Background: Asparagine synthetase A catalyzes the synthesis of asparagine from aspartate in the presence of ammonia as a nitrogen donor.
Results: L. donovani ASNA enzyme is of bacterial origin and is both ammonia- and glutamine-dependent.
Conclusion: LdASNA is essential for survival of the Leishmania parasite.
Significance: The absence of LdASNA homologs from humans validates Leishmania ASNA as a novel drug target.
Keywords: Aminoacyl-tRNA Synthetase; Drug Design; Drug Development; Drug Discovery; Drug Resistance; Leishmania; Parasite; Leishmania, Drug Target
Abstract
Asparagine is formed by two structurally distinct asparagine synthetases in prokaryotes. One is the ammonia-utilizing asparagine synthetase A (AsnA), and the other is asparagine synthetase B (AsnB) that uses glutamine or ammonia as a nitrogen source. In a previous investigation using sequence-based analysis, we had shown that Leishmania spp. possess asparagine-tRNA synthetase paralog asparagine synthetase A (LdASNA) that is ammonia-dependent. Here, we report the cloning, expression, and kinetic analysis of ASNA from Leishmania donovani. Interestingly, LdASNA was both ammonia- and glutamine-dependent. To study the physiological role of ASNA in Leishmania, gene deletion mutations were attempted via targeted gene replacement. Gene deletion of LdASNA showed a growth delay in mutants. However, chromosomal null mutants of LdASNA could not be obtained as the double transfectant mutants showed aneuploidy. These data suggest that LdASNA is essential for survival of the Leishmania parasite. LdASNA enzyme was recalcitrant toward crystallization so we instead crystallized and solved the atomic structure of its close homolog from Trypanosoma brucei (TbASNA) at 2.2 Å. A very significant conservation in active site residues is observed between TbASNA and Escherichia coli AsnA. It is evident that the absence of an LdASNA homolog from humans and its essentiality for the parasites make LdASNA a novel drug target.
Introduction
Leishmania donovani is a protozoan parasite that causes visceral leishmaniasis, a disease that is fatal if left untreated. Visceral leishmaniasis treatment primarily relies on chemotherapy due to problems related to vector control and lack of an effective vaccine to treat the disease (1). Because of the development of resistance against currently available antileishmanial drugs (2, 3), there is a growing need for discovering novel drug targets and developing new inhibitors.
Asparagine synthetase (l-aspartate:ammonia ligase (AMP-forming), EC 6.3.1.1) is an enzyme that catalyzes the synthesis of asparagine from aspartate using adenosine triphosphate (ATP) as the energy source in the presence of a nitrogen donor (4). The nitrogen donor can be glutamine or ammonia. Two families of asparagine synthetases are known to date. One is asparagine synthetase A (AsnA),6 which utilizes nitrogen only from an ammonia source (4). AsnA family members have been reported from prokaryotes such as Escherichia coli and Klebsiella aerogenes (5, 6) as well as from archaea such as Pyrococcus abyssi (7, 8). The other family is asparagine synthetase B (AsnB) whose members are found in prokaryotes and eukaryotes (9–11). Members of the AsnB family preferentially utilize glutamine as the nitrogen source, although they are capable of utilizing both glutamine and ammonia. E. coli and K. aerogenes have two asparagine synthetase genes, asnA and asnB, and the presence of either ensures sufficient asparagine biosynthesis, whereas inactivation of both causes asparagine auxotrophy (12). Although they are eukaryotes, Leishmania and Trypanosoma surprisingly possess both ASNA (LmjF.26.0830) and ASNB (LmjF.29.1490) (13).
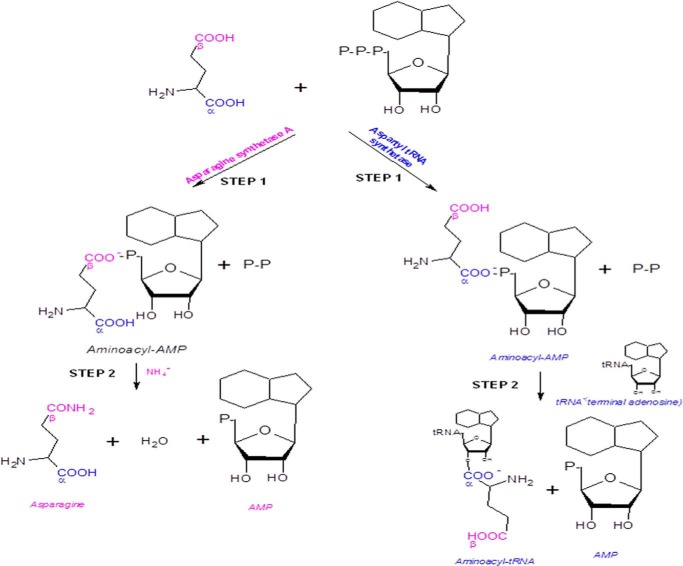
Asparagine is formed in two steps: the β-carboxylate group of aspartate is first activated by ATP to form an aminoacyl-AMP before its amidation by a nucleophilic attack with an ammonium ion. Interestingly, this mechanism of amino acid activation resembles that used by aminoacyl-tRNA synthetases, which first activate the α-carboxylate group of an amino acid to form also an aminoacyl-AMP before they transfer the activated amino acid onto cognate tRNA (Fig. 1) (4). We had previously reported a novel asparagine-tRNA synthetase paralog, asparagine synthetase A from Leishmania major (13). These paralogs retain the typical aminoacyl-tRNA synthetase catalytic domain with class II motifs but are devoid of the anticodon binding domain (14). Such paralogs have been reported only from prokaryotes and archaea so far (8, 15) where they participate in amino acid biosynthesis (e.g. AsnA and HisZ) and lysylation of a specific lysine in elongation factor P (GenX/PoxA) (8, 14, 16–20).
FIGURE 1.
Schematic representation of the reaction mechanisms of asparagine synthetase A and aspartyl-tRNA synthetase enzymes. Step 1 involves activation of the selective carboxyl groups by the respective enzymes to form aminoacyl-AMP. Step 2 involves amidation of the aminoacyl-AMP by asparagine synthetase A and transfer of the activated amino acid to the cognate tRNA by aspartyl-tRNA synthetase.
Asparagine biosynthesis is an unexplored area in kinetoplastids. Despite being eukaryotes, kinetoplastids and other parasites such as Trichomonas vaginalis, Entamoeba histolytica, and Cryptosporidium parvum possess an ammonia-dependent asparagine synthetase (AsnA-type) enzyme (13). Detailed sequence-based phylogenetic analysis reveals a close relationship of these parasitic ASNA enzymes to the ammonia-dependent AsnA-type enzymes of E. coli and other bacteria (13). A recent report indicates that asparagine synthetase A-knocked down T. brucei were auxotrophic to asparagine (21). To date, crystal structures of AsnA-type enzymes from E. coli and P. abyssi are known (8, 22). We report, for the first time, identification, molecular cloning, expression, and enzymatic characterization of a eukaryotic ASNA-type enzyme from Leishmania donovani. We additionally crystallized and solved the atomic structure of the L. donovani ASNA (LdASNA) homolog from Trypanosoma brucei (TbASNA). L. donovani and T. brucei share a sequence identity of ∼80%; therefore, the crystal structure of TbASNA provides a platform for structure-function studies on these enzymes. Gene replacement study indicates that the enzyme asparagine synthetase A (ASNA) from Leishmania donovani plays an essential role in the viability of this pathogenic organism.
EXPERIMENTAL PROCEDURES
Materials
All restriction enzymes and DNA-modifying enzymes were obtained from MBI Fermentas (Germany). Neomycin, paromomycin, blasticidin, and hygromycin were obtained from Sigma-Aldrich. Plasmid pET-30a was obtained from Novagen. Protein markers and DNA ladders were acquired from New England Biolabs. l-Aspartic acid, l-glutamine, lactate dehydrogenase, myokinase, and pyruvate kinase were obtained from Sigma-Aldrich. Other materials used in this study were of analytical grade and were commercially available.
Leishmania Strains and Culture Conditions
L. donovani Bob (LdBob strain/MHOM/SD/62/1SCL2D) promastigotes were cultured at 22 °C in M199 medium (Sigma) supplemented with 100 units/ml penicillin (Sigma), 100 μg/ml streptomycin (Sigma), and 5% heat-inactivated fetal bovine serum (Hyclone). Medium was additionally supplemented with 10 μg/ml hemin. Genetically manipulated parasites were derived from wild type L. donovani Bob originally obtained from Dr. Stephen Beverley (Washington University, St. Louis, MO). Wild type (WT) parasites were routinely cultured in medium with no drug supplementations, whereas the ASNA heterozygotes (ASNA/HYG and ASNA/NEO) and ASNA/NEO/HYG parasites were maintained in either 150 μg/ml hygromycin, 300 μg/ml paromomycin, or both, respectively. The ASNA overexpressor (ASNAOE) parasites were maintained in 10 μg/ml blasticidin. pSP72-α-neo-α-GFP-ASNA-transfected parasites were maintained in 40 μg/ml G418. For characterizing the single transfectant, double transfectant, and ASNA-overexpressing parasites phenotypically, cells were subcultured without the selection marker prior to experiments.
