Abstract
In eukaryotes, there are at least 60 members of the DExD/H helicase family, many of which are able to sense viral nucleic acids. By screening all known family members, we identified the helicase DHX33 as a novel double-stranded RNA (dsRNA) sensor in myeloid dendritic cells (mDCs). The knockdown of DHX33 using small heteroduplex RNA (shRNA) blocked the ability of mDCs to produce type I interferon (IFN) in response to poly I:C and reovirus. The HELICc domain of DHX33 was shown to bind poly I:C. The interaction between DHX33 and IPS-1 is mediated by the HELICc region of DHX33 and the C-terminal domain of IPS-1 (also referred to MAVS and VISA). The inhibition of DHX33 expression by RNA interference blocked the poly I:C-induced activation of MAP kinases, NF-κB and IRF3. The interaction between the helicase DHX33 and IPS-1 was independent of RIG-I/MDA5 and may be a novel pathway for sensing poly I:C and RNA viruses in mDCs.
Keywords: DHX33, helicase, innate immunity, IPS-1, myeloid dendritic cell, viral nucleic acid
INTRODUCTION
The innate immune system is able to recognize pathogenic microbes as foreign invaders and eliminate them. The key steps necessary for innate immune responses are the rapid detection of the invader and the induction of type I interferon (IFN) and/or pro-inflammatory cytokines. A growing number of receptors/sensors for pathogen RNA and poly I:C are currently being discovered. The Toll-like receptors (TLRs) are a class of membrane receptors that sense extracellular microbes and trigger anti-pathogen signaling cascades.1,2,3 The NOD-like receptors can sense intracellular microbes.4,5 The helicase-domain-containing antiviral proteins can sense intracellular viral RNA.6,7,8,9 In addition, there are cytoplasmic molecules that can sense microbial, as well as non-microbial, danger signals. There are multiple members of the DExD/H helicase family in eukaryotes. It has been reported that the RIG-I-like DExD/H helicases RIG-I, MDA5, LGP2 and DICER, as well as DDX3, are able to sense poly I:C or viral nucleic acids.6,7,8,10,11 Using biotinylated-poly I:C and biotinylated-CpG pulldown assays and mass spectrometry, we identified DHX36 and DHX9 as cytosolic CpG-DNA sensors that pair with MyD88 to trigger type I IFN responses in plasmacytoid dendritic cells.12 We also showed that DDX1–DDX21–DHX36 complexes pair with TRIF to sense cytosolic poly I:C in myeloid dendritic cells (mDCs).13 By screening proteins containing double-stranded RNA (dsRNA) binding motifs, we found that DHX9 pairs with IPS-1 to sense poly I:C in mDCs.14 DDX60 was recently identified as a novel DNA/RNA sensor that has the ability to bind both viral RNA and DNA and to promote RIG-I-dependent IFN production in response to dsRNA.15 DDX3 was found to bind IPS-1 and promote RIG-I-independent IFN production in response to dsRNA.11 These studies demonstrate the essential and broad role of the DExD/H helicase family in viral RNA/DNA sensing by serving either as direct sensors or positive regulators of the involved signal transduction pathways.
In this study, we investigated the potential roles of all 60 DExD/H helicase family members in poly I:C-induced IFN-β responses using small interfering RNA (siRNA) and small heteroduplex RNA (shRNA) screening. We report the identification of DHX33 as a novel dsRNA sensor in mDCs.
MATERIALS AND METHODS
Materials
Poly I:C and lipopolysaccharide (LPS) were purchased from Invivogen (San Diego, CA, USA). Lipofectamine 2000 was purchased from Invitrogen. The following antibodies were used for immunoprecipitation and/or immunoblotting: anti-DHX33 (Lifespan Biosciences, Seattle, WA, USA), anti-IRF3 (Santa Cruz, Dallas, TX, USA), anti-MDA5, anti-RIG-I, anti-IPS-1, anti-p65, anti-phospho-IRF3 and anti-phospho-p65 (Cell Signaling, Danvers, MA, USA), anti-β-actin, anti-HA-HRP and anti-Myc-HRP (Sigma, St Louis, MO, USA). Anti-HA and anti-Myc beads were purchased from Sigma (St Louis, MO, USA). NeutrAvidin-beads were purchased from Pierce (Pittsburgh, PA, USA).
Mouse embryonic fibroblast (MEF) cell culture and lentiviral infection
MEFs were maintained in Dulbecco's modified Eagle medium containing 15% heat-inactivated fetal calf serum and 1% penicillin-streptomycin (Invitrogen-GIBCO, Grand Island, NY, USA). MEFs were infected with a lentiviral vector carrying a DHX33 shRNA sequence, an IPS-1 shRNA sequence or a scrambled shRNA. Infected cells were selected by adding puromycin (1 ng/ml) to the medium 2 days post-infection. Cells were stimulated with poly I:C (2.5 µg/ml) plus Lipofectamine 2000 for 16 h. RNA was extracted from the stimulated cells and analyzed by real-time RT-PCR to determine the knockdown efficiency.
