Background: The molecular mechanisms by which Groucho/TLEs repress canonical Wnt signaling are incompletely understood.
Results: OGT interacts with TLEs and facilitates TLE-mediated transcriptional repression. Removal of O-GlcNAc is essential for gene activation from Wnt-responsive promoters.
Conclusion: OGT plays a vital role in TLE-mediated repression of Wnt signaling.
Significance: O-GlcNAc modification has a profound influence on diverse signaling pathways.
Keywords: Gene Silencing, O-GlcNAc, Signal Transduction, Transcription Repressor, Wnt Pathway, Groucho/TLE, OGT
Abstract
The Drosophila Groucho protein and its mammalian orthologues the transducin-like enhancers of split (TLEs) are critical transcriptional corepressors that repress Wnt and other signaling pathways. Although it is known that Groucho/TLEs are recruited to target genes by pathway-specific transcription factors, molecular events after the corepressor recruitment are largely unclear. We report that association of TLEs with O-GlcNAc transferase, an enzyme that catalyzes posttranslational modification of proteins by O-linked N-acetylglucosamine, is essential for TLE-mediated transcriptional repression. Removal of O-GlcNAc from Wnt-responsive gene promoters is critical for gene activation from Wnt-responsive promoters. Thus, these studies identify a molecular mechanism by which Groucho/TLEs repress gene transcription and provide a model whereby O-GlcNAc may control distinct intracellular signaling pathways.
Introduction
The roles of Groucho/TLEs2 have been implicated in many developmental processes, such as segmentation, neurogenesis, and sex determination of Drosophila and somitogenesis, neurogenesis, osteogenesis, and hematopoiesis in vertebrate homologs (1–5). Groucho/TLEs are global corepressors of Notch, Sonic hedgehog, and Wnt signaling through interaction with the Hes, Nkx, and TCF/LEF families of transcription factors, respectively (1, 6, 7). It has been shown that TLEs can directly interact with TCF/LEF during repression conditions, and the physical displacement of TLE by β-catenin occurs in response to Wnt activation (8). In the absence of nuclear β-catenin, transcriptional repression occurs as a result of the TCF/LEF nucleation of TLE. Upon Wnt signaling, β-catenin enters the nucleus to directly compete with Groucho/TLEs for TCF/LEF binding (8). Repression mediated by TLEs has been attributed to its interaction with Sin3A, histone deacetylases (HDACs), and other members of the global co-repression complex to alter local chromatin structure (9, 10) and Groucho/TLE oligomerization to promote long range chromatin condensation (11, 12).
Emerging evidence shows different regulatory modes for Groucho/TLE-mediated repression, including distribution of partner repressors, competition with coactivators, and posttranslational modifications of Groucho/TLE, such as phosphorylation and poly(ADP-ribosyl)ation (13–16). Our previous work suggests a general role for the posttranslational modification of the transcriptional apparatus by O-GlcNAc transferase (OGT) in gene repression (17, 18). These studies indicate that OGT, a tetratricopeptide repeat (TPR)-containing protein, may associate with general elements of the corepressor complex (17). It is well defined that Tup1, the yeast homologue of Groucho/TLEs, functions in complex with a TPR-containing protein, Cyc8 (19). However, such a Cyc8-like partner for Groucho/TLEs that functions in higher eukaryotes remains to be identified. Whereas Cyc8 has no known enzymatic activity associated with its TPR domains, OGT, which occurs in multicellular organisms, does. By placing O-GlcNAc modification on the transcription apparatus, OGT acts in concert with chromatin modification to inhibit transcriptional activators, such as Sp1 (18) and RNA polymerase II (20, 21), to maintain transcriptional repression. Nevertheless, specificity for OGT in regulating distinct intracellular pathways at the transcriptional level has not yet been defined. Because OGT contains the TPR structure that is reminiscent of Cyc8, we were prompted to explore the possibility that OGT is a partner for Groucho/TLEs in regulating expression of a distinct subset of developmental genes.
Herein, we provide evidence for pathway-specific regulation of the TCF/LEF locus by O-GlcNAc. We show that OGT is targeted to TCF/LEF sites on endogenous Wnt-responsive gene promoters by its interaction with Groucho/TLE transcriptional corepressors. In addition, these promoters and their downstream genes are specifically regulated by O-GlcNAc modification. Together, these data suggest that O-GlcNAc can function in regulating distinct intracellular signaling pathways. Moreover, these data serve as a model for how O-GlcNAc may specifically regulate other intracellular signaling pathways through targeting of OGT to distinct sites of transcriptional repression via its interaction with certain corepressor molecules.
MATERIALS AND METHODS
Plasmids
Vectors for bacterial expression of the full-length and deletion mutants of OGT in fusion with glutathione S-transferase (GST) were described previously (17). Catalytically dead OGT (D925N) was generated with the QuikChange site-directed mutagenesis kit (Stratagene). Full-length or fragments of human TLE1 and human TLE2 were produced by PCR and subcloned into pcDNA3 mammalian expression plasmid in fusion with the Gal4 DNA-binding domain. pcDNA3.1-His-TLE1 and -TLE2 were kindly provided by G. Stein.
GST Pull-down Assays
All GST fusion proteins were expressed in Escherichia coli and were purified and immobilized by batch affinity chromatography on glutathione-Sepharose 4B (Amersham Biosciences) as described previously (17). [35S]Methionine-labeled proteins were synthesized in vitro with a coupled transcription-translation system, TNT (Promega). 35S-Labeled proteins were incubated with equal amounts of immobilized GST fusion proteins in binding buffer (50 mm Tris (pH 7.5), 10% glycerol, 100 mm NaCl, 0.1% Nonidet P-40, 1 mm EDTA, 1 mm DTT, 1 mm PMSF, and protease inhibitors) for 2 h at 4 °C. Beads were washed five times with the binding buffer. Bound proteins were eluted with 1× SDS-PAGE buffer, separated by SDS-PAGE, and visualized by autoradiography.