Construction of Expression Vectors and Purification of the Proteins
A 1062-bp DNA fragment encompassing the whole open reading frame (ORF) of the LdASNA gene was amplified from genomic DNA using a sense primer with a flanking BamHI site (5′-CGGGATCCATGTCGTCCAGTCCGCAGGAGTACATT-3′) and an antisense primer with a flanking HindIII site (5′-CCCAAGCTTTTACAATAAGGAGTACTGCGTCGTGACCTC-3′). A single ∼1062-bp PCR product was obtained. The amplified product was cloned into BamHI and HindIII restriction sites of pET-30a vector using T4 DNA ligase (New England Biolabs). The fidelity of the PCR amplification of LdASNA was confirmed by automated DNA sequencing. Sequence comparison of ASNA was done using the NCBI search algorithm BLAST. Multiple amino acid sequence alignment was performed using the program ClustalW with default parameters (23). The T. brucei (TbASNA-pET28a) construct was kindly provided by Dr. Cordeiro da Silva (Institute for Molecular and Cell Biology, University of Porto, Portugal) (21). LdASNA-pET30a or TbASNA-pET28a constructs containing a His6 tag at the N terminus were transformed into E. coli BL-21 strain. Expression from the constructs LdASNA-pET30a or TbASNA-pET28a was induced at an A600 nm of 0.6 with 0.1 mm isopropyl 1-β-d-galactopyranoside at 16 °C for 16 h. Bacteria were then harvested by centrifugation at 5000 × g for 10 min, and the cell pellet was resuspended in lysis buffer (50 mm Tris-Cl, pH 7.4, 10 mm imidazole, 300 mm sodium chloride, 2 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture). The resulting cell suspension was sonicated six times for 30 s at 1-min intervals. The lysate was centrifuged at 10,000 × g for 30 min at 4 °C. The resulting supernatant, which contained the protein, was loaded onto pre-equilibrated Ni2+-nitrilotriacetic acid-agarose resin (Qiagen). The protein was eluted with increasing concentrations of imidazole. The purified protein was found to be >95% pure as judged by SDS-PAGE.
TbASNA was used for crystallization studies. The purest fractions were checked by SDS-PAGE and concentrated by using a 10-kDa-cutoff Centricon centrifugal device (Millipore). The protein was further purified by gel filtration chromatography on an S200 analytical gel filtration column (GE Biosciences) in buffer containing 50 mm Tris-Cl, 200 mm NaCl, and 10 mm β-mercaptoethanol, pH 7.5, for apoenzyme crystallization. The final protein was concentrated to ∼73 mg/ml (using A280 nm) and stored at −80 °C. The concentration of the purified protein was determined using Bradford reagent (Sigma).
Sequence Identification Decoding Analysis of ASNA Protein by MALDI-TOF
After protein visualization, the ASNA protein band was excised from SDS-PAGE gel and subjected to matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF/TOF). The MALDI-TOF mass spectra were recorded with an Autoflex II mass spectrometer (Bruker Daltonics, Germany) installed in the Advanced Instrumentation Facility, Jawaharlal Nehru University (New Delhi, India). The obtained spectra were calibrated externally with software available with the instrument. Initial sequences were subjected to flexControl and BioTools. Final sequence identification and scoring was performed using the Mascot search tool.
Asparagine Synthetase Assay
LdASNA-catalyzed asparagine synthesis activity was measured in a coupled enzyme system at 37 °C using a Varian Cary® 100 UV-visible spectrophotometer. The formed product (AMP) was coupled to an adenylate kinase, pyruvate kinase, and lactate dehydrogenase reaction for oxidation of NADH to NAD. The resultant decrease of NADH was monitored at 340 nm (4). The reaction mixture (1.0 ml) contained 100 mm Tris-HCl, pH 7.8, 1.5 mm l-aspartic acid, 20 mm NH4Cl, 3 mm ATP, 100 mm KCl, 10 mm (CH3COO)2Mg, 1 mm phosphoenolpyruvate, 0.24 mm of NADH, 5 mm DTT, 2 units of myokinase, 10 units of pyruvate kinase, and 25 units of lactate dehydrogenase. Kinetic analysis was done using 20 μg of the enzyme. Progress of the reaction was scanned against a buffer blank for 1200 s with an interval of 30 s at a wavelength of 340 nm. Determination of the Km and Vmax for l-aspartate, ATP, and NH4+ was achieved by varying the concentration of each component in the reaction mixture while maintaining others in excess. To confirm substrate specificity of the enzyme, l-glutamine was used as a nitrogen source instead of NH4Cl. The LdASNA activity in cell lysates of WT Bob, ASNAOE, and heterozygous (ASNA/HYG and ASNA/NEO) parasites was determined as reported above. Cell lysates of all the strains were prepared by repeated freeze/thaw. Statistical analyses were undertaken using GraphPad Prism 5.0 (GraphPad Software, Inc.).
Cloning of the pSP-α-blast-α-ASNA and pSP72-α-neo-α-GFP-ASNA Episome
For overexpression of ASNA gene in L. donovani, the 1062-bp ASNA ORF was amplified by PCR using the forward primer 5′-TTTTTCTAGAATGTCGTCCAGTCCGCAGGAGTACATT-3′ with a flanking XbaI site (underlined) and the reverse primer 5′CCCAAGCTTTTACAATAAGGAGTACTGCGTCGTGACCTC-3′ with a flanking HindIII site (underlined). The amplified DNA fragment LdASNA was cloned into the XbaI-HindIII site of pSP-α-blast-α vector (Leishmania-specific vector) containing a blasticidin acetyltransferase gene as the selection marker as reported earlier (24). The LdASNA-GFP construct was made by cloning LdASNA in pSP72-α-neo-α-GFP vector at BamHI and XbaI sites. The forward primer 5′-TTTGGATCCATGTCGTCCAGTCCGCAGGAGTACATT-3′ (BamHI site underlined) and the reverse primer 5′-TTTTTCTAGACAATAAAGAGTACTGCGCCGTGACCTC-3′ (XbaI site underlined) were used for amplification. Correct orientation and sequence fidelity of the inserts were verified by nucleotide sequence analysis.
Localization of ASNA in L. donovani
pSP72-α-neo-α-GFP-ASNA-transfected promastigotes were used for studying the localization of ASNA in L. donovani. Fluorescence imaging of stabilized culture was performed using a confocal laser scanning microscope (Zeiss LSM 510 META) equipped with a 63× objective at an excitation wavelength of 488 nm. Briefly, 106 promastigotes/ml were pelleted and resuspended in phosphate-buffered saline (PBS) containing 1% fetal bovine serum (FBS) with identical final cell concentration. Cells were pelleted and resuspended in 500 μl of ice-cold 4% paraformaldehyde and incubated on ice for 20 min. Promastigotes were then washed with PBS before treating them with 0.5% Triton X-100 in PBS for 5 min at room temperature. DAPI (0.5 μg/ml) (Invitrogen) was added and washed with PBS before microscopy. Parasites stained with DAPI were observed at an excitation wavelength of 405 nm. Furthermore, to investigate possible mitochondrial localization, 107 promastigotes/ml were pelleted and stained with MitoTracker Red-CMXRos (1 nm) (Molecular Probes, Eugene, OR) in fresh medium for 10–15 min to locate mitochondria. The cells were then washed twice with PBS containing 1% FBS and resuspended in the same PBS solution with identical final cell concentration. Promastigotes were then immobilized on poly-l-lysine-coated glass slides. For observing fluorescence of parasites stained with MitoTracker Red-CMXRos, the parasites were visualized using a confocal laser scanning microscope at an excitation wavelength of 568 nm.
Molecular Constructs for the Replacement of the ASNA Alleles
A targeted gene replacement strategy was used for inactivation of ASNA gene in L. donovani. A fusion PCR-based strategy was used as reported earlier (25). Briefly, ASNA-flanking regions were amplified from L. donovani wild type genomic DNA and ligated to hygromycin phosphotransferase gene (HYG) or neomycin phosphotransferase gene (NEO). The 5′-UTR (1000 bp) of L. donovani ASNA was obtained by PCR amplification with primers A and BHyg or primers A and BNeo (Table 1). The NEO gene was amplified from pX63-NEO with primers CNeo and DNeo. The HYG gene was amplified from pX63-HYG with primers CHyg and DHyg (Table 1). The 3′-UTR (750 bp) of L. donovani ASNA was obtained from L. donovani wild type genomic DNA by PCR amplification using primer EHyg and reverse primer F or with primer ENeo and reverse primer F (Table 1). The 5′-UTR of L. donovani ASNA was then ligated to the antibiotic resistance marker gene by PCR using primers A and DHyg or A and DNeo. This fragment (5′-UTR-marker gene) was fused with 3′-UTR of ASNA using primers A and F, yielding the fragment 5′-UTR-Hyg-3′-UTR or 5′-UTR-Neo-3′-UTR.
