Background: Mitochondrial health is difficult to assess in vivo.
Results: We have generated a reporter gene, MitoTimer, which targets mitochondria, and fluoresces green and shifts to red when oxidized, for assessment of mitochondrial content, structure, stress, and damage under physiological and pathological conditions.
Conclusion: MitoTimer is useful for assessment of mitochondrial health in vivo.
Significance: MitoTimer could advance mitochondrial research in multiple disciplines.
Keywords: Confocal Microscopy, Exercise, Mitochondria, Oxidative Stress, Skeletal Muscle, High-fat Diet, Reporter Gene
Abstract
Mitochondrial dysfunction plays important roles in many diseases, but there is no satisfactory method to assess mitochondrial health in vivo. Here, we engineered a MitoTimer reporter gene from the existing Timer reporter gene. MitoTimer encodes a mitochondria-targeted green fluorescent protein when newly synthesized, which shifts irreversibly to red fluorescence when oxidized. Confocal microscopy confirmed targeting of the MitoTimer protein to mitochondria in cultured cells, Caenorhabditis elegans touch receptor neurons, Drosophila melanogaster heart and indirect flight muscle, and mouse skeletal muscle. A ratiometric algorithm revealed that conditions that cause mitochondrial stress led to a significant shift toward red fluorescence as well as accumulation of pure red fluorescent puncta of damaged mitochondria targeted for mitophagy. Long term voluntary exercise resulted in a significant fluorescence shift toward green, in mice and D. melanogaster, as well as significantly improved structure and increased content in mouse FDB muscle. In contrast, high-fat feeding in mice resulted in a significant shift toward red fluorescence and accumulation of pure red puncta in skeletal muscle, which were completely ameliorated by voluntary wheel running. Hence, MitoTimer allows for robust analysis of multiple parameters of mitochondrial health under both physiological and pathological conditions and will be highly useful for future research of mitochondrial health in multiple disciplines in vivo.
Introduction
Mitochondrial dysfunction, e.g. reduced respiratory function and increased production of reactive oxygen species (ROS),3 features prominently in cardiovascular, metabolic, and neurodegenerative diseases, and other pathological conditions (1–9). Despite the importance of mitochondrial dysfunction in a myriad of pathological conditions, very few effective drugs and interventions are available for their prevention and treatment. One effective intervention to improve mitochondrial quality and quantity is healthy lifestyle interventions, in particular regular exercise (10–13). Our understanding of the mechanisms underlying the pathologies of the diseases and the benefits of exercise is obscure due to a lack of a reliable technology for a robust assessment of mitochondrial health in vivo.
Several fluorescence-based reporter genes have recently been developed for monitoring mitochondrial ROS production and redox state (14–17). Wolf et al. (14) developed mitochondria-targeted roGFP for assessing changes of redox state, which has been used in Drosophila (15). Two other reporter genes, HyPer and mt-cpYFP, have also been developed to detect H2O2 and superoxide in mitochondria, respectively (16, 17). These reagents are extremely useful for real-time measurement in cultured cells as they report instant changes in ROS production and redox state, but cannot be readily used for in vivo measurements because the readouts change instantaneously. Transmission electron microscopy is currently used for assessing mitochondrial quantity and structural integrity in whole organisms; however, the use of transmission electron microscopy is limited due to the high monetary and labor costs and the contingency on specialized equipment. It is therefore of great value to develop tools for assessing mitochondrial health with the ability to trace their history.
To this end we engineered a pMitoTimer reporter gene by targeting a fluorescent Timer protein to mitochondria by adding the mitochondrial targeting sequence of the cytochrome c oxidase subunit VIII gene to the N terminus of the coding region of Timer, under control of the constitutive CMV promoter. Timer encodes a DsRed mutant (DsRed1-E5) that fluoresces like green fluorescence protein when newly synthesized, and shifts the fluorescent spectrum irreversibly to red (18) following a form of oxidation (dehydrogenization) of the Tyr-67 residue (19, 20). This physical property in a biological environment makes it suitable for reporting the age of proteins and cells (18), and thus was named “Timer.” Recently, Ferree et al. (21) and Hernandez et al. (22) have used inducible MitoTimer reporter to show its usefulness in cell culture to report changes in mitochondrial turnover and transport. Because the fluorescence spectrum shifts upon oxidation, we predicted that MitoTimer driven by a constitutively active promoter would report cumulative redox history of the labeled mitochondria. We also provide additional value by using MitoTimer in vivo in transgenic Caenorhabditis elegans and Drosophila melanogaster as well as following somatic gene transfer in adult mouse skeletal muscle.
Here we have validated the reporter properties and confirmed that MitoTimer could be used to report mitochondrial content, structure, stress, and damage in vivo under physiological and pathological conditions. We also used MitoTimer to address a biological question of whether exercise training was protective against HFD-induced mitochondrial damage in mouse skeletal muscle. Our findings provide morphological evidence demonstrating HFD-induced mitochondrial defects in vivo using two independent methods (transmission electron microscopy and MitoTimer fluorescence), and the benefits of exercise training for mitochondrial health. This reporter gene can be potentially used in multiple disease models, and its applications, beyond this study, will likely lead to advancement in multiple disciplines of mitochondrial health research.
EXPERIMENTAL PROCEDURES
Plasmid DNA Construct
We constructed pMitoTimer by inserting the BamHI-NotI fragments of pTimer-1 (Clontech) into pDsRed2-Mito (Clontech) vector digested with BamHI and NotI using T4 ligase (New England Biolabs) following DNA isolation by using the Qiaex II gel extraction kit (Qiagen). The mitochondrial targeting sequence of the human cytochrome c oxidase subunit VIII gene from pDsRed2-Mito was then fused to the mutant DsRed1-E5 on the N terminus. Bacterial colonies were screened using a Qiaprep Spin Miniprep Kit (Qiagen) with restriction mapping and transfection in C2C12 myoblasts to confirm the correct construction of the plasmid DNA. Plasmid DNA pLamp1-YFP was purchased (Addgene).
Myoblast Transfection
Mouse C2C12 myoblasts were cultured on glass coverslips in normal growth medium (Dulbecco's modified Eagle's medium with high glucose containing 20% fetal bovine serum) and maintained at 37 °C with 5% CO2. The cells were transfected with pMitoTimer, pTimer, pDsRed-Mito, pGFP3, or the empty vector, pCI-neo (Promega). Briefly, the cells were incubated with a mixture of 1 μg of plasmid DNA and 3 μl of Lipofectamine (Invitrogen) in Opti-MEM media for 5 h at 37 °C before changing to normal growth medium with an overnight incubation. The cells were washed with PBS twice and fixed with 4% paraformaldhyde for 15 min on ice followed by 2 × 5-min washes with ice-cold phosphate-buffered saline (PBS). The coverslips were mounted on glass slides, and images were acquired by confocal microscopy (Olympus) for both green (excitation/emission 488/518 nm) and red (excitation/emission 543/572 nm) channels.
Western Blotting
Mitochondrial and cytosolic fractions were isolated from transfected C2C12 myoblasts using a mitochondrial isolation kit (Thermo Fisher Scientific Inc.). These fractions were subjected to SDS-PAGE and transferred onto PVDF membrane. The membrane was probed with the following primary antibodies: rabbit anti-DsRed (Clontech), mouse anti-COX IV (Invitrogen), and mouse anti-GAPDH (Cell Signaling). Secondary antibodies were goat anti-mouse IR680 and goat anti-rabbit IR800 (LICOR). Membranes were scanned using the Odyssey infrared imaging system (LICOR).