Coimmunoprecipitation and Immunohistochemical Assays
COS-7 cells were transfected using FuGENE6 (Roche Applied Science). The coimmunoprecipitation assay was performed as described (17). Antibodies used were α-OGT (v18), α-TLE1 (M101), α-TLE2 (H191), α-LEF (N-17), and α-Gal4 (DNA-binding domain; DBD) (all from Santa Cruz Biotechnology); α-active-β-catenin (clone 8E7, specific for the Ser-37/Thr-41 dephosphorylated form; Upstate Biotechnology); α-GST (clone 2) and α-β-actin (AC40) (Sigma); α-His6 and α-HA (clone 12CA5) (Roche Applied Science); and α-O-GlcNAc (RL2) (Abcam). All other methods generally adhered to protocols provided by Santa Cruz Biotechnology.
Cell-based Transcription Assays
HepG2 cells were transiently cotransfected with expression constructs for TLE1 or TLE2 (full-length and fragments) and OGT (wild-type or catalytically dead) plasmids either individually or in combination, together with the G5-Luc reporter construct as described previously (17). Transient transfection was performed using electroporation or Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. For luciferase assays, transfection efficiencies were normalized using a cotransfected β-galactosidase plasmid. β-Catenin/TCF-induced transcriptional activity was determined by using a β-catenin/TCF promoter-luciferase reporter construct, pTOPFLASH (22). Briefly, a dual luciferase reporter assay was performed whereby HEK293 cells were transfected with the experimental TCF promoter/luciferase reporter gene (TOPFLASH). A mutated TCF-luciferase reporter construct (pFOPFLASH) served as a negative control for TOPFLASH activity. A control reporter pRL-TK Renilla luciferase (Promega) was co-transfected in each sample to serve as an internal control for transfection efficiency. To promote Wnt signaling, Wnt1 conditioned medium was added 24 h after transfection, and luciferase activities were assayed 48 h after transfection. Experiments were performed in triplicate and repeated three times. Statistical analysis was performed using analysis of variance. A value of p < 0.05 was considered statistically significant.
Chromatin Immunoprecipitation Assay
ChIP assays were performed using previously described oligonucleotides and adhering generally to methods described previously (16). The oligonucleotide sequences used are listed in Table 1.
TABLE 1.
Primer sequences used in chromatin immunoprecipitation assay
| Target promoter | Sequence |
|---|---|
| Human c-MYC | 5′-AGGCAACCTCCCTCTCGCCCTA-3′ |
| 5′-AGCAGCAGATACCGCCCCTCCT-3′ | |
| Human cyclin D1 | 5′-CTGGAATTTTCGGGCATTTA-3′ |
| 5′-ACAACCCCTGTGCAAGTTTC-3′ | |
| Human COL2A1 | 5′-ACACCCCTCCTCTCCATCTT-3′ |
| 5′-TCATGAATGGGGCTTTTCTC-3′ |
RNA Interference
pSUPER-based vectors were constructed that contain DNA templates for the synthesis of siRNAs and transfected using Lipofectamine 2000. The sequences used are listed in Table 2.
TABLE 2.
Target sequences for design of pSUPER RNAi constructs
| RNAi target | RNAi target sequence |
|---|---|
| EGFP | 5′-GCGACGTAAACGGCCACAAGTTC-3′a |
| Human OGT-a | 5′-TGGCATCGACCTCAAAGCA-3′ |
| Human OGT-b | 5′-GGACAGATTCAAATAACAA-3′ |
| Human OGA-x | 5′-ACGCAAATTGGACCAGCTC-3′ |
| Human OGA-y | 5′-GACCTTGGGTTATGGAGCA-3′ |
| Human OGA-z | 5′-CATGAACGGAGTGAGGAAG-3′ |
| Scrambled control | 5′-GACATAGCGTAAGCCTATC-3′ |
a From Dharmacon.
Real-time RT-PCR
RNA was extracted using TRIzol reagent (Sigma) and purified with an RNase Easy Kit (Qiagen). cDNAs were synthesized from 0.2 μg of DNase-treated total RNA using the cDNA Archive Kit (Applied Biosystems). Relative RNA levels were quantified by real-time RT-PCR technology with an ABI PRISM 7700 detection system and SYBR Green reagent (Applied Biosystems). PCRs contained 1× SYBR Green Master Mix, 66 nm primers, and cDNA equivalent to 10 ng of total RNA in a 15-μl volume. Target mRNA levels were normalized against 36B4 or GAPDH mRNA level (as indicated) from the same total RNA sample. The primers used are listed in Table 3.
TABLE 3.
