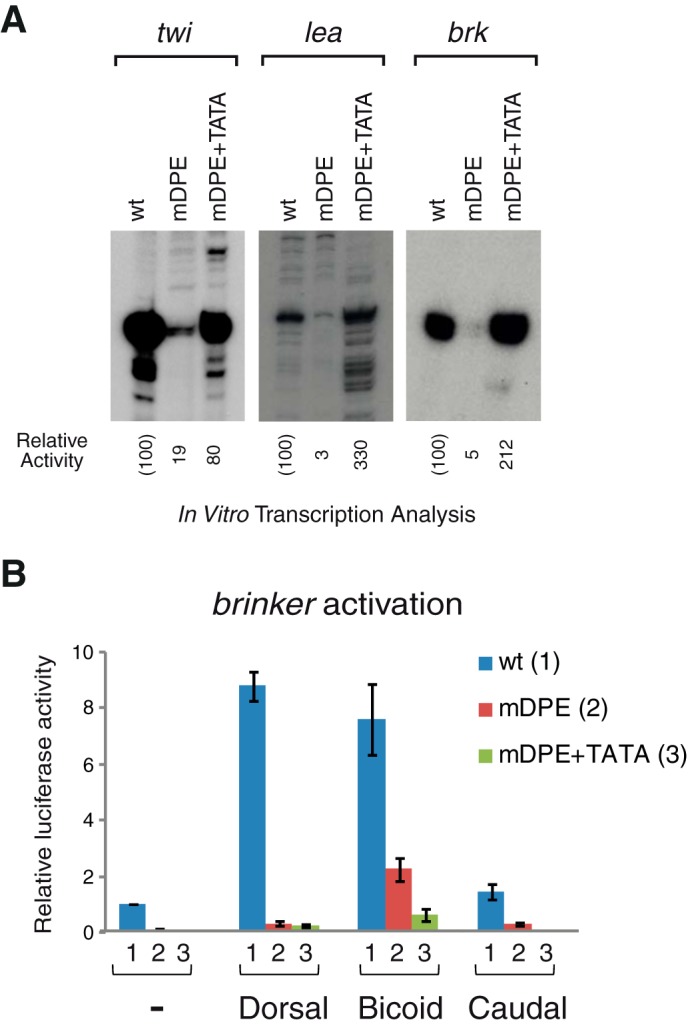
Transcription of the natural twist, leak, and brinker promoters is dependent on the DPE motif but could be restored via a TATA box using Drosophila embryo nuclear extracts, which contain transcription factors that are absent from Drosophila S2R+ cells. A, the natural enhancer-promoter constructs of twist, leak, and brinker containing either wt, mDPE, or mDPE + TATA motifs were subjected to in vitro transcription analysis with a Drosophila embryo nuclear extract. The resulting transcripts were detected by primer extension-reverse transcription analysis. B, unlike Dorsal and Caudal, Bicoid does activate the mDPE reporter and to some extent, also the mDPE+TATA brk reporter. Drosophila S2R+ cells were transfected with natural brinker promoters firefly luciferase reporter constructs containing wt (columns 1), mDPE (columns 2), or mDPE + TATA box promoter (columns 3), as well as expression vectors of either Dorsal, Bicoid, Caudal, or an empty vector, as indicated. To control for transfection efficiency variations, cells were co-transfected with a control Renilla luciferase reporter and assayed for dual luciferase. In both panels, n = 3 and error bars represent S.E.
FIGURE 6.
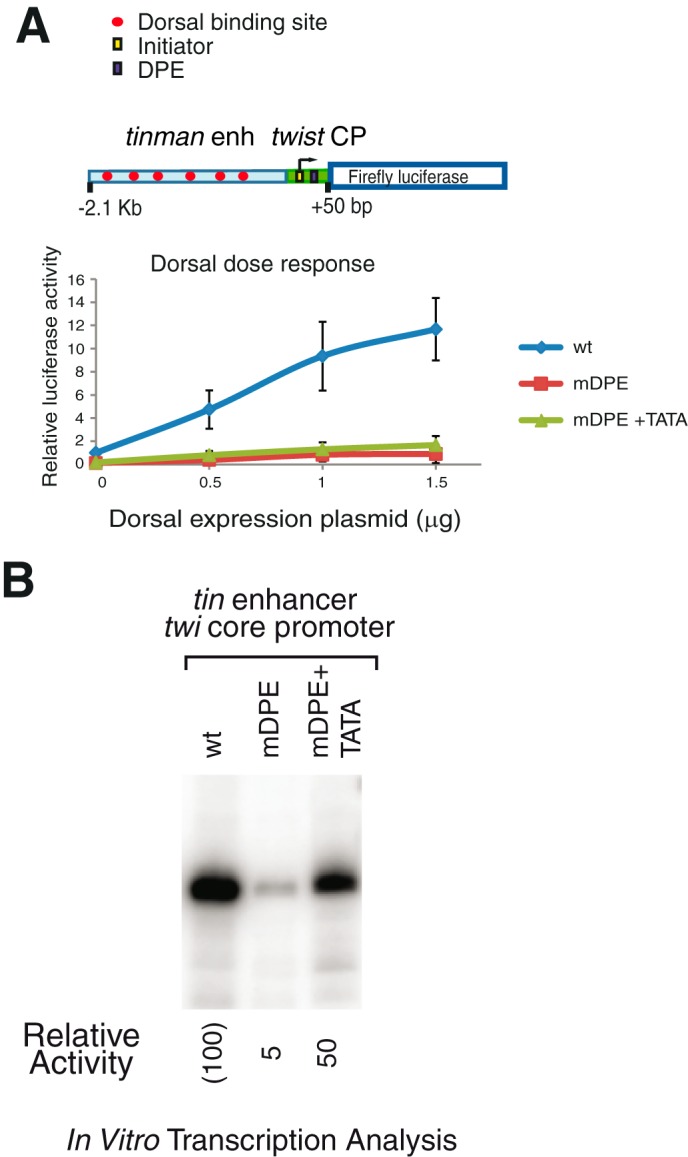
The core promoter is a key contributor to the transcriptional output. Drosophila S2R+ cells were transfected with firefly luciferase reporter constructs containing hybrid enhancer-promoters with either wt, mDPE, or MDPE + TATA core promoter, as well as varying amounts of a Dorsal expression plasmid, as indicated. To normalize for transfection efficiency, cells were co-transfected with a Pol III-Renilla luciferase control plasmid and assayed for dual luciferase activity. The activities are reported relative to the wild-type promoter in the absence of co-transfected Dorsal expression plasmid, which was defined to be 1. A schematic diagram of the hybrid genomic fragments is shown on top. A, transcriptional activation of the hybrid tinman enhancer-twist core promoter constructs by Dorsal (n = 3). B, the hybrid tinman enhancer-twist core promoter constructs containing either wt, mDPE, or mDPE + TATA motifs, were subjected to in vitro transcription analysis with a Drosophila embryo nuclear extract. The resulting transcripts were detected by primer extension-reverse transcription analysis.