RNA interference
D2SC mDC cell culture and the preparation of GM-CSF-derived DCs were performed as previously described.12 For siRNA knockdown, 0.5 million D2SC mDCs were transfected with 0.4 nmol of siRNAs (Dharmacon, Pittsburgh, PA, USA) using the Mouse Dendritic Cell Nucleofector Kit (Amaxa, Allendale, NJ, USA). The program number for electroporation was Y001. Twenty-four hours post-transfection, the cells were stimulated with poly I:C for 16 h. RNA was extracted from the collected cells and used in RT-PCR. For shRNA knockdown, shRNA lentiviral vectors were purchased from Open Biosystems (Pittsburgh, PA, USA) (target for IPS-1, clone TRCN0000124770; targets for DHX33, clone TRCN0000113057 (DHX33-a) and TRCN0000113059 (DHX33-b)). For the rescue experiment, the knockdown of DHX33 in D2SC cells was achieved using shRNA targeting the 3′ UTR of DHX33 (clone TRCN0000113058).
In vitro pulldown and immunoblotting assay
To prepare purified DHX33 and IPS-1 proteins, HEK293T cells were transfected with an expression plasmid encoding the full-length or truncated versions of HA-DHX33 or Myc-IPS-1. Lysates were prepared from the transfected cells and were incubated with anti-HA or anti-Myc beads. Proteins were eluted from the beads after washing with phosphate-buffered saline six times. For anti-HA bead or anti-Myc bead pulldowns, purified HA-DHX33 protein was incubated with purified Myc-IPS-1 protein for 1 h. Beads were added, and following a 1-h incubation, the bound complexes were pelleted by centrifugation. The proteins and beads were analyzed by immunoblotting with anti-HA-HRP or anti-Myc-HRP antibodies. For NA-bead pulldowns, the purified HA-DHX33 proteins were incubated for 2 h with biotin-labeled poly I:C. Following incubation with the NA-beads, the bound complexes were pelleted by centrifugation and analyzed by immunoblotting with an anti-HA antibody. To determine the DHX33 binding specificity, pulldown assays were repeated in the absence and presence of increasing amounts (0.5, 5 and 50 µg/ml) of unlabeled poly I:C or poly U.
RNA extraction and quantitative PCR
Total RNA was isolated from cells using the RNeasy kit (Qiagen, Valencia, CA, USA). Trace amounts of DNA in the RNA extract were digested with DNase (Qiagen). cDNA was prepared using SuperScript II (Invitrogen, Grand Island, NY, USA) and random hexamer primers. Quantitative PCR (QPCR) amplification was performed using the TaqMan universal master mix (Applied Biosystems, Carlsbad, CA, USA) and a 7500 Fast Real-Time PCR system (Applied Biosystems, Carlsbad, CA, USA). The following primers were used for QPCR: DHX33: forward 5′-TGCGTGAAGCAATTTCAGAC-3′ and reverse 5′-AGGTCGACATCCATGGTAGC-3′ IPS-1: forward 5′-AGAGCAACTCCTCCAGACCA-3′ and reverse 5′-AACGGTTGGAGACACAGGTC-3′ IFN-β: forward 5′-CCCTATGGAGATGACGGAGA-3′ and reverse 5′-TCCCACGTCAATCTTTCCTC-3′ and GAPDH: forward 5′-ACCCAGAAGACTGTGGATGG-3′ and reverse 5′-CAGTGAGCTTCCCGTTCAG-3′.
Signal transduction
D2SC cells were stimulated with 2.5 µg/ml poly I:C delivered by Lipofectamine 2000. The cells were lysed in RIPA buffer after stimulation for 0, 30, 60 or 120 min. The lysates were resolved by 4%–20% SDS–PAGE and blotted with antibodies recognizing the phosphorylated and unphosphorylated forms of p65, Erk1/2, p38 and IRF3.
RESULTS
DHX33 senses both poly I:C and RNA viruses in D2SC mDCs
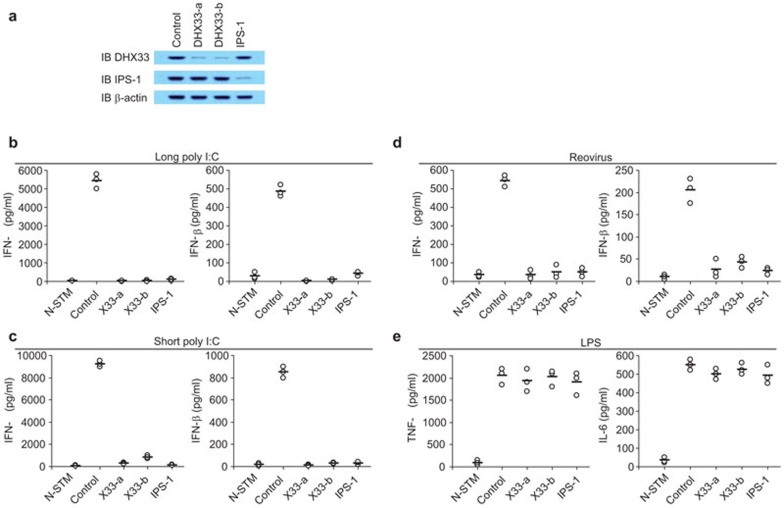
The DExD/H helicase family members have been shown to play important roles in sensing dsRNA and viral infection. We assessed the potential roles of all 60 DExD/H helicase family members by knocking down the expression of each helicase, followed by monitoring the IFN-β production levels in response to poly I:C stimulation. In addition to RIG-I, MDA5, LGP2, DDX1, DDX3, DHX9, DDX21, DHX36, DDX60 and DICER, we found that DHX33 is also involved in sensing poly I:C. We therefore established stable D2SC cell lines expressing shRNA to knockdown the expression of DHX33. Figure 1a shows that two distinct DHX33-targeting shRNAs (X33-a and X33-b) efficiently knocked down the expression of DHX33 at the protein level without affecting the expression of IPS-1. D2SC mDCs treated with the scrambled shRNA produced high levels of type I IFN (IFN-α and IFN-β) following stimulation with long or short poly I:C. This type I IFN response was strongly attenuated in IPS-1-knockdown D2SC mDCs, confirming previous reports showing that IPS-1 is the key adaptor for poly I:C sensors.16,17,18 This type I IFN response was also strongly attenuated in the two DHX33-knockdown D2SC cell lines (Figure 1b and c), indicating that DHX33 plays important roles in sensing both long and short poly I:C in D2SC mDCs. We next investigated whether DHX33 senses viral infection in D2SC mDCs. Type I IFN production was measured after culturing control and knockdown D2SC mDCs with reovirus. Both DHX33- and IPS-1-knockdown D2SC mDCs had a 90% reduction in type I IFN production in response to reovirus (Figure 1d). To determine whether DHX33 senses other stimuli, cytokine production was measured after control and knockdown D2SC cells were stimulated with LPS. As shown in Figure 1e, the knockdown of DHX33, as well as IPS-1, in D2SC mDCs, had little effect on TNF-α and IL-6 production in response to LPS. These data indicate that DHX33 plays an important role in mediating the responses of D2SC mDCs to poly I:C and an RNA virus, but not LPS.
