Background: Cycloisomaltooligosaccharide glucanotransferase catalyzes an intramolecular transglucosylation reaction and produces cycloisomaltooligosaccharides from dextran.
Results: The crystal structure of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase was determined.
Conclusion: The enzyme structures complexed with isomaltooligosaccharides and cycloisomaltooctaose revealed the molecular mechanism of action.
Significance: CBM35 functions in the product size determination and substrate recruitment.
Keywords: Carbohydrate-binding Protein, Carbohydrate Structure, Crystal Structure, Enzyme Catalysis, Enzyme Structure, Glycoside Hydrolases, X-ray Crystallography, Cycloisomaltooligosaccharide Glucanotransferase, Dextran
Abstract
Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase belongs to the glycoside hydrolase family 66 and catalyzes an intramolecular transglucosylation reaction that produces cycloisomaltooligosaccharides from dextran. The crystal structure of the core fragment from Ser-39 to Met-738 of B. circulans T-3040 cycloisomaltooligosaccharide glucanotransferase, devoid of its N-terminal signal peptide and C-terminal nonconserved regions, was determined. The structural model contained one catalytic (β/α)8-barrel domain and three β-domains. Domain N with an immunoglobulin-like β-sandwich fold was attached to the N terminus; domain C with a Greek key β-sandwich fold was located at the C terminus, and a carbohydrate-binding module family 35 (CBM35) β-jellyroll domain B was inserted between the 7th β-strand and the 7th α-helix of the catalytic domain A. The structures of the inactive catalytic nucleophile mutant enzyme complexed with isomaltohexaose, isomaltoheptaose, isomaltooctaose, and cycloisomaltooctaose revealed that the ligands bound in the catalytic cleft and the sugar-binding site of CBM35. Of these, isomaltooctaose bound in the catalytic site extended to the second sugar-binding site of CBM35, which acted as subsite −8, representing the enzyme·substrate complex when the enzyme produces cycloisomaltooctaose. The isomaltoheptaose and cycloisomaltooctaose bound in the catalytic cleft with a circular structure around Met-310, representing the enzyme·product complex. These structures collectively indicated that CBM35 functions in determining the size of the product, causing the predominant production of cycloisomaltooctaose by the enzyme. The canonical sugar-binding site of CBM35 bound the mid-part of isomaltooligosaccharides, indicating that the original function involved substrate binding required for efficient catalysis.
Introduction
Cyclodextrins are cyclic α-1,4-glucans composed of six or more glucose units (1) and are produced from starch by cyclodextrin glycosyltransferase (CGTase,7 EC 2.4.1.19 (2, 3)). The main enzymatic products are α-, β-, and γ-cyclodextrins consisting of six, seven, and eight glucose units, respectively. Cyclodextrins have a hydrophobic cavity at the center of the molecule, which enables it to form inclusion complexes with various hydrophobic molecules (3). Cyclodextrins have numerous applications in the pharmaceutical, food, and textile industries (4, 5). In particular, cyclodextrins are used to solubilize hydrophobic molecules in water because of the hydrophilic exterior and hydrophobic interior feature of these compounds. Thus, cyclodextrins aid drug delivery and protect compounds from degradation (6, 7). Cyclodextrins are also used as room refreshers.
Cycloisomaltooligosaccharides, also known as cyclodextrans (CIs), are similar cyclic oligomers of glucose molecules to cyclodextrins, but glucoses in CIs are linked with α-1,6-glucosidic bonds (8). CIs are produced from dextran via an intramolecular transglycosylation catalyzed by the enzyme cycloisomaltooligosaccharide glucanotransferase (CITase; EC 2.4.1.248). CIs of 7–17 glucose units (CI-7 to CI-17; named as per the number of glucose units present) are produced by Bacillus circulans T-3040 CITase (BcCITase (9–12)). The dominant product of BcCITase is cycloisomaltooctaose (CI-8), secondary CI-7, and the amount reduces in accordance with the higher degree of polymerization for CI-9 to CI-17. CIs are highly water-soluble and have a central hydrophobic cavity that is similar to cyclodextrins. Hence, the inclusion complex-forming ability is expected to be similar to that observed for cyclodextrins and that of CI-10 against Victoria blue B, as has been reported (10, 11). CIs are also known as strong inhibitors of glucansucrase, showing anti-plaque activity (13). Thus, CIs appear to be novel bio-nanomaterials applicable to various bio-industries as well as cyclodextrins.
The cit gene encoding BcCITase has been cloned, and its nucleotide sequence has been determined (14). According to amino acid sequence analysis, the enzyme belongs to the glycoside hydrolase family 66 (GH66). In the CAZy database (15), the following three CITases are listed: B. circulans T-3040; B. circulans U155 (9), and Paenibacillus sp. 598K (12). Most of the other enzymes in the family are dextranases, which hydrolyze dextran to produce linear isomaltooligosaccharides (IGs). Dextranases from streptococci, especially from Streptococcus mutans (SmDex), have been the most extensively studied among the GH66 proteins, and biochemical studies using site-directed mutagenesis revealed that Asp-385 in SmDex was essential for catalytic activity (16). Similarly, Asp-243 of dextranase from Thermotoga lettingae (17) and Asp-308 of BcCITase (18), both corresponding to Asp-385 in SmDex, are implicated as catalytic residues. We have solved the crystal structure of the fragment from Gln-100 to Ile-732 of SmDex, devoid of its N- and C-terminal variable regions, as the first structure of a GH66 protein, and we displayed that the conserved region in all GH66 proteins included three domains as follows: the domain N with an immunoglobulin fold, the catalytic domain A with a (β/α)8-barrel structure, and the domain C with tandemly repeated Greek key motifs (19). Structural analysis using the ligand-bound structures verified the catalytic nucleophile Asp-385 and also identified the catalytic acid/base Glu-453. Besides these GH66 core structural domains, BcCITase has two extra carbohydrate-binding module family 35 (CBM35) domains; one is inserted in the middle part of the conserved region, and this domain is common in CITases (BcCBM35-1); another is in the C-terminal region (BcCBM35-2) (20, 21). The detailed function of these domains remains to be elucidated, although the presence of the first domain has been shown to stabilize the protein and to enhance the production of CI-8 (21).
In this work, we present the crystal structure of a stable C-terminally truncated mutant of BcCITase and its ligand-bound structures, and we discuss the detailed reaction mechanism of the CITase by comparison with related structures. The novel function of the inserted domain of CBM35 in the product size determination is also addressed.
EXPERIMENTAL PROCEDURES
Sugar Ligands
CI-8 was produced from dextran 40 (GE Healthcare) by BcCITase (10) and purified to homogeneity by reversed-phase chromatography using an ODS column (Daisopak SP-120-5-ODS-BP, 20 × 250 mm, Daiso, Osaka, Japan) as described previously (22). Isomaltohexaose (IG-6), isomaltoheptaose (IG-7), and isomaltooctaose (IG-8) were produced from Fujioligo G67 (maltohexaose- and maltoheptaose-rich maltooligosaccharides, Nihon Shokuhin Kako Co Ltd., Tokyo, Japan) by Acetobacter capsulatum ATCC 11894 dextrin dextranase (23) and purified by gel permeation chromatography using a Toyopearl HW-40S column (5 × 62 cm, Tosoh Corp., Tokyo, Japan) (22).
Protein Expression and Crystallization
We constructed two enzyme forms containing His6 purification tags at either the N terminus (BcCITase-NH) or the C terminus (BcCITase-CH). BcCITase-NH was composed of a single polypeptide chain of 721 amino acids (18–738), where the N terminus 18MGSSHHHHHHSSGLVPRGSHM38 was derived from the expression vector, including the purification tag and thrombin cleavage site. BcCITase-CH was composed of a single polypeptide chain of 710 amino acids (37–746), where the N-terminal Met-37 and Gly-38 and the C-terminal 739LEHHHHHH746 were derived from the expression vector. Both constructs contained enzyme residues Ser-39–Met-738 (molecular mass of 78 kDa), and the N-terminal signal peptide and the C-terminal variable region were omitted. Both protein forms were expressed in Escherichia coli BL21(DE3) cells, purified by His6 tag nickel affinity chromatography and crystallized as reported previously (21, 24). BcCITase-NH with a concentration of 7 mg/ml was crystallized by the sitting drop vapor diffusion method using a precipitant solution composed of 1.0 m sodium acetate trihydrate, 0.1 m HEPES, pH 7.5, 0.05 m cadmium sulfate hydrate, and 12%(v/v) glycerol, at 293 K. BcCITase-CH with a concentration of 20 mg/ml was crystallized using a precipitant solution composed of 1.6 m ammonium sulfate, 0.1 m MES, pH 6.5, and 10% (v/v) 1,4-dioxane.