Primer sequences used in real-time RT-PCR analysis
| RT-PCR target | Primer sequence |
|---|---|
| Human OGT | |
| Forward | 5′-AGAAGGGCAGTGTTGCTGAAG-3′ |
| Reverse | 5′-TGATATTGGCTAGGTTATTCAGAGAGTCT-3′ |
| Human c-MYC | |
| Forward | 5′-CAGCTGCTTAGACGCTGGATTT-3′ |
| Reverse | 5′-ACCGAGTCGTAGTCGAGGTCAT-3′ |
| Human cyclin D1 | |
| Forward | 5′-CCGTCCATGCGGAAGATC-3′ |
| Reverse | 5′-ATGGCCAGCGGGAAGAC-3′ |
| Human ID2 | |
| Forward | 5′-TCAGCCTGCATCACCAGAGA-3′ |
| Reverse | 5′-GAATTCAGAAGCCTGCAAGGA-3′ |
| Human TLE1 | |
| Forward | 5′-GAGCCGGGCACAAGTAATTC-3′ |
| Reverse | 5′-TCATTGGAAAATTCAGGTCCATT-3′ |
| Human GAPDH | |
| Forward | 5′-GAAGGTGAAGGTCGGAGTC-3′ |
| Reverse | 5′-GAAGATGGTGATGGGATTTC-3′ |
| Human OGA | |
| Forward | 5′-GCGGTGTGGTGGAAGGATT-3′-3′ |
| Reverse | 5′-CCATTTCTGGAGCCTTCTAAAGAG-3′ |
RESULTS
TLEs Physically Interact with OGT
As a first step to assess a possible interaction between TLEs and OGT, we coexpressed hexahistidine-tagged TLE1 or TLE2 with hemagglutinin (HA)-tagged OGT in COS-7 cells. The result showed that TLE1 or TLE2 is coimmunoprecipitated with OGT, demonstrating that TLEs can physically associate with OGT (Fig. 1A). Direct protein-protein interactions between TLEs and OGT were confirmed by a glutathione S-transferase (GST) pull-down assay, in which TLE1 and TLE2 synthesized in vitro were able to bind to the purified GST-OGT fusion protein (Fig. 1B). In addition to multiple tandem TPRs at the N terminus, OGT contains two conserved domains (CDI and CDII) at the C terminus that contribute to its catalytic activity (23, 24). Deletion analysis reveals that a region spanning the first six TPR motifs and the CD I region in OGT independently bind to TLE1 and TLE2 (Fig. 1, B and C). As for the TLEs, both the conserved N-terminal glutamine-rich domain (Q domain) and the C-terminal WD-repeat domain interact with OGT (Fig. 1D). Interestingly, the Q domain of TLEs is able to bind to either TPR1–6 or CDI of OGT, whereas the WD domain mainly contacts OGT CDI (Fig. 1E). Together, the results suggest that there are multiple interfaces between an OGT molecule and a TLE molecule (Fig. 1F).
FIGURE 1.
Physical interaction between OGT and TLEs. A, coimmunoprecipitation analysis. COS-7 cells were transfected with HA-tagged OGT or His-tagged TLE1 or -2 expression vectors and immunoprecipitated with α-HA antibody, followed by immunoblotting with α-HA and α-His antibodies. B–E, mapping interaction domains in OGT and TLEs. TLE proteins and their deletion mutants were synthesized in reticulocyte lysate and were incubated with bacterially produced full-length OGT and deletion mutants. F, schematic representation of primary structures of TLE and OGT and the identified interactions. CcN, a region containing phosphorylation sites for Cdc2 and casein kinase 2; SP, serine/proline-rich domain; WD, WD40-repeat domains in tandem.
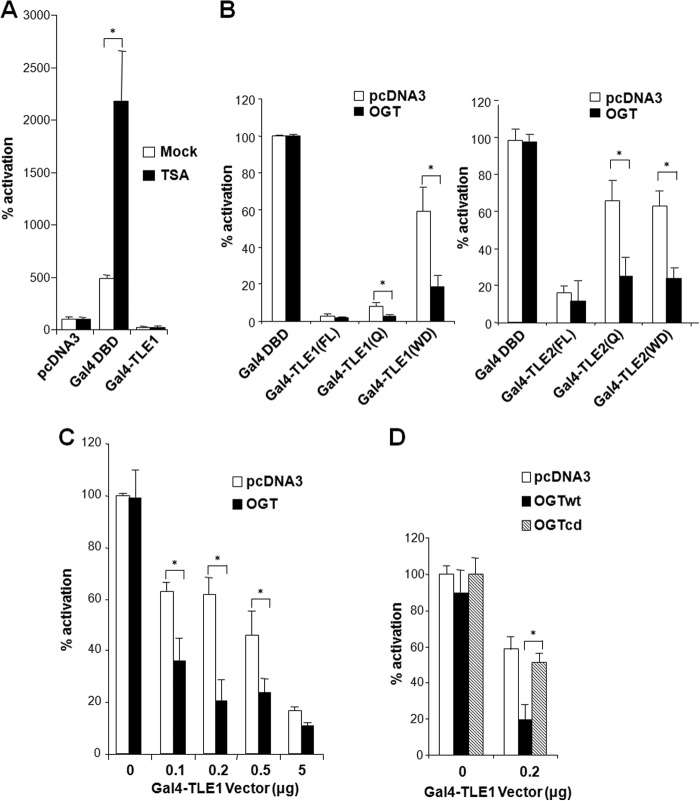
OGT Facilitates Transcriptional Repression of TLEs
Previous studies suggest that Groucho/TLEs mediate repression in part by recruiting HDACs (10, 25). Surprisingly, when TLE1 was tethered to a luciferase reporter plasmid by fusion to the Gal4 DBD, it retained potent repression activity irrespective of the presence of the HDAC inhibitor, TSA (Fig. 2A). In contrast, TSA was able to elevate Gal4 DBD-induced transcription from the reporter. These results suggest that HDACs are not sufficient to mediate TLE repression. It is known that both the Q and WD domains of the TLEs possess repression activity (26). These two domains physically interacted with OGT (Fig. 1, D and E), supporting the idea that their repression activity is dependent on OGT. Indeed, our results show that overexpression of OGT largely enhances transcriptional repression by either the Q or WD domain of TLE1 and -2, suggesting that both domains mediate repression by recruiting OGT (Fig. 2B). We next evaluated whether OGT could cooperate with TLE to inhibit transcription. The results indicate that although a moderate dose of exogenous OGT alone did not inhibit the reporter transcription by itself, OGT was capable of enhancing transcriptional repression by Gal4-TLE1 (Fig. 2C). OGT requires its endogenous catalytic activity to transfer O-GlcNAc moieties to proteins. We therefore used a catalytically dead OGT mutant lacking the ability to augment TLE1 repression by O-GlcNAc modification. Cotransfection of this inactive OGT mutant with TLE relieved TLE-mediated repression of the Gal4 reporter, suggesting that the enzymatic activity of OGT is necessary for TLE-mediated transcriptional repression (Fig. 2D).
FIGURE 2.
