Abstract
Controlling the density of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs) at synapses is essential for regulating the strength of excitatory neurotransmission. In particular, the phosphorylation of AMPARs is important for defining both synaptic expression and intracellular routing of receptors. Phosphorylation is a posttranslational modification known to regulate many cellular events and the C-termini of glutamate receptors are important targets. Recently, the first intracellular loop1 region of the GluA1 subunit of AMPARs was reported to regulate synaptic targeting through phosphorylation of S567 by Ca2+/calmodulin-dependent protein kinase II (CaMKII). Intriguingly, the loop1 region of all four AMPAR subunits contains many putative phosphorylation sites (S/T/Y), leaving the possibility that other kinases may regulate AMPAR surface expression via phosphorylation of the loop regions. To explore this hypothesis, we used in vitro phosphorylation assays with a small panel of purified kinases and found that casein kinase 2 (CK2) phosphorylates the GluA1 and GluA2 loop1 regions, but not GluA3 or GluA4. Interestingly, when we reduced the endogenous expression of CK2 using a specific shRNA against the regulatory subunit CK2β, we detected a reduction of GluA1 surface expression, whereas GluA2 was unchanged. Furthermore, we identified S579 of GluA1 as a substrate of CK2, and the expression of GluA1 phospho-deficient mutants in hippocampal neurons displayed reduced surface expression. Therefore, our study identifies CK2 as a regulator of GluA1 surface expression by phosphorylating the intracellular loop1 region.
Keywords: CK2, glutamate, AMPAR, synapse, trafficking
INTRODUCTION
The amino acid glutamate mediates most of the fast excitatory neurotransmission in the mammalian brain. After its release from the presynaptic terminal, glutamate binds principally to postsynaptic receptors. One subfamily of ionotropic glutamate receptors, the AMPA-type glutamate-gated receptors (AMPARs), mediates most of the fast neurotransmission. AMPARs play a crucial role in driving postsynaptic depolarization, and alterations in excitatory synaptic strength are considered to be the molecular basis for learning and memory. AMPAR subunits (GluA1–4) assemble as tetrameric channels, and hippocampal pyramidal cells mostly express GluA1A2 and GluA2A3 complexes (Lu et al., 2009; Wenthold et al., 1996). The dynamic and precise regulation of AMPARs is critical for synapse function, and their synaptic density is controlled by cell surface events such as lateral diffusion, endocytosis and exocytosis, but also by intracellular routing and trafficking (Carroll et al., 2001; Groc & Choquet, 2006; Hanley, 2010; Hirling, 2009).
Phosphorylation is a mechanism particularly important for the rapid regulation of channel function and trafficking of receptors. There are a variety of trafficking steps that a receptor goes through and some or all may be regulated by the coordinated action of kinases and phosphatases. Indeed, several kinases such as protein kinase A (PKA), protein kinase C (PKC) and Ca2+/calmodulin-dependent protein kinase II (CaMKII) phosphorylate various residues within the C-terminal tail of the AMPAR subunits GluA1–4. Moreover, the consequences of the phosphorylation state on AMPAR channel conductance, as well as trafficking to and from the postsynaptic membrane depend on the residue phosphorylated within a specific subunit (Banke et al., 2000; Barria et al., 1997; Derkach et al., 1999; Ehlers, 2000; Esteban et al., 2003; Kristensen et al., 2011; Lee et al., 2000, 2003; Lin et al., 2009; Lu & Roche, 2012; Roche et al., 1996; Serulle et al., 2007). Interestingly, although the phosphorylated sites in the GluA1 C-terminus are implicated in synaptic plasticity, they don’t appear to regulate basal synaptic transmission (Boehm et al., 2009; Hayashi et al., 2000; Lee et al., 2003; Lin et al., 2009). However, a molecular replacement approach was recently used to determine whether another region of the GluA1-containing AMPAR might function to regulate synaptic delivery and maintenance of synaptic transmission. Lu et al. (2010) reported that a CaMKII phosphorylation site, S567, in the first intracellular loop of GluA1 regulates the synaptic targeting of AMPARs. Because the function of the first intracellular region of AMPARs remains obscure, we hypothesized that other kinases might also phosphorylate this region of AMPARs to regulate their surface expression. In the present study, we used in vitro phosphorylation assays with a variety of purified kinases to identify new phosphorylation sites in the intracellular loop1 of AMPARs. We now report that casein kinase 2 (CK2) phosphorylates the loop1 of GluA1 and GluA2. There are at least two residues in GluA1 that are in vitro phosphorylated by CK2, with S579 being the major substrate. Furthermore, CK2 regulates the surface expression of the AMPAR subunit GluA1 by phosphorylating the intracellular loop1 region.