Figure 1.
DHX33 senses both poly I:C and an RNA virus in D2SC mDCs. (a) IB showing the knockdown efficiency of shRNAs targeting the indicated proteins in D2SC cells. Scrambled shRNA served as a control (left most lane). Actin blots are shown as loading controls (lower panel). ELISA of IFN-α and IFN-β production by D2SC cells treated with the indicated shRNAs after stimulation with (b) 2.5 µg/ml long poly I:C, (c) 5 µg/ml short poly I:C or (d) reovirus. Cells were stimulated with nucleic acids, delivered by Lipofectamine 2000, for 16 h. Virus was added to the cells at a MOI of 10 for 2 h. Cells were collected, washed with PBS and cultured for an additional 16 h. (e) ELISA of TNF-α and IL-6 production by D2SC cells treated with the indicated shRNAs after 16 h of stimulation with 1 µg/ml LPS. N-STM, unstimulated D2SC cells treated with scrambled shRNA. Individual circles represent the value from each independent experiment. Bars represent the average values from at least three independent experiments. IB, immunoblot; IFN, interferon; LPS, lipopolysaccharide; MOI, multiplicity of infection; PBS, phosphate-buffered saline; shRNA, small heteroduplex RNA.
DHX33 knockdown in bone marrow-derived DCs (BMDCs) abolishes their cytokine responses to poly I:C and an RNA virus
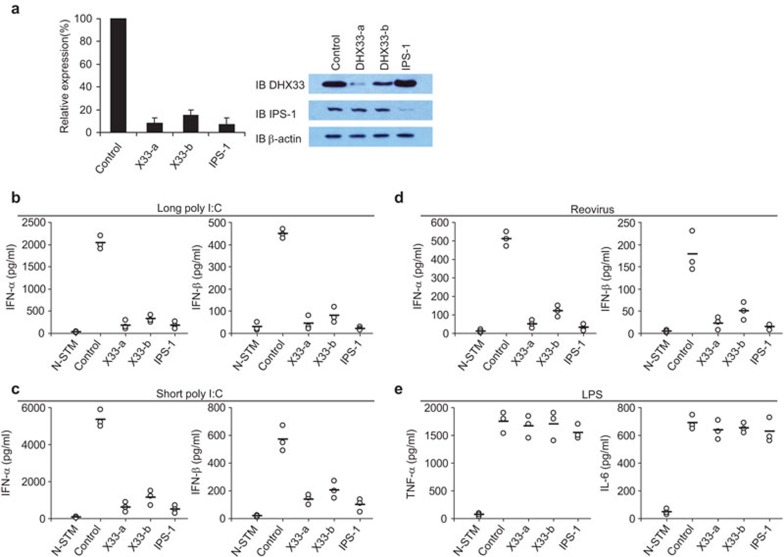
To determine whether DHX33 senses poly I:C or a dsRNA virus (reovirus) in primary cells, GM-CSF-derived mDCs were prepared and treated with shRNA to individually knock down the expression of DHX33 and IPS-1. Figure 2a shows that the shRNAs knocked down DHX33 and IPS-1 protein expression with an efficiency of 90%. The type I IFN responses of control and knockdown BMDCs were examined following stimulation with poly I:C, reovirus or LPS. As shown in Figure 2b–d, DHX33- and IPS-1-knockdown BMDCs produced much lower type I IFN levels in response to long poly I:C (Figure 2b), short poly I:C (Figure 2c) and reovirus (Figure 2d) than control cells stimulated under the same conditions. Compared with control cells stimulated with LPS, DHX33- and IPS-1-knockdown BMDCs produced similar levels of TNF-α and IL-6. These data indicate that both DHX33 and IPS-1 have critical functions in sensing both poly I:C and a dsRNA virus in BM DCs.
Figure 2.