OGT mediates transcriptional repression by TLEs. A, TLE can function independent of HDACs. HepG2 cells were transfected with a luciferase reporter with Gal4 DNA-binding sites and the indicated plasmids and were treated with 500 nm TSA. B, OGT acts via the Q or WD domains of TLE to repress transcription. HepG2 cells were transfected with a luciferase reporter with Gal4 DNA-binding sites and the indicated plasmids in the presence of 500 nm TSA. C, OGT overexpression potentiates TLE repression. Cells were transfected with the luciferase reporter and increasing amounts of the Gal4-TLE1/2 plasmids in the absence or presence of the OGT plasmid. D, TLE repression requires OGT catalytic activity. A catalytically dead OGT mutant (OGTcd) was compared with wild-type OGT (OGTwt) in a luciferase assay. Error bars, S.E.
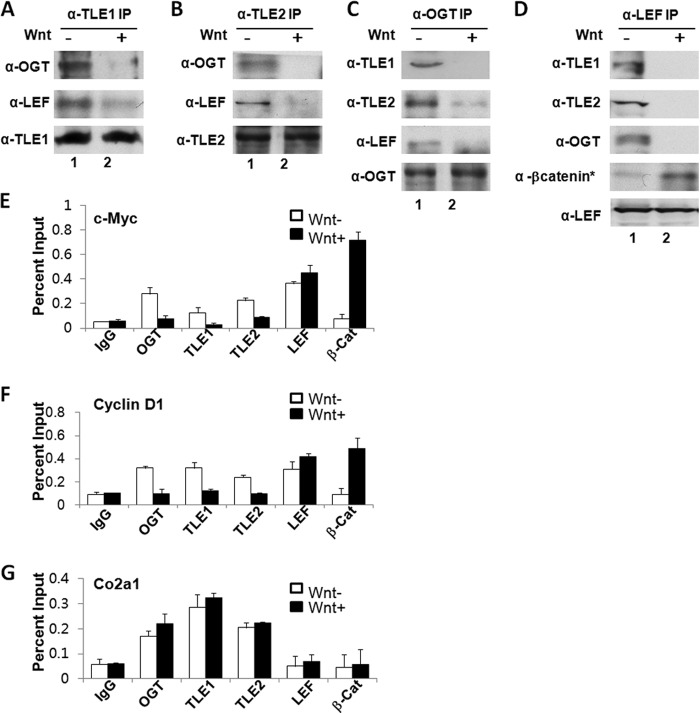
TLEs Associate with OGT in the Absence of Wnt Signaling
To evaluate the in vivo role for OGT in TLE-mediated repression, we analyzed the association of OGT with proteins known to form a complex with TCF/LEF during transcriptional inhibition and activation using Wnt1-conditioned media. OGT was immunoprecipitated with TLE1 or -2 in the absence of Wnt, and this association was disrupted in the presence of Wnt (Fig. 3, A and B). The reciprocal immunoprecipitation experiments further demonstrate that OGT is a component of the TLE corepressor complex on TCF/LEF in the absence of Wnt (Fig. 3, C and D) but not in the presence of Wnt when activated β-catenin (β-catenin*) associated with TCF/LEF (Fig. 3D). These data suggest that OGT interacts with TLE and TCF/LEF in a Wnt-responsive manner. To assess whether OGT associates with endogenous Wnt-responsive gene promoters, we used a ChIP assay to identify proteins present at consensus TCF/LEF-binding sites on the promoters of cyclin D1 and c-MYC genes (27, 28). In the absence of Wnt, OGT, TLE1, and TLE2 were associated with the LEF locus on cyclin D1 and c-MYC promoters, but Wnt activation displaces these repressor proteins from these promoters (Fig. 3E). Using an antibody against the activated form of β-catenin (β-catenin*), we found that β-catenin* replaced OGT/TLE on TCF/LEF in response to Wnt activation (Fig. 3E). As a control, we found that OGT, TLE1, and TLE2 were constitutively bound to the Notch-responsive HES-1 locus within the COL2A1 gene in the absence and presence of Wnt (Fig. 3E). These results indicate that OGT is an endogenous component of the TLE-LEF complex that represses Wnt signal.
FIGURE 3.
In vivo association of OGT with TLE at the TCF/LEF locus in the absence of Wnt activation. A and B, OGT associates with TLE1/2 in the absence of Wnt in 293 cells. C and D, OGT associates with LEF and TLEs in the absence of Wnt in 293 cells. In the presence of Wnt, activated β-catenin (β-catenin*) associates with TCF/LEF. E, ChIP demonstrates the association of OGT, TLE1/2, and LEF at TCF binding sites on Wnt-responsive promoters (cyclin D1 and c-MYC) in the absence of Wnt1 conditioned media in 293 cells. In the presence of Wnt conditioned media, activated β-catenin (B-cat*) and LEF are associated with the promoters. As a control, OGT, TLE1, and TLE2 were constitutively bound to the COl2A1 gene. IP, immunoprecipitation. Error bars, S.E.