Drug Treatment of Transfected Cells
pMitoTimer-transfected C2C12 myoblasts were treated with 10 μm rotenone (electron transport chain complex I inhibitor; Sigma), 100 nm antimycin A (complex III inhibitor; Sigma), or 100 μm paraquat (mitochondrial ROS producer primarily through complex I) in normal growth medium for 6 h. In experiments using diethylenetriamine-nitric oxide, cells were pre-treated with 200 μm diethylenetriamine-NO for 1 h prior to co-treatment with either antimycin A or dimethyl sulfoxide for 6 h. Cells were fixed and mounted immediately following drug treatment for confocal microscopy analysis. To assess the effect of the drug treatment to mitochondrial function, C2C12 myoblasts were treated with the same drugs for 6 h prior to measurement of the oxygen consumption rate (OCR) using a Seahorse XF-24 Flux Analyzer (Seahorse Biosciences, Billerica, MA). Briefly, prior to the assay, the media was changed to 5 mm HEPES-buffered Seahorse medium (Invitrogen) (pH 7.4) and equilibrated for 30 min at 37 °C. Oligomycin (Oligo, 1 μm), BAM15 (23) (2 μm), and rotenone and antimycin A (1 and 10 μm, respectively) were injected sequentially during the assay, whereas OCR was measured at 3-min intervals. OCR was normalized to α-Tubulin protein abundance in harvested cells following completion of the assay. Basal mitochondrial OCR, ATP-linked OCR, and reserve capacity were calculated.
D. melanogaster Strains and Drug Treatment
Wild type (w1118) and Mef2-Gal4 fly lines were obtained from the Bloomington Drosophila Stock Center (Indiana University, IN). Flies were housed in a humidified, temperature-controlled incubator at 25 °C on a 12:12-h light-dark cycle. To generate the MitoTimer transgenic fly line, the coding region of MitoTimer was subcloned into pUAS vector. A UAS-MitoTimer transgenic fly line was established commercially (Genetic Services, Inc.). UAS-MitoTimer flies were crossed with Mef2-Gal4 to obtain Mef2-Gal4>UAS-MitoTimer flies. Adult female flies were collected under light CO2 anesthesia and transferred to fresh medium every 2–3 days (20 per vial). To induce oxidative stress in Drosophila, we prepared the drugs as following. Antimycin A was dissolved in 100% ethanol at 1 mm and then added to 5% sucrose at 0.1 mm. Paraquat was dissolved in water at 100 mm and added to 5% sucrose at 10 mm. These two drugs were administered to adult flies (20 days old) 24 h after 6 h of fasting as described by Lee et al. (24). Rotenone was first dissolved in dimethyl sulfoxide at 4 mm, added to freshly made fly food at 60 °C at 0.4 mm, and used to feed the flies for 4 days after 6 h of fasting (25).
Exercise Training and Negative Geotaxis Assays in Flies
Exercise training of UAS-MitoTimer:Mef2-Gal4 flies were performed according to a previously reported method (26). Negative geotaxis was assessed in Rapid Negative Geotaxis (RING) assays in groups of 100 flies as described (27). Flies were tested 5 times per week for 5 weeks to assess the decline in negative geotaxis speed with age. Climbing endurance was measured using the fatigue assay as previously described (28).
Imaging Acquisition of Adult Fly Heart
Adult fly hearts were dissected according to an established method (29) and fixed in 4% paraformaldehyde for 20 min. Confocal Z-stack images were obtained within abdominal segment A1 with a set of fixed acquisition parameters. One set of images per heart, both green (excitation/emission 488/518 nm) and red (excitation/emission 543/572 nm) channels, were selected for analysis that was 6–8 nm from the top section.
C. elegans Experiments
Pmec-3MitoTimer was constructed by digesting pPD57.56 with BamHI and NheI and inserting a PCR fragment that contained the DsRed-2 mitochondrial portion of pTimer-1 also cut with BamHI and NheI. The sequence-confirmed plasmid was injected into wild type C. elegans to generate strain ZB4043 bzEx217[Pttx-3mCherry; Pmec-3MitoTimer] in which MitoTimer was expressed in 10 neurons including the touch receptor neurons. We imaged using a Zeiss Axoivert 200M upright microscope with deconvolution.
Mouse Model
Male C57BL/6J mice (7–8 weeks old) were obtained commercially (Jackson Laboratory) and housed in temperature-controlled (21 °C) quarters with a 12:12-h light-dark cycle and water and chow (Purina) provided ad libitum. All experimental protocols were approved by the University of Virginia, Institutional Animal Care and Use Committee. pMitoTimer and pLamp1-YFP transfection by somatic gene transfer in the adult mouse FDB muscle was performed as previously published (30). Briefly, the mice were anesthetized (isofluorane) and the base of each hind limb foot, where the flexor digitorum brevis (FDB) muscle is located, was injected with 10 μl of hyaluronidase (0.36 mg/ml; Sigma) subcutaneously. One h later, the mice were anesthetized a second time and the FDB injected with 20 μg of pMitoTimer in saline. In the case of co-transfection, pMitoTimer and pLamp1-YFP were mixed in a 1:1 ratio, and 20 μg of plasmid DNA was injected. Following another 10 min, a pair of gold-plated acupuncture needles were inserted under the skin at the heel of the foot and the base of the toes parallel to each other and perpendicular to the long axis of the foot. Ten pulses with 20 ms duration at 1 Hz and 75 V/cm were applied to each foot.
Diet and Exercise Interventions
Following somatic gene transfer, the mice were allowed to recover for 10 days and were then allocated to one of the following groups (n = 6); sedentary normal chow (NC-Sed), sedentary with 60% high-fat diet (HFD-Sed); Research Diets Inc.), voluntary wheel running with NC (NC-Ex), or voluntary wheel running with 60% HFD (HFD-Ex) for 3 weeks. In a separate experiment, the mice (n = 6) were allocated to either sedentary (Sed) or voluntary wheel running (Ex) for 6 weeks. All mice were housed individually in cages with running wheels, and the running wheels were locked for the mice allocated to the sedentary groups. At the end of the studies, all mice were humanely sacrificed under anesthesia, and FDB muscles were carefully excised and immediately fixed in 4% paraformaldehyde for 20 min.
Immunofluorescence
For detection of Cox4, pMitoTimer-transfected FDB muscles were processed for paraffin embedding as described (31). Briefly, harvested muscles were placed in 10% formalin at room temperature overnight. Samples were transferred to 70% ethanol before being paraffin-embedded and sectioned by a microtome. The sections were incubated in a dry 37 °C incubator overnight before being deparaffinized and rehydrated in a series of xylene and ethanol washes. Muscle sections (5 μm) were then washed for 10 min in Tris-buffered saline (TBS) with 0.1% Tween 20, followed by incubating with blocking buffer (10% normal goat serum in TBS) for 2 h at room temperature. Sections were incubated with mouse anti-Cox4 antibody (Invitrogen A21348, 1:100) in TBS overnight at 4 °C, washed for 10 min in 0.1% Tween 20 TBS, and incubated for 1 h at room temperature with goat anti-mouse Cy5 antibody (Jackson 115-175-146, 1:50) followed by mounting with Pro-Long Gold Antifade Reagent (Invitrogen) for confocal microscopy.
For detections of LC3, pMitoTimer-transfected FDB muscles were immediately fixed in 4% paraformaldehyde for 20 min and incubated overnight in PBS with 0.2% saponin at 4 °C. Muscles were then incubated in blocking solution (PBS with 0.5% BSA, 0.2% saponin, 3% normal goat serum, and 3% normal rat serum) for 1 h at room temperature, followed by overnight incubation with rabbit anit-LC3 (Novus Biologics, NB100-2220; 1:300) in blocking solution at 4 °C. Following 5 × 20-min washes in PBS with 0.2% saponin, muscles were incubated with goat anti-rabbit Cy5 antibody (Jackson 111-175-144, 1:200) for 2 h at room temperature and washes were repeated. Muscles were then whole mounted and imaged by confocal microscopy as described.