Site-directed mutagenesis, Asp-308 to alanine, of BcCITase-CH was performed using the QuikChange mutagenesis kit (Stratagene, Agilent Technologies, Inc., Santa Clara, CA). For incorporation of the selenomethionine (SeMet) enzyme, BcCITase-NH proteins were expressed in E. coli B834(DE3) cells grown in LeMaster medium supplemented with 25 mg/liter seleno-l-methionine (Wako Pure Chemical, Tokyo, Japan). Expression and purification were conducted in a similar manner to the native enzyme.
Data Collection and Structural Determination
Data for the native and its SeMet derivative crystals of BcCITase-NH protein were collected at 95 K to 2.3 Å resolution at beamline BL-6A and to 2.8 Å resolution at beamline BL-5A (Photon Factory, High Energy Accelerator Research Organization, Tsukuba, Japan). Data for BcCITase-CH, its D308A mutant, and ligand complexes were collected at 95 K at the Photon Factory. Data collection statistics are summarized in Table 1. Sugar-bound BcCITase-CH D308A mutant crystals were prepared by soaking the enzyme crystal into a precipitant drop containing 1–5% (w/v) sugar ligands for 10 min to 1 h and 10% ethylene glycol for about 10 min, before the diffraction experiments. All diffraction data were processed using HKL2000 (25). Initial structural determination was conducted by the multiple-wavelength anomalous dispersion method. Selenium atom positions in the SeMet derivative of BcCITase-NH were determined, and phases were calculated using the programs SOLVE/RESOLVE (26, 27). Further manual model building and refinement were performed for the native crystal using ARP/wARP (28), REFMAC5 (29), and COOT (30). Structural models included calcium and cadmium ions, which were distinguished by the anomalous signals and peak sizes. The crystal structures of the BcCITase-CH D308A mutant complexed with ligands were determined by the molecular replacement method using MOLREP (31) and refined with REFMAC5 and COOT. The structure of the ligand-free BcCITase-CH could not be refined because of crystal twinning, probably being transformation twins containing tetragonal and orthorhombic crystal systems. Structural refinement statistics are summarized in Table 2. Stereochemistry of the models was analyzed with the Rampage program (32), and structural drawings were prepared using the PyMOL program (DeLano Scientific LLC, Palo Alto, CA).
TABLE 1.
Data collection and structure refinement statistics of BcCITase
Values in parentheses refer to the highest resolution shell.
| Data | BcCITase-NH native | Se-Met (peak) | Se-Met (edge) | Se-Met (low remote) | Se-Met (high remote) | BcCITase·IG-6 | BcCITase·IG-7 | BcCITase·IG-8 | BcCITase·CI-8 |
|---|---|---|---|---|---|---|---|---|---|
| Space group | P3121 | P3121 | P3121 | P3121 | P3121 | P41212 | P41212 | P212121 | P212121 |
| Unit cell parameters | a = b = 106.4, c = 160.6 Å | a = b = 105.7, c = 160.6 Å | a = b = 172.1, c = 60.9 Å | a = b = 171.6, c = 61.3 Å | a = 61.3, b = 171.3, c = 173.3 Å | a = 61.9, b = 167.7, c = 174.9 Å | |||
| Beamline | BL-6A | BL-5A | BL-5A | BL-5A | BL-NE3A | BL-NW12A | |||
| Wavelength | 0.97800 Å | 0.97885 Å | 0.97931 Å | 0.98319 Å | 0.96408 Å | 1.0000 Å | 1.0000 Å | 1.0000 Å | 1.0000 Å |
| Resolution | 100 to 2.30 Å (2.38 to 2.30 Å) | 50.0 to 2.80 Å (2.90 to 2.80 Å) | 50.0 to 2.80 Å (2.90 to 2.80 Å) | 50.0 to 2.80 Å (2.90 to 2.80 Å) | 50.0 to 2.80 Å (2.90 to 2.80 Å) | 100.0 to 2.60 Å (2.64 to 2.60 Å) | 100.0 to 2.25 Å (2.29 to 2.25 Å) | 100.0 to 2.25 Å (2.33 to 2.25 Å) | 100.0 to 1.90 Å (1.93 to 1.90 Å) |
| Rsym | 0.079 (0.423) | 0.097 (0.281) | 0.103 (0.405) | 0.086 (0.295) | 0.119 (0.529) | 0.123 (0.610) | 0.099 (0.522) | 0.080 (0.566) | 0.077 (0.487) |
| Completeness | 100.0% (100.0%) | 100.0% (100.0%) | 100.0% (100.0%) | 100.0% (100.0%) | 100.0% (99.6%) | 99.7% (100.0%) | 99.6% (100.0%) | 98.9% (95.2%) | 99.6% (98.6%) |
| Multiplicity | 16.6 (16.7) | 22.0 (22.4) | 22.0 (21.5) | 22.0 (22.4) | 21.1 (14.7) | 9.6 (9.8) | 9.3 (9.2) | 5.7 (4.3) | 5.3 (5.0) |
| Average I/σ(I) | 47.1 (10.6) | 48.6 (11.7) | 36.9 (7.4) | 42.1 (10.7) | 27.7 (3.4) | 15.1 (4.0) | 17.4 (5.2) | 19.7 (2.3) | 17.9 (3.4) |
| Unique reflections | 47,410 (4667) | 26,226 (2558) | 26,276 (2567) | 26,159 (2547) | 26,350 (2559) | 28,917 (1441) | 44,028 (2161) | 87,986 (8336) | 143,236 (7058) |
| Observed reflections | 786,631 | 577,491 | 577,768 | 576,700 | 555,265 | 278,644 | 410,548 | 501,163 | 758,307 |
TABLE 2.
Structure refinement statistics of BcCITase
Values in parentheses refer to the highest resolution shell.
| Data | BcCITase-NH | BcCITase·IG-6 | BcCITase·IG-7 | BcCITase·IG-8 | BcCITase·CI-8 |
|---|---|---|---|---|---|
| PDB code | 3WNK | 3WNL | 3WNM | 3WNN | 3WNO |
| Structure refinement | |||||
| Resolution | 44.3 to 2.30 Å (2.36 to 2.30 Å) | 47.8 to 2.60 Å (2.67 to 2.60 Å) | 30.4 to 2.25 Å (2.05 to 2.25 Å) | 35.4 to 2.25 Å (2.16 to 2.10 Å) | 30.5 to 1.90 Å (1.95 to 1.90 Å) |
| R-factor | 0.158 (0.205) | 0.187 (0.263) | 0.172 (0.234) | 0.202 (0.310) | 0.170 (0.227) |
| Rfree-factor | 0.192 (0.269) | 0.247 (0.380) | 0.205 (0.283) | 0.244 (0.367) | 0.195 (0.267) |
| No. of reflections | 44,946 (3280) | 27,183 (2007) | 41,525 (3001) | 81,861 (5761) | 135,918 (9556) |
| No. of water molecules | 549 | 49 | 347 | 471 | 1226 |
| Average B-value | 37.7 Å2 | 69.2 Å2 | 42.5 Å2 | 48.5 Å2 | 28.4 Å2 |
| Root mean square deviations from ideal value | |||||
| Bond lengths | 0.008 Å | 0.015 Å | 0.010 Å | 0.013 Å | 0.006 Å |
| Bond angles | 1.235° | 1.684° | 1.320° | 1.618° | 1.176° |
| Ramachandran plot | |||||
| Favored region | 96.1% | 93.6% | 97.0% | 95.4% | 96.4% |
| Allowed region | 3.8% | 6.4% | 3.0% | 4.5% | 3.6% |
| Outlier region | 0.1% | 0.0% | 0.0% | 0.1% | 0.0% |
RESULTS
Overall Structure
The crystal structure of BcCITase-NH was determined by the multiwavelength anomalous dispersion method at 2.3 Å resolution. The BcCITase-NH model includes a single protein molecule, as well as the surrounding water and glycerol molecules and sodium, calcium, cadmium, and acetate ions. The N-terminal nine residues Met-18–His-26 were not identified because of a lack of electron density. Successively, four ligand complex structures of the BcCITase-CH D308A mutant with IG-6 (BcCITase·IG-6), IG-7 (BcCITase·IG-7), IG-8 (BcCITase·IG-8), and CI-8 (BcCITase·CI-8) were determined (Table 2). The crystals of BcCITase·IG-6 and BcCITase·IG-7 belong to the tetragonal space group of P41212, whereas the BcCITase·IG-8 and BcCITase·CI-8 crystals belong to the less symmetrical orthogonal space group of P212121. This difference might have been caused by the structural rearrangement upon binding of relatively large ligands at the catalytic cleft and the substrate-binding site in BcCBM35-1, although the crystals were obtained from identical conditions. The tetragonal and orthorhombic crystals contained one and two protein molecules in the asymmetric unit, respectively. The N-terminal five residues Met-37–Ser-41 and the C-terminal five residues His-742–His-746 were not identified because of a lack of electron density. Besides the sugar and water molecules, the structural model contained sodium, calcium, and sulfate ions, as well as MES and ethylene glycol molecules.