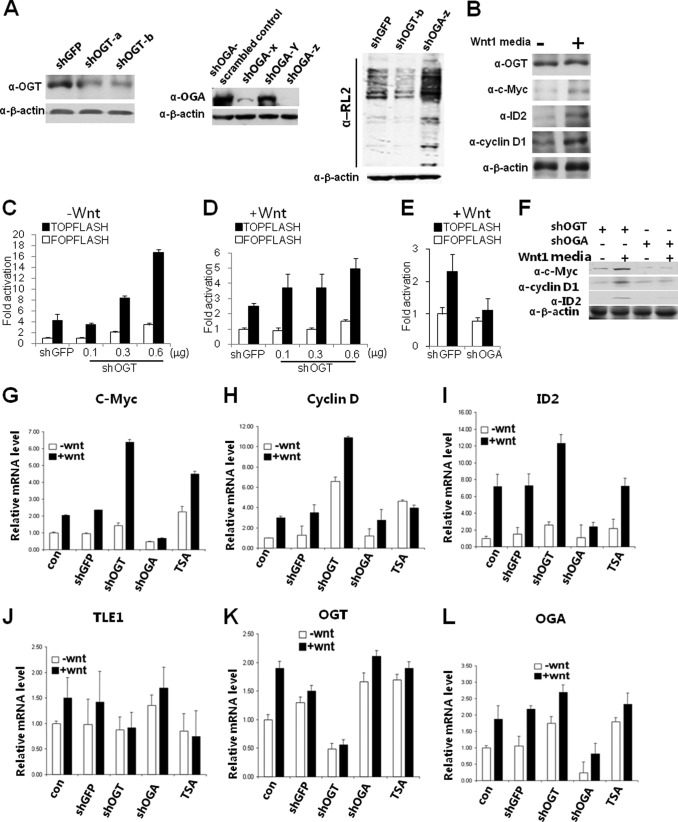
O-GlcNAc Is Required for Gene Repression by TLEs in the Wnt Pathway
The TOPFLASH system is commonly used to assess responsiveness at the TCF/LEF locus (29). This synthetic system compares expression of luciferase in response to TCF/LEF-binding sites (TOPFLASH) with that of mutant TCF/LEF-binding sites (FOPLASH). To evaluate the role of endogenous OGT in this system, we used several short hairpin RNAs combined with Wnt1-conditioned media, which was shown to activate expression of downstream genes (Fig. 4, A and B). Using shRNA to knock down levels of OGT (shOGT) caused a decrease in the O-GlcNAc level (Fig. 4A). Knockdown of OGT resulted in a dose-dependent derepression of the TCF/LEF locus on the TOPFLASH reporter (Fig. 4C). Additionally, luciferase expression on the FOPFLASH reporter was also increased to a certain extent, which is probably related to global derepression by reduction of O-GlcNAc levels (Fig. 4C). In the presence of Wnt, we observed a less striking difference between the TOPFLASH and FOPFLASH reporters (Fig. 4D). These results suggest that OGT represses transcription via TCF/LEF- dependent as well as -independent pathways.
FIGURE 4.
Gene repression in the Wnt pathway requires O-GlcNAc. A, Western blot analysis of knockdown of OGT and OGA in HEK293 cells using pSUPER expressing short hairpin RNA sequences listed in Table 2 and corresponding changes in global O-GlcNAc levels. B, Western blot analysis of Wnt-responsive genes in the absence and presence of Wnt1 conditioned media. C, knocking down OGT expression by shRNA significantly increases Wnt-responsive promoter activity. The pSUPER vector expressing shRNA against OGT was transfected into HEK293 cells, together with a Wnt-responsive luciferase reporter (TOPFLASH) or a non-responsive promoter (FOPFLASH). For a control, pSUPER encoding a shRNA against EGFP sequence was used. The β-galactosidase assay was used as a control for transfection efficiency. D, OGT shRNA elevates expression of endogenous Wnt target genes in the presence of Wnt. HEK293 cells were transfected with OGT shRNA or control shRNA. E, shRNA against OGA prevents activation by Wnt using the TOP/FOPFLASH reporter system. F, shRNA knockdown of OGT enhances and shRNA knockdown of OGA prevents expression of Wnt target genes analyzed by Western blot. G–L, shRNA knockdown of OGT enhances and shRNA knockdown of OGA prevents expression of Wnt target genes analyzed by quantitative real-time RT-PCR. Gene expression was normalized to GAPDH. Error bars, S.E.
O-GlcNAc moieties are removed from proteins enzymatically by O-GlcNAcase (OGA) (30). We examined whether OGA is essential for activation of the TCF/LEF locus. Knockdown of OGA by shOGA causes an increase in intracellular O-GlcNAc levels (Fig. 4A). OGA knockdown inhibited activation of the TCF/LEF locus by Wnt (Fig. 4E). Hence, OGA is required for Wnt activation and suppressed expression of Wnt-responsive genes at both the translational (Fig. 4F) and transcriptional levels (Fig. 4, G–L). Thus, OGA is a distinct requirement for canonical Wnt signaling.
Amino Enhancer of Split (AES) Antagonizes TLE Repression by Competition with OGT
The AES protein is homologous to the N-terminal Q and GP domains of TLEs, and it can partially antagonize TLE-mediated repression in a dominant negative fashion (31, 32). We employed AES as a tool to evaluate the structure-function relationship for the OGT-TLE interaction. To this end, we determined whether AES function as a derepressor lies in its ability to interfere with the TLE-OGT interaction via the Q-domain of AES. The results show that the addition of AES inhibits TLE1 binding to OGT in a pull-down assay (Fig. 5A). In doing so, AES might bind directly to OGT through its Q domains and prevent this enzyme from associating with TLEs. Alternatively, AES might bind to TLEs and compete off OGT. In an attempt to distinguish these two possibilities, we show that, although AES contains the highly conserved Q domain, it fails to bind to OGT (Fig. 5B). In contrast, it directly interacts with TLE1 (Fig. 5C), which can be explained by the intrinsic affinity between the Q domains (33). Hence, the dominant negative function of AES is probably due to its competition with OGT for binding to the Q domains of TLEs.
FIGURE 5.
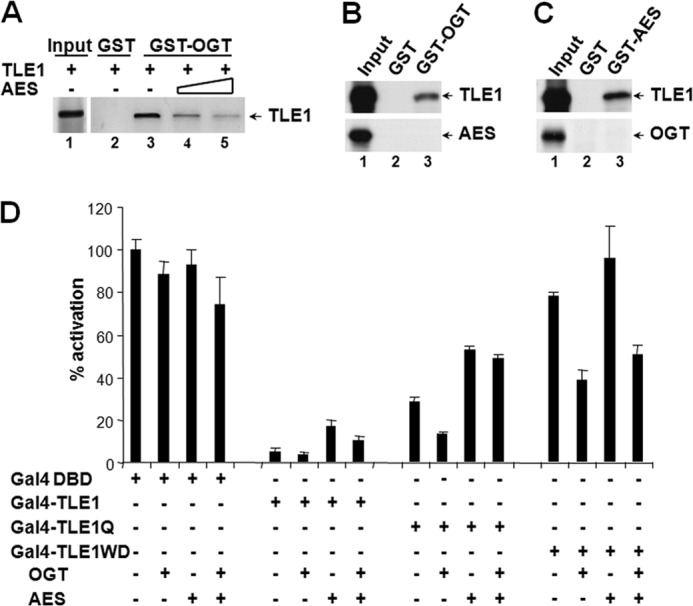
AES relieves TLE repression by inhibiting TLE-OGT interactions. A, AES competes with OGT for TLE1 binding. TLE1 and increasing amounts of AES were co-incubated with GST-OGT fusion protein. B and C, GST pull-down analyses revealed the interaction of AES with TLE1 but not OGT. D, AES specifically relieves Q domain-mediated repression. Cells were transfected with the plasmids for Gal4-TLE1 or its deletion mutants, together with OGT and AES either individually or in combination. Error bars, S.E.