MATERIALS AND METHODS
ANIMALS
The National Institute of Neurological Disorders and Stroke Animal Care and Use Committee approved our use of experimental animals (protocol #1171). All animals were handled and the experiments performed according to the guidelines of the National Institutes of Health Office of Intramural Animal Care and Use of Laboratory Animals. In addition, efforts were made to minimize the use of animals. Rats and mice were housed in a temperature- and humidity-controlled room under a 12 hours light/dark cycle with food and water freely available. Timed-pregnant rats (Harlan, Indianapolis, IN, USA) were received at least one day prior to dissection, and were euthanized in carbon dioxide prior to decapitation. We also used a GluA1 knockout strain (Zamanillo et al., 1999) generated by Rolf Sprengel (Department of Molecular Neurobiology, Max Planck Institute for Medical Research, Heidelberg, Germany). Heterozygous mouse line was used to generate a first generation of knockouts, which were then mated to obtain the pups required for dissection on postnatal day 0 or 1.
RAT NEURONAL CULTURES
Primary rat hippocampal and cortical neuronal cultures were prepared from embryonic day 18–19 Sprague-Dawley embryos of either sex. The hippocampi and cortices were isolated, the tissue was dissociated for 12 mins at 37°C by 0.05% trypsin in 10 mM HEPES-supplemented HBSS and containing penicillin-streptomycin (Life Technologies, Frederick MD, USA) with 1.37 mg/ mL DNAse (Sigma, St-Louis, MO, USA). Cells were triturated using fire-polished glass Pasteur pipettes. Hippocampal cells were plated on poly-D-lysine (Sigma) -coated 12 mm glass coverslips to ~ 75,000 cells per well of a 24-well plate for immunocytochemistry. For biochemistry and immunoblotting, cortical neurons were plated at ~1.25 × 106 cells per well on poly-D-lysine-coated 6-well plate or at ~7.5 × 106 cells on 100 mm poly-D-lysine-coated dishes. Neurons were maintained in serum-free Neurobasal Medium supplemented with 2% B-27 (Life Technologies) and 2 mM L-glutamine (Sigma). The cultures were maintained at 37°C in 5% CO2/ 95% air mixture and used at 15–19 days in vitro (DIV).
MOUSE NEURONAL CULTURES
Primary mouse hippocampal cultures were prepared from postnatal day 0–1 pups of either sex, which were sacrificed by decapitation. The hippocampi were isolated, and the tissue was dissociated for 45 mins at 37°C using a papain dissociation kit (Worthington Biochemical Corp, Lakewood, NJ, USA) in HBSS (Life Technologies). Cells were triturated using fire-polished glass Pasteur pipettes. Hippocampal cells were plated on poly-D-lysine-coated glass coverslips at ~ 175,000 cells per well of a 24-well plate for immunocytochemistry and at ~ 300,000 cells per well of a 12-well plate for electrophysiology. Neurons were maintained in serum-free Neurobasal Medium supplemented with 2% B-27 (Life Technologies) and 2 mM L-glutamine (Sigma). The mouse hippocampal neurons were transfected at DIV10 using 1.5 µg of pIRES2-EGFP plasmid DNA with 0.75 µL LipofecAMINE 2000 (Life Technologies) per well, and were maintained at 37°C in 5% CO2/ 95% air mixture until used for electrophysiology at 14–16 DIV.
MOLECULAR BIOLOGY
The GluA1 C-terminus (amino acids 827–907), or loop1 regions of GluA1 (amino acids 558–585), GluA2 (amino acids 565–592), GluA3 (amino acids 568–597), or GluA4 (amino acids 566–593) were cloned in frame into pGEX-4T-1 (GE Healthcare). The Flag-tagged GluA1 “flip” cloned in the pRK5 mammalian expression vector served as a template for creating an shGluA1 resistant molecular replacement construct harboring silent mutations (uppercased: ggaatTcgGaaAatCggtt). The shGluA1-resistant-construct was used for creating S579A and D580Q mutations in the loop1-region. Mutations in the different vectors pRK5 or pGEX-4T-1 were introduced using the QuikChange site-directed mutagenesis system (Agilent Technologies). For electrophysiology, the shGluA1-resistant Flag-GluA1 WT or D580Q in pRK5 vector was PCR amplified then inserted into pIRES2-EGFP (Clontech). Specific shRNA targeting GluA1 (shGluA1, target sequence: 5’-ggaatccgaaagattggtt-3’) and CK2β (shCK2β, target sequence: 5’-tggtttccctcacatgctct-3’ from Pallares et al., 2009; shCTL, scrambled shCK2β sequence: 5’-gccttctcacgtcgatttct-3’) were inserted after the H1 promotor in pSUPER vector according to the manufacturer’s instructions (OligoEngine, Seatle, WA, USA), and then the H1-shRNA DNA cassette was excised and subcloned into a modified FUGW lentiviral vector (kindly provided by Robert C. Malenka, Stanford School of Medicine, Stanford, CA, USA). This vector expresses EGFP under the expression of the human Ubiquitin C promoter and allows the monitoring of transfected or infected cells. All engineered constructs were subsequently analyzed from sequences provided by the NINDS DNA Sequencing Facility.