TABLE 1.
Primers used for generation of the hygromycin (Hyg)- and neomycin (Neo)-specific linear replacement cassette fragments
| L. donovani primers | Sequences |
|---|---|
| A | 5′-ACAGCGAACGAAATCGAGCG-3′ |
| BHyg | 5′-GGTGAGTTCAGGCTTTTTCATGGCTGGCAGTGAAAGAAAAAGGGATGAATGGA-3′ |
| CHyg | 5′-TCCATTCATCCCTTTTTTCTTTCACTGCCAGCCATGAAAAAGCCTGAACTCAC-3′ |
| DHyg | 5′-GTTCCACCACCCTCCCCCGTCTTTCTATTCCTTTGCCCTCGGACGAG-3′ |
| EHyg | 5′-CTCGTCCGAGGGCAAAGGAATAGAAAGACGGGGGAGGGTGGGTGGGAACGCCT-3′ |
| BNeo | 5′-AATCCATCTTGTTCAATCATGGCTGGCAGTGAAAGAAAAAGGGATGAATGGA-3′ |
| CNeo | 5′-TCCATTCATCCCTTTTTTCTTTCACTGCCAGCCATGATTGAACAAGATGGATT-3′ |
| DNeo | 5′-GTTCCACCACCCTCCCCCGTCTTTTCAGAAGAACTCGTCAAGAAG-3′ |
| ENeo | 5′-CTTCTGACGAGTTCTTCTGAAAAGACGGGGGAGGGTGGGTGGGAACGCCT-3′ |
| F | 5′-AGGCGTCGCTCCGCTCTGCACTTTC-3′ |
Generation of Genetically Manipulated Parasites
After PCR amplification and purification, ∼2–3 μg of linear replacement cassette fragment 5′-UTR-Hyg-3′-UTR or 5′-UTR-Neo-3′-UTR were individually transfected by electroporation in wild type L. donovani promastigotes (26). Depending on the marker gene, transfectants were selected in the presence of 150 μg/ml hygromycin (Sigma) or 300 μg/ml paromomycin (Sigma). After three to four passages, cells resistant to antibiotic selection were subjected to PCR-based analysis to check for the correct integration of the replacement cassettes using primers shown in Table 2. Thereafter, a second round of transfection was initiated to knock out the other copy of ASNA gene. The replacement cassette fragment 5′-UTR-Hyg-3′-UTR was used to transfect ASNA/NEO heterozygous parasites, and 5′-UTR-Neo-3′-UTR was used to transfect ASNA/HYG heterozygous parasites to generate homozygous ASNA gene deletion mutants. A similar PCR-based analysis was carried out to check for the correct integration of the replacement cassettes as was done in the case of ASNA heterozygotes. Furthermore, the genotypes of the ASNA mutants were confirmed by Southern analysis using standard protocols (27).
TABLE 2.
Primers used for the molecular characterization of the genetically manipulated parasites by PCR-based analysis
| L. donovani primers | Sequences |
|---|---|
| Primer 1 | 5′-TGTAGAAGTACTCGCCGATAGTGG-3′ |
| Primer 2 | 5′-CGACACAATACTCTGGCGCTGGAGC-3′ |
| Primer 3 | 5′-CGCAGCTATTTACCCGCAGGACAT-3′ |
| Primer 4 | 5′-ATGACGGAGGCCCTCTCCTTTTTGC-3′ |
| Primer 5 | 5′-ATAGCGTTGGCTACCCGTGATATTGC-3′ |
| Primer 6 | 5′-AACACGGCGGCATCAGAGCAGCCGATTG-3′ |
| Primer 7 | 5′-CGAGGTCACGGCGCAGTACTCTTTA-3′ |
| Primer 8 | 5′-GGTCTGGAGATCAATGTACTCCTGC-3′ |
The L. donovani strain overexpressing ASNA was made by transfecting wild type L. donovani promastigotes with the pSP-α-blast-α-ASNA construct. The transfected cells were maintained in 10 μg/ml blasticidin. Wild type L. donovani transfected with pSP-α-blast-α vector was used for comparison. 20 μg of pSP72-α-neo-α-GFP-ASNA plasmid was transfected into the wild type L. donovani promastigotes for localization studies. The transfected cells were maintained in 40 μg/ml G418. A control transfection with vector alone was used for comparison.
Growth Studies
Growth rate experiments were conducted by inoculating stationary phase parasites at a density of 1 × 106 cells/ml in M199 medium with 5% FBS in 25-cm2 flasks without respective selection drug and culturing at 22 °C. Growth rates of each of the cultures were determined at 24-h intervals by using a Neubauer hemocytometer. Growth studies with each individual cell line were done at least three times, and similar results were obtained each time.
Crystallization, Data Collection, and Structure Determination
TbASNA crystals (apo form) were obtained at 20 °C by the hanging drop vapor diffusion method using 1 μl of TbASNA and 1 μl of 10% (w/v) PEG 20000, 20% (v/v) PEG monomethyl ether 550, 0.03 m NPS (sodium nitrate, disodium hydrogen phosphate, ammonium sulfate), and 0.1 m MOPS/HEPES-Na, pH 7.5. A plate-shaped single crystal was directly mounted in cooled nitrogen gas at 100 K. X-ray diffraction data were collected on a MAR CCD x-ray detector at BM14 beam line of the European Synchrotron Radiation Facility at Grenoble, France. A total of 360 images were collected with 12-s exposure each and 0.5° oscillations per frame. The diffraction images were processed and scaled with HKL2000 (28). The initial model was built by AutoBuild in PHENIX (29), which was subsequently rebuilt manually using Coot (30). Model refinement was performed using phenix.refine in PHENIX. Structural superimposition and figures were generated using Chimera (31). Coordinates and structure factors for TbASNA have been deposited in the RCSB Protein Data Bank under accession code 4LNS.
Statistical Analysis
Data are represented as mean ± S.D. A value of p < 0.05 was accepted as an indication of statistical significance. Results for enzyme activity in the cell lysates were entered as column data in GraphPad Prism 5.0 (GraphPad Software, Inc.) and analyzed by Student's t test.
RESULTS
Sequence Analysis and Genomic Organization
ASNA sequences have been identified in each of the Leishmania infantum and L. major genomes (see the European Molecular Biology Laboratory-European Bioinformatics Institute web site). ASNA sequences have also been identified in the T. brucei 927 (Tb927.7.1110) and Trypanosoma cruzi Es genomes (Tc00.1047053503899.90). In addition to kinetoplastids, BLAST searches against EuPathDB (32) suggest that parasites such as T. vaginalis (A2EKG5), E. histolytica (C4LT17), and C. parvum (Q5CPD9) possess a copy of ASNA gene. BLAST analysis of L. major and L. infantum genomes revealed that the ASNA-encoding gene is present on chromosome 26. Trypanosoma and Leishmania contain a single copy of ASNA gene (33). Sequence analysis, database search, and alignment of the LdASNA amino acid sequence were performed. The LdASNA amino acid sequence had a single open reading frame consisting of 1062 bp. The ORF encoded a putative polypeptide of 353 amino acids with a predicted molecular mass of 40 kDa. Sequence analysis suggests that asparagine synthetase A enzymes from kinetoplastids share reasonably high sequence identity (∼60%). E. coli AsnA is 58% identical to LdASNA and 60% identical to TbASNA. Lactobacillus delbrueckii and E. histolytica share sequence identity of ∼45% with LdASNA and TbASNA. Archaeal sequences are distant homologs with low (<15%) identity to LdASNA and TbASNA sequences. Phylogenetic analysis reveals a close relationship of kinetoplastid enzymes to E. coli and other bacterial ammonia-dependent AsnA-type enzymes (13).