DHX33 knockdown in BMDCs abolishes their cytokine responses to poly I:C and reovirus. (a) QPCR showing the knockdown efficiency of shRNA targeting the indicated genes in BMDCs (left panel). IB showing the knockdown efficiency of shRNAs targeting the indicated proteins in BMDCs (right panel). Scrambled shRNA served as a control. ELISA of IFN-α and IFN-β production by BMDCs treated with the indicated shRNAs after stimulation with (b) 2.5 µg/ml long poly I:C, (c) 5 µg/ml short poly I:C or (d) reovirus at a MOI of 10. Cells were stimulated with nucleic acids, delivered by Lipofectamine 2000, for 16 h. Virus was added to the cells for 2 h. Cells were collected, washed with PBS and cultured for an additional 16 h. (e) ELISA of TNF-α and IL-6 production by BMDCs treated with the indicated shRNAs after 16 h of stimulation with 1 µg/ml LPS. N-STM, unstimulated D2SC cells treated with the scrambled shRNA. Individual circles represent the values from each independent experiment. Bars represent the average values from at least three independent experiments. BMDC, bone marrow-derived dendritic cell; IB, immunoblot; IFN, interferon; MOI, multiplicity of infection; LPS, lipopolysaccharide; PBS, phosphate-buffered saline; QPCR, quantitative PCR; shRNA, small heteroduplex RNA.
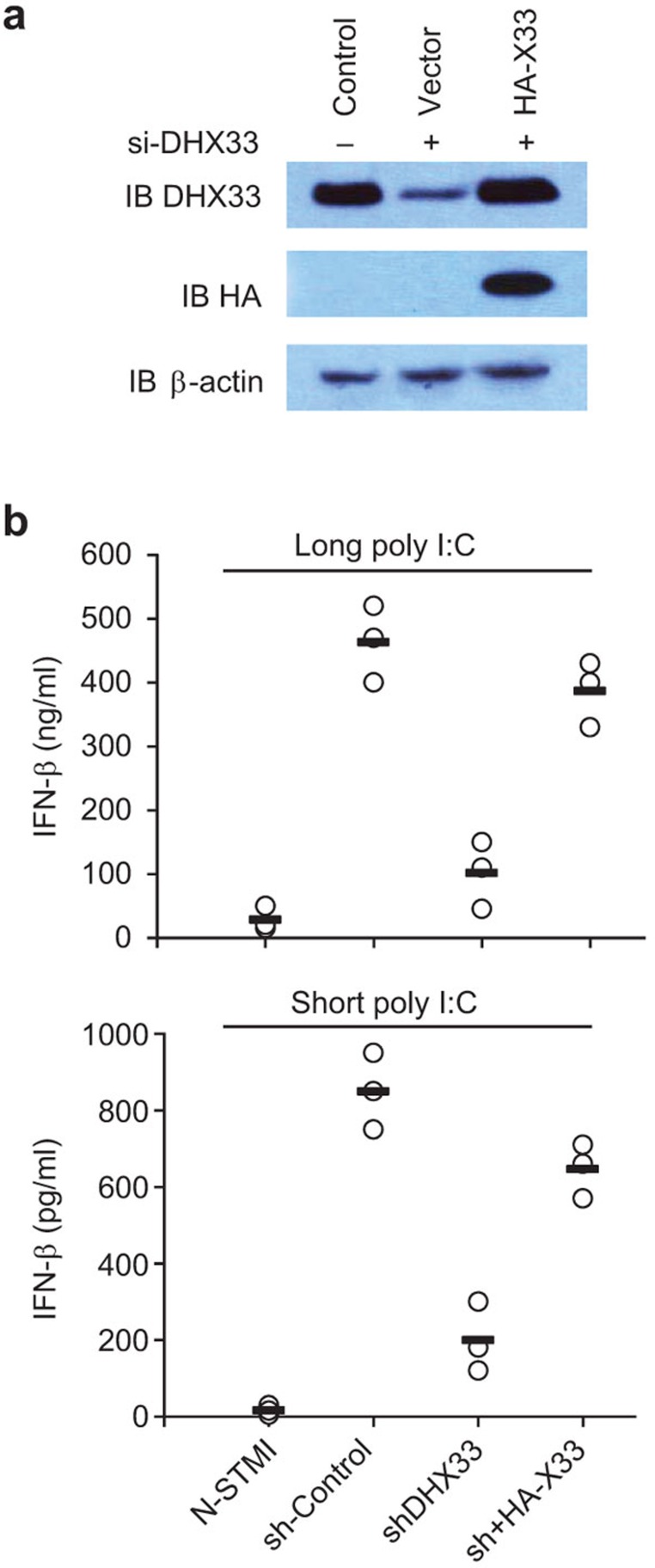
Recombinant DHX33 rescues the DHX33 shRNA-induced defect
To determine whether recombinant DHX33 could rescue the defective IFN production in response to poly I:C caused by DHX33 shRNA knockdown, we reconstituted DHX33-knockdown D2SC cells with HA-tagged DHX33 (Figure 3a). The shRNA used in this study selectively targets the 3′ UTR of the DHX33 mRNA. Therefore, only the expression of endogenous DHX33 was knocked down. The reconstitution of DHX33-knockdown cells with HA-tagged DHX33 rescued the IFN-β production that was rendered defective in DHX33-knockdown cells in response to stimulation with both long poly I:C and short poly I:C (Figure 3b).
Figure 3.
Recombinant DHX33 can rescue the defect in poly I:C-activated IFN production caused by DHX33 shRNA knockdown. (a) Immunoblot of endogenous DHX33 in D2SC mDCs (Control) and recombinant HA-DHX33 (sh+HA-X33) in D2SC mDCs in which endogenous DHX33 was selectively knocked down (shDHX33). (b) ELISA of IFN-β production by D2SC mDCs. IFN, interferon; mDC, myeloid dendritic cell; shRNA, small heteroduplex RNA.