LENTIVIRAL PARTICLE PACKAGING
To knockdown CK2β or GluA1 in cultured neurons, lentiviral particles were produced in HEK293T/17 cells (ATCC [CRL-11268], Manassas, VA, USA) co-transfected with the lentiviral vector harboring the shRNA sequence, the packaging vector Δ8.9, and the VSVG envelope glycoprotein vector by using FUGENE HD (Promega, Madison, WI, USA) in UltraCULTURE (Lonza, Walkersville, MD, USA) containing 1 mM sodium pyruvate, 0.075% sodium bicarbonate (Corning Life Sciences, Manassas, VA, USA) and 2 mM L-glutamine (Sigma). Culture media containing the lentiviral particles were collected 48–72h post-transfection and centrifuged at 100,000 × g for 2h at 4°C. The lentiviral particle pellet was resuspended in PBS, aliquoted and frozen at −80°C until use. Typically, 1 µL of lentiviruses was used per well of 24-well plate, 5 µL for 35 mm plates, and 10–15 µL for 100 mm plates.
GST-FUSION PROTEIN PURIFICATION
GST-fusion proteins were purified from E. coli BL21 (pLys). The expression of GST-fusion protein was induced from an overnight culture (10 mL) by incubating the cells with 0.5 mM IPTG at 37°C for 3h. The cells were collected by centrifugation (4,000 × g, 10 mins, 4°C), lysed by sonication on ice in 1.5mL of GST buffer (20 mM TrisHCl, pH 8.0, 1% Triton X-100, 150 mM NaCl, 5 mM ethylenediaminetetraacetic acid (EDTA), 1× complete protease inhibitor cocktail [Roche]) containing 1 mM dithiothreitol (DTT), then incubated with agitation at 4°C for 30 mins. The clarified lysate (16,000 × g, 20 mins, 4°C) was then incubated with glutathione-sepharose beads (GE Healthcare) for 30 mins at room temperature, or overnight at 4°C. Beads were then washed 4 times in GST lysis buffer, the proteins verified on 10% SDS-PAGE by staining with GelCode Blue Stain Reagent (Thermo Scientific).
IN VITRO KINASE ASSAY
~1 ug of GST-fusion protein on beads (usually in final volume of 20 µL of slurry beads) were washed with the appropriate kinase buffer containing 100 µM ATP. Enzymatic reactions were performed at 30°C for 30 mins with a specific kinase and buffer (described below) containing 100 µM ATP, 1 pmol of [γ-32P]-ATP (BLU002A, 3000Ci/mmol, Perkin Elmer, Waltham, MA, USA). At the end of the reactions, the beads were cautiously washed with 100 µL of PBS containing 1× of phosphatases inhibitors cocktails II and III. The proteins eluted in 2× Laemmli buffer were resolved by SDS-PAGE (10% gel), transferred on PVDF membranes (0.45 µm pore, Millipore), and visualized by autoradiography using Kodak BioMax MR or MS films. Some experiments were also visualized using Fuji Super RX-N Blue films.
CaMKII: 50 units (CaMKIIα subunit, Calbiochem) in 20 mM Tris-HCl pH 7.5, 10 mM MgCl2, 0.1 mM EDTA, 0.5 mM DTT, 2 mM CaCl2, 1.2 µM Calmodulin. CK2: 100 units (116 ng, NEB) in 20 mM Tris-HCl pH 7.5, 50 mM KCl, 10 mM MgCl2. CK1: 250 units (125 ng, NEB) in 50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 5 mM DTT. PKA: 50 ng (catalytic subunit, Promega) in 10 mM HEPES pH 7.0, 20 mM MgCl2. PKC: 10 ng (Promega) in 20 mM HEPES pH 7.0, 10 mM MgCl2, 1.67 mM CaCl2, 1 mM DTT.