Cloning, Overexpression, and Purification of LdASNA
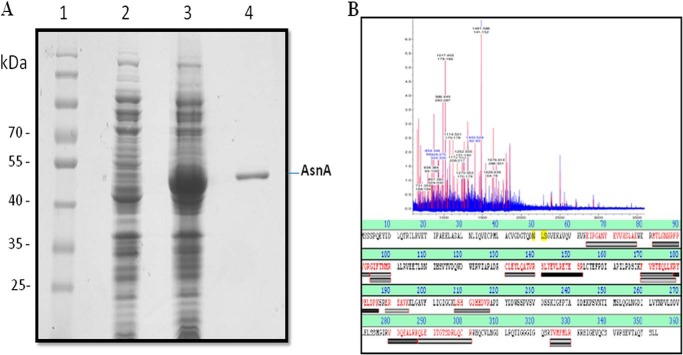
LdASNA is predicted to be of ∼40-kDa molecular mass with a theoretical pI of 5.4 using the ExPASy bioinformatics tool ProtParam. To characterize the recombinant LdASNA protein, the full-length gene was cloned into the backbone fragment of pET-30a expression vector. The resultant construct pET30a-LdASNA with an N-terminal His6 tag and thrombin protease cleavage site was transformed into E. coli (BL21). A single band of homogenously purified LdASNA protein with an N-terminal His6 tag was identified with a molecular mass of ∼44 kDa (Fig. 2A). Purification yielded ∼3 mg of purified protein/liter of bacterial culture. To further characterize LdASNA, the purified protein was analyzed by MALDI-TOF/TOF mass spectroscopy (Fig. 2B). The spectrum of the protein analyzed by BioTools version 2.2 showed an intensity coverage of 35.8% for putative asparagine synthetase A (L. infantum JPCM5). The polypeptide sequence fragments covered in the MS/MS data for each peak are colored red in Fig. 2B. A sequence search analysis by a Mascot search revealed top scores of 99 and 74 with putative asparagine synthetase A of Leishmania spp. with accession numbers gi|146089599 and gi|398016957, respectively. These results confirm the presence of a putative asparagine synthetase A in L. donovani.
FIGURE 2.
A, purification of recombinant LdASNA protein on Ni2+-nitrilotriacetic acid affinity resin. Lane 1, molecular mass marker; lane 2, uninduced cell lysate; lane 3, induced cell lysate; lane 4, eluted fraction with 200 mm imidazole showing purified LdASNA. B, MALDI-TOF peptide mass fingerprinting and alignment of the identified peptides of a tryptic digest of LdASNA protein obtained after nickel-nitrilotriacetic acid purification.
Enzymatic Activity and Kinetic Parameters for LdASNA
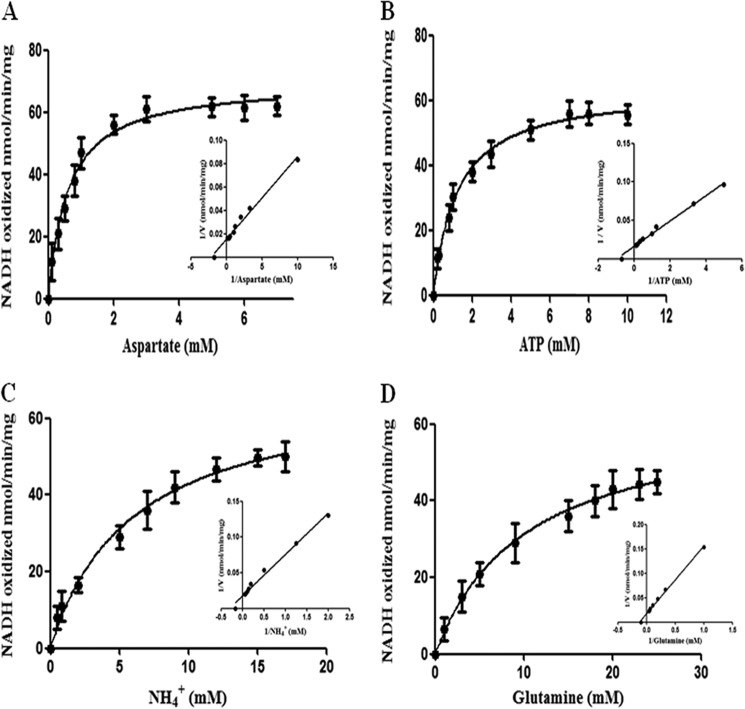
Asparagine synthetase A catalyzes the synthesis of asparagine, AMP, and PPi in the presence of ATP, l-aspartate, and NH4+ (4). To determine any ammonium-dependent asparagine synthesis activity of LdASNA, the product AMP was coupled with an NADH oxidation reaction (4). In the case of prokaryotic organisms, ammonium- and glutamine-dependent asparagine synthesis activity, each corresponding to a nitrogen source of NH4+ or glutamine, respectively, has been reported (10, 15). In archaea bacteria, e.g. P. abyssi, asparagine formation occurs only by the process of amidation of aspartate (34). To establish the kinetic parameters and specificity of LdASNA using l-aspartate, ATP, and N2 source, we examined the effect of various concentrations of substrate l-aspartate while other components were kept constant (Fig. 3A). The Vmax value for l-aspartate was determined to be 69.6 ± 2 nmol min−1 mg of protein−1. The Km value of LdASNA for l-aspartate was measured to be 0.6 ± 0.08 mm, which is closer to that reported in the case of K. aerogenes (0.91 mm) (6). ATP is an essential component for substrate amidation during the asparagine synthesis reaction. Therefore, we performed ATP-dependent enzyme kinetic studies (Fig. 3B). In the case of LdASNA, the estimated Vmax value was 63.5 ± 1.3 nmol min−1 mg of protein−1, and the Km for ATP was 1.2 ± 0.09 mm. To establish the N2 source specificity, we measured LdASNA activity with various concentrations of NH4+ and l-glutamine (Fig. 3, C and D). An LdASNA Vmax value of 68.4 ± 3.6 nmol min−1 mg of protein−1 and Km value of 5.95 ± 0.8 mm was obtained for NH4+, which is closer to that reported for Streptococcus bovis (4.0 mm) and T. brucei (5.55 mm) (21, 35). Interestingly, LdASNA was found to be capable of utilizing l-glutamine as the nitrogen source. This feature never reported before appears to be unique for kinetoplastid ASNAs as has also been recently reported in case of T. brucei and T. cruzi (21). The kinetic parameters obtained for the utilization of l-glutamine for LdASNA were a Vmax value of 63.4 ± 1.8 nmol min−1 mg of protein−1 and Km value of 10.3 ± 0.7 mm. Hence, we conclude that LdASNA is active and preferentially utilizes ammonia, although it is also capable of utilizing glutamine as a nitrogen source (Fig. 3, C and D).
FIGURE 3.
Michaelis-Menten and Lineweaver-Burk plots (inset) for conversion of l-aspartic acid to asparagine by LdASNA enzyme. The enzyme assay was carried out as described under “Experimental Procedures.” A–D, kinetic constants were computed using the Michaelis-Menten algorithm within GraphPad Prism 5.0 for utilization of l-aspartate, ATP, NH4Cl, and l-glutamine by LdASNA enzyme. Results are representative data from three separate experiments and are represented as mean ± S.E. (error bars).
Subcellular Localization of LdASNA
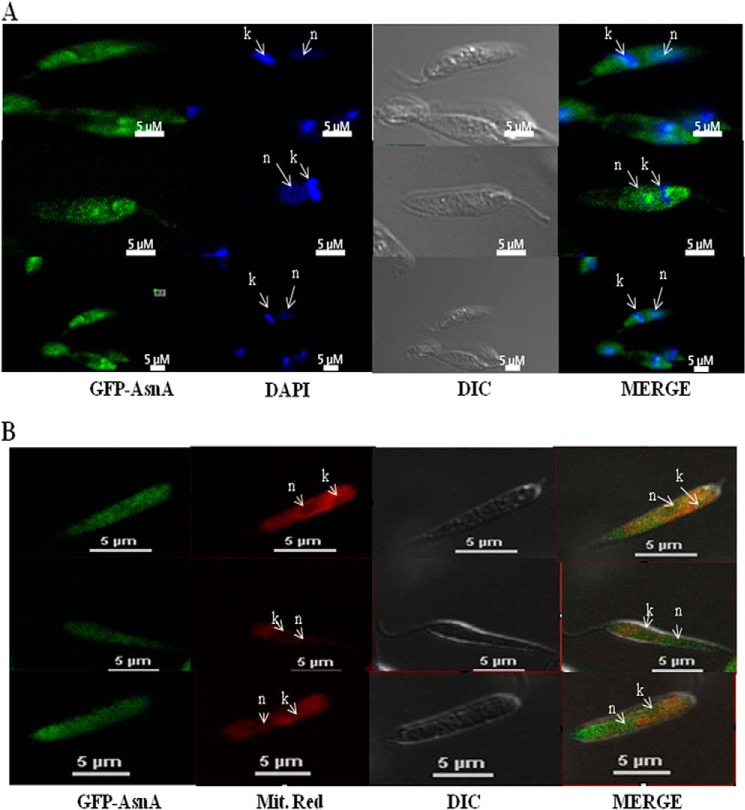
The GFP-LdASNA construct was designed for episomal expression in L. donovani parasites as described under “Experimental Procedures.” This construct was electrotransformed in parasites and selected as reported earlier (36). L. donovani parasites transfected with the GFP-LdASNA fusion constructs were fixed and analyzed by fluorescence microscopy. GFP fluorescence was detectable in ∼60% of the cells transfected with the fusion constructs. The kinetoplast and nuclear DNA in these cells were readily identified by their bright staining with DAPI (Fig. 4A). GFP-LdASNA was found to be present in the cytoplasm of the parasite (Fig. 4A). We repeated localization studies several times and viewed ∼90 different parasites. The parasites transfected with the GFP vector alone (lacking an insert) showed GFP fluorescence in the entire promastigote (data not shown). L. major ASNA appears to have predicted mitochondrial (PSORT II) and cytosolic (MARSPred) localization. However, the mitochondrion-targeting signal peptide sequences and the cleavage sites could not be predicted by any of the currently available computational tools. Therefore, we wanted to probe mitochondrial localization of LdASNA, so GFP-LdASNA construct-transfected parasites were stained with MitoTracker Red (Fig. 4B). Our data indicate localization of ASNA in both the cytosol and mitochondria of the parasites.