DHX33 signaling is independent of MDA5 signaling and RIG-I signaling
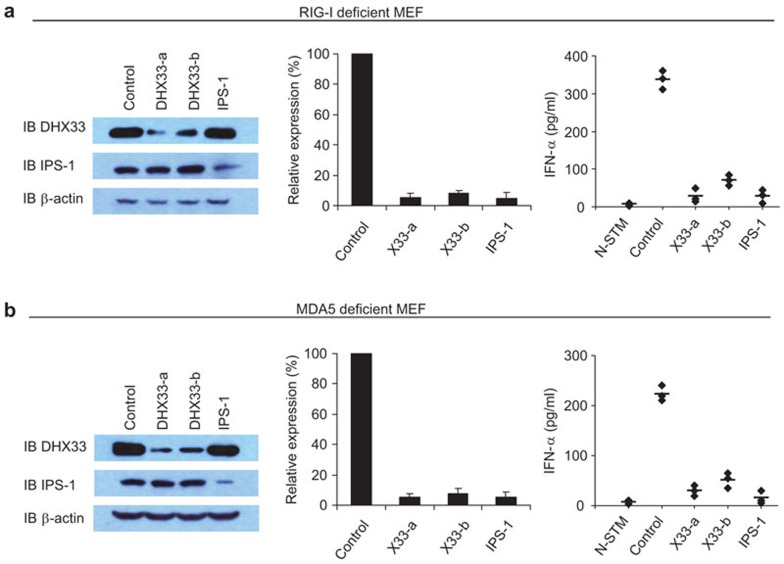
Because IPS-1 is the key adaptor molecule that mediates RIG-I and MDA5 signaling in the sensing of poly I:C, we sought to determine whether DHX33 signaling is independent of MDA5 and RIG-I signaling. We performed experiments using MEFs derived from MDA5- and RIG-I-deficient mice. The production of IFN-α in response to poly I:C in MDA5- and RIG-I-deficient MEFs remained intact, agreeing with a previous report.6 However, IFN-α responses induced by poly I:C were blocked in MDA5- and RIG-I-deficient MEFs after the expression of DHX33 was knocked down by shRNA with 90% efficiency (Figure 4), suggesting that the ability of DHX33 to sense poly I:C is independent of RIG-I and MDA5.
Figure 4.
DHX33 signaling is independent of MDA5 and RIG-I signaling. (a) IB showing the knockdown efficiency of shRNAs targeting the indicated proteins in RIG-I-deficient MEFs (left panel). QPCR showing the knockdown efficiency of shRNAs targeting the indicated genes in RIG-I-deficient MEFs (middle panel). Scrambled shRNA served as a control. ELISA of IFN-α production by RIG-I-deficient MEFs treated with the indicated shRNAs after 16 h of stimulation with 5 µg/ml short poly I:C delivered to the cells by Lipofectamine 2000 (right panel). (b) IB showing the knockdown efficiency of shRNAs targeting the indicated proteins in MDA5-deficient MEFs (left panel). QPCR analysis showing the knockdown efficiency of shRNAs targeting the indicated genes in MDA5-deficient MEFs (middle panel). Scrambled shRNA served as a control. ELISA of IFN-α production by MDA5-deficient MEFs treated with the indicated shRNAs after 16 h of stimulation with 2.5 µg/ml long poly I:C delivered to the cells by Lipofectamine 2000 (right panel). N-STM, unstimulated RIG-1- or MDA5-deficient MEFs treated with scrambled shRNA. The data represent the mean±s.d. of triplicate or quadruplicate measurements. Individual diamonds represent the values from each independent experiment. Bars represent the average values from at least three independent experiments. IB, immunoblot; IFN, interferon; MEF, mouse embryonic fibroblast; QPCR, quantitative PCR; shRNA, small heteroduplex RNA.
DHX33 forms a complex with IPS-1 in D2SC mDCs
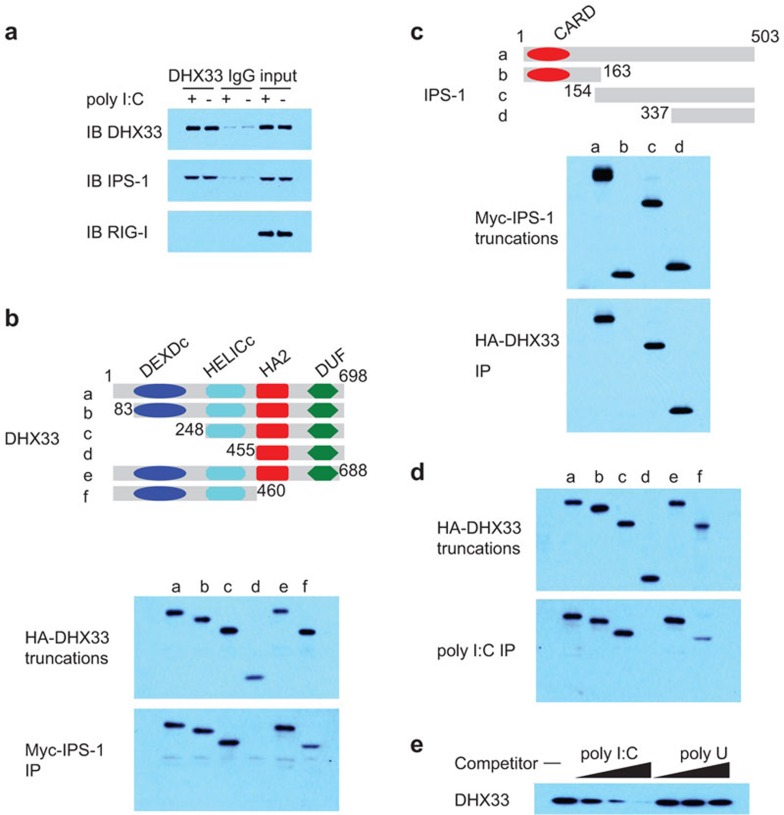
D2SC mDCs and BMDCs in which DHX33 or IPS-1 had been knocked down displayed similar cytokine responses to poly I:C and a dsRNA virus. We next determined whether IPS-1 is the adaptor molecule for DHX33 signal transduction. To investigate the potential interaction between DHX33 and IPS-1, we performed anti-DHX33 immunoprecipitation experiments with resting and poly I:C-activated D2SC mDCs. As shown in Figure 5a, endogenous DHX33 was associated with IPS-1 in both resting and poly I:C-stimulated D2SC mDCs. By contrast, an association between DHX33 and RIG-I was undetectable.