Genetic and biochemical studies suggest that AES does not abrogate TLE repressive function entirely but rather fine tunes its function. Consistent with this notion, overexpression of AES only partially diminishes repression activity of the full-length TLE1 fused to Gal4 DBD in a luciferase reporter assay (Fig. 5D). Further analysis shows that AES specifically relieves repression imposed by the interaction between the Q domain of TLE1 and OGT, whereas it has no effect on repression by the WD domain via OGT (Fig. 5D). These results lead to a mechanistic interpretation for AES function as a moderate or partial “derepressor” (Fig. 5D).
DISCUSSION
OGT is a ubiquitous transcriptional regulator that plays important roles in gene repression (34–36). This work provides evidence that O-GlcNAc modification can specifically regulate the canonical Wnt locus. We show that the enzyme OGT can directly interact with Groucho/TLE transcription factors. OGT associates with a LEF/TLE repression complex in vivo (Fig. 3). This repression complex is disrupted upon Wnt stimulation (Fig. 4). We provide a framework for the role of O-GlcNAc in regulating the canonical Wnt locus (Fig. 6).
FIGURE 6.
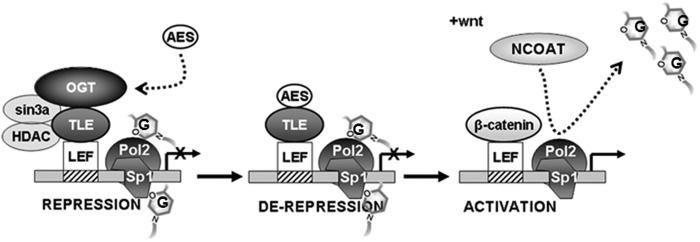
Proposed model representing the role of OGT at the TCF/LEF locus. OGT contributes to transcriptional repression through interaction with Groucho/TLEs. Human AES may displace OGT to evoke derepression. An activating signal is necessary for binding of β-catenin to LEF, and the removal of O-GlcNAc residues from transcriptional coactivators and other proteins involved in activation is essential for activation of the canonical Wnt locus.
We also show that genetic manipulation of OGT and OGA is sufficient to modulate a TCF/LEF reporter activity (TOPFLASH) (Fig. 4, C–E). Activation of the canonical Wnt locus is linked to expression of target genes, including c-MYC, ID2, and cyclin D1, whose expression levels are enhanced by lowering O-GlcNAc levels (Fig. 4, G–L). However, although O-GlcNAc modification appears to repress the canonical Wnt locus and prevent expression of Wnt-responsive genes, removal of O-GlcNAc moieties from proteins is not by itself indicative of activation of the canonical Wnt locus (Fig. 4) (i.e. shRNA against OGT causes a similar activation of mutant TCF reporter versus wild-type TCF-reporter in the absence of Wnt). In contrast, for TCF/LEF activation, the removal of O-GlcNAc modification from proteins seems to be essential along with the presence of β-catenin for the Wnt conditioned media to achieve full activation. It should be pointed out that the studies by Olivier-Van Stichelen et al. (37, 38) show that OGT enhances the transcriptional capability of β-catenin through stabilizing the protein in the cytosol. The contrary effect of OGT on β-catenin activity may reflect the spatiotemporal difference in OGT regulation in distinct cellular contexts.
AES has been hypothesized to antagnonize repression at the canonical Wnt locus (31, 32). Herein, we use AES to demonstrate critical features of the structure-function relationship that exists between OGT and TLE. AES contains high homology with the Q domain of full-length TLE. We observed that multiple interaction motifs may exist between OGT and TLE (Fig. 1) but that the Q domain of TLE, in particular, was more important for repression via OGT (Fig. 2A). Using AES, we showed that AES may antagonize the ability of OGT to associate with TLE, specifically by relieving repression through the Q domain (Fig. 5D). This supports the notion that a physical interaction between OGT and TLE is functionally important in mediating repression by TLE.
Groucho/TLE-mediated transcriptional repression may involve multiple mechanisms, such as expression of partner repressors, competition with coactivators, and posttranslational modifications (13). The experiments presented here support a model in which OGT is a component of the TLE-LEF repression complex in the nucleus. OGT associates directly with the TLEs, thereby specifically repressing canonical Wnt signals. There are multiple interaction surfaces between the TLE and OGT proteins. Thus, the TLEs are able to effectively recruit OGT and form a versatile corepressor pair. It has been shown that mSin3A interacts with HCF-1 to enhance transcriptional repression (39). We and others demonstrate that OGT physically interacts with mSin3A and HCF-1 (17, 40, 41). The association of OGT with the general corepressor mSin3A (17) suggests that the covalent modification of the transcriptional apparatus by O-GlcNAc represents a general mechanism for gene repression in diverse signaling pathways. However, some degree of specificity must be required to maintain transcriptional control. Based on the TPR domain of OGT (42), it has been hypothesized that specificity for O-GlcNAc modification is derived from the association of OGT with specific repressor molecules. The current work supports this notion. Although TLEs may also recruit mSin3A, HDACs, and thereby OGT, the direct association of the TLEs with OGT may provide more specificity to the repression complex. Thus, our evidence supports a distinct role for O-GlcNAc modification regulating the canonical Wnt locus.