The core structure of BcCITase is composed of four domains. The ribbon model of the BcCITase structure for chain B of the BcCITase·CI-8 complex is shown in Fig. 1. As three of them are conserved in the C-terminal truncated mutant of SmDex belonging to GH66 (Fig. 2) (19), the domain names are designated in accordance with those of SmDex; the N-terminal domain N, the catalytic domain A, and the C-terminal domain C. BcCITase contained an additional domain B comprising a CBM35 β-jellyroll fold designated as BcCBM35-1.
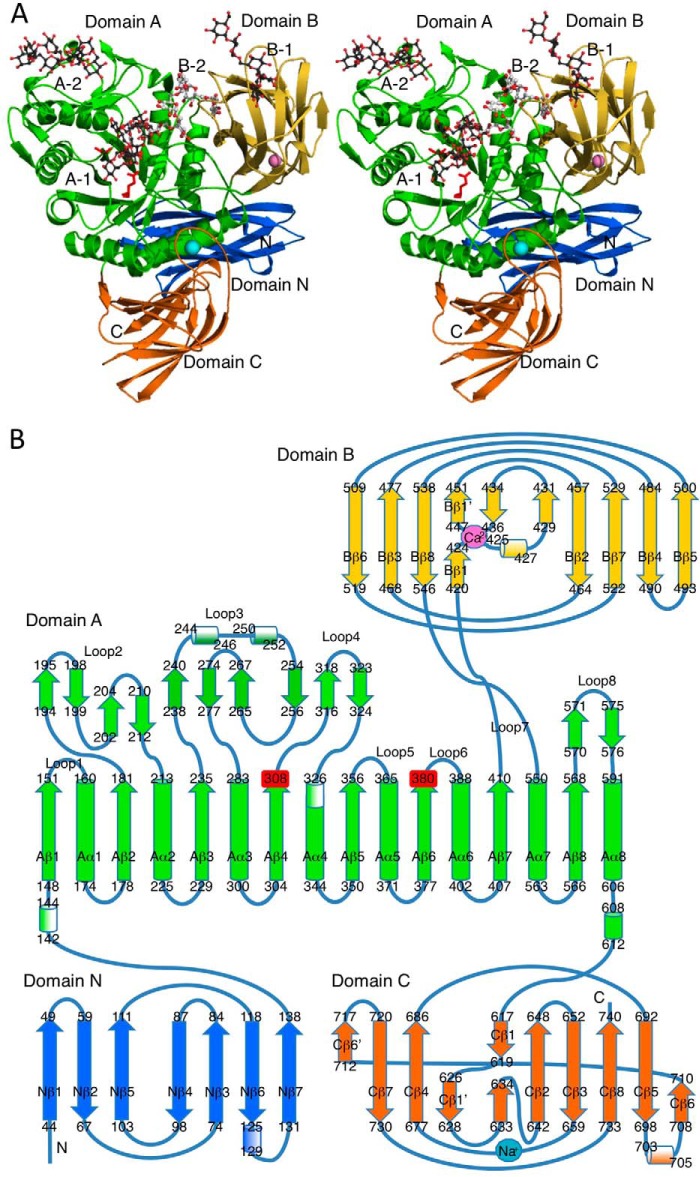
FIGURE 1.
Structure of BcCITase. A, stereoview of the BcCITase·CI-8 complex ribbon model. The model was drawn for chain B. IG-8 molecule bound in the catalytic site of chain B in the BcCITase·IG-8 complex was superimposed. Each domain is shown in different colors as follows: domains N, A, B, and C, are colored blue, green, yellow, and orange, respectively; two catalytic residues, red; bound CI-8 molecules, gray; superimposed IG-8 molecule, white; calcium ion, pink; sodium ion, cyan. Four sugar-binding sites are labeled as follows: catalytic site, A-1; the surface-binding site in domain A, A-2, canonical sugar-binding site in BcCBM35-1, B-1; the second sugar-binding site (subsite −8) in BcCBM35-1, B-2. B, topological diagram of BcCITase as follows: α-helices, 310-helices, and β-strands, as filled cylinders, shaded cylinders, and filled arrows, respectively. Catalytic residues, bound calcium, and sodium ions were placed.
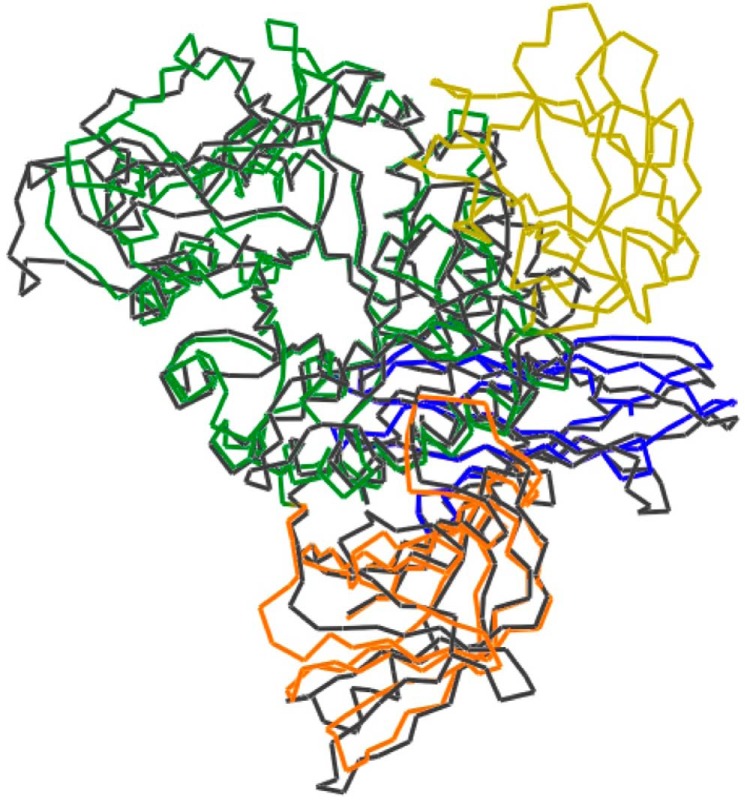
FIGURE 2.
Superimposed models of BcCITase and SmDex. BcCITase was colored by domains, and SmDex is presented in gray, from PDB code 3VMN (19).
Domain N (Ser-39–Ser-139), composed of seven β-strands, formed a more complete C2 type immunoglobulin fold than that of SmDex (Fig. 2) (19), in which the second and fifth strands are both split into two strands.
Domain A (Ser-140–Ala-419 and Thr-547–Asp-614) folded into a (β/α)8-barrel. The loop3, located between the 3rd β-strand Aβ3 and the 3rd α-helix Aα3 (see the names in the secondary structures of Fig. 1B), is the longest with 47 residues, and it was found to be compactly folded into a small block with the loop4 in a manner similar to that observed for the SmDex structure. The structure of the barrel was also almost identical with that of SmDex.
Domain C (Ser-615–Met-738) was composed of a tandemly repeated Greek key motifs. The overall fold of domain C was similar to that of SmDex (Fig. 2) (19), except that BcCITase lacked the α-helix located between Cβ3 and Cβ4. The electron density at the loop region between Cβ1′ and Cβ1 was poor, and this region had variable structures between the models.
Domain arrangement of BcCITase was similar to that of SmDex but different from that of CGTases that consisted of five domains and belonged to GH13 (33). Besides CGTases, significant similarities were not observed with the structures of amylomaltase belonging to GH77 (34–37) or 4-α-glucanotransferase belonging to GH57 (38) that produce larger cyclic α-1,4-glucan or cycloamylose.