FIGURE 4.
A, confocal microscopy of wild type L. donovani transfected with pSP72-α-neo-α-GFP-ASNA. First panel, wild type L. donovani expressing LdASNA as a GFP translational fusion protein; second panel, wild type L. donovani stained with DAPI; third panel, phase-contrast image; fourth panel, merged micrograph. B, co-localization of LdASNA in mitochondria. First panel, wild type L. donovani expressing LdASNA as a GFP translational fusion protein; second panel, wild type L. donovani stained with MitoTracker Red; third panel, phase-contrast image; fourth panel, merged micrograph. “k” and “n” indicate kinetoplastid and nuclear DNA, respectively. Results are representative data from three separate experiments. DIC, differential interference contrast.
Gene Deletion Studies of Asparagine Synthetase A
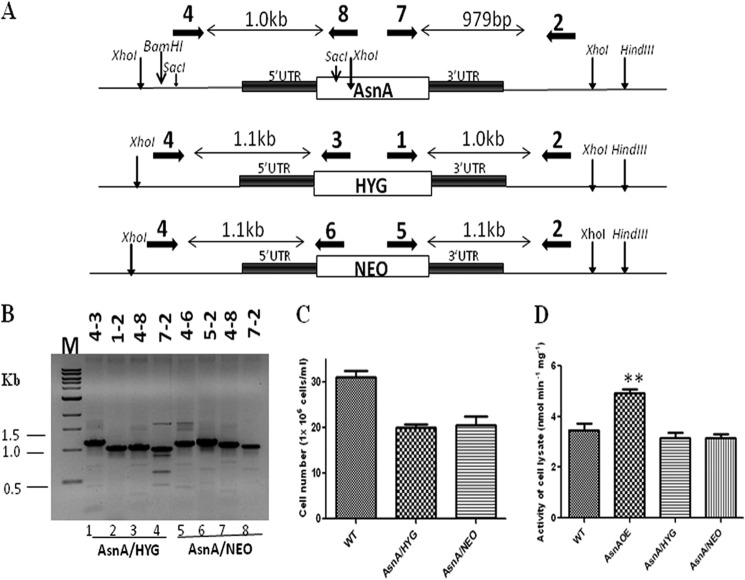
The essentiality of LdASNA was assessed by classical gene replacement experiments where attempts were made to replace both copies of LdASNA by drug resistance genes. Cassettes having HYG or NEO as selection markers along with the flanking 5′-UTR and 3′-UTR of LdASNA gene were used to replace LdASNA as described under “Experimental Procedures” (Fig. 5A). Linear replacement cassettes made by fusion PCR were electrotransfected into wild type L. donovani promastigotes, leading to generation of heterozygous parasites in which one copy of LdASNA gene was replaced with either the hygromycin or neomycin drug resistance gene. Replacement of a single copy of LdASNA gene from the chromosome by the hygromycin or neomycin drug resistance gene cassette was confirmed by PCR-based analysis by using primers external to the inactivation cassette of LdASNA gene (Fig. 5A). Cells transfected with the replacement cassette were selected after three to four passages. DNA from the transfected L. donovani promastigotes was isolated and subjected to PCR-based analysis. 1.1- and 1.0-kb bands in the case of the HYG cassette and 1.1- and 1.1-kb bands in the case of the NEO cassette along with the 1.0-kb and 979-bp bands corresponding to the WT LdASNA gene were also obtained (Fig. 5B). This confirmed that a single copy of LdASNA gene had been replaced in heterozygotes parasites. Interestingly, the heterozygous parasites consistently showed a growth delay compared with wild type parasites (Fig. 5C). The ASNA activity was lower in heterozygous parasites in comparison with the wild type strain (Fig. 5D). It is reasonable to assume that a gene dosage effect resulted in the production of lesser ASNA protein, and such a conjecture would suggest that ASNA is involved in optimal cell proliferation.
FIGURE 5.
A, restriction map of the LdASNA genomic locus and the location of the primers used for confirmation by PCR analysis along with the expected band sizes. Primer 4 was designed as a forward primer to match the upstream region of LdASNA gene, and primers 8, 3, and 6 were designed to be internal to ASNA, HYG, and NEO coding regions, respectively. Primer 2 was designed as a reverse primer to match the downstream region of LdASNA gene, and primers 7, 1, and 5 were designed as forward primers internal to ASNA, HYG, and NEO coding regions, respectively. B, genomic DNA from ASNA/HYG and ASNA/NEO heterozygous parasites was used as a template for PCR analysis. The specific integration of the replacement cassette was checked with LdASNA- and HYG- or NEO-specific primers. M indicates the molecular size marker in kb. C, growth analysis of L. donovani WT and single transfectants ASNA/HYG and ASNA/NEO. The parasites were counted after 4 days on a hemocytometer. These results represent mean ± S.D. (error bars) with n = 3. D, comparison of the LdASNA enzyme activity (nmol min−1 mg−1) of WT Bob, ASNAOE, ASNA/HYG, and ASNA/NEO in the respective cell lysates by an NADH-ATP coupled reaction. These results represent mean ± S.D. (error bars) with n = 3. **, p < 0.01.
LdASNA heterozygotes were subsequently transfected with a second cassette to replace the second copy of the gene. PCR analysis demonstrated that cells selected on the double antibiotic medium showed HYG and NEO replacement cassettes at the LdASNA gene locus similar to the heterozygotes, but these transfectants also showed 1.0-kb and 979-bp bands corresponding to the WT LdASNA gene (data not shown). This result indicates the presence of wild type gene in the double transfectants.
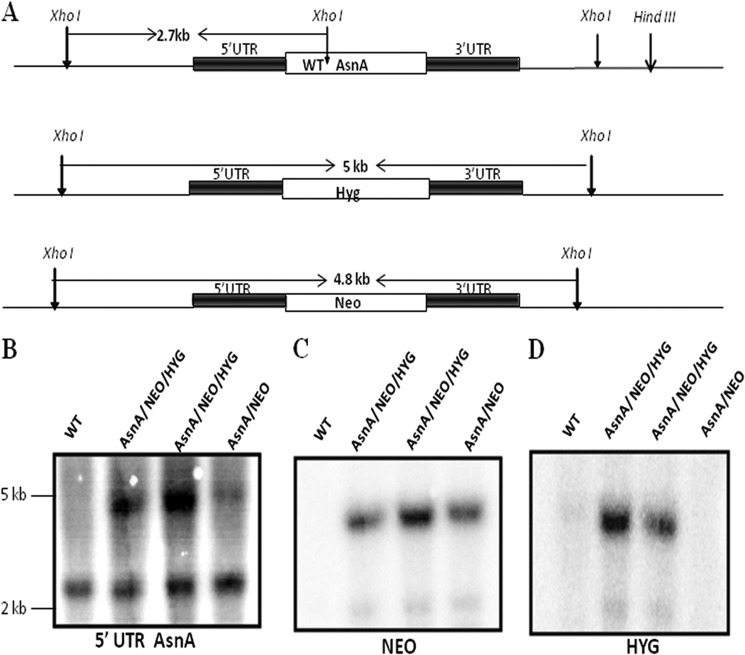
The presence of WT LdASNA gene along with HYG and NEO replacement cassettes in the LdASNA gene locus was further checked by Southern blot analysis by digesting the genomic DNA with XhoI. In the WT cells, digestion of WT LdASNA gene locus with XhoI was expected to yield a 2.7-kb band after probing with the 5′-UTR of LdASNA gene as probe (Fig. 6A). Integration of the HYG or NEO cassette in single transfectants was expected to yield 5- and 4.8-kb 5′-UTR-hybridizing bands, respectively, in addition to the 2.7-kb wild type band (Fig. 6A). Bands of the expected sizes hybridizing with the L. donovani ASNA 5′-UTR as probe were obtained in the case of the heterozygous parasites (Fig. 6B), but in the case of double transfectants, a band corresponding to the WT LdASNA was found, indicating the presence of WT gene in these transfectants. The same blot probed with the HYG and NEO genes as probes showed the presence of both HYG and NEO in double transfectants (Fig. 6, C and D). At least five clones of each of the transfectant cell lines were picked and screened for phenotypic characterization. All attempts to generate LdASNA homozygous null mutant were unsuccessful. Southern blot analyses demonstrated that the LdASNA gene was still present in the genome of these parasite lines, indicating that LdASNA may be an essential gene.