Confocal Microscopy
Immediately following fixation (described above) the muscles were whole mounted on gelatin-coated glass slides with 50% glycerol in PBS with a coverslip. MitoTimer images were acquired at ×100 magnification under a confocal microscope (Olympus Fluoview FV1000) using the green (excitation/emission 488/518 nm) and red (excitation/emission 543/572 nm) channels with identical predetermined acquisition parameters for all samples to ensure no saturation of the signals and similar intensity of the green and red channels in control samples. Lamp1-YFP images were acquired using the yellow channel (excitation/emission 515/527 nm), whereas LC3 and Cox4 immunofluorescence (Cy5) was detected in the far-red spectrum (excitation/emission 635/672 nm).
Ratiometric Analysis
MitoTimer signals were analyzed using our custom-designed Matlab-based algorithm. Briefly, positive pixels were thresholded at a value of 1.5 times the mean gray value of the above background pixels for each channel. This made the threshold robust against differences in fiber size between images. Saturated pixels (gray level = 255) were removed from analysis. In addition, the pixels with a red to green ratio of >2.5 were removed for the ratiometric analysis. The ratio of red to green was calculated as a mean value of red to green ratio for all the remaining positive pixels in the image (Fig. 2c). The number of red dots in the images was calculated by counting areas with clusters of ≥5 pixels with high fluorescence signal (>175) and a red to green ratio of >2.5. The red dot counting was done on images including saturated pixels (Fig. 2d).
FIGURE 2.
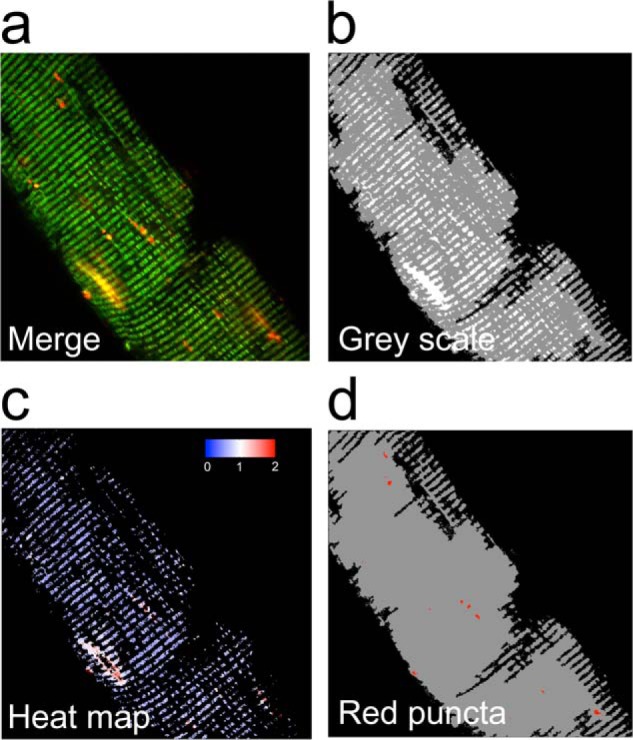
Computational analysis of MitoTimer fluorescence. A Matlab-based computer algorithm was designed for identification of MitoTimer positive pixels, quantification of MitoTimer fluorescence spectrum shift between green and red, and accumulation of pure red puncta. a, a representative confocal image (merged image of green and red) of a mouse FDB muscle fiber; b, the processed confocal image showing background (black), fiber (gray), and MitoTimer (white) pixels; c, corresponding heat map of the red:green ratio of MitoTimer positive pixels; and d, pure red puncta identified by the MitoTimer image analysis algorithm.
Transmission Electron Microscopy
Transmission electron microscopy was performed as previously described (32).
Statistical Analysis
The data were analyzed using a t test or one-way analysis of variance with Student's Newman-Kuels post hoc where appropriate. Data are presented as mean ± S.E. and statistical significance was set at p < 0.05.
RESULTS
MitoTimer Protein Targets to Mitochondria and Displays Heterogeneous Green and Red Fluorescence
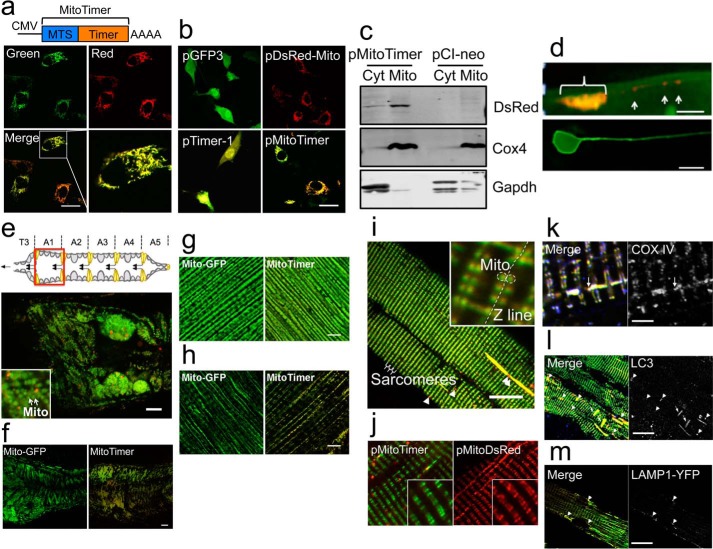
We engineered the MitoTimer reporter gene to target the oxidation-sensitive Timer protein to mitochondria. To confirm the subcellular location, we transiently transfected C2C12 myoblasts with plasmid pMitoTimer (Fig. 1a) and compared the expression pattern with those of pGFP3, pDsRed-Mito, and pTimer-1 (Fig. 1b). Cells transfected with pMitoTimer showed typical mitochondrial network structures (Fig. 1a) identical to those transfected with pDsRed-Mito (Fig. 1b). Cells transfected with pMitoTimer also showed heterogeneous green and red fluorescence (Fig. 1a, panel labeled Merge), similar to that of pTimer-1 (Fig. 1b). Mitochondrial targeting of the protein was further confirmed by Western blot of the mitochondrial and cytosolic fractions of C2C12 myoblasts transiently transfected with pMitoTimer compared with cells transfected with an empty vector, pCI-neo (Fig. 1c). MitoTimer protein was highly enriched in the mitochondrial fraction as probed by anti-DsRed antibodies (Fig. 1c). To validate these findings in vivo, we generated transgenic C. elegans with touch neuron-specific expression under control of the neuron-specific promoter mec-4 and observed green and red mitochondria in the soma and distributed through the sensory neuron process (Fig. 1d), which is similar to mitochondrial targeted standard GFP as reported previously (33). We also generated UAS-MitoTimer transgenic flies and crossed with Mef2-Gal4 transgenic flies to generate Mef2-Gal4>UAS-MitoTimer flies with muscle expression (34). Confocal microscopy showed mitochondrial structure identical to that obtained in Mef2-Gal4>UAS-Mito-GFP flies not only in adult heart tube (Fig. 1, e and f), but also in other muscles, such as indirect flight muscle and larval body wall muscle (Fig. 1, g and h). There were variable green and red fluorescence in different regions of the heart tube. In addition, we detected pure red fluorescent puncta, which were often disconnected from, but in close proximity to, the mitochondrial reticulum (Fig. 1e).
FIGURE 1.