Structure of BcCBM35-1
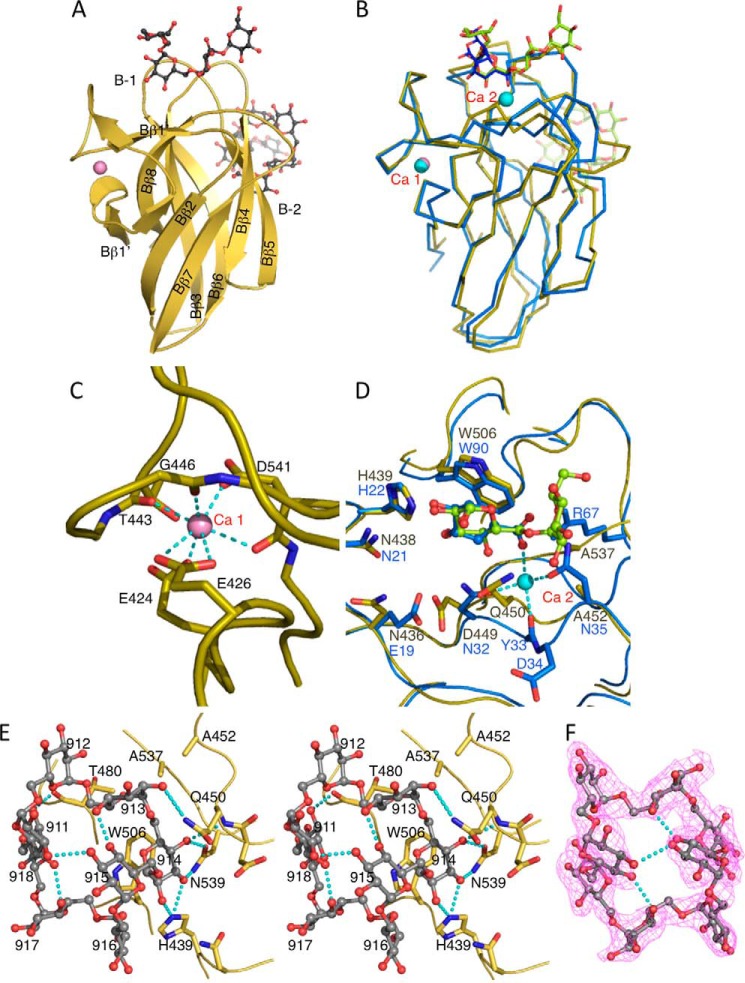
Domain B (Gly-420–Gly-546) folded into a β-jellyroll, comprising BcCBM35-1, and this structure was inserted between Aβ7 and Aα7 of the catalytic domain, where the corresponding region in SmDex was loop 7 with an α-helix fold (Figs. 1 and 3A). BcCBM35-1 protruded from the catalytic domain on the catalytic cleft side. Structural comparison using the Dali server showed that this domain had the highest similarity with CBM35 structures of Amycolatopsis orientalis exo-β-d-glucosaminidase CsxA (Chi-CBM35, PDB code 2VZP) and Clostridium thermocellum rhamnogalacturonan acetyl esterase Rgae12A (Cthe_3141) (Rhe-CBM35, PDB code 2W1W), with a root mean square difference of 1.4 Å (39). These CBM35 structures have two calcium ion-binding sites (Fig. 3B). One of them is located between the end of Bβ1 and the beginning of Bβ1′. This site is conserved among CBM35s, except for Cellvibrio japonicus endo-β-1,4-mannanase (Mac5C-CBM35, PDB code 2BGO (40)) and Rhe-CBM35 (39). In BcCBM35-1, this calcium ion-binding site is conserved and formed by Glu-424, Glu-426, Thr-443, Gly-446, and Asp-541 (Fig. 3, A and C). In the BcCITase-NH structure, the calcium ion was replaced by a cadmium ion, which was derived from the crystallization conditions. The second calcium ion-binding site of CBM35s is located at the tip of the fold on two loops between Bβ1′ and Bβ2 and between Bβ7 and Bβ8, which is conserved in all CBM35 structures determined and contributes to substrate recognition. In BcCBM35-1, however, the main chain of these loops was structured differently from those of the other CBM35s, and the asparagine residue on the first loop was replaced by Ala-414 (Fig. 3D). Concomitantly, the second calcium-binding site was not conserved, and the metal ion was not observed. Instead, this site acted as one of the sugar-binding sites with affinity toward isomaltooligosaccharides, as described later (Fig. 3E).
FIGURE 3.
Structure of BcCBM35-1. A, ribbon diagram of BcCBM35-1 from the BcCITase·IG-8 complex. Bound IG-8, gray ball-and-stick model; calcium ion, pink. Two sugar-binding sites, B-1 and B-2, are labeled. B, superimposed model of BcCBM35-1 (yellow, calcium ion in pink) on A. orientalis exo-β-d-glucosaminidase Chi-CBM35 (blue, calcium ions in cyan, PDB code 2VZP, Ref. 39). C, calcium-binding structure of BcCBM35-1. D, superimposed model of the sugar-binding sites of BcCBM35-1 and Chi-CBM35. Two of four bound glucoses in BcCBM35-1 and glucuronic acid in Chi-CBM35 are shown in ball-and-stick models. E, stereoview of BcCBM35-1 conserved sugar-binding site B-1 from the BcCITase·CI-8 complex chain A; bound CI-8, gray ball-and-stick model; estimated hydrogen bonds, cyan dashed lines. F, 2Fobs − Fcalc electron density map of the bound CI-8; contour level, 1σ.
Sugar Complex Structures of BcCITase
In the crystal structure of BcCITase-NH, the peptide tag used for protein expression and purification was maintained without protease digestion and stacked into the catalytic cleft of the adjacent molecule in the crystal, preventing ligands from accessing the active site in the soaking experiments. To obtain stable ligand·protein complex structures, the catalytic site mutant recombinant BcCITase-CH was used. Simultaneously, for the purpose of the soaking experiments of the protein·sugar complex, a point mutation of nucleophile Asp-308 to alanine was introduced, because the CITase mutants in which the nucleophile was mutated showed no enzyme activity (11, 21). The ligands were observed mainly at two sites, the catalytic cleft in domain A (Fig. 1A, site A-1) and the sugar-binding site of BcCBM35-1 (Fig. 1A, site B-1). BcCITase-CI-8 contained an additional CI-8 molecule in the intermolecular cavity surrounded by loop 3 of domain A and the bound CI-8 molecule at the catalytic cleft (Fig. 1A, site A-2). This CI-8 bound loosely, as judged from the B-factor of the bound CI-8 molecule and appeared to be an artifact due to crystal packing. However, it was embedded within the exposed hydrophobic residues of Tyr-243, Tyr-255, Tyr-274, Tyr-276, and Tyr-319 on loop3 and loop4 (Fig. 1B), and thus on the opposite side of the catalytic cleft. Therefore, these hydrophobic residues might function as a surface-binding site, as has been observed for barley α-amylases (41).
In the catalytic cleft, four glucose residues out of six were observed in the BcCITase·IG-6 complex structure, whereas in the other complex structures, the whole molecules were observed (Fig. 4A). In BcCBM35-1, four or five glucose moieties are modeled in an almost identical conformation in the sugar-binding site of BcCITase·IG-6, BcCITase·IG-7, and BcCITase·IG-8 complex structures (Fig. 1A, site B-1). In the BcCITase·CI-8 complex structure, the bound CI-8 molecules were observed differently between two nonsymmetrically related molecules. Five glucose moieties were observed at the same positions with the BcCITase·IG-8 complex structure in chain B, whereas all eight glucoses of the CI-8 molecule were observed in chain A (Fig. 3, E and F).
FIGURE 4.
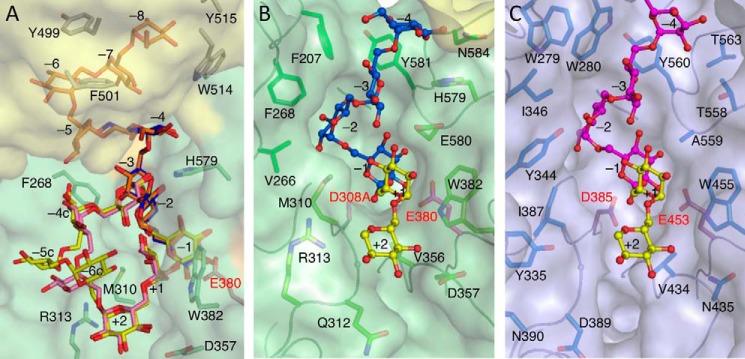
Ligand-binding surface structures of BcCITase and SmDex in the catalytic cleft. A, bound IG-6, IG-7, IG-8, and CI-8 models in the catalytic cleft are superimposed on the surface model of the BcCITase·IG-8 complex chain A. Surface is colored green for domain A and yellow for BcCBM35-1. B, surface structure in the catalytic cleft of the BcCITase·IG-6 complex. Two glucoses at subsite +1 and +2 derived from the BcCITase·CI-8 complex are superimposed for clarity in yellow. C, surface structure in the catalytic cleft of the SmDex·isomaltotriose derivative complex from PDB code 3VMO (19).