Our proposed model (Fig. 6) suggests that during transcriptional repression, OGT is targeted to the canonical Wnt locus by its interaction with the TLEs, where it may modify critical elements of the transcriptional apparatus, such as Sp1 and RNA polymerase II (17, 21). AES may fine tune the system by antagonizing the interaction of OGT with TLEs. For activation of the TCF/LEF locus, stabilization of β-catenin and removal of O-GlcNAc residues by OGA are both necessary. Considering the wide array of proteins modified by O-GlcNAc, including transcriptional coactivators (43) and the proteasome (44), identification of a mammalian phenotype resultant from specific O-GlcNAc regulation of the canonical Wnt pathway may not be possible. Nevertheless, these data indicate that OGT activity may play a role in a variety of developmental processes regulated through the TCF/LEF locus.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK089098 and P01 DK057751 (to X. Y.) and DK043652 (to A. J. P.). This work was also supported by a grant from the Ellison Medical Foundation (to X. Y.) and National Natural Science Foundation of China Grant 81120108005/81225003 (to Y. N.).
This paper is dedicated to the memory of Dr. Jeffrey E. Kudlow (1947–2009), who trained and inspired many of us.
- TLE
- transducin-like enhancer of split
- Q
- WD, and GP domain, glutamine-rich, WD-repeat, and glycine/proline-rich domain, respectively
- TCF
- T cell factor
- LEF
- lymphoid enhancer factor
- TPR
- tetratricopeptide repeat
- CDI and CDII
- catalytic domain I and II, respectively
- OGT
- O-GlcNAc transferase
- HDAC
- histone deacetylase
- DBD
- DNA-binding domain
- OGA
- O-GlcNAcase
- AES
- amino enhancer of split
- TSA
- trichostatin.
REFERENCES
- 1. Jennings B. H., Ish-Horowicz D. (2008) The Groucho/TLE/Grg family of transcriptional co-repressors. Genome Biol. 9, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dehni G., Liu Y., Husain J., Stifani S. (1995) TLE expression correlates with mouse embryonic segmentation, neurogenesis, and epithelial determination. Mech. Dev. 53, 369–381 [DOI] [PubMed] [Google Scholar]
- 3. Li S. S. (2000) Structure and function of the Groucho gene family and encoded transcriptional corepressor proteins from human, mouse, rat, Xenopus, Drosophila and nematode. Proc. Natl. Sci. Counc. Repub. China B 24, 47–55 [PubMed] [Google Scholar]
- 4. Grbavec D., Lo R., Liu Y., Stifani S. (1998) Transducin-like Enhancer of split 2, a mammalian homologue of Drosophila Groucho, acts as a transcriptional repressor, interacts with Hairy/Enhancer of split proteins, and is expressed during neuronal development. Eur. J. Biochem. 258, 339–349 [DOI] [PubMed] [Google Scholar]
- 5. Gasperowicz M., Otto F. (2005) Mammalian Groucho homologs: redundancy or specificity? J. Cell. Biochem. 95, 670–687 [DOI] [PubMed] [Google Scholar]
- 6. Grbavec D., Stifani S. (1996) Molecular interaction between TLE1 and the carboxyl-terminal domain of HES-1 containing the WRPW motif. Biochem. Biophys. Res. Commun. 223, 701–705 [DOI] [PubMed] [Google Scholar]
- 7. Jennings B. H., Pickles L. M., Wainwright S. M., Roe S. M., Pearl L. H., Ish-Horowicz D. (2006) Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol. Cell 22, 645–655 [DOI] [PubMed] [Google Scholar]
- 8. Daniels D. L., Weis W. I. (2005) Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol. 12, 364–371 [DOI] [PubMed] [Google Scholar]
- 9. Yochum G. S., Ayer D. E. (2001) Pf1, a novel PHD zinc finger protein that links the TLE corepressor to the mSin3A-histone deacetylase complex. Mol. Cell. Biol. 21, 4110–4118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arce L., Pate K. T., Waterman M. L. (2009) Groucho binds two conserved regions of LEF-1 for HDAC-dependent repression. BMC Cancer 9, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jennings B. H., Wainwright S. M., Ish-Horowicz D. (2008) Differential in vivo requirements for oligomerization during Groucho-mediated repression. EMBO Rep. 9, 76–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buscarlet M., Stifani S. (2007) The “Marx” of Groucho on development and disease. Trends Cell Biol. 17, 353–361 [DOI] [PubMed] [Google Scholar]
- 13. Cinnamon E., Paroush Z. (2008) Context-dependent regulation of Groucho/TLE-mediated repression. Curr. Opin. Genet. Dev. 18, 435–440 [DOI] [PubMed] [Google Scholar]
- 14. Hanson A. J., Wallace H. A., Freeman T. J., Beauchamp R. D., Lee L. A., Lee E. (2012) XIAP monoubiquitylates Groucho/TLE to promote canonical Wnt signaling. Mol. Cell 45, 619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nuthall H. N., Joachim K., Stifani S. (2004) Phosphorylation of serine 239 of Groucho/TLE1 by protein kinase CK2 is important for inhibition of neuronal differentiation. Mol. Cell. Biol. 24, 8395–8407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ju B. G., Solum D., Song E. J., Lee K. J., Rose D. W., Glass C. K., Rosenfeld M. G. (2004) Activating the PARP-1 sensor component of the groucho/TLE1 corepressor complex mediates a CaMKinase IIδ-dependent neurogenic gene activation pathway. Cell 119, 815–829 [DOI] [PubMed] [Google Scholar]
- 17. Yang X., Zhang F., Kudlow J. E. (2002) Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell 110, 69–80 [DOI] [PubMed] [Google Scholar]
- 18. Yang X., Su K., Roos M. D., Chang Q., Paterson A. J., Kudlow J. E. (2001) O-Linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc. Natl. Acad. Sci. U.S.A. 98, 6611–6616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wong K. H., Struhl K. (2011) The Cyc8-Tup1 complex inhibits transcription primarily by masking the activation domain of the recruiting protein. Genes Dev. 25, 2525–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Comer F. I., Hart G. W. (2001) Reciprocity between O-GlcNAc and O-phosphate on the carboxyl terminal domain of RNA polymerase II. Biochemistry 40, 7845–7852 [DOI] [PubMed] [Google Scholar]