Genetic engineering and expression of the MitoTimer reporter gene and mitochondrial targeting of the MitoTimer protein. Confocal microscopy analysis of MitoTimer reporter gene expression was performed for transiently transfected C2C12 myoblasts, transgenic C. elegans, Drosophila, and transfected adult mouse FDB muscle by somatic gene transfer. a, diagram of the pMitoTimer construct (top) and its expression (bottom) by transient transfection of C2C12 myoblasts showing heterogeneous intensities of green and red fluorescence. An enlarged image (right lower corner) shows a cell with mitochondrial network-like structure. MTS, mitochondrial targeting sequence. Scale bar = 10 μm. b, expression patterns in C2C12 cells transfected with pGFP3, pDsRed-Mito, pTimer-1, and pMitoTimer. Note the cytosolic expression pattern of pGFP3 and pTimer-1 and mitochondrial expression pattern of pDsRed-Mito and pMitoTimer. Scale bar = 10 μm. c, Western blot of cytosolic (Cyt) and mitochondrial (Mito) fractions of cells transfected with pMitoTimer or pCI-neo probed with anti-DsRed showing mitochondria-specific expression in cells transfected with pMitoTimer. Cox4 enrichment in the mitochondrial fraction and Gapdh in the cytosolic fraction indicate successful cell fractionation. d, expression of MitoTimer in a C. elegans touch neuron with mitochondria concentrated in the cell body (bracket) and individual mitochondria along the process (arrows) in ZB4043 bzEx217[Pttx-3mCherry; Pmec-3MitoTimer] transgenic C. elegans (top panel). The cell body has higher green content of mitochondria (yellow composite color for MitoTimer signal). The bottom panel shows the neuronal cell body and its process visualized by the cytoplasmic GFP fluorescence in zdIs5[pmec-4GFP] transgenic nematodes. Scale bar = 4 μm. e, expression of MitoTimer in A1 segment of heart tube (a schematic illustration shown above) of Mef2-Gal4>UAS-MitoTimer transgenic Drosophila (merged image of green and red). Scale bar = 10 μm. Mitochondria are indicated by arrows in the enlarged image as an inset. Expression patterns of Mito-GFP and MitoTimer in the A1 segment of adult heart tube (f), adult indirect flight muscle (g), and larval body wall muscle (h) in transgenic Drosophila. Scale bar = 10 μm. i, whole mount-fixed FDB fibers in adult mice 11 weeks after electric pulse-mediated gene transfer of pMitoTimer (merged image of green and red). Sarcomeres are marked, and pure red puncta are indicated by arrows. Scale bar = 20 μm. Mitochondria are labeled by dashed circles in the enlarged image as an insert on either side of the Z-line. j, somatic gene transfer of pDsRed-Mito and pMitoTimer in adult mouse FDB fibers. Scale bar = 10 μm. k, Cox4 immunofluorescence of FDB muscle transfected with pMitoTimer showing co-localization of MitoTimer and Cox4 in some (arrow) but not all red puncta. Scale bar = 2 μm. l, LC3 whole mount immunofluorescence of FDB muscle transfected with pMitoTimer showing co-localization of MitoTimer and LC3 in some (large arrowheads) but not all red puncta and yellow subsarcolemmal mitochondrial (small arrows). Scale bar = 20 μm. m, somatic gene transfer of pLamp1-YFP with pMitoTimer in adult mouse FDB muscle showing co-localization of MitoTimer and Lamp1 in red puncta (arrowheads). Scale bar = 20 μm.
To further validate the use of the MitoTimer reporter gene in vivo, we performed electric pulse-mediated gene transfer in adult mouse FDB muscle followed by whole mount confocal microscopy. FDB muscle fibers transfected with pMitoTimer showed a striking sarcomeric striation pattern at low magnification (Fig. 1i) identical to that in FDB muscle transfected with pDsRed-Mito (Fig. 1j). High magnification images showed labeling of inter myofibrillar mitochondria on each side of the Z-line as well as subsarcolemmal mitochondria with streaking patterns (Fig. 1i). These patterns have also been shown by the mitochondria-targeted reporter gene mt-cpYFP (35). Mitochondria targeting of the MitoTimer protein was further confirmed by co-localization of fluorescence with immunofluorescence staining of Cox4 (Fig. 1k). Similar to the fly heart tube, we also detected pure red puncta in mouse skeletal muscle transfected with pMitoTimer and confirmed that they were often positive for Cox4, suggesting that they are of mitochondrial origin and are likely degenerative mitochondria (Fig. 1k). In addition we found that some red puncta and yellow fluorescent subsarcolemmal mitochondria co-localized with LC3 immunofluorescence, a marker for autophagasome (Fig. 1l), as well as a YFP signal from pLamp1-YFP transfection (Fig. 1m), a maker of lysosome. The findings strongly suggest that red puncta were mitochondria targeted for removal from the cell via autophagy.
Quantification of Fluorescence Spectrum Shift and Pure Red Puncta
We developed a Matlab-based automated image analysis platform for quantification of fluorescence of each channel that was acquired with the same parameters of laser intensity and scanning speed for all samples under the confocal microscope. In the case of transfected FDB muscles, this Matlab-based computer algorithm segmented the muscle fiber and identified MitoTimer positive pixels within the fiber using a threshold value of 1.5 times the mean gray value above background pixels for each channel (Fig. 2, a and b). This processing controlled for differences in fiber number and size between images. The shift of the fluorescence spectrum was calculated by taking the ratio of red to green fluorescence intensity of each of the positive pixels (Fig. 2c). The algorithm also quantified the number of pure red puncta (Fig. 2d), which were defined as more than 5 pixels in contact with each other with a high fluorescence signal (>175) and a red to green ratio of >2.5. With this automated image analysis algorithm we were able to quantitatively characterize the fluorescent properties of MitoTimer signals in various model systems quickly and reproducibly. The algorithm can process 100 pairs of images in ∼30 s.
MitoTimer Protein Changes Fluorescence from Green to Red under Conditions of Mitochondrial Stress
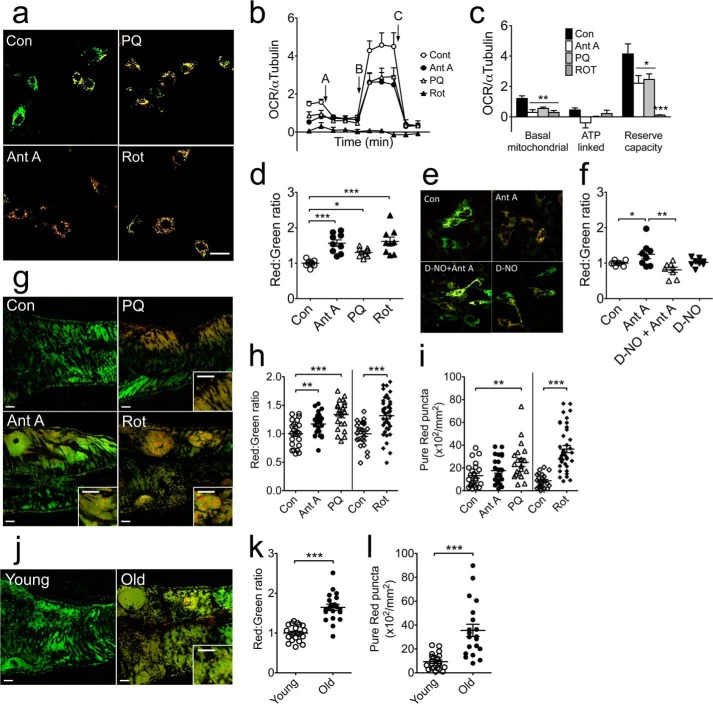
To determine whether mitochondrial stress promotes a shift of the fluorescent spectrum from green to red, we treated pMitoTimer-transfected C2C12 myoblasts with rotenone (10 μm), antimycin A (100 nm), or paraquat (100 μm) for 6 h. These drugs are known to induce mitochondrial ROS production from complex I or complex III of the electron transport chain. A ratiometric algorithm showed that treatment with any of these drugs resulted in a significant (p < 0.05) shift of mitochondrial fluorescence toward red (Fig. 3, a and d). The drug treatments significantly reduced mitochondrial respiratory function as measured by a mitochondrial stress test using a Seahorse oxygen flux analyzer. Specifically, these three drug treatments all resulted in reduced basal mitochondrial respiration and reserve capacity along with a trend of reduced ATP-linked respiration (Fig. 3, b and c). To further confirm that the change in MitoTimer fluorescence in cells is due to ROS production we showed that pre-treatment with diethylenetriamine-NO (200 μm) was sufficient to prevent the antimycin A-induced fluorescence shift (Fig. 3, e and f). When adult Mef2-Gal4>UAS-MitoTimer transgenic flies (20 days of age) were treated with these drugs supplemented in feed, there was also a significant shift toward red fluorescence in the heart tube concurrent with increases of pure red puncta (Fig. 3, g–i). Similar changes were observed in aged (70 days old) compared with young (7 days old) flies (Fig. 3, j–l). These findings support the use of the MitoTimer reporter gene to report mitochondrial stress and damage in vitro and in vivo.