FIGURE 2.
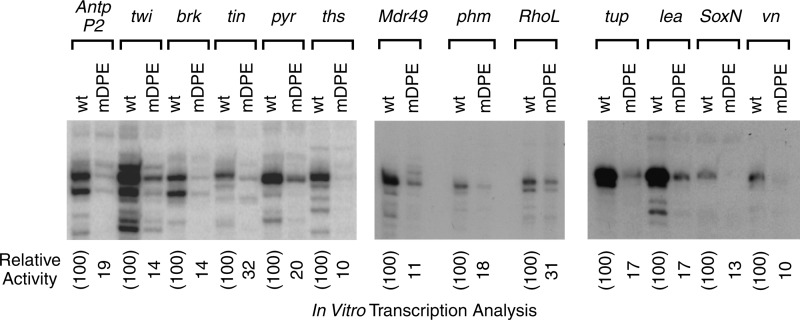
The DPE is functional in multiple Dorsal target genes. The wt and mDPE versions of the indicated core promoters (from −10 to +40 relative to the A + 1 start site) were subjected to in vitro transcription analysis with a Drosophila embryo nuclear extract. The resulting transcripts were detected by primer extension-reverse transcription analysis. The previously characterized Drosophila Antp P2 (Antennapedia downstream promoter) served as a control.
Expression Plasmids and Luciferase Reporter Genes
pAcDorsal expression plasmid was kindly provided by Dr. Albert Courey (UCLA) For the construction of a Relish expression vector, the active N terminus of Relish (amino acids 1–532; not including the ankyrin repeat-containing inhibitory domain) (44) was cloned into a pAc expression vector. An N-terminally FLAG-tagged full-length Caudal was cloned into a pAc expression vector. A C-terminally V5-His-tagged full-length Bicoid was cloned into a pAc expression vector. The natural enhancer-promoter tin reporter construct encompasses a genomic fragment from −2089 to +80 relative to the A+1 of the Inr (containing either a wt, or a mDPE), upstream of a firefly luciferase reporter gene. The natural enhancer-promoter brk reporter construct encompasses a genomic fragment from −3531 to +60 relative to the A+1 of the Inr (containing either a wt, a mDPE, or a mDPE with a TATA box), upstream of a firefly luciferase reporter gene. The natural enhancer-promoter twi reporter constructs encompasses a genomic fragment from −2041 to +50 relative to the A + 1 of the Inr (containing either a wt, a mDPE or a mDPE with a TATA box), upstream of a firefly luciferase reporter gene. The natural enhancer-promoter lea reporter construct encompasses a genomic fragment from −3931 to +69 relative to the A+1 of the Inr (containing either a wt, a mDPE, or a mDPE with a TATA box), upstream of a firefly luciferase reporter gene. The tin enhancer-twi core promoter hybrid promoter was constructed using the tin enhancer from −2089 to −138 followed by the twi promoter from −134 to +50 (with a 3-nucleotide spacer in between), upstream of a firefly luciferase reporter gene.
Luciferase reporter constructs driven by either the tin, brk, lea, or twi genomic region with a mutated DPE (in which nucleotides at positions +28 to +34 relative to A+1 in the Inr were mutated to CTCATGT) or an additional TATA box (TATAAAAA, where the upstream T is at position −31 relative to the A + 1 of the Inr) were generated using a QuikChange II site-directed mutagenesis kit (Stratagene). DNA sequence-verified fragments that encompass the mutated nucleotides were subcloned into their corresponding locations in the wild-type vectors. Pol III-Renilla luciferase reporter was kindly provided by Dr. Norbert Perrimon (Harvard Medical School).
Transient Transfection and Reporter Gene Assays
Drosophila melanogaster Schneider S2R+ adherent cells were cultured in Schneider's Drosophila medium (Biological Industries) that was supplemented with 10% heat-inactivated FBS. Cells were transfected in 24-well plates by using the Escort IV reagent (Sigma). For Dorsal dose-response dual luciferase assays, cells were transfected with the firefly luciferase reporter constructs (80 ng) and the indicated amounts of a Dorsal expression vector that was supplemented, where necessary, with pAc control expression vector to give a total of 1.5 μg of DNA of expression vector. For assaying the tin, brk, twi, or lea reporter constructs with mDPE and an added TATA box, cells were co-transfected with either 0.5 μg of Dorsal, Relish, Bicoid, or Caudal expression vector that was supplemented, where necessary, with pAc control expression vector to give a total of 1.0 μg of DNA of expression vector, in addition to either tin (80 ng), brk (80 ng), lea (80 ng), or twi reporter (20 ng).
To normalize for transfection efficiency, cells were co-transfected with a Pol III-Renilla luciferase control plasmid (10 ng). Cells were harvested 36–48 h post-transfection and assayed for dual luciferase activities, as specified by the manufacturer (Promega). The graphs represent averages of three to four independent experiments.