SUBCELLULAR FRACTIONATION OF CULTURED NEURONS
Cultured cortical neurons (DIV17–19), infected at DIV10 with lentiviruses expressing shCK2β or a control shCTL, were washed and harvested in ice-cold ACSF (in mM; 10 HEPES, 150 NaCl, 3 KCl, 10 glucose, 2 CaCl2, 1 MgCl2, pH adjusted with CsOH to 7.35) on ice. After centrifugation (1,000 × g, 4°C, 10 mins), the cell pellet was homogenized on ice using a 23 gauge needle (25 strokes) in 20 mM Tris-HCL pH 7.5, 320 mM sucrose, 5 mM EDTA, 1× complete protease inhibitor cocktail, 1× phosphatase inhibitor cocktails II and III. Lysate was centrifuged (1,000 × g, 4°C, 10 mins), resulting in a pellet (discarded) and a post-nuclear supernatant (PNS) from which the protein content was estimated using Pierce BCA protein assay (Thermo Scientific). Equal amount of PNS proteins (400–600 µg) were centrifuged (10,000 × g, 4°C, 20 mins), resulting in a pellet (P2 / crude synaptosomes) and a discarded supernatant. Pellets were resuspended on ice in 20 mM Tris-HCL pH7.5, 5 mM EDTA, 1× complete protease inhibitor cocktail, 1× of phosphatase inhibitor cocktails II and III. An aliquot of total P2 fraction was kept, and Triton X-100 added to final concentration of 1% before incubating with gentle agitation (4°C, 30 mins). Lysates were centrifuged (33,000 × g, 4°C, 30 mins) to obtain a soluble fraction and a pellet (Triton X-100 insoluble / PSD fraction) which was solubilized by heating at 37°C for 30 mins in 1% sodium dodecyl sulfate (SDS), 20 mM Tris-HCL pH7.5, 5 mM EDTA, 1× complete protease inhibitor cocktail, 1× of phosphatase inhibitor cocktails II and III. An equal percentage of each fraction was separated by SDS-PAGE followed by specific immunoblotting.
HEK293T TRANSFECTION AND GLUA1 MUTANT ANALYSIS
Each well of a poly-D-lysine-coated 6-well dish contained 1 µg of GluA1 plasmid DNA diluted in 250 µL Opti-MEM I (Life Technology), to which 2 µL of LipofecAMINE 2000 (Life Technologies) diluted in 250 µL Opti-MEM I (Life Technologies) was added and mixed thoroughly. After 20 mins incubation at room temperature, 1 × 106 HEK293T/17 cells (ATCC) in 1.5 mL culture media without antibiotics was added in each well, before incubating at 37°C in 5% CO2/ 95% air mixture for ~40h until processed for immunoblotting.
IMMUNOBLOTTING
Transfected HEK293T/17 cells or cultured cortical neurons infected with lentiviruses for 7 days were cooled on ice, washed in ice-cold ACSF, and then lysed in ice-cold RIPA buffer (150 mM NaCl, 1% Triton X-100, 0.5% deoxycholic acid, 0.1% SDS, 5 mM EDTA, 20 mM Tris-HCl, pH 8.0, 1× complete protease inhibitor cocktail). After brief sonication and centrifugation (16,000 × g, 4°C, 20 mins), the protein content of the soluble material was estimated using Pierce BCA protein assay (Thermo Scientific), equal amount of proteins were separated by SDS-PAGE and transferred to a 0.45 µm PVDF membrane. Membranes were blocked in TBS-T (TBS with 0.1% Tween 20) containing 5% [w/v] nonfat skim milk before incubation with primary antibodies in TBS-T. Quality Controlled Biochemicals (Hopkinton, MA, USA) generated the affinity purified rabbit polyclonal GluA1 antibody (SHSSGMPLGATGL, 1 µg/ mL). The affinity purified rabbit polyclonal GluA2 (VAKNPQNINPSSSQNS, 0.5 µg/ mL) antibody was a gift of the late Robert J. Wenthold (National Institute of Deafness and Other Communication Disorders, Bethesda, MD, USA). The following antibodies are commercially available: rabbit anti-GST (cat. A190–122A; 1:25,000, Bethyl Laboratories, Montgomery, TX, USA), mouse anti-β-actin (cat. G043; clone 8H10, 1:10,000; Applied Biological Materials, Richmond, BC, Canada), mouse anti-CK2β (cat. C3617; clone 6D5, 1:1,500; Sigma), rabbit monoclonal anti-CK2α (cat. 2079-1; clone EP1963Y, 1:500; Epitomics, Burlingame, CA, USA), mouse anti-synaptophysin (cat. S5768; clone SVP-38, 1:10,000; Sigma), mouse monoclonal anti-PSD-95 (cat. 75-028; clone K28/43; 1:50 000; UC Davis / NIH NeuroMab Facility, Davis, CA, USA). Peroxidase-conjugated sheep anti-mouse or donkey anti-rabbit secondary antibodies (1:10,000, GE Healthcare), diluted in TBS-T, were detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) and Fuji Super RX-N Blue films. Quantitation was performed using ImageJ software (NIH) on subsaturated films. The experimental value (shCK2β) was normalized to the control sample (shCTL, set to 100%) from the corresponding experiment, pooled together, and the statistical analysis was performed using an unpaired two-tailed t-test using GraphPad Prism 6 (GraphPad Prism Software, La Jolla, CA, USA). Statistical difference was significant when P < 0.05. A representative immunoblot from at least three experiments is shown.