FIGURE 3.
MitoTimer shifts fluorescence spectrum in response to mitochondrial stress in C2C12 myoblasts and Drosophila heart tube. Confocal microscopy analysis of MitoTimer reporter gene expression was performed for transiently transfected C2C12 myoblasts and transgenic Drosophila heart tube following treatment or feeding, respectively, with drugs that are known to cause mitochondrial stress. a, representative merged images of pMitoTimer-transfected C2C12 myoblasts (scale bar = 10 μm). b, OCR normalized to α-tubulin during a Seahorse mitochondrial stress test. Baseline OCR was measured twice prior to addition of oligomycin (A) to determine ATP-dependent OCR followed by addition of the mitochondrial uncoupler BAM15 (B) for measurement of maximal capacity and finally addition of antimycin A and rotenone (C) for non-mitochondrial OCR. c, quantification of basal mitochondrial, ATP linked, and reserve capacity OCR normalized to α-Tubulin protein during a Seahorse mitochondrial stress test. d, quantification of the red:green fluorescence intensities (n = 9) in transfected C2C12 cells following treatment with rotenone (Rot, 10 μm), antimycin A (Ant A, 100 nm), or paraquat (PQ, 100 μm) for 6 h. e, representative merged images of pMitoTimer-transfected C2C12 myoblasts (scale bar = 10 μm); and f, quantification of red:green fluorescence intensities following treatment with dimethyl sulfoxide or diethylenetriamine-NO (D-NO, 200 μm) for 1 h followed by either dimethyl sulfoxide (Con), antimycin A (Ant A, 100 μm), D-NO + Ant A, or D-NO alone for 6 h. g, representative merged images of Drosophila heart tube MitoTimer signals (scale bar = 10 μm). h, quantification of the red:green fluorescence intensity. i, quantification of number of pure red puncta in the A1 heart tube segment of Drosophila following treatment with rotenone (0.4 mm in food for 4 days, n = 39), paraquat (10 mm in 5% sucrose for 24 h, n = 21), or antimycin A (0.1 mm in 5% sucrose for 24 h, n = 25). Controls were either normal food for 4 days (n = 24) or sucrose for 24 h (n = 23). j, representative images of MitoTimer in Drosophila heart tube (scale bar = 10 μm). k, quantification of the red:green fluorescence intensity. l, quantification of pure red puncta from 7-day-old flies (n = 21) compared with 70-day-old flies (n = 20). Data represent mean ± S.E. *, **, and *** denote p < 0.05, 0.01, and 0.001, respectively.
MitoTimer Reports the Beneficial Impact of Exercise on Mitochondria in Vivo
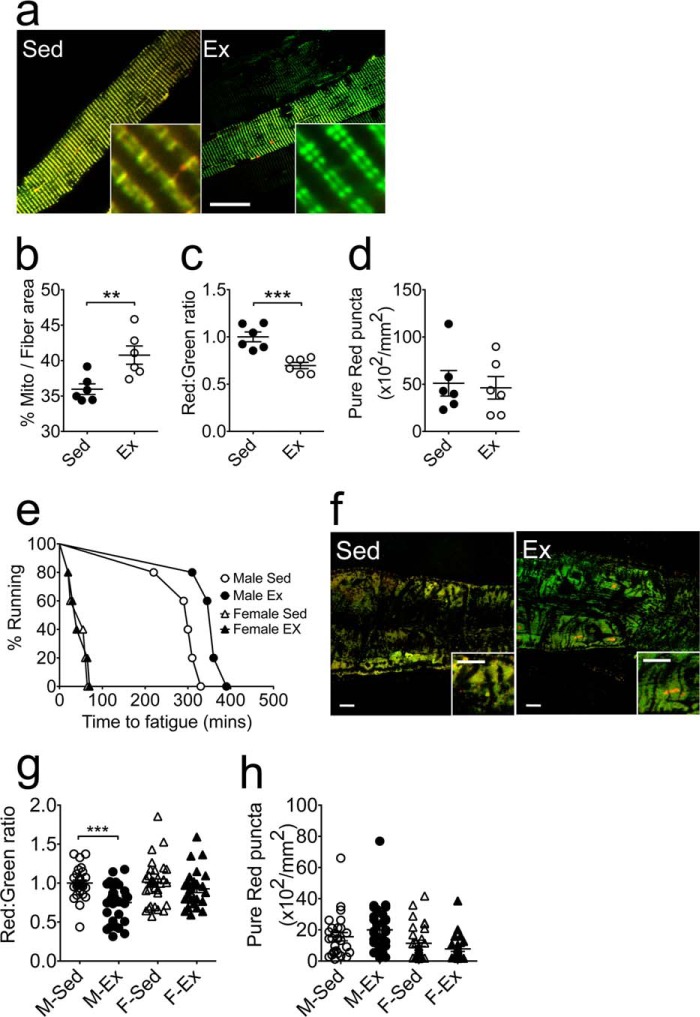
It is known that endurance exercise training increases mitochondrial quantity and improves mitochondrial quality in skeletal muscle (36). To determine whether the MitoTimer reporter gene could be used for this purpose, we performed somatic gene transfer of pMitoTimer in mouse FDB muscles followed by voluntary wheel running for 6 weeks. Whole mount confocal microscopy showed that exercise training promoted a significant increase in mitochondrial content per fiber area (Fig. 4, a and b), as well as a shift of the fluorescence toward green (Fig. 4, a and c), indicative of reduced mitochondrial stress. There was no significant change in the number of pure red puncta (Fig. 4d). Under higher magnification, we also observed improved mitochondrial alignment and uniformity of mitochondrial pairs along the Z-line in FDB fibers from exercised mice compared with those of sedentary mice (Fig. 4a). We then employed exercise training in flies using a unique training device known as the power tower (26). Exercise training in male flies resulted in improved exercise capacity (Fig. 4e) associated with a significant shift of the MitoTimer fluorescence toward green (Fig. 4, f and g) with no significant changes in pure red puncta in the heart tube (Fig. 4, f and h).
FIGURE 4.
MitoTimer reports a beneficial impact of exercise training on mitochondria in vivo. Confocal microscopy analysis of MitoTimer reporter gene expression was performed for somatic gene transfer-transfected FDB muscle and transgenic Drosophila heart tube following exercise training by voluntary running exercise and induced negative geotaxis, respectively. a, representative merged images of pMitoTimer-transfected adult mouse FDB fibers following 6 weeks of voluntary wheel running (Ex) with sedentary mice as control (Sed). Scale bar = 20 μm. b, quantification of the percent of MitoTimer positive pixels per fiber area. c, quantification of the red:green fluorescence intensity under each condition (n = 6). d, quantification of the number of pure red puncta. Each value is the average of 10–15 images per mouse; e, time to fatigue curve of male and female flies following 3 weeks of exercise training on the Power Tower compared with sedentary controls. f, representative merged images of MitoTimer in Drosophila heart tubes from male flies following exercise training. Scale bar = 10 μm. g, quantification of the red:green fluorescence intensity in male (Sed, n = 27; Ex, n = 32) and female (Sed, n = 30; Ex, n = 26) Drosophila heart tubes; and h, quantification of pure red puncta. Data represent mean ± S.E. ** and *** denote p < 0.01 and 0.001, respectively.