RESULTS
Identification of DPE Motifs in Core Promoters of Dorsal Target Genes
To discover distinct gene regulatory networks (GRNs) and pathways that are regulated via the core promoter, we have examined the core promoter composition of genes that are involved in early embryonic fly development. This analysis led us to focus on the dorsal-ventral GRN and in particular on Dorsal target genes. The dorsal-ventral GRN is critical for early embryonic development. The network includes multiple genes that are activated by different nuclear concentrations of the Dorsal transcription factor along the dorsal-ventral axis (45–54). Activation of Dorsal target genes is achieved by the recruitment of Dorsal to the enhancers of these genes, which contain Dorsal-binding sites hundreds or even thousands of base pairs upstream of the transcription start site.
To examine whether core promoter elements play a role in the regulation of Dorsal-target gene expression, we first analyzed the core promoter sequences of Dorsal-target genes. Using the RefSeq database, we have carried out a comprehensive analysis of the core promoter composition of previously identified Dorsal target genes (Refs. 47 and 51 and references therein) (Table 1). Strikingly, the majority of known Dorsal target genes appear to contain Inr and DPE motifs. Specifically, in the mesoderm, where Dorsal nuclear concentration is highest, of a total 30 mesodermal genes analyzed, 17 genes contain potential Inr and DPE motifs with high probability (Table 1, Inr and DPE +++ and Inr and DPE ++), including Dorsal target genes critical for mesodermal development such as tin, twi, Hbr, and Mef2, and 3 genes contain TATA box, Inr, and DPE motifs. In the neuroectoderm, where Dorsal nuclear concentration is intermediate, 17 genes contain Inr and DPE motifs with high probability, including well characterized Dorsal target genes such as brk, ths, and pyr, and 7 genes contain TATA box, Inr, and DPE motifs of a total 35 neuroectodermal genes analyzed. In the dorsal ectoderm, where Dorsal nuclear concentration is lowest, 10 genes contain Inr and DPE motifs with high probability, including well characterized dorsoectodermal targets such as dpp and tup, and 9 genes contain TATA box, Inr, and DPE motifs of a total 26 dorsal ectoderm genes analyzed. Overall, 63 of 91 genes analyzed (over 69%) are likely to contain functional DPE motifs.
Computational analyses of the frequencies of Drosophila core promoter elements had previously indicated that the DPE is present in ∼2.1–22% of Drosophila core promoters (19, 25, 29). We have calculated the frequencies of core promoter elements among Drosophila transcripts that initiate at different chromosomal positions using strict Inr-DPE spacing criteria. We have discovered that 23% of Drosophila promoters contain Inr and DPE motifs, whereas 4% contain Inr, DPE, and TATA box motifs (Table 2). The frequency of DPE-containing promoters among Dorsal target genes (69%) is significantly higher than the calculated (23%) and the previously reported frequencies (2.1% to 22%) of DPE in Drosophila core promoters (Table 2).
The overall frequency of a TATA box among Drosophila transcripts is 18% (Table 2). Interestingly, only a small subset of the Dorsal target genes appear to contain a TATA box and no DPE. In the mesoderm there are no genes that contain only a TATA box, 2 genes that contain a TATA box and an Inr motif, and 3 more genes that contain TATA box, Inr, and DPE motifs. In the neuroectoderm, 1 gene contains a TATA box, there are no genes that contain a TATA box and Inr, and 7 genes contain TATA box, Inr, and DPE motifs. In the dorsal ectoderm, 1 gene contains a TATA box, 3 genes contain a TATA box and Inr, and 9 genes contain TATA box, Inr, and DPE. The overall frequency of TATA box without a DPE motif among Dorsal target genes is less than 8% (7 of 91), and only a few (3 of 65) of the Dorsal targets that are regulated by high or intermediate nuclear concentrations of Dorsal contain a TATA box without a DPE motif, which is significantly lower than the overall frequency of Drosophila core promoters containing a TATA box without a DPE motif (13%; Table 2). Taken together, the Inr and DPE motifs appear to be overrepresented in the core promoters of multiple Dorsal target genes.
The DPE Motif in Multiple Drosophila Dorsal Target Genes Is Conserved
To examine whether core promoter elements play a role in the regulation of Dorsal-target gene expression, we have analyzed the sequence conservation of Dorsal target genes that are activated by different nuclear concentration of Dorsal: mesodermal genes that are activated by the highest concentrations of Dorsal, namely tinman (tin), twist (twi), phantom (phm), rho-like (RhoL; Mes1), and Multi drug resistance 49 (Mdr49; Mes5) (45, 47, 55–58), neurogenic ectoderm genes that are activated by intermediate levels of Dorsal, namely brinker (brk), leak (lea), SoxNeuro (SoxN), and vein (vn) (59–63), and genes that are activated by low levels of Dorsal: thisbe (ths), pyramus (pyr) in the neuroectoderm, and tailup (tup) in the dorsalectoderm (64, 65). In particular, we have searched these genes for both an Inr motif and a DPE motif that matches the functional DPE range set (DSWYVY) (33). Additionally, we have verified that the DPE motif is precisely located at +28 relative to the A + 1 of the Inr. Such spacing was previously shown to be critical for the function of DPE-dependent promoters (28, 33). Importantly, none of these genes contain a TATA box.
To assess the potential biological significance of the DPE motifs in the core promoters of these Dorsal target genes, we have examined the sequence conservation of these core promoters in the 12 sequenced Drosophila species. The Inr and DPE consensus sequences, as well as the Inr-to-DPE spacing in the core promoters of tin, brk, twi, pyr, ths, Mdr49 (Mes5), phm, RhoL (Mes1), tup, lea, and SoxN are conserved across 12 Drosophila species (Fig. 1 and supplemental Fig. S1). vn is highly conserved throughout the core promoter sequence but in only five Drosophila species (supplemental Fig. S1). Hence, the core promoters of multiple Dorsal target genes appear to contain conserved Inr and DPE motifs that match the DPE functional range set (33).