Sugar-binding Structure in the Catalytic Site
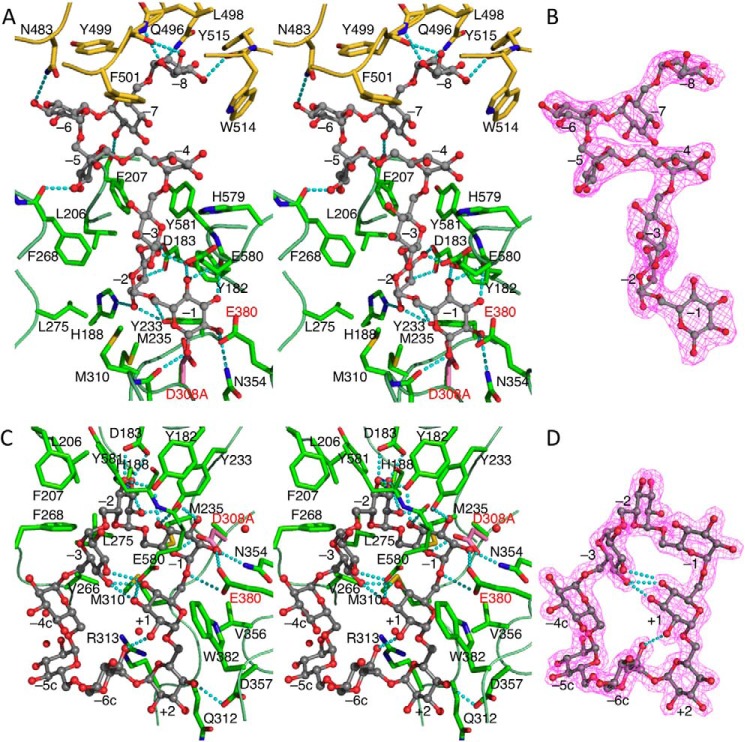
In the BcCITase·IG-6 complex structure, four glucose residues (Glc-1 to Glc-4 from the reducing end) were observed at four subsites (−1 to −4 from the catalytic site) only in one side of the active site, similar to those in SmDex (Fig. 4, B and C) (19), and the other two glucose moieties (Glc-5 and Glc-6) were disordered. In the BcCITase·IG-8 complex, the entire IG-8 molecule (Glc-1 to Glc-8 from the reducing end) was observed, and the four glucose moieties from the reducing end were located at the same position with those of the BcCITase·IG-6 complex (Fig. 5, A and B). The glucose moieties at the subsites −1 (Glc-1) and −2 (Glc-2) were recognized via hydrogen bonds by the enzyme, and Glc-2 was buried in the pocket made by hydrophobic residues Leu-206, Phe-207, Tyr-233, Met-235, Phe-268, Leu-275, Met-310, and Tyr-581, in a manner similar to SmDex (Fig. 2) (19). The anomeric hydroxyl group of Glc-1 adopted a β-configuration in both complexes. The differences from SmDex are found in His-188 and Glu-580 of BcCITase, both of which were replaced by alanine in SmDex. Glc-3 and Glc-4 were mainly recognized by the hydrophobic residues Tyr-581 and Phe-207. The other four glucose moieties Glc-5 to Glc-8 in the BcCITase·IG-8 complex were observed to extend outwards from the catalytic cleft (Fig. 1A, site B-2). Glc-5 to Glc-7 were located in the hydrophobic straight between domains A and B, each had one direct hydrogen bond to protein residues but did not show tight binding architectures. Glc-8 was buried in the pocket made by aromatic residues Tyr-499, Phe-501, Trp-514, and Tyr-515 in domain B with its O6 atom at the bottom and recognized directly via the formation of four hydrogen bonds with Gln-496, Tyr-499, and Trp-514. This glucose-binding pocket was located at the side of BcCBM35-1, involving the above residues on Bβ5 and Bβ6. This second sugar-binding site was not conserved in the other CBM35 structures reported.
FIGURE 5.
Ligand-binding structures of BcCITase in the catalytic cleft. A, stereoview of the BcCITase·IG-8 complex. Bound IG-8, gray ball-and-stick model; estimated hydrogen bonds, cyan dashed lines. Asp-308 side chain of the wild-type enzyme is superimposed in pink. B, 2Fobs − Fcalc electron density map of the bound IG-8 in the catalytic cleft; contour level, 1σ. C, stereoview of the BcCITase·CI-8 complex. D, 2Fobs − Fcalc electron density map of the bound CI-8; contour level, 1.2σ.
In the BcCITase·CI-8 complex, CI-8 (Glc-1 to Glc-8) was bound to the catalytic cleft (Fig. 5, C and D). The bound CI-8 took a distorted cyclic structure along the catalytic surface that consisted of loops 3 and 4 encircling the side chain of Met-310. Glc-1 and Glc-2 were located at the same position with those of the bound IG-6 and IG-8. Glc-8 of CI-8 was situated adjacent to Glc-1 and over the catalytic site. A covalent α-1,6-bond was also observed between the Glc-1 O1 atom and the Glc-8 O6 atom. Glc-3 was located at subsite −3, as observed for the bound IG-6 and IG-8, but the orientation of the sugar ring was different, and it was found to have formed a hydrogen bond between Glu-580 and the O3 atom. Glc-4, Glc-5, and Glc-6 had no direct hydrogen bonds with the protein and were situated on the hydrophobic surface that included residues Val-266, Phe-268, Met-310, and Arg-313. Glc-7 was located on the main chain of Gly-311–Gln-312 and bound by one hydrogen bond with the side chain of Asp-357. Glc-8 was located on the Trp-382 side chain, half-covered by Met-310 and bound by one hydrogen bond with the side chain of Glu-580. These two glucoses, Glc-7 and Glc-8, were considered to lie at subsites +1 and +2, respectively. The subsites for the bound CI-8 were designated after the nomenclature of the subsites for CGTase (42) as subsites −1, −2, −3, −4c, −5c, −6c, +2, and +1 (Figs. 4A and 5C).
In the BcCITase·IG-7 complex, IG-7 (Glc-1 to Glc-7) was bound to the catalytic cleft in a similar manner to CI-8 in the BcCITase·CI-8 complex (Fig. 4, A and D). Glc-1 to Glc-4 were located at the same position with CI-8, and Glc-6 and Glc-7 were located at the positions of Glc-7 and Glc-8 of CI-8. Glc-5 was located on the side chain plane of Arg-313 that corresponded to the mid-point of Glc-5 and Glc-6 of CI-8, forming the shorter α-1,6-glucan chain between Glc-4 and Glc-6. The anomeric hydroxyl group of Glc-1 adopted a β-configuration, and no covalent bond was observed with the O6 atom of Glc-7, a distance of 3.3 Å apart.
Sugar-binding Structure in BcCBM35-1
Domain B, BcCBM35-1, bound isomaltooligosaccharides at two sites. The second site was located at the side of the domain and recognized the nonreducing end glucose of IG-8, which extended from the catalytic site and worked as subsite −8, as mentioned above. Another site was the canonical sugar-binding site located on the top of the domain, where most CBM35s recognize their cognate sugars, involving the second calcium ion (Fig. 3D). This first site mainly recognizes one glucose moiety through stacking interactions with Trp-506 and hydrogen bonds by the side chains of His-439 and Asn-539, and the main chain N atom of Gln-450 (Fig. 3E). His-439, Trp-506, and Asn-539 are highly conserved among CBM35s, including Chi-CBM35 and Xyl-CBM35 (39), and are engaged in uronic sugar recognition in an identical manner. In these structures, an arginine residue on loop6 (the loop between β6 and β7) and the second conserved calcium ion interact with the carboxylic group of glucuronic acid. Isomaltooligosaccharides do not have a carboxylic group, and this position was occupied by the α-1,6-glucosyl bond to the adjacent glucose moiety. Thus, the arginine residue and calcium ion are not present in BcCBM35-1, because of the structural changes of the loops. Because of these structural changes, the first site of BcCBM35-1 could bind to the midpoint of the linear isomaltooligosaccharides. One glucose moiety on the reducing end side and two glucoses on the nonreducing end side of the bound glucose moiety (Glc-914, Fig. 3E) were commonly observed to link through α-1,6-glucosyl bonds in all ligand·complex structures. Intriguingly, the entire CI-8 molecule was observed in chain A of the BcCITase·CI-8 complex. The bound CI-8 took a constricted but symmetric cyclic structure, like a teacup, containing three inner hydrogen bonds that formed the bottom of the cup (Fig. 3F). The CI-8 molecule was found to wrap up the side chain of Trp-506, as if CI-8 had formed an inclusion complex.
DISCUSSION
Molecular Mechanism of Action for BcCBM35-1
A previous deletion mutant study (21) revealed that the presence of BcCBM35-1 is relevant to the predominant production of CI-8 by BcCITase, and our present ligand complex structures proved that BcCBM35-1 was involved in CI-8 production as shown in Fig. 6. The BcCITase·IG-8 complex structure showed that the IG-8 molecules bound in the catalytic cleft extending from subsite −1 at the catalytic site to subsite −8 in BcCBM35-1, whereas the BcCITase·CI-8 complex structure showed that the bound CI-8 was located in the catalytic cleft over the catalytic site. This implies that these structures correspond to the enzyme·substrate complex (Fig. 6, step B) and the enzyme·product complex (Fig. 6, step D) when this enzyme produced the product CI-8. These observations provide the CI-8 producing mechanism with the aid of BcCBM35-1 as follows. BcCBM35-1 binds the nonreducing end glucose of the substrate at the second sugar-binding site (Fig. 6, step A). The bound α-1,6-glucan folds into the catalytic cleft along the subsites −8 to −1 with eight glucose moieties occupied (Fig. 6, step B). The α-glucosidic linkage between Glc −1 and Glc +1 is cleaved at the catalytic site, and the glucose at subsite −1 of the rest is held by the catalytic nucleophile as an enzyme-substrate intermediate, at the same moment the nonreducing end glucose is released from subsite −8 and occupies subsite +1 by folding into a circular conformation around Met-310 (Fig. 6, step C). The glucose at subsite +1 then functions as an acceptor of the transglucosylation to complete the cyclization reaction (Fig. 6, step D). The mutational experiments at subsite −8 (Y515A or Y515G), in which the mutants showed reduced production of CI-8 (data not shown), also support the finding that BcCBM35-1 enhanced CI-8 production. Similar observations that the carbohydrate-binding module is involved with the product size have been reported for CGTases. The small domain E, belonging to CBM20, adjacent to the active site of CGTase has two sugar-binding sites, and a substrate-binding site with lower affinity guides the substrate to the active site (43). Mutational studies of CGTase showed that the amino acid residues at donor subsites −3, −6, and −7 influence the product sizes (3, 44).