Figure 5.
DHX33 interacts with both poly I:C and IPS-1. (a) IB of the indicated proteins precipitated with an anti-DHX33 antibody from whole-cell lysates of D2SC cells that were unstimulated (−) or stimulated with long poly I:C (+). An IgG antibody served as a control. (b) Top: schematic representation of DHX33 and its serial truncations. DExDc: Asp–Glu–Ala–Asp box motif; Helic C: helicase C-terminal domain. HA2: helicase-associated domain. DUF: domain of unknown function. Numbers denote amino-acid residues. Middle panel: immunoblot of purified HA-DHX33 truncated proteins using an anti-HA antibody (a–f). Bottom: immunoblot using an anti-HA antibody of proteins precipitated (IP) with an anti-Myc antibody from a mixture of an HA-DHX33 protein (a–f) individually incubated with a Myc-IPS-1 fusion protein. (c) Top: schematic representation of IPS-1 and its serial truncations. CARD: the caspase activation and recruitment domain. Numbers denote amino-acid residues. Middle panel: immunoblot using an anti-Myc antibody of purified Myc-IPS-1 truncated proteins (a–d). Bottom: immunoblot using an anti-Myc antibody of proteins precipitated (IP) from a mixture of a Myc-IPS-1 protein (a–d) individually incubated with an HA-DHX33 fusion protein. (d) Immunoblot using an anti-HA antibody of pulldown assays in which purified, serial truncations of HA-DHX33 (a–f) were individually incubated with biotinylated poly I:C, followed by the addition of NeutrAvidin beads. (e) Immunoblot using an anti-HA antibody of pulldown competition assay products in which 0.5, 5 or 50 µg/ml poly I:C or poly U was added to a mixture of HA-DHX33 plus biotinylated poly I:C, followed by the addition of NeutrAvidin beads. IB, immunoblot; IP, immunoprecipitation.
DHX33 and IPS-1 interact via the HELICc domain and the C-terminal domain, respectively
To map the regions of DHX33 and IPS-1 that mediate their interaction, serial truncations of DHX33 and IPS-1 were prepared. A Myc fusion of IPS-1 was incubated with either full-length or truncated HA fusions of DHX33, followed by pulldown with anti-Myc beads. As shown in Figure 5b, the HELICc domain of DHX33 was required for the interaction with IPS-1. Likewise, we conducted reciprocal experiments using truncated versions of Myc-IPS-1 and full-length HA-DHX33 to identify the site(s) in IPS-1 bound by DHX33. We found that the C-terminal domain of IPS-1 was required for the interaction between IPS-1 and DHX33 (Figure 5c).
DHX33 binds poly I:C through its HELICc region
To determine whether DHX33 directly binds poly I:C, HA fusions of full-length DHX33 or truncated DHX33 were incubated with biotin-labeled poly I:C, followed by pulldown with NeutrAvidin beads. The HELICc region of DHX33 was required for binding to poly I:C, suggesting that DHX33 binds dsRNA directly (Figure 5d). To confirm that DHX33 specifically binds to poly I:C, we performed competition experiments by adding increasing amounts of unlabeled poly I:C or poly U to a mixture of an HA fusion of DHX33 plus biotin-labeled poly I:C, followed by pulldown with NeutrAvidin beads. We found that only unlabeled poly I:C could block the binding of biotin-labeled poly I:C to DHX33 (Figure 5e), supporting the hypothesis that DHX33 directly associates with poly I:C.