The DPE Motifs in Multiple Dorsal Target Genes Are Functionally Important for Transcription
To examine whether the conserved DPE motifs are functionally important for transcription of Dorsal target genes, we have cloned both wild-type (wt) and mutant DPE (mDPE) core promoters (−10 to +40 relative to the A + 1 of the Inr) of 12 putative DPE-containing Dorsal target genes (namely, twi, brk, tin, pyr, ths, Mdr49, phm, RhoL, tup, lea, soxN, and vn) into the pUC119 vector. The activity of wt and mDPE core promoters was tested by in vitro transcription with Drosophila embryo nuclear extracts followed by primer extension analysis. Mutations in the DPE motif of each of the core promoters of the aforementioned Dorsal target genes resulted in reduced transcription levels (Fig. 2). Hence, transcription of the core promoters of these 12 Dorsal target genes is highly dependent on the DPE.
Activation of Dorsal Target Genes via the DPE
To examine the effect of core promoter composition on activation of Dorsal target genes in the context of their natural enhancers, we mapped the putative Dorsal DNA binding sites within the enhancers of tin, brk, and twi. Consensus Dorsal DNA-binding sites (GGG(W)nCCM, where (W)n is four or five repeats of T/A (48)) were identified using TFsearch and JASPAR.
To test the activation of the tin, brk, and twi genes by Dorsal and their dependence on the DPE motif, genomic fragments encompassing either 2.2 kb of the natural tin enhancer and promoter, 3.6 kb of the natural brk enhancer and promoter (each containing 6 putative Dorsal binding sites), or 2.1 kb of the natural twi enhancer and promoter (which contains 15 putative Dorsal binding sites) were subcloned upstream of the firefly luciferase reporter gene (illustrated in Fig. 3). D. melanogaster Schneider S2R+ cells were co-transfected, and the transcriptional activity of wt and mDPE reporter genes in the presence of varying amounts of transfected Dorsal expression plasmid was assayed. The transcriptional activity of both tin and brk is higher in the presence of increasing amounts of transfected Dorsal expression plasmid, as expected (Fig. 3, A and B). The levels of transcription of the mDPE tin and brk do not increase in response to increasing amounts of transfected Dorsal, suggesting that the transcription of tin and brk is highly dependent on the presence of the DPE motif.
The transcriptional activity of twi is augmented with increasing amounts of transfected Dorsal expression plasmid, as expected (Fig. 3C). The levels of transcription are dependent on the presence of the DPE motif, because the transcriptional activity of mDPE twi is significantly lower. There is, however, an increase in the mDPE twi reporter activity with increasing amounts of transfected Dorsal expression vector. The basal transcription levels (in the absence of transfected Dorsal) of the twi reporter constructs are also dependent on the DPE motif (Fig. 3D). The basal activity of the wt twi promoter was 19-fold higher than that of the mDPE twi, whereas the basal activities of both wt tin and wt brk promoters were 6-fold higher than that of their corresponding mDPE reporters (data not shown).
The TATA Box Motif Cannot Substitute for the Loss of a DPE Motif in brk Transcription and Can Only Partially Substitute for the Loss of a DPE Motif in twi in Drosophila S2R+ Cells
Transcription of TATA-dependent genes differs from transcription of DPE-dependent genes in many respects. We demonstrated that mutation of the DPE reduces activation of brk and twi (Fig. 3). Thus, to examine whether the activation of these genes by Dorsal could occur via a TATA box motif, we have generated reporter constructs containing either brk or twi natural enhancer-promoter that, in addition to a mutation in the DPE motif, contain a TATA box. Addition of a TATA box to the brk natural enhancer-promoter with a mutation in the DPE motif could not restore transcription levels in the absence or in the presence of transfected Dorsal (Fig. 4A).
Interestingly, transfection of Relish, another Rel family transcription factor, which binds the same or nearly identical DNA sequence motifs as Dorsal (66, 67), was able to activate the wt brk, albeit to lower levels as compared with Dorsal. Addition of a TATA box to the brk natural enhancer-promoter with a mutation in the DPE motif could not restore transcription levels in the presence of transfected Relish (Fig. 4A).
Addition of a TATA box to the mDPE twi reporter was only able to partially restore activation by transfected Dorsal in the context of the natural enhancer (less than 30%) (Fig. 4B). Similarly, addition of a TATA box to the mDPE twi reporter was only able to partially restore activation by transfected Relish in the context of the natural enhancer. Interestingly, addition of a TATA box to a twi reporter containing a mDPE motif was unable to restore the basal transcription activity in the absence of transfected Dorsal (Fig. 4C). The twi enhancer contains 15 putative Dorsal binding sites, whereas the brk enhancer contains 6 putative Dorsal binding sites. To examine whether twi is less dependent on the DPE because it contains a higher number of putative Dorsal binding sites, we have cloned a genomic fragment encompassing 4.0 kb of the natural leak (lea) enhancer and promoter, which contains 13 putative Dorsal DNA binding sites. We have also generated lea reporter constructs containing either a mDPE motif or a mDPE with a TATA box and have tested their activation by Dorsal and Relish (Fig. 4D). A mutation of the DPE reduces activation of lea by both Dorsal and Relish. Interestingly, addition of a TATA box to the lea reporter containing a mDPE motif was able to restore the basal transcription activity, as well as the activation by transfected Dorsal or Relish (Fig. 4D).
Taken together, in Drosophila S2R+ cells, brk, twi, and lea promoters are dependent on the DPE motif. Dorsal activates transcription of its targets brk and twi via the DPE, whereas it activates lea via the DPE as well as the TATA box. Furthermore, Relish, which binds the same or nearly identical DNA sequence motifs as Dorsal, is able to activate transcription of Dorsal targets with similar core promoter preference as Dorsal.