FIGURE 6.
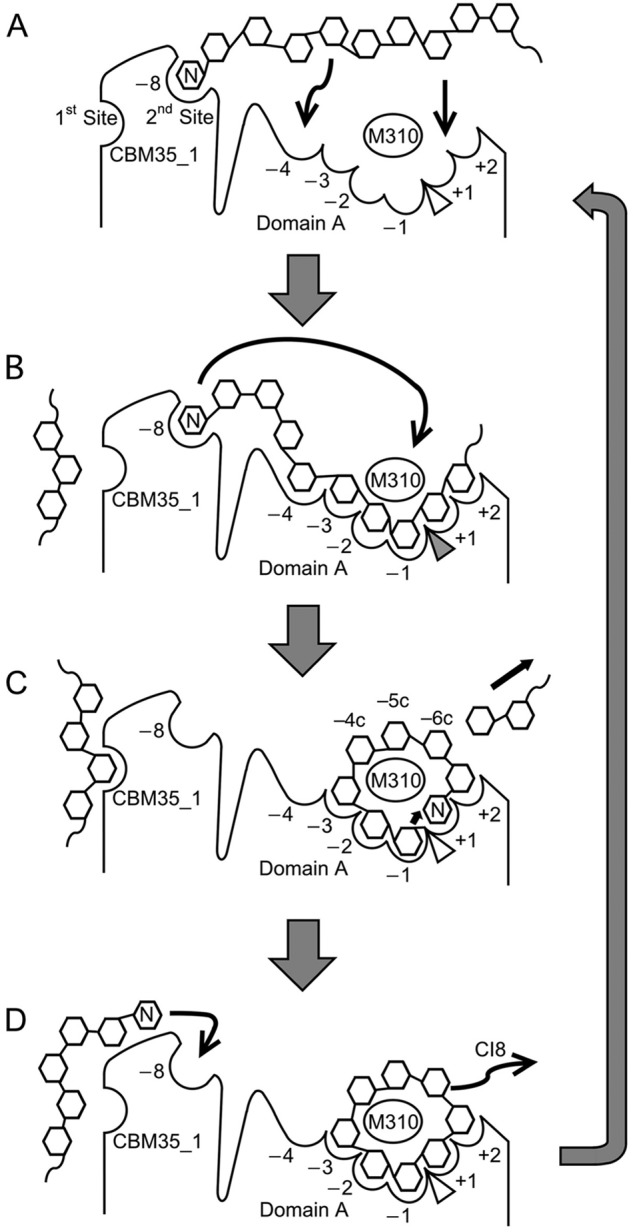
Schematic drawing of the CI-8-producing mechanism of BcCITase. A, substrate binding at the second sugar-binding site of BcCBM35-1. B, enzyme·substrate complex with eight glucose moieties occupied in the minus subsites. C, enzyme-substrate intermediate. D, enzyme·product complex. Incidentally, substrate recruitment at the conserved sugar-binding site is drawn.
The BcCITase·IG-7 complex structure showed that the bound IG-7 was located in the catalytic cleft with a circular structure, and IG-7 appeared to be the snapshot just before the cyclization reaction when this enzyme produced CI-7, with the nonreducing end glucoses Glc-6 and Glc-7 docked at subsites +2 and +1, respectively. The O6 atom of Glc-7 at subsite +1 is the closest transglucosylation acceptor with a proximity of 3.3 Å from the C1 atom at subsite −1. It also implied that IG-7 could not reach subsite −8 from the catalytic site, and that CI-7 was produced without the aid of subsite −8 in BcCBM35-1. The inability to produce CI-6 or smaller CIs can also be attributed to the structure of catalytic subsites. In the BcCITase·IG-7 and BcCITase·CI-8 complex structures, the catalytic site bound the circular sugar ligand with at least four subsites, namely −2, −1, +1, and +2, occupied, although in the BcCITase-IG-6 structure, bound IG-6 was observed only in −1 to −4 subsites. This indicates that the cyclization reaction involves at least four subsites −2, −1, +1, and +2, and IG-6 appears to be too short to occupy these subsites in a circular structure.
Sugar Binding in BcCBM35-1
The original function of BcCBM35-1 appears to be the binding of the substrate for efficient recruitment, as the canonical sugar-binding site (Figs. 1A, site B-1, and 3E)) was revealed to bind the mid-part of α-1,6-glucan, showing the type B glycan chain-binding architecture among the three sugar-binding types of carbohydrate-binding modules (45). This first binding site was located on the opposite side from the catalytic site in the three-dimensional structure of BcCITase, and hence its ligand binding appears to occur independently from the catalytic machinery. It may work to select the long substrate that could reach to the catalytic site so that CIs could be produced by the enzyme. Conversely, the second binding site, subsite −8 (Figs. 1A, site B-2, and 5A), showed the type C small sugar-binding architecture. BcCBM35-1 is the first example among the solved CBM35 structures that has two sugar-binding sites, although the presence of two sugar-binding sites has been reported for CBM6s, a related family of CBM35 (46, 47). More interestingly, this site is unique to BcCBM35-1 among CBM35s and is not conserved in BcCBM35-2 in BcCITase or the CBM35s of CITase from Paenibacillus sp. 598K strain (12). Paenibacillus CITase does not predominantly produce CI-8, but CI-7 instead, and this observation is consistent with the concept that this enzyme does not possess the second sugar-binding site.
Structural Determinant for Cyclization Activity
GH66 enzymes contain mainly two enzymes, endo-dextranase and CITase. Because all CITases have the inserted CBM35 (20) compared with dextranases, it had been considered that CITase activity might be attributed to CBM35, but the relationship remains unclear. Recently, both Bacteroides thetaiotaomicron dextranase lacking this inserted domain and Paenibacillus sp. endo-dextranases containing the CBM35 domain have been reported to show CI producing activity, although the activity of these two enzymes is reduced when compared with the activity of BcCITase (48, 49). Furthermore, deletion mutants, in which BcCBM35-1 is removed from BcCITase, still retain approximately one-twentieth of the CI producing activity of the wild-type enzyme (21). Therefore, CITase activity cannot be attributed wholly to CBM35, although CBM35 enhances the CITase activity. The crystal structures of BcCITase showed that BcCBM35-1 might contribute to the determination of product size preferences, as well as the usual task of CBMs in substrate recognition for efficient catalysis. BcCBM35-1 is distal from the cleavage site, and it would be difficult for the domain to affect the cyclization reaction.
Structural comparison of the catalytic cleft between BcCITase and SmDex displayed an obvious difference in the widths of the subsites +1 and +2, showing that the cleft of BcCITase is narrower than that of SmDex (Fig. 3, A and B). At subsite +1, Met-310 and Glu-580 in BcCITase, corresponding to Ile-387 and Ala-559 in SmDex, are involved in acceptor recognition and are possible candidates that enhance transglycosylation. At subsite +2, Gly-311, Gln-312, and Arg-313 provided van der Waals contact with the glucose in BcCITase, whereas the main chain trace of Gly-388–Asn-390 had shifted to make the cleft wider in SmDex.
Met-310, which is located at the center of the bound IG-7 or CI-8 molecules in these complex structures, appears to play a role similar to that of Tyr-195 of B. circulans 251 CGTases (50, 51). In this enzyme, substrate oligosaccharides are wrapped around this tyrosine residue upon the cyclization reaction, and mutational experiments of this residue cause a change in product profiles (50). In contrast with CGTases, GH13 α-amylases, which hydrolyze linear chains of α-1,4-glucans, have smaller residues at this position (52). In GH66, methionine is conserved only in CITases, whereas this residue is replaced by isoleucine in streptococcal dextranases and a variety of hydrophobic residues are found in other enzymes. Paenibacillus dextranase has tryptophan at this position and synthesizes relatively large CI molecules (48). Thus, the amino acid residues at this position might function in the determination of product size.