IMMUNOCYTOCHEMISTRY
Hippocampal cells in 24-well plates were transfected at DIV14–15 using 1.5 µg of plasmid DNA with 0.75 µL LipofecAMINE 2000 (Life Technologies) per well. When required to knockdown GluA1, 1 µL of lentiviruses was added to each well at DIV5–6 previous to transfection at DIV14–15 with a molecular replacement construct (described under molecular biology section). 3–4 days post-transfection (DIV 17–19), neurons were used for specific assays. To measure surface expression of endogenous AMPARs, neurons were labeled by incubating the live neurons (37°C, 12 mins) in culture media containing mouse monoclonal antibody against an extracellular epitope of GluA1 (cat. MAB2263; clone RH95, 5 µg/ mL, Millipore) or GluA2 (cat. MAB397; clone 6C4, 10 µg/mL; Millipore). Cells expressing Flag-tagged GluA1 were surface labeled by incubating the live neurons (37°C, 8 mins) in culture media containing mouse M2 monoclonal antibody against Flag (cat. F1804; 1 µg/ mL, Sigma). After washing the cells in cold ACSF, the neurons were fixed with ice-cold 4% paraformaldehyde / 4% sucrose in 1× PBS for 15 mins. Without permeabilization, cells were blocked for 30 mins in 1× PBS containing 10% normal goat serum (NGS) (Vector Laboratories, Burlingame, CA, USA). Endogenous surface AMPARs were labeled using Alexa Fluor 647 conjugated goat anti-mouse IgG2a secondary antibody, while recombinant Flag-GluA1 was labeled with Alexa Fluor 555-conjugated goat anti-mouse secondary antibody (Molecular Probe / Life Technologies, Carlsbad, CA, USA). After permeabilization using 0.1% Triton X-100 in 1× PBS for 15 mins at room temperature, and blocking for 30 mins in 1× PBS containing 10% NGS, internal recombinant Flag-GluA1 was labeled using our custom affinity purified GluA1C antibody (0.25 µg/ mL) in 1× PBS / 3% NGS for 30 mins at room temperature before labeling with Alexa Fluor 647-conjugated goat anti-rabbit secondary antibody (Molecular Probe). To evaluate the total expression of endogenous AMPARs, neurons were washed in cold ACSF, fixed, permeabilized with 0.1% Triton X-100, and blocked in 1× PBS containing 10% NGS. Labeling was performed at room temperature with our affinity purified rabbit polyclonal GluA1C antibody (1 µg/ mL) in 1× PBS / 3% NGS or with a mouse monoclonal anti-GluA2 (cat. 75-002; clone L21/32; 1:100; NeuroMab). A chicken anti-GFP and an Alexa Fluor 488-conjugated goat anti-chicken secondary antibody (Molecular Probe) on permeabilized cells were used to intensify the EGFP signal.
CONFOCAL MICROSCOPY, IMAGE ACQUISITION AND STATISTICAL ANALYSIS
Coverslips were mounted in ProLong Gold Antifade reagent (Life Technologies) and all immunochemistry images were acquired on a Zeiss LSM 710 laser scanning confocal microscope using a 63× oil objective (1.4 numerical aperture). Image acquisition for a particular experiment was performed using identical settings (gain, laser power, pinhole size, etc). All confocal images were averaged 4 times, obtained at a resolution of 1024 × 1024, dwell time of 0.79 µsec, and with step intervals of 0.37 µm in the Z direction. For quantitative analysis, maximal projection images were created with the ZEN software (Zeiss) from 4–6 serial optical sections. For analysis, the fluorescence signal from surface (red channel) and total (blue channel) was thresholded separately to levels distinguishing dendritic morphology from background, not only the fluorescence from puncta. The values were kept identical for all experimental and control images within an experiment, and only slightly adjusted between independent experiments. Metamorph (Universal Imaging Corp., Downingtown, PA, USA) was used to measure the integrated intensity of 3 sections of dendrites selected and averaged for each neuron from 3 experiments generated from independent dissections. For every section measured, a ratio of surface divided by total fluorescence was calculated, and then normalized to the average ratio value obtained for all cells (expressing the GluA1 WT construct) in one specific experiment. The three segments for one cell were averaged, and the cells from independent experiments were pooled. By using GraphPad Prism 6, we identified one outlier under a very stringent condition (ROUT method, Q=0.1%). Only one cell was identified and removed from the dataset (S579A, surface/total value of 5.35 which represented more than 14 fold the standard error). The statistical significance between condition was calculated using Kruskal-Wallis one-way ANOVA with Dunn’s multiple comparison test, and was considered significant when P < 0.05.