The DPE Motif, Which Is Important for the Activity of twi, lea, and brk in Drosophila Nuclear Extracts, Can Be Replaced by a TATA Box
We next sought to examine the dependence of the twi, lea, and brk on the DPE motif in Drosophila embryo nuclear extracts, which provide a different experimental system. To this end we have performed in vitro transcription reactions of the enhancer-promoter reporters containing wt, mDPE, or mDPE + TATA with Drosophila embryo nuclear extracts followed by primer extension analysis. Mutations in the DPE motifs of the core promoters of twi, lea, and brk result in reduced transcription levels of the enhancer-promoter constructs (Fig. 5A). Hence, transcription of these Dorsal target genes is dependent on the DPE.
The addition of a TATA box to mDPE-containing twi and brk promoters cannot fully restore the reporter activity of the mDPE constructs in transfected S2R+ cells (Fig. 4). The addition of a TATA box to mDPE-containing twi, lea, and brk promoters can, however, restore in vitro transcription using Drosophila embryo nuclear extracts (Fig. 5A). This demonstrates both the strength of the TATA box in general as well as the apparent absence of transcription factors from Drosophila S2R+ cells. We have analyzed the enhancer of brk for putative binding sites of transcription factors that are expressed in 0–12-h embryos but are not expressed in S2R+ cells. The brk enhancer contains 19 putative Bicoid binding sites and 60 putative Caudal bindings sites (JASPAR). To examine whether Caudal or Bicoid could activate transcription of a mDPE + TATA-containing brk reporter, we co-transfected S2R+ cells with natural brk reporter constructs containing wt, mDPE, or mDPE +TATA box promoter, as well as expression vectors of either Dorsal, Bicoid, or Caudal (Fig. 5B). As expected, Caudal, which has previously been shown to be a preferential DPE-specific transcription factor (68), was able to activate the wt brk promoter but was unable to activate transcription of mDPE or mDPE + TATA-containing promoters. Interestingly, Bicoid was able to activate transcription of the wt brk reporter. Unlike Dorsal and Caudal, Bicoid does not have such core promoter preference because it activates the mDPE reporter and to some extent, also the mDPE + TATA brk reporter. It is of note that Bicoid activates its natural target gene giant, which contains functional TATA box, Inr, and DPE motifs, regardless of its core promoter composition (data not shown). Hence, a factor other than Caudal or Bicoid accounts for the transcription of the brk reporter containing a mDPE + TATA box using embryo nuclear extracts.
The Core Promoter Composition Is an Important Contributor to the Transcriptional Output
To determine the contribution of the core promoter to the transcriptional output, we have generated hybrid enhancer-promoter constructs with the tin enhancer and twi core promoter and have tested their activities. Co-transfection of a tin enhancer-twi core promoter hybrid reporter and increasing amounts of Dorsal expression vector to S2R+ cells resulted in an overall Dorsal activation that is typical for the tin enhancer (compare Figs. 3A and 6A). The tin enhancer-twi core promoter hybrid reporters containing mDPE or mDPE + TATA can be somewhat activated by Dorsal, as we have observed for twi enhancer-promoter reporters (Figs. 3, 4, and 6A). Hence, the ability of Dorsal to activate the mDPE or mDPE + TATA hybrid promoters in S2R+ cells relies on the core promoter in addition to the enhancer.
We next compared the activity of the hybrid reporters using Drosophila embryo nuclear extracts. As can be seen in Fig. 6B, the transcription activities of the hybrid tin enhancer-twi core promoter reporter constructs using embryo nuclear extracts are very similar to the activities of the natural twi enhancer-promoter constructs (Fig. 5A), suggesting that even though the enhancers are present in these constructs, the activity of the constructs in the embryo nuclear extracts is dictated by the core promoters. Collectively, the analysis of the hybrid enhancer-promoter constructs in both S2R+ cells and nuclear extracts derived from Drosophila embryos highlights the contribution of the core promoter to the transcriptional output.
DISCUSSION
The DPE Is a Transcriptional Element Shared by Many Dorsal Target Genes
In this study we demonstrate that the DPE is an important, conserved transcription element shared by multiple Dorsal target genes, which comprise the dorsal-ventral gene regulatory network. Specifically, we have shown that over two-thirds of the known Dorsal target genes contain DPE motifs, which is significantly higher than the percentage of DPE promoters in Drosophila genes (19, 25, 29). Remarkably, only less than 8% of the Dorsal target genes contain TATA box elements without DPE motifs. The DPE is most prevalent in mesodermal Dorsal targets. The number of DPE containing genes decreases in the neuroectoderm and further decreases in the dorsal ectoderm, where the Dorsal nuclear concentration is lowest. On the other hand, the number of genes containing a TATA box is higher in regions where Dorsal nuclear concentration is decreased. The occurrence of the DPE in many developmentally regulated genes (68), as well as our analysis of the frequencies of core promoter elements in Drosophila genes (Table 2) and the identification of DPE motifs in the majority of Dorsal targets, imply that the DPE is not randomly distributed in ∼23% of the genes; rather it is enriched in specific GRNs and pathways.
Regulation of twi, lea, and brk via the Core Promoter
We have examined the transcription of the natural enhancers and promoters of twi, lea, tin, and brk in Drosophila Schneider S2R+ cells and have discovered that the basal transcription levels of twi, lea, tin, and brk in the absence of ectopically expressed Dorsal are highly dependent on the DPE motif (Figs. 3 and 4).
We show that the DPE core promoter motif is an important regulatory component in Dorsal target genes in S2R+ cells. The brk and twi core promoters are dependent on the DPE motif and could not be fully activated in S2R+ cells by an added TATA box, whereas the lea core promoter is functionally dependent on the DPE, but its activity could be restored via an added TATA box.
Unlike the results obtained using Drosophila S2R+ cells, in vitro transcription analysis using nuclear extracts derived from Drosophila embryos has demonstrated that the DPE motif, which is important for the transcriptional activity of twi, lea, and brk, can be replaced by a TATA box. This demonstrates the strength of the TATA box, which can restore transcription of some mDPE-containing promoters. Nevertheless, the TATA box is only naturally used in a minority of Dorsal target genes.