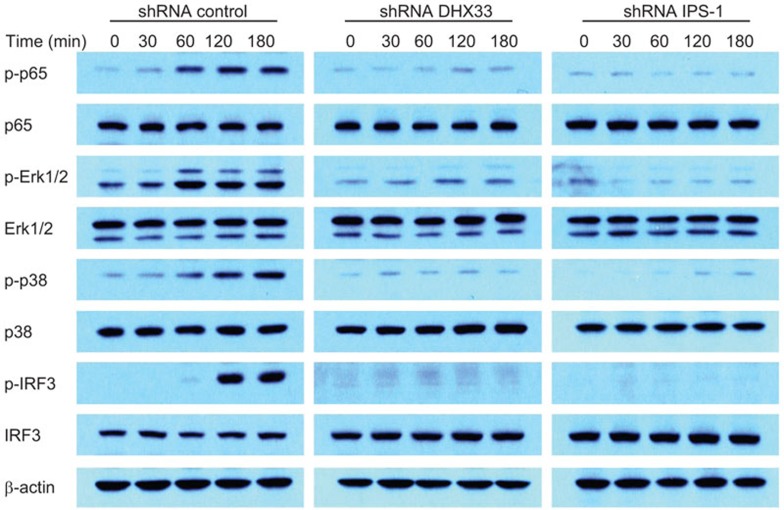
DHX33 is required to activate MAP kinases, NF-κB and IRF3 in mDCs
Studies with IPS-1-deficient mice have shown that the loss of IPS-1 prevents the activation of NF-κB and IRF3 in multiple cell types.19 We next sought to determine whether the downstream signaling of DHX33 in D2SC mDCs induced by poly I:C involves MAP kinases and NF-κB and IRF3 activation. Control, IPS-1-knockdown and DHX33-knockdown D2SC mDCs were stimulated with poly I:C, and total cell extracts were prepared and separated by SDS–PAGE, followed by immunoblotting (Figure 6). In control D2SC mDCs, the phosphorylation of p65, Erk1/2, p38 and IRF3 was detected between 60 and 120 min. In contrast, the phosphorylation of p65, Erk1/2, p38 and IRF3 was barely detectable in DHX33- and IPS-1-knockdown D2SC mDCs.
Figure 6.
DHX33 is required for the activation of MAP kinases, NF-κB and IRF3 in mDCs. Immunoblot of the indicated proteins from the lysates of D2SC mDCs treated with a scrambled shRNA (control), a DHX33-targeting shRNA (DHX33) or an IPS-1-targeting shRNA (IPS-1) after stimulation for the indicated time with 2.5 µg/ml long poly I:C delivered by Lipofectamine 2000. β-Actin served as a loading control. mDC, myeloid dendritic cell; shRNA, small heteroduplex RNA.
DISCUSSION
The TLRs, especially TLR3, are a class of membrane receptors that sense poly I:C and viral RNA.20 However, results from in vivo experiments indicate that TLR3-deficient and wild-type mice have similar responses to several viruses, including vesicular stomatitis virus, lymphocytic choriomeningitis virus, reovirus and murine cytomegalovirus.21 Likewise, cellular responses to poly I:C in vitro are rendered TLR3 independent when the cells are incubated with sufficient levels of poly I:C.20 In addition to TLRs, other cytoplasmic dsRNA-binding proteins, such as double-stranded RNA-dependent protein kinase and 2′,5′-oligoadenylate synthetase, are important antiviral molecules.22,23 However, knockout mouse data indicate that RNA-dependent protein kinase is not directly essential for cellular responses to poly I:C,24,25,26 suggesting that it acts in a second, IFN-dependent phase.27 The RIG-I-like DExD/H helicases (RIG-I, MDA5 and LGP2) were identified as cytoplasmic sensors of poly I:C and virally derived dsRNA.6,7,8 Interestingly, RIG-I was found to preferentially sense 5′-triphosphate dsRNA, ssRNA and short poly I:C (>300 bp and <1000 bp). MDA5 primarily senses long poly I:C (>1000 bp). Furthermore, residual responses to poly I:C in MDA5- and RIG-I-deficient MEFs still exist,6 suggesting the presence of additional RNA sensors that function as part of the innate immune system. Recently, we found that these residual responses in MDA5- and RIG-I-deficient MEFs were blocked after the knockdown of DDX1, DDX21, DHX36 or TRIF expression by shRNA.12 In addition, a residual response to short poly I:C in RIG-I-deficient D2SC mDCs also exists. This residual response was decreased dramatically after the knockdown of DHX9 expression.14 A previous study reported that RIG-I and MDA-5 are type I IFN-induced genes in response to poly I:C,28 suggesting that these RIG-I-like receptors need to be upregulated to a sufficient level secondary to IFN stimulation. Collectively, these findings indicate that there are other molecules involved in the initial sensing of viral RNA.
Our bioinformatics analysis indicated that there are at least 60 members of the (DExD/H)-box helicase superfamily, including the following subfamilies: RIG-I-like receptor (RIG-I, MDA5 and LGP-2), DEAH/RHA, aspartate–glutamate–alanine–aspartate (DEAD)-box and Snf1-related kinase interacting protein 2-like.29 Indeed, a recent study demonstrated that DICER-2, another RIG-I-like DExD/H helicase, senses viral nucleic acids as part of the Drosophila innate immune system.10 DDX3, a member of the DEAD-Box subfamily, is able to recognize poly I:C in 293T and HeLa cells, and it induces an IFN-β response mediated by IPS-1. Members of the DEAD-Box subfamily, as well as DDX1, DDX21, DHX9 and DHX36, play critical roles in sensing both short and long poly I:C in mDCs. All of these studies suggest that DExD/H helicases play very important and broad roles in antiviral innate immune responses. Other members of the (DExD/H)-box helicase superfamily may be missing the initial poly I:C and viral RNA sensors.
Different helicases may use different adaptor molecules as well. The DDX1–DDX21–DHX36 complex uses TRIF as an adaptor molecule.12 RIG-I and MDA5, as well as the NOD-like receptors, contain a CARD domain that interacts with another CARD-containing protein, IPS-1. IPS-1 transduces the signal through IκB kinase-related kinases, such as TNF receptor-associated factor family member-associated NF-κB activator-binding kinase-1 and inducible IκB kinase, culminating in the activation of IRF3 and the induction of the transcription of type I IFNs.30 We identified DHX33 as a protein that binds IPS-1. Although DHX33 has DEXDc and HELICc type domains that are similar to those of RIG-I and MDA5, it does not have a CARD domain.