COLOCALIZATION
3–4 days post-transfection (DIV 17–19), hippocampal neurons were washed in cold ACSF, then fixed with ice-cold 4% paraformaldehyde / 4% sucrose in 1× PBS for 15 mins. Following permeabilization with 100% methanol (−20°C) for 10 mins on ice, the methanol was gradually removed and replaced with 1× PBS, then cells were blocked for 30 mins in 1× PBS containing 10% NGS. Endogenous GluA1 (custom affinity purified GluA1C antibody, 1 µg/ mL) and Shank (pan-Shank antibody; cat. 75-089; clone N23B/49; 1:100; NeuroMab) were labeled in 1× PBS / 3% NGS overnight at 4°C. After washes, cells were incubated with Alexa Fluor 647-conjugated goat anti-rabbit secondary and Alexa Fluor 555-conjugated goat anti-mouse IgG1 secondary antibodies (Molecular Probe). A chicken anti-GFP and an Alexa Fluor 488-conjugated goat anti-chicken secondary antibody (Molecular Probe) on permeabilized cells were used to intensify the EGFP signal. Unsaturated single plan confocal images (parameters mentioned previously in the confocal microscopy section) of a Z-section with good signal for both Shank and GluA1 were acquired. For analysis, the Shank and GluA1 signal from untransfected cells (GFP negative) was removed using Adobe Photoshop CS5.1 (Adobe System Incorporated, San Jose, CA, USA), generating the signal specific to the transfected cells (GFP positive). The Manders’ coefficient of colocalization was then calculated using the JACoP plugin (Bolte & Cordelieres, 2006) in ImageJ (NIH). All the cells from 5 independent experiments were pooled, and the statistical significance between condition was calculated using Mann-Whitney t-test in GraphPad Prism 6, and was considered significant when P < 0.05.
ELECTROPHYSIOLOGICAL RECORDINGS
GluA1 knockout mouse hippocampal neurons grown on coverslips were transferred to a submersion chamber for whole-cell recording. The chamber was perfused with recording solution (in mM: NaCl 119, KCl 2.5, NaHCO3 26, Na2PO4 1, glucose 11, CaCl2 2.5, MgCl2 1.3, and saturated with 95% O2/5% CO2) and GFP positive neurons (from pIRES2-EGFP GluA1 constructs) were identified by epifluorescence microscopy. Non transfected neurons next to GFP positive ones were recorded as the control. The intracellular solution contained (in mM) CsMeSO4 135, NaCl 8, HEPES 10, Na3GTP 0.3, MgATP 4, EGTA 0.3, QX-314 5, and spermine 0.1. Cells were recorded with 3- to 5- MΩ borosilicate glass pipettes at −70 mV. mEPSCs were acquired in the presence of 0.5 µM TTX and 100 µM Picrotoxin and semiautomatically detected by offline analysis using in-house software in Igor Pro (Wavemetrics). Series resistance was monitored and not compensated, and cells in which series resistance varied by 25% during a recording session were discarded. Synaptic responses were collected with a Multiclamp 700A amplifier (Axon Instruments, Foster City, CA), filtered at 2 kHz, digitized at 10 Hz. An unpaired t-test was used to determine the statistical significance. All error bars represent standard error measurement.