Importantly, the mDPE and mDPE + TATA reporter constructs of twi, brk, and lea have comparable strength in vitro, yet they do not have the same strength in S2R+ cells. This suggests that there are additional DPE specificity factors in the cells. The identification of such DPE specificity factor(s) that act in the activation of twi and brk, but not in lea, awaits future investigation. Taken together, our data indicate that the core promoter contributes to the overall transcription levels and adds an important regulatory dimension to the complex dorsal-ventral gene network.
Not All Sequence-specific Transcription Factors Activate Their Target Genes in a Core Promoter Preferential Manner
In this study we have demonstrated that multiple Dorsal target genes are dependent on the DPE and that Dorsal has the ability to preferentially activate some of its targets via the DPE. It is interesting to note that Relish, another Rel family transcription factor, which is important for the activation of the Imd (Immune deficiency) pathway of innate immunity and binds the same or nearly identical DNA sequence motifs as Dorsal, activates transcription in a similar manner to Dorsal, albeit to a lower extent, presumably because of the lack of additional factors that might be missing in the S2R+ cells in which the Imd pathway has not been activated (Fig. 4).
In a separate study, we have observed that the maternal sequence-specific transcription factor Bicoid activates its natural target gene giant, which contains functional TATA box, Inr, and DPE motifs, regardless of its core promoter composition (data not shown). In this study we show that Bicoid activates the mDPE brk reporter (Fig. 5B), highlighting the fact that the ability of Dorsal, Relish, and Caudal to activate transcription of some of their target genes with a preference for the core promoter composition is not a general property of sequence-specific transcription factors; rather, it is a unique feature of these specific transcription factors.
DPE Transcription May Provide a Specialized Transcription System Directed toward Development
The present study demonstrates that the DPE is broadly used in the regulation of genes that mediate the formation of the dorsal-ventral axis during early embryonic development. Furthermore, our findings highlight the importance of the core promoter, in addition to sequence-specific DNA binding motifs in enhancers, in the regulation of gene expression. We have previously demonstrated that the core promoters of the majority of the Hox genes, which are key regulators of the development of the embryonic body plan, contain functional DPE motifs (68). We have also shown that Caudal, a key regulator of the Hox gene network and a sequence-specific enhancer binding transcription factor, activates transcription with a distinct preference for the DPE over the TATA box (68). We now propose that the prevalence of the DPE in developmentally regulated genes provides these genes with certain advantages in the complex regulation of gene expression (e.g. kinetics of responsiveness to specific signals).
The concept of a specialized transcription system has been articulated with regards to the TCT core promoter element, which has been shown to play a key role in a system that is directed toward the synthesis of ribosomal proteins (20). Transcription of TATA-dependent genes is different from transcription of DPE-dependent genes in many respects, e.g. the necessary basal transcription factors, the existence of core promoter-specific enhancers, and the necessity for TBP for TATA transcription as opposed to DPE transcription, where TBP exerts inhibitory effects. Taken together, DPE transcription may be regarded as a specialized transcription system that is directed toward development.
Supplementary Material
Acknowledgments
We thank Jim Kadonaga, Ze'ev Paroush, Doron Ginsberg, Lilach Gilboa, Diana Ideses, Sascha Duttke, Adi Kedmi, Hila Shir, Yehuda Danino, Shani Basch, Anna Sloutskin, and Julia Sharabany for critical reading of the manuscript. Initial experiments were performed in the lab of Jim Kadonaga. We are indebted to Jim for generous support and invaluable suggestions and comments. We thank Chris Benner and Chris K. Glass for sharing the Drosophila transcription start site data. We thank Albert Courey, Norbert Perrimon, and Christos Samakovlis for the generous gift of reagents. We thank Lilach Gilboa, Eileen Furlong, Ze'ev Paroush, Rona Grossman, Offer Gerlitz, Alexandra Lusser, Ron Wides and his lab members, Benny Shilo and his lab members, Joshua Theisen, and Tirza Doniger for invaluable advice and assistance.
This work was supported, in whole or in part, by Israel Science Foundation Grant 798/10 (to T. J.-G.), European Union Seventh Framework Programme Marie Curie International Reintegration Grant 256491 (to T. J.-G.) and National Institutes of Health Grant GM041249 (to James T. Kadonaga). The analysis of the transcriptional activity of Biocoid was supported by United States-Israel Binational Science Foundation Grant 2009428 (to T. J.-G. and James T. Kadonaga).

This article contains supplemental Fig. S1.
- Inr
- initiator
- DPE
- downstream core promoter element
- MTE
- motif 10 element
- TBP
- TATA box-binding protein
- Pol
- polymerase
- GRN
- gene regulatory network
- mDPE
- mutant DPE.