By screening 60 members of the DExD/H helicase family by siRNA-mediated knockdown of protein expression followed by the detection of type I IFN responses to poly I:C, we identified RIG-I, MDA5, LGP2, DDX1, DDX3, DHX9, DDX21, DHX33, DHX36 and DDX60 as sensors of poly I:C. It has been reported previously that RIG-I, MDA5, LGP2, DDX1, DDX3, DHX9, DDX21, DHX36 and DDX60 play roles in sensing poly I:C.6,7,8,12,14,15 In this study, we discovered the important function of DHX33 in sensing poly I:C and an RNA virus. The knockdown of DHX33 by shRNA in the mouse DC cell line D2SC strongly attenuated type I IFN production in response to poly I:C and reovirus. DHX33 can bind poly I:C and uses IPS-1 as an adaptor molecule. Importantly, we demonstrated that the observed association between DHX33 and IPS-1 in overexpression studies also occurs endogenously. Taken together, our results suggest that the DExD/H helicases play critical roles in innate immunity by sensing dsRNA and an RNA virus.
Acknowledgments
We thank M Wentz for critical reading. We thank all of our colleagues in our laboratory.
The authors declare that they have no competing financial interests.
References
- Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Recognition of viruses by innate immunity. Immunol Rev. 2007;220:214–224. doi: 10.1111/j.1600-065X.2007.00562.x. [DOI] [PubMed] [Google Scholar]
- Blasius AL, Beutler B. Intracellular Toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Martinon F, Tschopp J. NLRs join TLRs as innate sensors of pathogens. Trends Immunol. 2005;26:447–454. doi: 10.1016/j.it.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Kufer TA, Fritz JH, Philpott DJ. NACHT-LRR proteins (NLRs) in bacterial infection and immunity. Trends Microbiol. 2005;13:381–388. doi: 10.1016/j.tim.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, et al. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pippig DA, Hellmuth JC, Cui S, Kirchhofer A, Lammens K, Lammens A, et al. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 2009;37:2014–2025. doi: 10.1093/nar/gkp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deddouche S, Matt N, Budd A, Mueller S, Kemp C, Galiana-Arnoux D, et al. The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat Immunol. 2008;9:1425–1432. doi: 10.1038/ni.1664. [DOI] [PubMed] [Google Scholar]
- Oshiumi H, Sakai k, Matsumoto M, Seya T. 2010. DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential. Eur J Immunol. 2010;40:940–948. doi: 10.1002/eji.200940203. [DOI] [PubMed] [Google Scholar]
- Kim T, Pazhoor S, Bao M, Zhang Z, Hanabuchi S, Facchinetti V, et al. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci USA. 2010;107:15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kim T, Bao M, Facchinetti V, Jung SY, Ghaffari AA, et al. Helicases pair with TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34:866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yuan B, Lu N, Facchinetti V, Liu YJ. DHX9 pairs with IPS-1 to sense dsRNA in myeloid dendritic cells. J Immunol. 2011;187:4501–4508. doi: 10.4049/jimmunol.1101307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita M, Oshiumi H, Matsumoto M, Seya T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol Cell Biol. 2011;31:3802–3819. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, et al. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-B by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Edelmann KH, Richardson-Burns S, Alexopoulou L, Tyler KL, Flavell RA, Oldstone MB. Does Toll-like receptor 3 play a biological role in virus infections. Virology. 2004;322:231–238. doi: 10.1016/j.virol.2004.01.033. [DOI] [PubMed] [Google Scholar]
- Balachandran S, Roberts PC, Brown LE, Truong H, Pattnaik AK, Archer DR, et al. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity. 2000;13:129–141. doi: 10.1016/s1074-7613(00)00014-5. [DOI] [PubMed] [Google Scholar]
- Schröder HC, Suhadolnik RJ, Pfleiderer W, Charubala R, Müller WE. (2′–5′) Oligoadenylate and intracellular immunity against retrovirus infection. Int J Biochem. 1992;24:55–63. doi: 10.1016/0020-711x(92)90229-t. [DOI] [PubMed] [Google Scholar]
- Yang YL, Reis LF, Pavlovic J, Aguzzi A, Schäfer R, Kumar A, et al. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi LB, Jr, Heitmeier MR, Scheuner D, Kaufman RJ, Buller RM, Corbett JA. Potential role of PKR in double-stranded RNA-induced macrophage activation. EMBO J. 2000;19:3630–3638. doi: 10.1093/emboj/19.14.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WM, Ostertag D, Li ZW, Chang L, Chen Y, Hu Y, et al. JNK2 and IKKbeta are required for activating the innate response to viral infection. Immunity. 1999;11:721–731. doi: 10.1016/s1074-7613(00)80146-6. [DOI] [PubMed] [Google Scholar]
- Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta M, Kawai T, Yokoi N, Akira S, Kinoshita S. Contribution of IPS-1 to polyI:C-induced cytokine production in conjunctival epithelial cells. Biochem Biophys Res Commun. 2011;404:419–423. doi: 10.1016/j.bbrc.2010.11.136. [DOI] [PubMed] [Google Scholar]
- Linder P. Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Kim YJ. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu Rev Biochem. 2007;76:447–480. doi: 10.1146/annurev.biochem.76.060605.122847. [DOI] [PubMed] [Google Scholar]