RESULTS
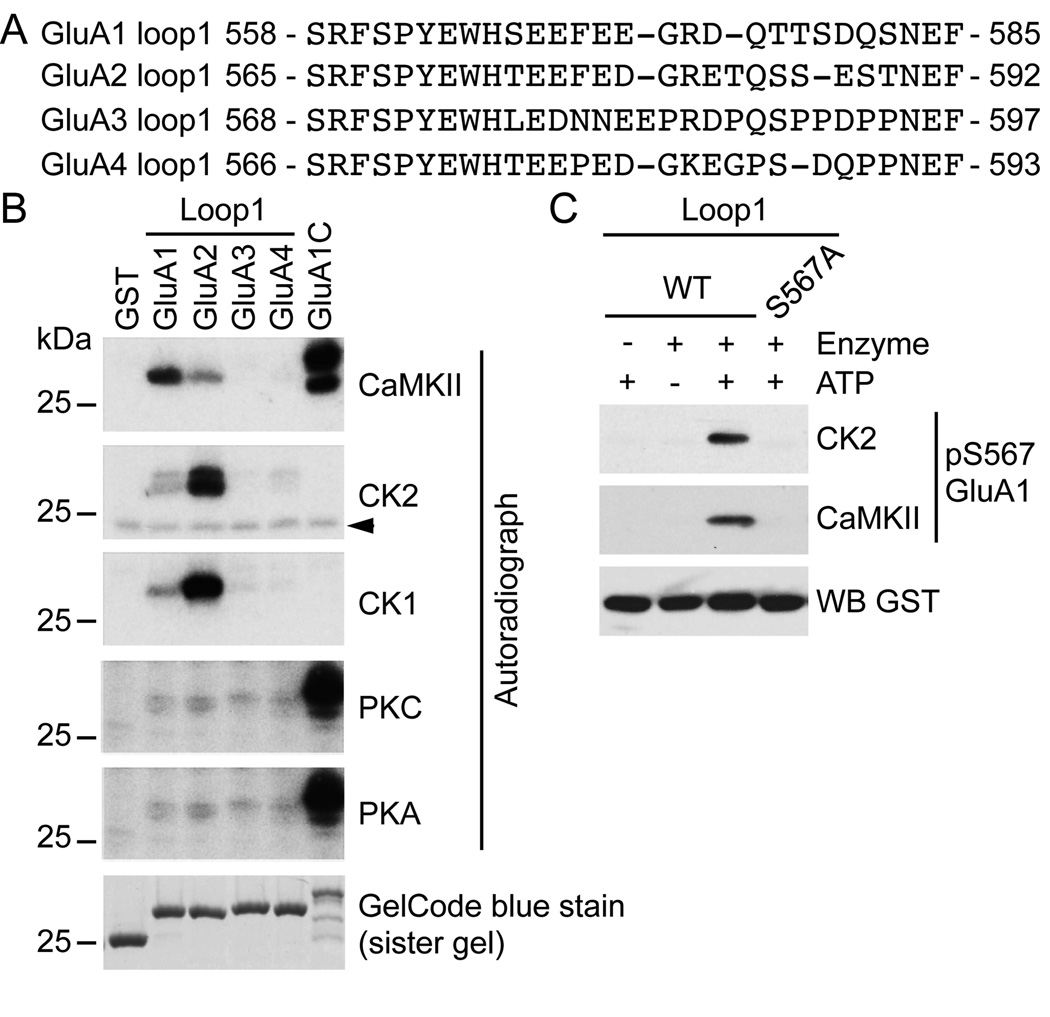
We previously demonstrated that CaMKII phosphorylates the intracellular loop1 region of GluA1 on S567, and that neither PKA nor PKC efficiently phosphorylated the loop1 of GluA1 or GluA2 (Lu et al., 2010). The intracellular loop1 region of GluA1–4 is a segment of approximately 30 amino acids containing several residues that could be phosphorylated [e.g. serine (S), threonine (T) and tyrosine (Y)] (Fig. 1A). As shown by the sequence alignment, the first portion of the loop1 region of GluA1–4 is well conserved, whereas the remaining portion is highly divergent. This observation suggests that some of these residues might be phosphorylated in all AMPAR subunits, whereas the phosphorylation of other residues might be subunit-specific. This observation led us to investigate more completely the phosphorylation of the loop1 region by several kinases. Interestingly, the loop1 region contains several acidic residues that could confer a prefential phosphorylation site for casein kinase 1 (CK1) and casein kinase 2 (CK2) (Fig. 1A). However, despite the similarity in their names, CK1 is totally unrelated to CK2 in the phylogenetic tree and has a different preferential consensus phosphorylation motif (Hanks & Hunter, 1995; Pinna & Ruzzene, 1996). Since these acidic residues are present in all AMPAR subunits, we performed our analysis of the loop1 region on AMPAR subunits GluA1–4.