- 21. Ranuncolo S. M., Ghosh S., Hanover J. A., Hart G. W., Lewis B. A. (2012) Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription in vitro and in vivo. J. Biol. Chem. 287, 23549–23561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Korinek V., Barker N., Morin P. J., van Wichen D., de Weger R., Kinzler K. W., Vogelstein B., Clevers H. (1997) Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science 275, 1784–1787 [DOI] [PubMed] [Google Scholar]
- 23. Kreppel L. K., Blomberg M. A., Hart G. W. (1997) Dynamic glycosylation of nuclear and cytosolic proteins: cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 272, 9308–9315 [DOI] [PubMed] [Google Scholar]
- 24. Lubas W. A., Hanover J. A. (2000) Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J. Biol. Chem. 275, 10983–10988 [DOI] [PubMed] [Google Scholar]
- 25. Chen G., Fernandez J., Mische S., Courey A. J. (1999) A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 13, 2218–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fisher A. L., Ohsako S., Caudy M. (1996) The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol. Cell. Biol. 16, 2670–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sasaki T., Suzuki H., Yagi K., Furuhashi M., Yao R., Susa S., Noda T., Arai Y., Miyazono K., Kato M. (2003) Lymphoid enhancer factor 1 makes cells resistant to transforming growth factor β-induced repression of c-myc. Cancer Res. 63, 801–806 [PubMed] [Google Scholar]
- 28. Sierra J., Yoshida T., Joazeiro C. A., Jones K. A. (2006) The APC tumor suppressor counteracts β-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 20, 586–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Korinek V., Barker N., Willert K., Molenaar M., Roose J., Wagenaar G., Markman M., Lamers W., Destree O., Clevers H. (1998) Two members of the Tcf family implicated in Wnt/β-catenin signaling during embryogenesis in the mouse. Mol. Cell. Biol. 18, 1248–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Toleman C., Paterson A. J., Whisenhunt T. R., Kudlow J. E. (2004) Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J. Biol. Chem. 279, 53665–53673 [DOI] [PubMed] [Google Scholar]
- 31. Roose J., Molenaar M., Peterson J., Hurenkamp J., Brantjes H., Moerer P., van de Wetering M., Destrée O., Clevers H. (1998) The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature 395, 608–612 [DOI] [PubMed] [Google Scholar]
- 32. Muhr J., Andersson E., Persson M., Jessell T. M., Ericson J. (2001) Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell 104, 861–873 [DOI] [PubMed] [Google Scholar]
- 33. Pinto M., Lobe C. G. (1996) Products of the grg (Groucho-related gene) family can dimerize through the amino-terminal Q domain. J. Biol. Chem. 271, 33026–33031 [DOI] [PubMed] [Google Scholar]
- 34. Li M. D., Ruan H. B., Singh J. P., Zhao L., Zhao T., Azarhoush S., Wu J., Evans R. M., Yang X. (2012) O-GlcNAc transferase is involved in glucocorticoid receptor-mediated transrepression. J. Biol. Chem. 287, 12904–12912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gambetta M. C., Oktaba K., Müller J. (2009) Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 325, 93–96 [DOI] [PubMed] [Google Scholar]
- 36. Ozcan S., Andrali S. S., Cantrell J. E. (2010) Modulation of transcription factor function by O-GlcNAc modification. Biochim. Biophys. Acta 1799, 353–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Olivier-Van Stichelen S., Guinez C., Mir A. M., Perez-Cervera Y., Liu C., Michalski J. C., Lefebvre T. (2012) The hexosamine biosynthetic pathway and O-GlcNAcylation drive the expression of β-catenin and cell proliferation. Am. J. Physiol. Endocrinol. Metab. 302, E417–424 [DOI] [PubMed] [Google Scholar]
- 38. Olivier-Van Stichelen S., Drougat L., Dehennaut V., El Yazidi-Belkoura I., Guinez C., Mir A. M., Michalski J. C., Vercoutter-Edouart A. S., Lefebvre T. (2012) Serum-stimulated cell cycle entry promotes ncOGT synthesis required for cyclin D expression. Oncogenesis 1, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wysocka J., Myers M. P., Laherty C. D., Eisenman R. N., Herr W. (2003) Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17, 896–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ruan H. B., Han X., Li M. D., Singh J. P., Qian K., Azarhoush S., Zhao L., Bennett A. M., Samuel V. T., Wu J., Yates J. R., 3rd, Yang X. (2012) O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1α stability. Cell Metab. 16, 226–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Capotosti F., Guernier S., Lammers F., Waridel P., Cai Y., Jin J., Conaway J. W., Conaway R. C., Herr W. (2011) O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell 144, 376–388 [DOI] [PubMed] [Google Scholar]
- 42. Iyer S. P., Hart G. W. (2003) Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J. Biol. Chem. 278, 24608–24616 [DOI] [PubMed] [Google Scholar]
- 43. Bowe D. B., Sadlonova A., Toleman C. A., Novak Z., Hu Y., Huang P., Mukherjee S., Whitsett T., Frost A. R., Paterson A. J., Kudlow J. E. (2006) O-GlcNAc integrates the proteasome and transcriptome to regulate nuclear hormone receptors. Mol. Cell. Biol. 26, 8539–8550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu J., Wang S., Viollet B., Zou M. H. (2012) Regulation of the proteasome by AMPK in endothelial cells: the role of O-GlcNAc transferase (OGT). PloS One 7, e36717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]