Glu-580 hydrogen bonded to the glucose at subsites +1 in the BcCITase-CI-8 and BcCITase·IG-7 complex structures. Interestingly, Glu-580 of BcCITase is also conserved in CITases and Paenibacillus dextranase, but this residue is replaced by alanine in streptococcal dextranases. Amino acid residues at the acceptor subsites +1 and +2 in CGTases have been shown to play important roles in the cyclization reaction. Mutational studies cause the conversion of enzymatic activities, from CGTases to maltogenic amylase (53) or from Novamyl, a maltose-forming maltogenic amylase, to a cyclodextrin-forming enzyme (54). With regard to BcCITase, the significance of residues at subsites +1 and +2 to their enzymatic activities requires further clarification, and our biochemical studies have not yet provided a clear solution on this issue. Thus, more careful and detailed structural and biochemical studies on BcCITase are necessary to clarify the factors determining the CI producing activity.
Only a few bacteria are known to possess CITases that produce CIs from dextran, which is not an abundant natural resource synthesized from sucrose by some lactic acid bacteria (55). CIs appear to be by-products that finally degraded to glucoses by CITase for energy use, and the biological function of CIs in bacteria remains unclear. CIs, however, show profitable properties for human life. They inhibit glucan synthesis by glucansucrase, applicable for dental caries prevention (13), and show inclusion complex-forming abilities similar to that observed for cyclodextrins that are used in various ways (10, 11). Furthermore, CIs show higher water solubility and a wider range of sizes from 7 to 17 degrees of polymerization (21). This is larger than that found for cyclodextrins that have three forms, and CIs are therefore expected to expand the use of cyclic glucans. Here, we have determined the crystal structure of CITase, elucidating its catalytic mechanism and providing the structural basis for enzymatic improvement. We hope that the elucidation of biological, chemical, and physical properties of CIs is accelerated and widespread use of CIs is developed.
Acknowledgments
We thank the beamline researchers and staff at the Photon Factory.
This work was supported in part by the Program for Promotion of Basic and Applied Research for Innovations in the Bio-oriented Industry (BRAIN, Japan) and by the scientific technique research promotion program for agriculture, forestry, fisheries and food industry (Japan).
- CGTase
- cyclodextrin glycosyltransferase
- BcCITase
- B. circulans T-3040 cycloisomaltooligosaccharide glucanotransferase
- BcCITase-CH
- recombinant BcCITase attached by a C-terminal His6 purification tag
- BcCITase-NH
- recombinant BcCITase attached by an N-terminal His6 purification tag
- Chi-CBM35
- carbohydrate-binding module family 35 domain of A. orientalis exo-β-d-glucosaminidase
- CI
- cycloisomaltooligosaccharide
- CITase
- cycloisomaltooligosaccharide glucanotransferase
- SeMet
- selenomethionine
- SmDex
- Streptococcus mutans endodextranase
- PDB
- Protein Data Bank.
REFERENCES
- 1. Qi Q., Zimmermann W. (2005) Cyclodextrin glucanotransferase: from gene to applications. Appl. Microbiol. Biotechnol. 66, 475–485 [DOI] [PubMed] [Google Scholar]
- 2. Tilden E. B., Hudson C. S. (1939) The conversion of starch to crystalline dextrins by the action of a new type of amylase separated from cultures of Aerobacillus macerans. J. Am. Chem. Soc. 61, 2900–2902 [Google Scholar]
- 3. van der Veen B. A., Uitdehaag J. C., Dijkstra B. W., Dijkhuizen L. (2000) Engineering of cyclodextrin glycosyltransferase reaction and product specificity. Biochim. Biophys. Acta 1543, 336–360 [DOI] [PubMed] [Google Scholar]
- 4. Li Z., Wang M., Wang F., Gu Z., Du G., Wu J., Chen J. (2007) γ-Cyclodextrin: a review on enzymatic production and applications. Appl. Microbiol. Biotechnol. 77, 245–255 [DOI] [PubMed] [Google Scholar]
- 5. Biwer A., Antranikian G., Heinzle E. (2002) Enzymatic production of cyclodextrins. Appl. Microbiol. Biotechnol. 59, 609–617 [DOI] [PubMed] [Google Scholar]
- 6. Loftsson T., Hreinsdóttir D., Stefánsson E. (2007) Cyclodextrin microparticles for drug delivery to the posterior segment of the eye: aqueous dexamethasone eye drops. J. Pharm. Pharmacol. 59, 629–635 [DOI] [PubMed] [Google Scholar]
- 7. Astray G., Gonzalez-Barreiro C., Mejuto J. C., Rial-Otero R., Simal-Gándara J. (2009) A review on the use of cyclodextrins in foods. Food Hydrocoll. 23, 1631–1640 [Google Scholar]
- 8. Oguma T., Horiuchi T., Kobayashi M. (1993) Novel cyclic dextrans, cycloisomaltooligosaccharides, from Bacillus sp. T-3040 culture. Biosci. Biotechnol. Biochem. 57, 1225–1227 [DOI] [PubMed] [Google Scholar]
- 9. Oguma T., Tobe K., Kobayashi M. (1994) Purification and properties of a novel enzyme from Bacillus spp. T-3040, which catalyzes the conversion of dextran to cyclic isomaltooligosaccharides. FEBS Lett. 345, 135–138 [DOI] [PubMed] [Google Scholar]
- 10. Funane K., Terasawa K., Mizuno Y., Ono H., Miyagi T., Gibu S., Tokashiki T., Kawabata Y., Kim Y. M., Kimura A., Kobayashi M. (2007) A novel cyclic isomaltooligosaccharide (cycloisomaltodecaose, CI-10) produced by Bacillus circulans T-3040 displays remarkable inclusion ability compared with cyclodextrins. J. Biotechnol. 130, 188–192 [DOI] [PubMed] [Google Scholar]
- 11. Funane K., Terasawa K., Mizuno Y., Ono H., Gibu S., Tokashiki T., Kawabata Y., Kim Y. M., Kimura A., Kobayashi M. (2008) Isolation of Bacillus and Paenibacillus bacterial strains that produce large molecules of cyclic isomaltooligosaccharides. Biosci. Biotechnol. Biochem. 72, 3277–3280 [DOI] [PubMed] [Google Scholar]
- 12. Suzuki R., Terasawa K., Kimura K., Fujimoto Z., Momma M., Kobayashi M., Kimura A., Funane K. (2012) Biochemical characterization of a novel cycloisomaltooligosaccharide glucanotransferase from Paenibacillus sp. 598K. Biochim. Biophys. Acta 1824, 919–924 [DOI] [PubMed] [Google Scholar]
- 13. Kobayashi M., Funane K., Oguma T. (1995) Inhibition of dextran and mutan synthesis by cycloisomaltooligosaccharides. Biosci. Biotechnol. Biochem. 59, 1861–1865 [DOI] [PubMed] [Google Scholar]
- 14. Oguma T., Kurokawa T., Tobe K., Kobayashi M. (1995) Cloning and sequence analysis of the cycloisomaltooligosaccharide glucanotransferase gene from Bacillus ciyculans T-3040 and expression in Escherichia coli cells. J. Appl. Glycosci. 42, 415–419 [Google Scholar]
- 15. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The carbohydrate-active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Igarashi T., Morisaki H., Yamamoto A., Goto N. (2002) An essential amino acid residue for catalytic activity of the dextranase of Streptococcus mutans. Oral Microbiol. Immunol. 17, 193–196 [DOI] [PubMed] [Google Scholar]
- 17. Kim Y. M., Kim D. (2010) Characterization of novel thermostable dextranase from Thermotoga lettingae TMO. Appl. Microbiol. Biotechnol. 85, 581–587 [DOI] [PubMed] [Google Scholar]
- 18. Yamamoto T., Terasawa K., Kim Y. M., Kimura A., Kitamura Y., Kobayashi M., Funane K. (2006) Identification of catalytic amino acids of cyclodextran glucanotransferase from Bacillus circulans T-3040. Biosci. Biotechnol. Biochem. 70, 1947–1953 [DOI] [PubMed] [Google Scholar]
- 19. Suzuki N., Kim Y. M., Fujimoto Z., Momma M., Okuyama M., Mori H., Funane K., Kimura A. (2012) Structural elucidation of dextran degradation mechanism by Streptococcus mutans dextranase belonging to glycoside hydrolase family 66. J. Biol. Chem. 287, 19916–19926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aoki H., Sakano Y. (1997) A classification of dextran-hydrolysing enzymes based on amino-acid-sequence similarities. Biochem. J. 323, 859–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Funane K., Kawabata Y., Suzuki R., Kim Y. M., Kang H. K., Suzuki N., Fujimoto Z., Kimura A., Kobayashi M. (2011) Deletion analysis of regions at the C-terminal part of cycloisomaltooligosaccharide glucanotransferase from Bacillus circulans T-3040. Biochim. Biophys. Acta 1814, 428–434 [DOI] [PubMed] [Google Scholar]