FIGURE 6.
A, restriction map of the LdASNA locus and the predicted chromosomal rearrangements occurring during HYG and NEO replacement of ASNA in the chromosomal locus. B, Southern blot analysis of WT Bob, single transfectant ASNA/NEO, and double transfectant ASNA/NEO/HYG. Genomic DNA was digested with XhoI, separated on a 0.6% agarose gel, and probed with the 5′-UTR of LdASNA. Molecular size markers are indicated to the right of the blot. C and D, blot in B was striped and reprobed with the NEO or HYG gene, respectively, to indicate the presence of NEO and HYG in single and double transfectants.
Because the double transfectant mutant clones (ASNA/HYG/NEO) were found to contain 2.7-kb wild type ASNA allele as well as HYG and NEO integrations, DNA content-based analysis was performed to check the ploidy levels of the cells. Fluorescence-activated cell sorting (FACS) analysis of fixed cells stained with propidium iodide was performed to determine the ploidy levels of WT and single and double transfectants. In the wild type cells, two peaks corresponding to 2N (G1) and 4N (G2) DNA content were observed as expected. An identical pattern was observed with the heterozygous mutants ASNA/HYG and ASNA/NEO, thus indicating that these heterozygous parasites contain normal diploid DNA content similar to wild type cells. Interestingly, FACS analysis of the double transfected parasite clone (ASNA/HYG/NEO) showed peaks that corresponded to 3N and 6N DNA content (data not shown). This suggests a difference in the ploidy level in the cell population of the double transfectant parasites when compared with the single transfectants and wild type cells. The G1 and G2 peaks of the double transfected parasite indicated that these cells are aneuploid (data not shown). All the single and double transfected clones screened showed similar results.
Crystal Structure Analysis
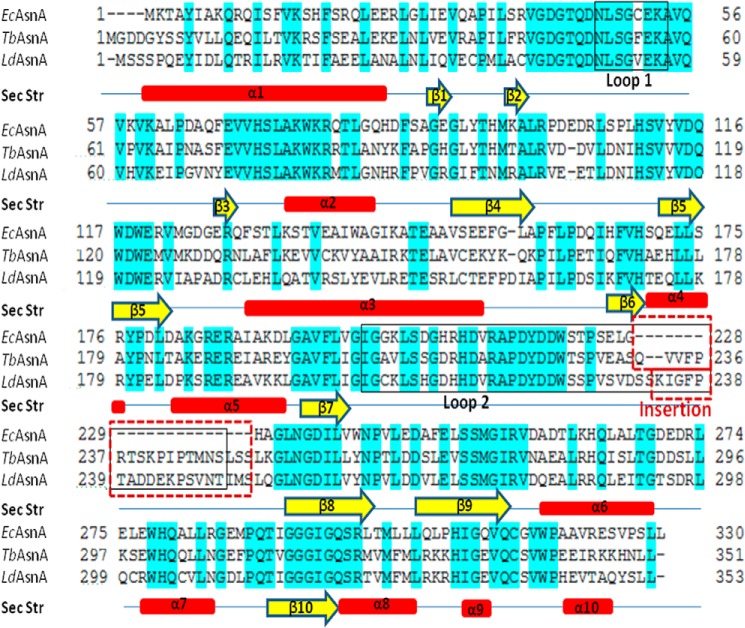
The TbASNA crystal structure was solved by molecular replacement using EcAsnA (Protein Data Bank code 11AS) as the search model in apo form. The structure was refined using native diffraction data at 2.2-Å resolution. TbASNA crystals belong to space group (P6122) with one monomer of TbASNA in the asymmetric unit. TbASNA exists as a homodimer inside the crystals (based on packing considerations) (Table 3). The final electron density for the TbASNA model is well defined except for loop 1 (residues 51–57) and loop 2 (residues 210–250; Figs. 7–9). The structure of TbASNA (monomer) consists of 10 β-strands and 10 α-helices (Fig. 7). Sequence identity between Leishmania and the E. coli enzyme counterpart is ∼57%, and the overall fold of TbASNA is hence very similar to that of EcAsnA. TbASNA and EcAsnA monomers superimpose with a root mean square deviation of ∼1.2 Å for 304 Cα atoms. However, residues 210–250 in TbASNA have a unique ∼19-residue insertion (in the TbASNA sequence, this insertion is QVVFPRTSKPIPTMNSLSS; in LdASNA, it is KIGFPTADDEKPSVNTIMS) as part of loop 2 (Fig. 8). The significance of this insertion at this location in the structure of TbASNA is unclear because in the crystal structure of apo-TbASNA this loop is completely disordered. Furthermore, these ∼19-residue insertions seem to have little sequence conservation between TbASNA and LdASNA. In contrast, a structure-based multiple sequence alignment between T. brucei and L. donovani ASNA shows very few regions of sequence divergence (Fig. 7). Therefore, the role of insertion sequences in ASNAs may need to be explored separately. From the structure of TbASNA, it is evident that α-helices 6 and 7 are distinctly displaced from their E. coli AsnA counterparts by up to ∼9 Å. Furthermore, in EcAsnA, there are no major conformational changes between the apo and holo forms of the enzyme. Indeed, the apo form of TbASNA shows no significant deviation from its E. coli counterpart except in the noted ∼9-Å displacement near α-helices 6 and 7 (Fig. 8B). Using visualization software (Chimera), we cataloged active site amino acid residues (a total of 15) that interact with l-Asn and AMP in the crystal structure of EcAsnA (Protein Data Bank code 12AS). We then mapped these l-Asn- and AMP-binding active site residues onto the TbASNA structure (Fig. 9). Interestingly, all 15 active site residues (of which seven recognize l-Asn and eight bind AMP) are conserved among E. coli, TbASNA, and LdASNA. Residue Tyr-218 in EcAsnA is part of loop 2, and although it remains conserved, it is disordered in TbASNA. These constraints within the active sites of ASNAs, therefore, can be exploited to screen specific inhibitors that can selectively target these enzymes. The high resolution atomic structure presented here can serve as a platform for development of specific inhibitors against both TbASNA and Leishmania ASNA.
TABLE 3.
Data collection and refinement statistics
Values in parentheses are for the highest resolution shell. r.m.s.d., root mean square deviation.
| Data collection | |
| Wavelength (Å) | 0.976 |
| Crystal-to-detector distance (mm) | 232.1 |
| Exposure time (s) | 12 |
| Number of frames | 360 |
| Unit cell parameters (Å; °) | a = 71.55, b = 71.55, c = 268.63; α = 90, β = 90, γ = 120 |
| Space group | P6122 |
| Resolution (Å) | 25.0-2.20 (2.24-2.20) |
| Unique reflections | 21721 (1082) |
| Multiplicity | 8.0 (8.5) |
| Completeness (%) | 99.6 (100) |
| I/σ(I) | 27.14 (3.2) |
| Rmerge | 0.076 (0.482) |
| Refinement | |
| Resolution (Å) | 24.6-2.20 |
| Reflections in work set/test set | 20,543/1,105 |
| Rwork/Rfree (%) | 20.5/25.0 |
| Stereochemistry | |
| r.m.s.d. in bond lengths (Å) | 0.008 |
| r.m.s.d. in bond angles (°) | 1.17 |
| Ramachandran plot | |
| Residues in most favored regions (%) | 93.7 |
| Residues in additionally allowed regions (%) | 6.3 |
FIGURE 7.
Sequence alignment of TbASNA and LdASNA with EcAsnA. The secondary structure (Sec Str) elements derived from the crystal structure of TbASNA are placed at the bottom of the alignment. The loop in TbASNA and LdASNA (herein called loop 2) contains a ∼19-residue insertion (red box) that is absent in E. coli and archaeal AsnAs.
FIGURE 8.
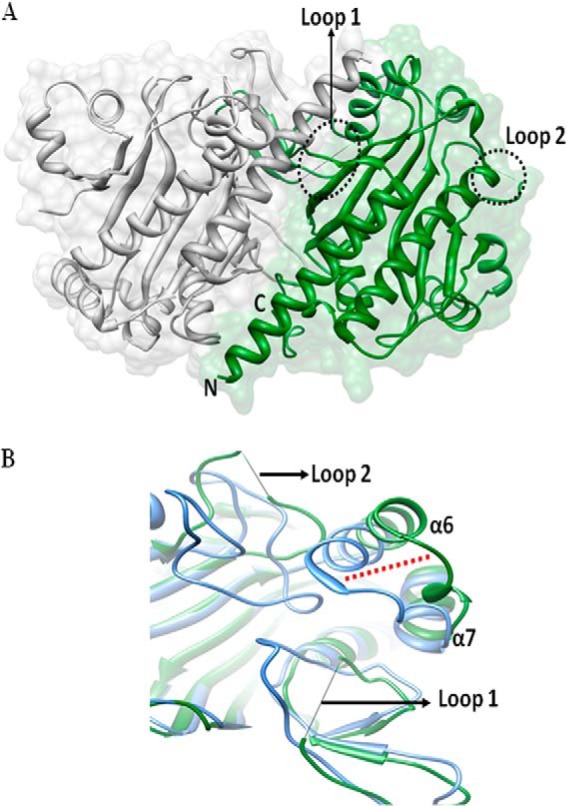
A, overall structure of TbASNA in apo form. The monomers that form ASNA dimer are shown with loop 1 and loop 2 highlighted in black dots. Loops 1 and 2 are disordered in TbASNA, although in the E. coli apoenzyme, they are ordered. B, superposition of TbASNA and EcAsnA monomer showing α-helices 6 and 7 that are displaced from their EcAsnA counterparts by ∼9 Å.