MitoTimer Reports Mitochondrial Stress and Damage under Physiological and Pathological Conditions in Vivo
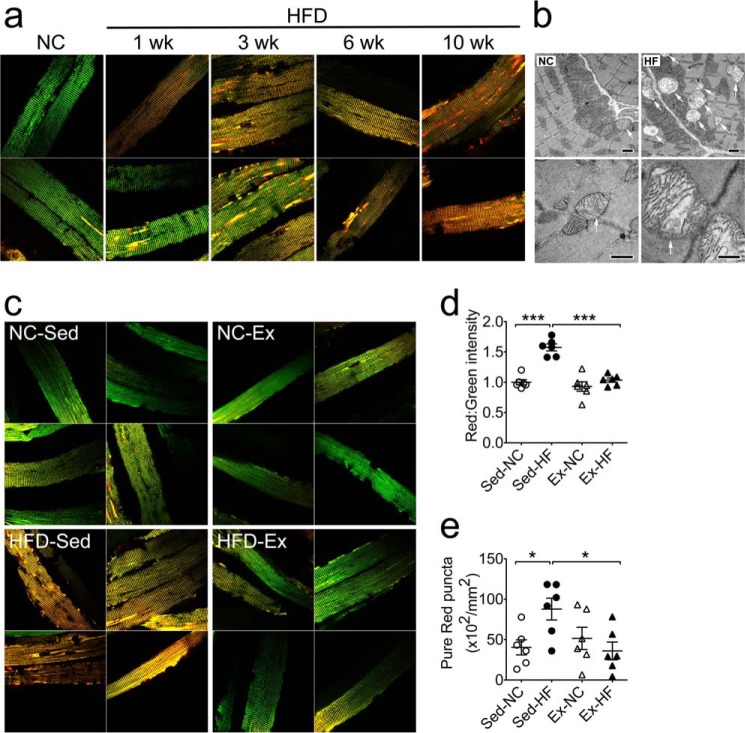
To determine the utility of the MitoTimer reporter gene in a mammalian model for mitochondrial stress and damage under pathological conditions, we subjected mice transfected with pMitoTimer in the FDB muscles to a 60% HFD for up to 10 weeks. Confocal microscopy of whole mounted fixed muscles showed a considerable shift in fluorescence toward red in mice on the HFD along with a substantial increase of pure red puncta compared with mice on normal chow diet (Fig. 5a). These changes appear to be time-dependent as shown by increasing severity in a time course at 1, 3, 6, and 10 weeks (Fig. 5a). Transmission electron microscopy confirmed that the HFD resulted in accumulation of damaged mitochondria with various degrees of enlargement and vacuolization in skeletal muscles (Fig. 5b). We then set to determine whether exercise training could prevent these changes and found that the shift of the fluorescence toward red with increased pure red puncta following 3 weeks of HFD were completely prevented by voluntary wheel running (Fig. 5, c–e). Taken together, we have shown that the MitoTimer reporter gene encodes an oxidation-sensitive protein that targets to mitochondria and reports in vitro and in vivo mitochondrial health under physiological and pathological conditions.
FIGURE 5.
MitoTimer reports mitochondrial stress and damage under physiological and pathological conditions in vivo. Confocal microscopy analysis of the MitoTimer reporter gene was performed for somatic gene transfer-transfected FDB muscle following high-fat diet feeding with/without voluntary running exercise in mice. a, representative merged images of pMitoTimer-transfected adult mouse FDB fibers following 10 weeks of feeding on normal chow (NC), 1, 3, 6, and 10 weeks of feeding on 60% HFD (n = 2 per time point). Scale bar = 20 μm. b, transmission electron microscope images of longitudinal sections of mouse plantaris muscle following 12 weeks of a high-fat diet showing enlargement and accumulation of damaged mitochondria (arrows), which were occasionally detected in normal chow fed mice. Scale bar = 1.0 μm. c, representative merged images of pMitoTimer-transfected adult mouse FDB fibers following normal chow with sedentary cage activity (NC-Sed), NC with 3 weeks of exercise training (NC-Ex), 60% HFD with sedentary cage activity (HFD-Sed), or 60% HFD with exercise training (HFD-Ex). Scale bar = 20 μm. d, quantification of the red:green fluorescence intensity under each condition (n = 6). Each value is the average of 10–15 images per mouse. e, quantification of number of pure red puncta. Data represent mean ± S.E. * and *** denote p < 0.05 and 0.001, respectively.
DISCUSSION
Mitochondrial dysfunction is one of the most important features of many prevalent diseases (1–9). For example, reduced mitochondrial content and electron transport chain activity in skeletal muscles have been reported both in obese, diabetic patients (37, 38) and in lean, insulin-resistant offspring of type 2 diabetic patients (2, 39), suggesting its importance in the pathogenesis. Accumulation of damaged mitochondria, due to lipotoxicity, in cardiac myocytes has been reported in a rat genetic model of obesity (4). Mitochondrial DNA damage and dysfunction are associated with heart failure induced by myocardial infarction (40). Abnormal regulation of genes essential for mitochondrial function is sufficient to induce heart failure in animals (5). Finally, there is accumulating evidence of loss of the mitochondrial function in the central nervous system in neurodegenerative diseases (3, 6, 41). It is clear that mitochondrial damage and dysfunction is detrimental, if not causal, to the aforementioned disease conditions. More importantly, mitochondrial abnormalities promote excess production of reactive oxygen species in a vicious cycle leading to further mitochondrial damage (42). In light of the importance of mitochondria in health and disease, there is an urgent need to develop appropriate animal models to improve our understanding of mitochondrial pathology and develop interventions to improve mitochondrial function.
This study has taken critical steps to advance our ability to measure mitochondrial stress and damage through validation of MitoTimer under physiological and pathological conditions in vitro and in vivo. We believe that the MitoTimer reporter gene will be highly useful for future in vivo research because it allows for robust measurement of multiple mitochondrial parameters. The most significant changes that we observed with the MitoTimer reporter gene in whole animals were the overall change of fluorescence spectrum and the abundance of pure red puncta. These two parameters may reflect two different features of mitochondrial health. Within a given region of mitochondrial network, the MitoTimer fluorescence spectrum is likely to be determined by a balance between the import of the newly synthesized protein and its oxidation. Because MitoTimer protein in this study is expressed under control of constitutively active promoters, an overall increase of red:green ratio of the mitochondrial network under the conditions of mitochondrial stress are likely due to increased oxidation of the protein rather than altered transcription/translation. This could be caused by increased reactive oxygen species production and/or reduced antioxidant defense, i.e. oxidative stress. Because MitoTimer is translated in the cytosol and transported to mitochondria, increased oxidative stress in the cytosol, decreased import (of newly synthesized green MitoTimer), and/or reduced degradation (of old red MitoTimer) could all potentially lead to the finding.