REFERENCES
- 1. Nüsslein-Volhard C., Wieschaus E. (1980) Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801 [DOI] [PubMed] [Google Scholar]
- 2. Pearson J. C., Lemons D., McGinnis W. (2005) Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 6, 893–904 [DOI] [PubMed] [Google Scholar]
- 3. Levine M., Tjian R. (2003) Transcription regulation and animal diversity. Nature 424, 147–151 [DOI] [PubMed] [Google Scholar]
- 4. Muse G. W., Gilchrist D. A., Nechaev S., Shah R., Parker J. S., Grissom S. F., Zeitlinger J., Adelman K. (2007) RNA polymerase is poised for activation across the genome. Nat. Genet. 39, 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zeitlinger J., Stark A., Kellis M., Hong J. W., Nechaev S., Adelman K., Levine M., Young R. A. (2007) RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 39, 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lagha M., Bothma J. P., Levine M. (2012) Mechanisms of transcriptional precision in animal development. Trends Genet. 28, 409–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Spitz F., Furlong E. E. (2012) Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626 [DOI] [PubMed] [Google Scholar]
- 8. Ochoa-Espinosa A., Small S. (2006) Developmental mechanisms and cis-regulatory codes. Curr. Opin. Genet. Dev. 16, 165–170 [DOI] [PubMed] [Google Scholar]
- 9. Malik S., Roeder R. G. (2010) The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bulger M., Groudine M. (2011) Functional and mechanistic diversity of distal transcription enhancers. Cell 144, 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smale S. T., Kadonaga J. T. (2003) The RNA polymerase II core promoter. Annu. Rev. Biochem. 72, 449–479 [DOI] [PubMed] [Google Scholar]
- 12. Juven-Gershon T., Hsu J.-Y., Theisen J. W., Kadonaga J. T. (2008) The RNA polymerase II core promoter: the gateway to transcription. Curr. Opin. Cell Biol. 20, 253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Juven-Gershon T., Kadonaga J. T. (2010) Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 339, 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thomas M. C., Chiang C. M. (2006) The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41, 105–178 [DOI] [PubMed] [Google Scholar]
- 15. Heintzman N. D., Ren B. (2007) The gateway to transcription: identifying, characterizing and understanding promoters in the eukaryotic genome. Cell. Mol. Life Sci. 64, 386–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smale S. T. (2001) Core promoters: active contributors to combinatorial gene regulation. Genes Dev. 15, 2503–2508 [DOI] [PubMed] [Google Scholar]
- 17. Lenhard B., Sandelin A., Carninci P. (2012) Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 13, 233–245 [DOI] [PubMed] [Google Scholar]
- 18. Ohler U., Wassarman D. A. (2010) Promoting developmental transcription. Development 137, 15–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gershenzon N. I., Trifonov E. N., Ioshikhes I. P. (2006) The features of Drosophila core promoters revealed by statistical analysis. BMC Genomics 7, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parry T. J., Theisen J. W., Hsu J.-Y., Wang Y.-L., Corcoran D. L., Eustice M., Ohler U., Kadonaga J. T. (2010) The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery. Genes Dev. 24, 2013–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dikstein R. (2011) The unexpected traits associated with core promoter elements. Transcription 2, 201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goldberg M. L. (1979) Sequence analysis of Drosophila histone genes. Ph.D. Thesis, Stanford University [Google Scholar]
- 23. Reeve J. N. (2003) Archaeal chromatin and transcription. Mol. Microbiol. 48, 587–598 [DOI] [PubMed] [Google Scholar]
- 24. Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C. A., Taylor M. S., Engström P. G., Frith M. C., Forrest A. R., Alkema W. B., Tan S. L., Plessy C., Kodzius R., Ravasi T., Kasukawa T., Fukuda S., Kanamori-Katayama M., Kitazume Y., Kawaji H., Kai C., Nakamura M., Konno H., Nakano K., Mottagui-Tabar S., Arner P., Chesi A., Gustincich S., Persichetti F., Suzuki H., Grimmond S. M., Wells C. A., Orlando V., Wahlestedt C., Liu E. T., Harbers M., Kawai J., Bajic V. B., Hume D. A., Hayashizaki Y. (2006) Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38, 626–635 [DOI] [PubMed] [Google Scholar]
- 25. FitzGerald P. C., Sturgill D., Shyakhtenko A., Oliver B., Vinson C. (2006) Comparative genomics of Drosophila and human core promoters. Genome Biol. 7, R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chalkley G. E., Verrijzer C. P. (1999) DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the Initiator. EMBO J. 18, 4835–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Burke T. W., Kadonaga J. T. (1996) Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 10, 711–724 [DOI] [PubMed] [Google Scholar]
- 28. Burke T. W., Kadonaga J. T. (1997) The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAF(II)60 of Drosophila. Genes Dev. 11, 3020–3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ohler U., Liao G. C., Niemann H., Rubin G. M. (2002) Computational analysis of core promoters in the Drosophila genome. Genome Biol. 3, RESEARCH0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lim C. Y., Santoso B., Boulay T., Dong E., Ohler U., Kadonaga J. T. (2004) The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 18, 1606–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Juven-Gershon T., Cheng S., Kadonaga J. T. (2006) Rational design of a super core promoter that enhances gene expression. Nat. Methods 3, 917–922 [DOI] [PubMed] [Google Scholar]
- 32. Theisen J. W., Lim C. Y., Kadonaga J. T. (2010) Three key subregions contribute to the function of the downstream RNA polymerase II core promoter. Mol. Cell Biol. 30, 3471–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kutach A. K., Kadonaga J. T. (2000) The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell Biol. 20, 4754–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lewis B. A., Sims R. J., 3rd, Lane W. S., Reinberg D. (2005) Functional characterization of core promoter elements: DPE-specific transcription requires the protein kinase CK2 and the PC4 coactivator. Mol. Cell 18, 471–481 [DOI] [PubMed] [Google Scholar]