FIGURE 9.
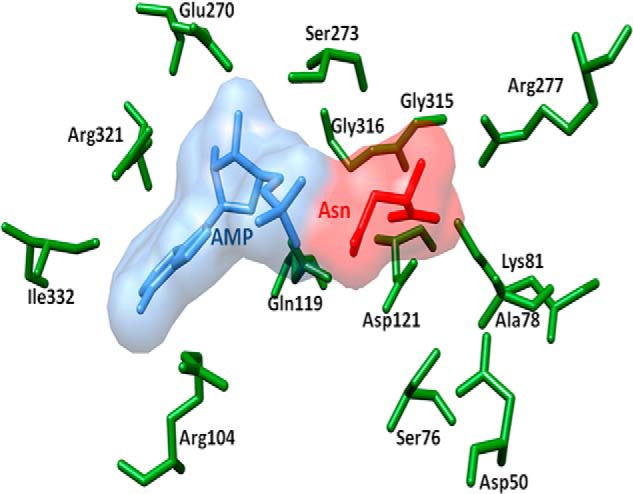
Superposition of EcAsnA complexed with Asn and AMP and TbASNA. Ligands are shown as space-filling models. l-Asn (red), AMP (blue), and side chains of TbASNA are shown (green). The corresponding EcAsnA side chains have been removed for simplicity. All the l-Asn- and AMP-binding side chains are conserved between E. coli and TbASNA and LdASNA.
DISCUSSION
Aminoacyl-tRNA synthetase paralogs involved in amino acid biosynthesis have been known for many years (8, 14, 16–19). These activities include AsnA (asparagine-tRNA synthetase paralog), HisZ (histidyl-tRNA synthetase paralog), and lysylation of a specific lysine in bacterial elongation factor P (GenX/PoxA). Among these, E. coli AsnA and GenX/PoxA are well characterized (8, 14, 16–20, 22). Because they are absent in humans but present in pathogenic organisms like Leishmania, these paralogs form a novel category of drug targets. Our earlier detailed bioinformatics analysis revealed the presence of ASNA paralogs in certain eukaryotic pathogens (13, 37). Recently, characterization of ASNA from T. brucei and T. cruzi has been reported (21). Although the asparagine-tRNA synthetases contain a synthetase domain and an anticodon binding domain, these paralogs contain only the synthetase domain to perform asparagine biosynthesis (13). We had reported earlier that although aminoacyl-tRNA synthetases of L. major are suggested to be of archaeal/eukaryotic origins, L. major asparagine synthetase A is likely to be of bacterial origin (13).
Two structurally distinct groups of asparagine synthetases are known in prokaryotes: the ammonia-utilizing asparagine synthetases (AsnA) and glutamine-utilizing asparagine synthetases (AsnB). Although asnB genes are reported from all three domains of life, asnA genes have been reported from prokaryotes (15), archaea (8), and very recently eukaryotic pathogens (13, 21, 37). AsnA is present in certain anaerobic Gram-positive bacteria such as Clostridium perfringens, Bacillus anthracis, Streptococcus pneumoniae, and Clostridium botulinum. AsnA also exists in certain facultative anaerobic Gram-negative bacteria such as E. coli, Yersinia enterocolitica, Salmonella enterica, Yersinia pestis, and Shigella flexneri (33). Enterocytozoon bieneusi is the only fungal pathogen that has an ASNA gene (33). It is also the only eukaryotic pathogen with 45 copies of ASNA genes of variable amino acid lengths ranging from 50 to 300 amino acids long (33). Interestingly, ASNA is present only in C. parvum and Cryptosporidium hominis and is absent from other apicomplexan parasites (33). Among Viridiplantae, a single copy of ASNA gene is present in Thalassiosira pseudonana CCMP 1335 (33). T. vaginalis is the other eukaryotic pathogen that carries a copy of ASNA gene (13).
In this study, we show that LdASNA is an ammonia-dependent AsnA-type enzyme also capable of utilizing glutamine as the nitrogen source. The overall Km values deduced by kinetic analysis for LdASNA appear to be closer to those reported earlier in the case of prokaryotic organisms (6, 35) Attempts to delete the ASNA gene from the parasite genome failed, and these parasites showed aneuploidy. Thus, L. donovani ASNA appears to be an essential gene. However, ASNA knockdown in T. brucei did not affect in vitro growth of bloodstream forms of the parasite (21).
Previous crystal structure analysis of EcAsnA suggested a class II catalytic core of aminoacyl-tRNA synthetases for these enzymes (22). Structure-based sequence comparison of EcAsnA with the catalytic domain of yeast aspartyl-tRNA synthetase showed a high conservation of catalytic residues (22). Although the substrates of these two enzymes are similar, the activation of carboxyl groups on the aspartyl residues is different. Yeast aspartyl-tRNA synthetase activates the α-carboxyl group of the substrate, whereas EcAsnA activates the β-carboxyl group (22). The recent crystal structure of an archaeal AsnA from P. abyssi in various substrate-bound forms helped to decode the plausible mechanism of asparagine biosynthesis by AsnA in archaea (8). Sequence-based phylogenetic analysis of the LdASNA followed by structural modeling and comparison with the E. coli AsnA suggested an evolutionary origin of kinetoplastid ASNA genes closer to that of the prokaryotic AsnA rather than the archaeal enzyme (13). The LdASNA is also expected to follow a similar catalytic mechanism as EcAsnA based on its structural similarity to archaeal AsnA (13). Our extensive attempts to crystallize recombinant LdASNA failed, but we were able to solve the high resolution crystal structure of the highly homologous TbASNA. Sequence identity between LdASNA and TbASNA is ∼80%. As expected, the overall fold of TbASNA is similar to that of EcAsnA, and their respective monomers superimpose with a root mean square deviation of ∼1.2 Å for 304 Cα atoms (Figs. 7–9). The loop in TbASNA and LdASNA (herein called loop 2) contains a unique 19-residue insertion of variable sequence that is absent from E. coli and archaeal AsnAs. In both the apo and holo structures of EcAsnA, loop 2 is ordered, suggesting that ligand engagement is not necessary for ordering of loop 2. However, in the TbASNA structure of the apoenzyme, the corresponding loop 2 with its unique 19-residue insertion remains fully disordered. We also observed very high sequence conservation in 15 residues that line the active site pockets of these enzymes (Fig. 9). Hence, the presented crystal structure of TbASNA can serve as a model for structure-based inhibitor design against not only LdASNA but, given the extensive active site conservation (residues that interact with l-Asn and AMP are identical) noted previously, possibly also for other homologs of ASNA from various pathogens that harbor this enzyme.
Asparagine synthetase is a potential target for cancer chemotherapy (38) because asparagine depletion caused by the administration of l-asparaginase is presently being used as a protocol for the treatment of acute lymphoblastic leukemia (39). Therefore, it is highly desirable to develop inhibitors of ASNA that could be used as chemotherapeutic agents. Koizumi et al. (40) synthesized a transition state analog of cysteine sulfoximine, N-adenylated S-methyl-l-cysteine sulfoximine. This inhibitor was found to be an extremely potent slow binding inhibitor of E. coli AsnA. This inhibitor was also found to inhibit AsnB effectively and irreversibly (41). The crystal structure of E. coli asparagine synthetase A in complex with its potent slow binding inhibitor has revealed the structural basis for transition state stabilization as well as for substrate recognition (40). Our analyses of ASNA from Leishmania parasites along with the structure of its homolog from Trypanosoma together provide a rationale for targeting this enzyme in drug discovery efforts. The absence of ASNA from humans and its essentiality in Leishmania together provide a new window for exploiting ASNA for drug development to treat visceral leishmaniasis.
Acknowledgments
We thank the Advance Instrumentation Research Facility at Jawaharlal Nehru University for providing the imaging facility and for MALDI-TOF analysis. We also thank Marc Ouellette, University of Laval, Quebec, Canada for providing Leishmania shuttle vectors.
This work was supported by a grant from the Department of Biotechnology, Government of India and a Department of Science and Technology Promotion of University Research and Scientific Excellence grant (to R. M.).