- 22. Suzuki S., Yukiyama T., Ishikawa A., Yuguchi Y., Funane K., Kitamura S. (2014) Conformation and physical properties of cycloisomaltooligosaccharides in aqueous solution. Carbohydr. Polym. 99, 432–437 [DOI] [PubMed] [Google Scholar]
- 23. Suzuki M., Unno T., Okada G. (1999) Simple purification and characterization of an extracellular dextrin dextranase from Acetobacter capsulatum ATCC 11894. J. Appl. Glycosci. 46, 469–473 [Google Scholar]
- 24. Suzuki N., Kim Y. M., Momma M., Fujimoto Z., Kobayashi M., Kimura A., Funane K. (2013) Crystallization and preliminary x-ray crystallographic analysis of cycloisomaltooligosaccharide glucanotransferase from Bacillus circulans T-3040. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 69, 946–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26. Terwilliger T. C. (2003) Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Terwilliger T. C. (2003) SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374, 22–37 [DOI] [PubMed] [Google Scholar]
- 28. Perrakis A., Morris R., Lamzin V. (1999) Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 29. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31. Vagin A., Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 32. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 33. Lawson C. L., van Montfort R., Strokopytov B., Rozeboom H. J., Kalk K. H., de Vries G. E., Penninga D., Dijkhuizen L., Dijkstra B. W. (1994) Nucleotide sequence and x-ray structure of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 in a maltose-dependent crystal form. J. Mol. Biol. 236, 590–600 [DOI] [PubMed] [Google Scholar]
- 34. Barends T. R., Bultema J. B., Kaper T., van der Maarel M. J., Dijkhuizen L., Dijkstra B. W. (2007) Three-way stabilization of the covalent intermediate in amylomaltase, an α-amylase-like transglycosylase. J. Biol. Chem. 282, 17242–17249 [DOI] [PubMed] [Google Scholar]
- 35. Jung J. H., Jung T. Y., Seo D. H., Yoon S. M., Choi H. C., Park B. C., Park C. S., Woo E. J. (2011) Structural and functional analysis of substrate recognition by the 250s loop in amylomaltase from Thermus brockianus. Proteins 79, 633–644 [DOI] [PubMed] [Google Scholar]
- 36. Przylas I., Tomoo K., Terada Y., Takaha T., Fujii K., Saenger W., Sträter N. (2000) Crystal structure of amylomaltase from Thermus aquaticus, a glycosyltransferase catalysing the production of large cyclic glucans. J. Mol. Biol. 296, 873–886 [DOI] [PubMed] [Google Scholar]
- 37. Imamura K., Matsuura T., Ye Z., Takaha T., Fujii K., Kusunoki M., Nitta Y. (2005) Crystallization and preliminary x-ray crystallographic study of disproportionating enzyme from potato. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61, 109–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Imamura H., Fushinobu S., Yamamoto M., Kumasaka T., Jeon B. S., Wakagi T., Matsuzawa H. (2003) Crystal structures of 4-α-glucanotransferase from Thermococcus litoralis and its complex with an inhibitor. J. Biol. Chem. 278, 19378–19386 [DOI] [PubMed] [Google Scholar]
- 39. Montanier C., van Bueren A. L., Dumon C., Flint J. E., Correia M. A., Prates J. A., Firbank S. J., Lewis R. J., Grondin G. G., Ghinet M. G., Gloster T. M., Herve C., Knox J. P., Talbot B. G., Turkenburg J. P., Kerovuo J., Brzezinski R., Fontes C. M., Davies G. J., Boraston A. B., Gilbert H. J. (2009) Evidence that family 35 carbohydrate binding modules display conserved specificity but divergent function. Proc. Natl. Acad. Sci. U.S.A. 106, 3065–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tunnicliffe R. B., Bolam D. N., Pell G., Gilbert H. J., Williamson M. P. (2005) Structure of a mannan-specific family 35 carbohydrate-binding module: evidence for significant conformational changes upon ligand binding. J. Mol. Biol. 347, 287–296 [DOI] [PubMed] [Google Scholar]
- 41. Nielsen M. M., Bozonnet S., Seo E. S., Mótyán J. A., Andersen J. M., Dilokpimol A., Abou Hachem M., Gyémánt G., Naested H., Kandra L., Sigurskjold B. W., Svensson B. (2009) Two secondary carbohydrate binding sites on the surface of barley α-amylase 1 have distinct functions and display synergy in hydrolysis of starch granules. Biochemistry 48, 7686–7697 [DOI] [PubMed] [Google Scholar]
- 42. Uitdehaag J. C., Kalk K. H., van Der Veen B. A., Dijkhuizen L., Dijkstra B. W. (1999) The cyclization mechanism of cyclodextrin glycosyltransferase (CGTase) as revealed by a γ-cyclodextrin-CGTase complex at 1.8-Å resolution. J. Biol. Chem. 274, 34868–34876 [DOI] [PubMed] [Google Scholar]
- 43. Penninga D., van der Veen B. A., Knegtel R. M., van Hijum S. A., Rozeboom H. J., Kalk K. H., Dijkstra B. W., Dijkhuizen L. (1996) The raw starch binding domain of cyclodextrin glycosyltransferase from Bacillus circulans strain 251. J. Biol. Chem. 271, 32777–32784 [DOI] [PubMed] [Google Scholar]
- 44. Leemhuis H., Kelly R. M., Dijkhuizen L. (2010) Engineering of cyclodextrin glucanotransferases and the impact for biotechnological applications. Appl. Microbiol. Biotechnol. 85, 823–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pires V. M., Henshaw J. L., Prates J. A., Bolam D. N., Ferreira L. M., Fontes C. M., Henrissat B., Planas A., Gilbert H. J., Czjzek M. (2004) The crystal structure of the family 6 carbohydrate binding module from Cellvibrio mixtus endoglucanase 5a in complex with oligosaccharides reveals two distinct binding sites with different ligand specificities. J. Biol. Chem. 279, 21560–21568 [DOI] [PubMed] [Google Scholar]
- 47. Henshaw J. L., Bolam D. N., Pires V. M., Czjzek M., Henrissat B., Ferreira L. M., Fontes C. M., Gilbert H. J. (2004) The family 6 carbohydrate binding module CmCBM6–2 contains two ligand-binding sites with distinct specificities. J. Biol. Chem. 279, 21552–21559 [DOI] [PubMed] [Google Scholar]
- 48. Kim Y. M., Kiso Y., Muraki T., Kang M. S., Nakai H., Saburi W., Lang W., Kang H.K., Okuyama M., Mori H., Suzuki R., Funane K., Suzuki N., Momma M., Fujimoto Z., Oguma T., Kobayashi M., Kim D., Kimura A. (2012) Novel dextranase catalyzing cycloisomaltooligosaccharide formation and identification of catalytic amino acids and their functions using chemical rescue approach. J. Biol. Chem. 287, 19927–19935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim Y. M., Yamamoto E., Kang M. S., Nakai H., Saburi W., Okuyama M., Mori H., Funane K., Momma M., Fujimoto Z., Kobayashi M., Kim D., Kimura A. (2012) Bacteroides thetaiotaomicron VPI-5482 glycoside hydrolase family 66 homolog catalyzes dextranolytic and cyclization reactions. FEBS J. 279, 3185–3191 [DOI] [PubMed] [Google Scholar]
- 50. Penninga D., Strokopytov B., Rozeboom H. J., Lawson C. L., Dijkstra B. W., Bergsma J., Dijkhuizen L. (1995) Site-directed mutations in tyrosine 195 of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 affect activity and product specificity. Biochemistry 34, 3368–3376 [DOI] [PubMed] [Google Scholar]
- 51. Wind R. D., Buitelaar R. M., Dijkhuizen L. (1998) Engineering of factors determining α-amylase and cyclodextrin glycosyltransferase specificity in the cyclodextrin glycosyltransferase from Thermoanaerobacterium thermosulfurigenes EM1. Eur. J. Biochem. 253, 598–605 [DOI] [PubMed] [Google Scholar]
- 52. Nakajima R., Imanaka T., Aiba S. (1986) Comparison of amino acid sequence of eleven different α-amylases. Appl. Microbiol. Biotechnol. 23, 355–360 [Google Scholar]
- 53. Leemhuis H., Rozeboom H. J., Wilbrink M., Euverink G. J., Dijkstra B. W., Dijkhuizen L. (2003) Conversion of cyclodextrin glycosyltransferase into a starch hydrolase by directed evolution: the role of alanine 230 in acceptor subsite +1. Biochemistry 42, 7518–7526 [DOI] [PubMed] [Google Scholar]
- 54. Beier L., Svendsen A., Andersen C., Frandsen T. P., Borchert T. V., Cherry J. R. (2000) Conversion of the maltogenic alpha-amylase Novamyl into a CGTase. Protein Eng. 13, 509–513 [DOI] [PubMed] [Google Scholar]
- 55. Robyt J. F., Yoon S. H., Mukerjea R. (2008) Dextransucrase and the mechanism for dextran biosynthesis. Carbohydr. Res. 343, 3039–3048 [DOI] [PubMed] [Google Scholar]