In adult fly heart tubes and adult mouse FDB muscles, we readily detected pure red puncta that is approximately the size of enlarged mitochondria. We suspected that these pure red puncta were severely damaged mitochondria, as they were often disconnected from, but close to, normal healthy mitochondria, consistent with a phenomenon of mitochondrial fission after damage. Furthermore, we found that the pure red puncta were similar in size and location to those of enlarged, partially vacuolated mitochondria with remnant cristae structures detected by transmission electron microscopy of muscle sections from mice on a HFD. The direct proof of this speculation came as evidence of co-localization of Cox4 immunofluorescence with the pure red puncta. Some pure red puncta did not show detectable Cox4 immunofluorescence possibly due to complete degradation of electron transport chain proteins. It is likely that severely damaged mitochondria no longer import new MitoTimer protein, thus appearing as pure red puncta once the remaining protein is oxidized. Another possibility is that some of the autophagasome containing degenerating mitochondria were fused with lysosome to form autolysosome and display only red fluorescence due to acidic pH as demonstrated similarly by pmCherry-GFP-LC3B previously (43). In supporting this notion, we found that some yellow and red puncta were positive for LC3 and Lamp1, suggesting that they were part of autophagosome and autolysosome, respectively, targeted for degradation and removal from the cell.
Morphological analysis of the total fluorescent signal of MitoTimer allows for estimation of mitochondrial content and examination of structure. As shown in Fig. 4b, the % area occupied by the mitochondrial fluorescence signal of myofibers increased significantly in response to voluntary wheel running exercise training. Furthermore, high magnification of the MitoTimer images showed a typical structure of paired mitochondria on either side of the Z-line of the sarcomere. Exercise training not only increased the density of mitochondria, but also improved the size uniformity of the mitochondrial pair. Taken together, we have shown that the MitoTimer reporter could be used for robust, simultaneous analysis of multiple parameters, including mitochondrial content, structure, stress, and damage in vivo. These unique features of MitoTimer will prove to be valuable for mitochondrial research.
While the current studies were underway, Hernandez et al. (22) and Ferree et al. (21) published findings using a similar structural design of MitoTimer with the reporter gene under control of a doxycycline-inducible promoter for assessment of mitochondrial turnover in cultured cells. Hernandez et al. (22) provided evidence in HEK293 cells that MitoTimer targeted to the mitochondrial matrix and transitioned to red fluorescence over time demonstrating the usefulness in assessment of mitochondrial turnover and protein import. Ferree et al. (21) expanded on these findings in other cell types to show that mitochondrial dynamics and motility could also alter MitoTimer fluorescence and would thus be a useful reporter under conditions where mitochondria turnover and transport is affected. These findings set an important stage for this novel reporter gene being used for assessment of mitochondrial dynamics and health. In the current study, our application of MitoTimer in various animal models ranging from worms, flies to mice, allows analysis of mitochondrial content, structure, stress, and damage. This robust in vivo analysis of mitochondrial health adds significant value to the application of this novel reporter gene.
In summary, we took advantage of cultured cells, transgenic flies, transgenic worms, and somatic gene transfer in adult mice to show that the MitoTimer reporter gene in combination with computer-assisted imaging analysis is an excellent tool for robust, simultaneous measurements of mitochondrial content, structure, stress, and damage in vitro and in vivo. We anticipate that a combination of this technology with other cell and animal models will aid innovative research and facilitate the development of effective interventions to prevent and treat many of the prevalent diseases related to abnormal mitochondrial structure and function in various tissue/organ systems.
Acknowledgments
We thank Dr. Weiping Han for the conceptual contribution to the project, and Drs. Evelyn Ralston and Sarah Oddoux for help in setting up somatic gene transfer procedure for FDB muscles. We thank Dr. Norbert Leitinger for careful review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant AR050429 (to Z. Y.).
- ROS
- reactive oxygen species
- Cox4
- cytochrome c oxidase subunit IV
- FDB
- flexor digitorum bevis
- HFD
- high-fat diet
- OCR
- oxygen consumption rate
- DsRed
- Discosoma red fluorescent protein.
REFERENCES
- 1. Lowell B. B., Shulman G. I. (2005) Mitochondrial dysfunction and type 2 diabetes. Science 307, 384–387 [DOI] [PubMed] [Google Scholar]
- 2. Morino K., Petersen K. F., Dufour S., Befroy D., Frattini J., Shatzkes N., Neschen S., White M. F., Bilz S., Sono S., Pypaert M., Shulman G. I. (2005) Reduced mitochondrial density and increased IRS-1 serine phosphorylation in muscle of insulin-resistant offspring of type 2 diabetic parents. J. Clin. Invest. 115, 3587–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chang L., Cornford M., Miller B. L., Itabashi H., Mena I. (1995) Neuronal ultrastructural abnormalities in a patient with frontotemporal dementia and motor neuron disease. Dementia 6, 1–8 [DOI] [PubMed] [Google Scholar]
- 4. Zhou Y. T., Grayburn P., Karim A., Shimabukuro M., Higa M., Baetens D., Orci L., Unger R. H. (2000) Lipotoxic heart disease in obese rats: implications for human obesity. Proc. Natl. Acad. Sci. U.S.A. 97, 1784–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Czubryt M. P., McAnally J., Fishman G. I., Olson E. N. (2003) Regulation of peroxisome proliferator-activated receptor gamma coactivator 1 α (PGC-1α) and mitochondrial function by MEF2 and HDAC5. Proc. Natl. Acad. Sci. U.S.A. 100, 1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ravid R., Ferrer I. (2012) Brain banks as key part of biochemical and molecular studies on cerebral cortex involvement in Parkinson's disease. FEBS J. 279, 1167–1176 [DOI] [PubMed] [Google Scholar]
- 7. Woo D. K., Green P. D., Santos J. H., D'Souza A. D., Walther Z., Martin W. D., Christian B. E., Chandel N. S., Shadel G. S. (2012) Mitochondrial genome instability and ROS enhance intestinal tumorigenesis in APC(Min/+) mice. Am. J. Pathol. 180, 24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Müller W. E., Eckert A., Kurz C., Eckert G. P., Leuner K. (2010) Mitochondrial dysfunction: common final pathway in brain aging and Alzheimer's disease: therapeutic aspects. Mol. Neurobiol. 41, 159–171 [DOI] [PubMed] [Google Scholar]
- 9. Shen X., Zheng S., Thongboonkerd V., Xu M., Pierce W. M., Jr., Klein J. B., Epstein P. N. (2004) Cardiac mitochondrial damage and biogenesis in a chronic model of type 1 diabetes. Am. J. Physiol. Endocrinol. Metab. 287, E896–E905 [DOI] [PubMed] [Google Scholar]
- 10. Broderick T. L., Belke T., Driedzic W. R. (2002) Effects of chronic caloric restriction on mitochondrial respiration in the ischemic reperfused rat heart. Mol. Cell Biochem. 233, 119–125 [DOI] [PubMed] [Google Scholar]
- 11. Minamiyama Y., Bito Y., Takemura S., Takahashi Y., Kodai S., Mizuguchi S., Nishikawa Y., Suehiro S., Okada S. (2007) Calorie restriction improves cardiovascular risk factors via reduction of mitochondrial reactive oxygen species in type II diabetic rats. J. Pharmacol. Exp. Ther. 320, 535–543 [DOI] [PubMed] [Google Scholar]
- 12. Ponsot E., Dufour S. P., Zoll J., Doutrelau S., N′Guessan B., Geny B., Hoppeler H., Lampert E., Mettauer B., Ventura-Clapier R., Richard R. (2006) Exercise training in normobaric hypoxia in endurance runners. II. Improvement of mitochondrial properties in skeletal muscle. J. Appl. Physiol. 100, 1249–1257 [DOI] [PubMed] [Google Scholar]