The atomic coordinates and structure factors (code 4LNS) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- AsnA
- asparagine synthetase A
- AsnB
- asparagine synthetase B
- LdASNA
- L. donovani asparagine synthetase A
- TbASNA
- T. brucei asparagine synthetase A
- ASNAOE
- ASNA overexpressor
- CMXRos
- chloromethyl-X-rosamine
- HYG
- hygromycin phosphotransferase gene
- NEO
- neomycin phosphotransferase gene
- EcAsnA
- E. coli asparagine synthetase A.
REFERENCES
- 1. Freitas-Junior L. H., Chatelain E., Kim H. A., Siqueira-Neto J. L. (2012) Visceral leishmaniasis treatment: what do we have, what do we need and how to deliver it? Int. J. Parasitol. Drugs Drug Resist. 2, 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maltezou H. C. (2010) Drug resistance in visceral leishmaniasis. J. Biomed. Biotechnol. 2010, 617521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. García-Hernández R., Manzano J. I., Castanys S., Gamarro F. (2012) Leishmania donovani develops resistance to drug combinations. PLoS Negl. Trop. Dis. 6, e1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sugiyama A., Kato H., Nishioka T., Oda J. (1992) Overexpression and purification of asparagine synthetase from Escherichia coli. Biosci. Biotechnol. Biochem. 56, 376–379 [DOI] [PubMed] [Google Scholar]
- 5. Humbert R., Simoni R. D. (1980) Genetic and biomedical studies demonstrating a second gene coding for asparagine synthetase in Escherichia coli. J. Bacteriol. 142, 212–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reitzer L. J., Magasanik B. (1982) Asparagine synthetases of Klebsiella aerogenes: properties and regulation of synthesis. J. Bacteriol. 151, 1299–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Charron C., Roy H., Blaise M., Giegé R., Kern D. (2004) Crystallization and preliminary x-ray diffraction data of an archaeal asparagine synthetase related to asparaginyl-tRNA synthetase. Acta Crystallogr. D Biol. Crystallogr. 60, 767–769 [DOI] [PubMed] [Google Scholar]
- 8. Blaise M., Fréchin M., Oliéric V., Charron C., Sauter C., Lorber B., Roy H., Kern D. (2011) Crystal structure of the archaeal asparagine synthetase: interrelation with aspartyl-tRNA and asparaginyl-tRNA synthetases. J. Mol. Biol. 412, 437–452 [DOI] [PubMed] [Google Scholar]
- 9. Hughes C. A., Beard H. S., Matthews B. F. (1997) Molecular cloning and expression of two cDNAs encoding asparagine synthetase in soybean. Plant Mol. Biol. 33, 301–311 [DOI] [PubMed] [Google Scholar]
- 10. Scofield M. A., Lewis W. S., Schuster S. M. (1990) Nucleotide sequence of Escherichia coli asnB and deduced amino acid sequence of asparagine synthetase B. J. Biol. Chem. 265, 12895–12902 [PubMed] [Google Scholar]
- 11. Van Heeke G., Schuster S. M. (1989) Expression of human asparagine synthetase in Escherichia coli. J. Biol. Chem. 264, 5503–5509 [PubMed] [Google Scholar]
- 12. Yoshida K., Fujita Y., Ehrlich S. D. (1999) Three asparagine synthetase genes of Bacillus subtilis. J. Bacteriol. 181, 6081–6091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gowri V. S., Ghosh I., Sharma A., Madhubala R. (2012) Unusual domain architecture of aminoacyl tRNA synthetases and their paralogs from Leishmania major. BMC Genomics 13, 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Francklyn C. (2003) tRNA synthetase paralogs: evolutionary links in the transition from tRNA-dependent amino acid biosynthesis to de novo biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 100, 9650–9652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakamura M., Yamada M., Hirota Y., Sugimoto K., Oka A., Takanami M. (1981) Nucleotide sequence of the asnA gene coding for asparagine synthetase of E. coli K-12. Nucleic Acids Res. 9, 4669–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sissler M., Delorme C., Bond J., Ehrlich S. D., Renault P., Francklyn C. (1999) An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 96, 8985–8990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gilreath M. S., Roy H., Bullwinkle T. J., Katz A., Navarre W. W., Ibba M. (2011) β-Lysine discrimination by lysyl-tRNA synthetase. FEBS Lett. 585, 3284–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ambrogelly A., O'Donoghue P., Söll D., Moses S. (2010) A bacterial ortholog of class II lysyl-tRNA synthetase activates lysine. FEBS Lett. 584, 3055–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yanagisawa T., Sumida T., Ishii R., Takemoto C., Yokoyama S. (2010) A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P. Nat. Struct. Mol. Biol. 17, 1136–1143 [DOI] [PubMed] [Google Scholar]
- 20. Roy H., Zou S. B., Bullwinkle T. J., Wolfe B. S., Gilreath M. S., Forsyth C. J., Navarre W. W., Ibba M. (2011) The tRNA synthetase paralog PoxA modifies elongation factor-P with (R)-β-lysine. Nat. Chem. Biol. 7, 667–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Loureiro I., Faria J., Clayton C., Ribeiro S. M., Roy N., Santarém N., Tavares J., Cordeiro-da-Silva A. (2013) Knockdown of asparagine synthetase A renders Trypanosoma brucei auxotrophic to asparagine. PLoS Negl. Trop. Dis. 7, e2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakatsu T., Kato H., Oda J. (1998) Crystal structure of asparagine synthetase reveals a close evolutionary relationship to class II aminoacyl-tRNA synthetase. Nat. Struct. Biol. 5, 15–19 [DOI] [PubMed] [Google Scholar]
- 23. Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pérez-González J. A., Ruiz D., Esteban J. A., Jiménez A. (1990) Cloning and characterization of the gene encoding a blasticidin S acetyltransferase from Streptoverticillum sp. Gene 86, 129–134 [DOI] [PubMed] [Google Scholar]
- 25. Darveau A., Pelletier A., Perreault J. (1995) PCR-mediated synthesis of chimeric molecules. Methods Neurosci. 26, 77–85 [Google Scholar]
- 26. Kapler G. M., Coburn C. M., Beverley S. M. (1990) Stable transfection of the human parasite Leishmania major delineates a 30-kilobase region sufficient for extrachromosomal replication and expression. Mol. Cell. Biol. 10, 1084–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 28. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 29. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 32. Aurrecoechea C., Brestelli J., Brunk B. P., Fischer S., Gajria B., Gao X., Gingle A., Grant G., Harb O. S., Heiges M., Innamorato F., Iodice J., Kissinger J. C., Kraemer E. T., Li W., Miller J. A., Nayak V., Pennington C., Pinney D. F., Roos D. S., Ross C., Srinivasamoorthy G., Stoeckert C. J., Jr., Thibodeau R., Treatman C., Wang H. (2010) EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res. 38, D415–D419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen F., Mackey A. J., Stoeckert C. J., Jr., Roos D. S. (2006) OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 34, D363–D368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roy H., Becker H. D., Reinbolt J., Kern D. (2003) When contemporary aminoacyl-tRNA synthetases invent their cognate amino acid metabolism. Proc. Natl. Acad. Sci. U.S.A. 100, 9837–9842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Burchall J. J., Reichelt E. C., Wolin M. J. (1964) Purification and properties of the asparagine synthetase of Streptococcus bovis. J. Biol. Chem. 239, 1794–1798 [PubMed] [Google Scholar]
- 36. Chauhan S. C., Madhubala R. (2009) Glyoxalase I gene deletion mutants of Leishmania donovani exhibit reduced methylglyoxal detoxification. PLoS One 4, e6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Heinz E., Williams T. A., Nakjang S., Noël C. J., Swan D. C., Goldberg A. V., Harris S. R., Weinmaier T., Markert S., Becher D., Bernhardt J., Dagan T., Hacker C., Lucocq J. M., Schweder T., Rattei T., Hall N., Hirt R. P., Embley T. M. (2012) The genome of the obligate intracellular parasite Trachipleistophora hominis: new insights into microsporidian genome dynamics and reductive evolution. PLoS Pathog. 8, e1002979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Richards N. G., Kilberg M. S. (2006) Asparagine synthetase chemotherapy. Annu. Rev. Biochem. 75, 629–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pieters R., Hunger S. P., Boos J., Rizzari C., Silverman L., Baruchel A., Goekbuget N., Schrappe M., Pui C. H. (2011) L-Asparaginase treatment in acute lymphoblastic leukemia: a focus on Erwinia asparaginase. Cancer 117, 238–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koizumi M., Hiratake J., Nakatsu T., Kato H., Oda J. (1999) A potent transition-state analogue inhibitor of Escherichia coli asparagine synthetase A. J. Am. Chem. Soc. 121, 5799–5800 [Google Scholar]
- 41. Boehlein S. K., Nakatsu T., Hiratake J., Thirumoorthy R., Stewart J. D., Richards N. G., Schuster S. M. (2001) Characterization of inhibitors acting at the synthetase site of Escherichia coli asparagine synthetase B. Biochemistry 40, 11168–11175 [DOI] [PubMed] [Google Scholar]