- 35. Goodrich J. A., Tjian R. (2010) Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat. Rev. Genet. 11, 549–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ohtsuki S., Levine M., Cai H. N. (1998) Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 12, 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Butler J. E., Kadonaga J. T. (2001) Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15, 2515–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Butler J. E., Kadonaga J. T. (2002) The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 16, 2583–2592 [DOI] [PubMed] [Google Scholar]
- 39. Hsu J.-Y., Juven-Gershon T., Marr M. T., 2nd, Wright K. J., Tjian R., Kadonaga J. T. (2008) TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev. 22, 2353–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Willy P. J., Kobayashi R., Kadonaga J. T. (2000) A basal transcription factor that activates or represses transcription. Science 290, 982–985 [DOI] [PubMed] [Google Scholar]
- 41. van Werven F. J., van Bakel H., van Teeffelen H. A., Altelaar A. F., Koerkamp M. G., Heck A. J., Holstege F. C., Timmers H. T. (2008) Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev. 22, 2359–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wampler S. L., Tyree C. M., Kadonaga J. T. (1990) Fractionation of the general RNA polymerase II transcription factors from Drosophila embryos. J. Biol. Chem. 265, 21223–21231 [PubMed] [Google Scholar]
- 43. Soeller W. C., Poole S. J., Kornberg T. (1988) In vitro transcription of the Drosophila engrailed gene. Genes Dev. 2, 68–81 [DOI] [PubMed] [Google Scholar]
- 44. Dushay M. S., Asling B., Hultmark D. (1996) Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc. Natl. Acad. Sci. U.S.A. 93, 10343–10347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pan D. J., Huang J. D., Courey A. J. (1991) Functional analysis of the Drosophila twist promoter reveals a dorsal-binding ventral activator region. Genes Dev. 5, 1892–1901 [DOI] [PubMed] [Google Scholar]
- 46. Jiang J., Levine M. (1993) Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell 72, 741–752 [DOI] [PubMed] [Google Scholar]
- 47. Stathopoulos A., Van Drenth M., Erives A., Markstein M., Levine M. (2002) Whole-genome analysis of dorsal-ventral patterning in the Drosophila embryo. Cell 111, 687–701 [DOI] [PubMed] [Google Scholar]
- 48. Markstein M., Markstein P., Markstein V., Levine M. S. (2002) Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl. Acad. Sci. U.S.A. 99, 763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Levine M., Davidson E. H. (2005) Gene regulatory networks for development. Proc. Natl. Acad. Sci. U.S.A. 102, 4936–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Papatsenko D., Levine M. (2005) Quantitative analysis of binding motifs mediating diverse spatial readouts of the Dorsal gradient in the Drosophila embryo. Proc. Natl. Acad. Sci. U.S.A. 102, 4966–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zeitlinger J., Zinzen R. P., Stark A., Kellis M., Zhang H., Young R. A., Levine M. (2007) Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 21, 385–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bonn S., Furlong E. E. (2008) cis-Regulatory networks during development: a view of Drosophila. Curr. Opin. Genet. Dev. 18, 513–520 [DOI] [PubMed] [Google Scholar]
- 53. Hong J. W., Hendrix D. A., Papatsenko D., Levine M. S. (2008) How the Dorsal gradient works: insights from postgenome technologies. Proc. Natl. Acad. Sci. U.S.A. 105, 20072–20076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Reeves G. T., Stathopoulos A. (2009) Graded dorsal and differential gene regulation in the Drosophila embryo. Cold Spring Harb. Perspect. Biol. 1, a000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thisse C., Perrin-Schmitt F., Stoetzel C., Thisse B. (1991) Sequence-specific transactivation of the Drosophila twist gene by the dorsal gene product. Cell 65, 1191–1201 [DOI] [PubMed] [Google Scholar]
- 56. Jiang J., Kosman D., Ip Y. T., Levine M. (1991) The dorsal morphogen gradient regulates the mesoderm determinant twist in early Drosophila embryos. Genes Dev. 5, 1881–1891 [DOI] [PubMed] [Google Scholar]
- 57. Bodmer R., Jan L. Y., Jan Y. N. (1990) A new homeobox-containing gene, msh-2, is transiently expressed early during mesoderm formation of Drosophila. Development 110, 661–669 [DOI] [PubMed] [Google Scholar]
- 58. Warren J. T., Petryk A., Marqués G., Parvy J. P., Shinoda T., Itoyama K., Kobayashi J., Jarcho M., Li Y., O'Connor M. B., Dauphin-Villemant C., Gilbert L. I. (2004) Phantom encodes the 25-hydroxylase of Drosophila melanogaster and Bombyx mori: a P450 enzyme critical in ecdysone biosynthesis. Insect Biochem. Mol. Biol. 34, 991–1010 [DOI] [PubMed] [Google Scholar]
- 59. Campbell G., Tomlinson A. (1999) Transducing the Dpp morphogen gradient in the wing of Drosophila: regulation of Dpp targets by brinker. Cell 96, 553–562 [DOI] [PubMed] [Google Scholar]
- 60. Jaźwińska A., Kirov N., Wieschaus E., Roth S., Rushlow C. (1999) The Drosophila gene brinker reveals a novel mechanism of Dpp target gene regulation. Cell 96, 563–573 [DOI] [PubMed] [Google Scholar]
- 61. Schimmelpfeng K., Gögel S., Klämbt C. (2001) The function of leak and kuzbanian during growth cone and cell migration. Mech. Dev. 106, 25–36 [DOI] [PubMed] [Google Scholar]
- 62. Crémazy F., Berta P., Girard F. (2000) Sox neuro, a new Drosophila Sox gene expressed in the developing central nervous system. Mech. Dev. 93, 215–219 [DOI] [PubMed] [Google Scholar]
- 63. Schnepp B., Grumbling G., Donaldson T., Simcox A. (1996) Vein is a novel component in the Drosophila epidermal growth factor receptor pathway with similarity to the neuregulins. Genes Dev. 10, 2302–2313 [DOI] [PubMed] [Google Scholar]
- 64. Stathopoulos A., Tam B., Ronshaugen M., Frasch M., Levine M. (2004) pyramus and thisbe: FGF genes that pattern the mesoderm of Drosophila embryos. Genes Dev. 18, 687–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. de Navascués J., Modolell J. (2007) tailup, a LIM-HD gene, and Iro-C cooperate in Drosophila dorsal mesothorax specification. Development 134, 1779–1788 [DOI] [PubMed] [Google Scholar]
- 66. Senger K., Armstrong G. W., Rowell W. J., Kwan J. M., Markstein M., Levine M. (2004) Immunity regulatory DNAs share common organizational features in Drosophila. Mol. Cell 13, 19–32 [DOI] [PubMed] [Google Scholar]
- 67. Copley R. R., Totrov M., Linnell J., Field S., Ragoussis J., Udalova I. A. (2007) Functional conservation of Rel binding sites in drosophilid genomes. Genome Res. 17, 1327–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Juven-Gershon T., Hsu J.-Y., Kadonaga J. T. (2008) Caudal, a key developmental regulator, is a DPE-specific transcriptional factor. Genes Dev. 22, 2823–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.