- 13. Phielix E., Meex R., Moonen-Kornips E., Hesselink M. K., Schrauwen P. (2010) Exercise training increases mitochondrial content and ex vivo mitochondrial function similarly in patients with type 2 diabetes and in control individuals. Diabetologia 53, 1714–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wolf A. M., Asoh S., Ohsawa I., Ohta S. (2008) Imaging mitochondrial redox environment and oxidative stress using a redox-sensitive fluorescent protein. J. Nippon Med. Sch. 75, 66–67 [DOI] [PubMed] [Google Scholar]
- 15. Liu Z., Celotto A. M., Romero G., Wipf P., Palladino M. J. (2012) Genetically encoded redox sensor identifies the role of ROS in degenerative and mitochondrial disease pathogenesis. Neurobiol. Dis. 45, 362–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roma L. P., Duprez J., Takahashi H. K., Gilon P., Wiederkehr A., Jonas J. C. (2012) Dynamic measurements of mitochondrial hydrogen peroxide concentration and glutathione redox state in rat pancreatic β-cells using ratiometric fluorescent proteins: confounding effects of pH with HyPer but not roGFP1. Biochem. J. 441, 971–978 [DOI] [PubMed] [Google Scholar]
- 17. Wang W., Fang H., Groom L., Cheng A., Zhang W., Liu J., Wang X., Li K., Han P., Zheng M., Yin J., Wang W., Mattson M. P., Kao J. P., Lakatta E. G., Sheu S. S., Ouyang K., Chen J., Dirksen R. T., Cheng H. (2008) Superoxide flashes in single mitochondria. Cell 134, 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Terskikh A., Fradkov A., Ermakova G., Zaraisky A., Tan P., Kajava A. V., Zhao X., Lukyanov S., Matz M., Kim S., Weissman I., Siebert P. (2000) “Fluorescent timer”: protein that changes color with time. Science 290, 1585–1588 [DOI] [PubMed] [Google Scholar]
- 19. Yarbrough D., Wachter R. M., Kallio K., Matz M. V., Remington S. J. (2001) Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 98, 462–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Verkhusha V. V., Chudakov D. M., Gurskaya N. G., Lukyanov S., Lukyanov K. A. (2004) Common pathway for the red chromophore formation in fluorescent proteins and chromoproteins. Chem. Biol 11, 845–854 [DOI] [PubMed] [Google Scholar]
- 21. Ferree A. W., Trudeau K., Zik E., Benador I. Y., Twig G., Gottlieb R. A., Shirihai O. S. (2013) MitoTimer probe reveals the impact of autophagy, fusion, and motility on subcellular distribution of young and old mitochondrial protein and on relative mitochondrial protein age. Autophagy 9, 1887–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hernandez G., Thornton C., Stotland A., Lui D., Sin J., Ramil J., Magee N., Andres A., Quarato G., Carreira R. S., Sayen M. R., Wolkowicz R., Gottlieb R. A. (2013) MitoTimer: a novel tool for monitoring mitochondrial turnover. Autophagy 9, 1852–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kenwood B. M., Weaver J. L., Bajwa A., Poon I. K., Byrne F. L., Murrow B. A., Calderone J. A., Huang L., Divakaruni A. S., Tomsig J. L., Okabe K., Lo R. H., Cameron Coleman G., Columbus L., Yan Z., Saucerman J. J., Smith J. S., Holmes J. W., Lynch K. R., Ravichandran K. S., Uchiyama S., Santos W. L., Rogers G. W., Okusa M. D., Bayliss D. A., Hoehn K. L. (2014) Identification of a novel mitochondrial uncoupler that does not depolarize the plasma membrane. Mol. Metab. 3, 114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee K. S., Iijima-Ando K., Iijima K., Lee W. J., Lee J. H., Yu K., Lee D. S. (2009) JNK/FOXO-mediated neuronal expression of fly homologue of peroxiredoxin II reduces oxidative stress and extends life span. J. Biol. Chem. 284, 29454–29461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Coulom H., Birman S. (2004) Chronic exposure to rotenone models sporadic Parkinson's disease in Drosophila melanogaster. J. Neurosci. 24, 10993–10998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Piazza N., Gosangi B., Devilla S., Arking R., Wessells R. (2009) Exercise-training in young Drosophila melanogaster reduces age-related decline in mobility and cardiac performance. PLoS One 4, e5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gargano J. W., Martin I., Bhandari P., Grotewiel M. S. (2005) Rapid iterative negative geotaxis (RING): a new method for assessing age-related locomotor decline in Drosophila. Exp. Gerontol. 40, 386–395 [DOI] [PubMed] [Google Scholar]
- 28. Tinkerhess M. J., Ginzberg S., Piazza N., Wessells R. J. (2012) Endurance training protocol and longitudinal performance assays for Drosophila melanogaster. J. Vis. Exp. 61, e3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vogler G., Ocorr K. (2009) Visualizing the beating heart in Drosophila. J. Vis. Exp. 31, e1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. DiFranco M., Quinonez M., Capote J., Vergara J. (2009) DNA transfection of mammalian skeletal muscles using in vivo electroporation. J. Vis. Exp. 32, e1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gomez D., Shankman L. S., Nguyen A. T., Owens G. K. (2013) Detection of histone modifications at specific gene loci in single cells in histological sections. Nat. Methods 10, 171–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miller S. E., Howell D. N. (1988) Viral infections in the acquired immunodeficiency syndrome. J. Electron Microsc. Tech. 8, 41–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Toth M. L., Melentijevic I., Shah L., Bhatia A., Lu K., Talwar A., Naji H., Ibanez-Ventoso C., Ghose P., Jevince A., Xue J., Herndon L. A., Bhanot G., Rongo C., Hall D. H., Driscoll M. (2012) Neurite sprouting and synapse deterioration in the aging Caenorhabditis elegans nervous system. J. Neurosci. 32, 8778–8790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ranganayakulu G., Schulz R. A., Olson E. N. (1996) Wingless signaling induces nautilus expression in the ventral mesoderm of the Drosophila embryo. Dev. Biol. 176, 143–148 [DOI] [PubMed] [Google Scholar]
- 35. Fang H., Chen M., Ding Y., Shang W., Xu J., Zhang X., Zhang W., Li K., Xiao Y., Gao F., Shang S., Li J. C., Tian X. L., Wang S. Q., Zhou J., Weisleder N., Ma J., Ouyang K., Chen J., Wang X., Zheng M., Wang W., Zhang X., Cheng H. (2011) Imaging superoxide flash and metabolism-coupled mitochondrial permeability transition in living animals. Cell Res. 21, 1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krieger D. A., Tate C. A., McMillin-Wood J., Booth F. W. (1980) Populations of rat skeletal muscle mitochondria after exercise and immobilization. J. Appl. Physiol. 48, 23–28 [DOI] [PubMed] [Google Scholar]
- 37. Ritov V. B., Menshikova E. V., He J., Ferrell R. E., Goodpaster B. H., Kelley D. E. (2005) Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes 54, 8–14 [DOI] [PubMed] [Google Scholar]
- 38. Kelley D. E., He J., Menshikova E. V., Ritov V. B. (2002) Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51, 2944–2950 [DOI] [PubMed] [Google Scholar]
- 39. Petersen K. F., Dufour S., Befroy D., Garcia R., Shulman G. I. (2004) Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N. Engl. J. Med. 350, 664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ide T., Tsutsui H., Hayashidani S., Kang D., Suematsu N., Nakamura K., Utsumi H., Hamasaki N., Takeshita A. (2001) Mitochondrial DNA damage and dysfunction associated with oxidative stress in failing hearts after myocardial infarction. Circ. Res. 88, 529–535 [DOI] [PubMed] [Google Scholar]
- 41. Wang J., Xiong S., Xie C., Markesbery W. R., Lovell M. A. (2005) Increased oxidative damage in nuclear and mitochondrial DNA in Alzheimer's disease. J. Neurochem. 93, 953–962 [DOI] [PubMed] [Google Scholar]
- 42. Bonawitz N. D., Rodeheffer M. S., Shadel G. S. (2006) Defective mitochondrial gene expression results in reactive oxygen species-mediated inhibition of respiration and reduction of yeast life span. Mol. Cell Biol. 26, 4818–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]