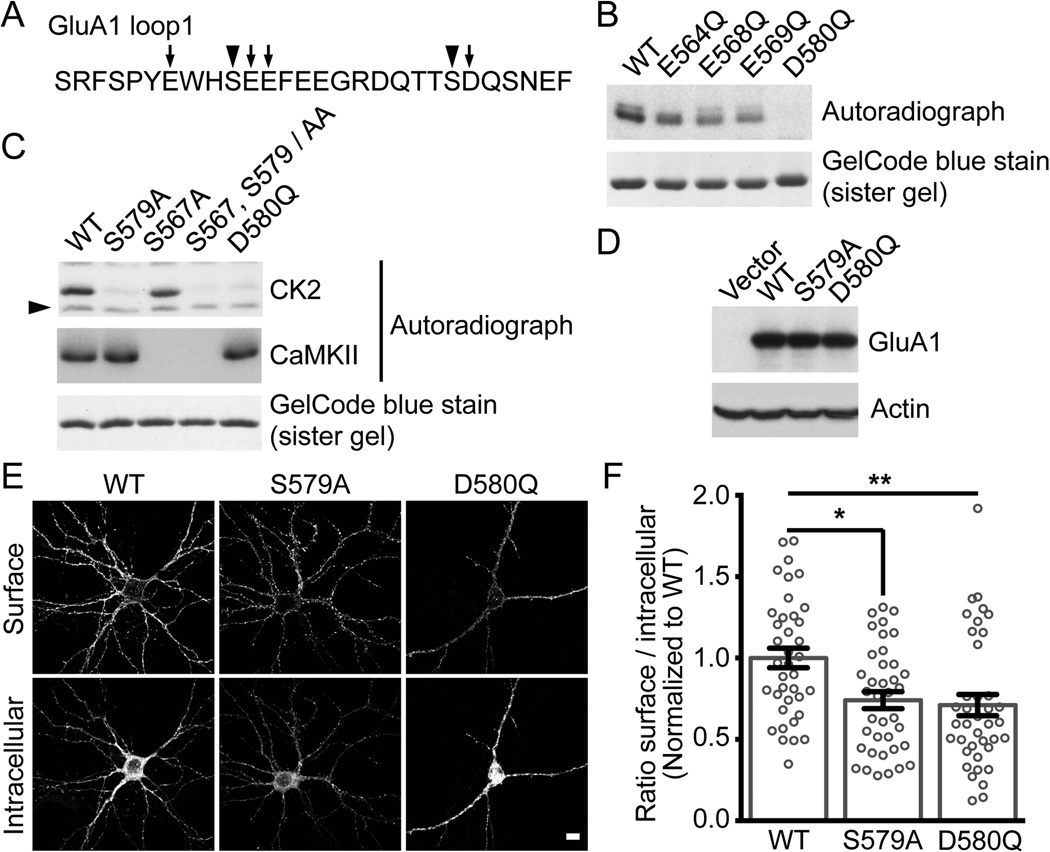
Figure 1. Phosphorylation of the intracellular loop1 region of AMPARs by a variety of kinases.
A) Sequence alignment of the intracellular loop1 region of the AMPAR subunits GluA1–4. B) GST, GST-fusion proteins of the loop1 region or the C-terminus of GluA1 were incubated with the indicated kinase and [γ-32P]-ATP, and analyzed by autoradiography. GelCode blue staining shows the amount of protein used for the assays. A typical result for each condition is shown. The arrowhead in (B) represents a non-specific band. C) As revealed by a phospho-specific immunoblot, purified GST-fusion protein of GluA1 loop1 is in vitro phosphorylated by CK2 and CaMKII on S567. An immunoblot against GST demonstrates that similar amount of protein was used for each condition. The specificity of our phospho-specific antibody against GluA1 pS567 for both CK2 and CaMKII is demonstrated, as GluA1 S567A shows no immunoreactivity.
We performed in vitro kinase assays on purified GST-fusion protein of the GluA1–4 loop1 regions, using the GluA1 C-tail as a control (Fig. 1B). Our results show that CaMKII phosphorylates the GluA2 loop1, although the efficiency was significantly less than the GluA1 C-terminus and GluA1 loop1, which was previously reported to be a substrate (Lu et al., 2010). In contrast, GluA2 loop1 was a better in vitro substrate for CK2 and CK1 than GluA1 loop1. Also, CaMKII, CK2 or CK1 do not phosphorylate the loop1 regions of GluA3 and GluA4. Furthermore, the loop1 regions of the AMPAR subunits are not efficiently phosphorylated by PKA or PKC, although these kinases robustly phosphorylated the GluA1 C-terminus. Taken together, our results demonstrate that the loop1 region of AMPAR subunits is a good substrate for several kinases, which may act at one or all subunits.
The region adjacent to S567 contains a cluster of acidic residues (Fig. 1A). Therefore, we examined if the GluA1 loop1 region was phosphorylated at S567 by CK2. We incubated GST-GluA1 loop1 with ATP and either CK2 or CaMKII, and immunoblotted using our phospho-specific antibody that recognizes phosphorylated GluA1 S567 (Fig.1B). Strikingly, both CK2 and CaMKII phosphorylate GluA1 S567, and importantly no signal is detected upon phosphorylation of the GluA1 S567A mutant demonstrating the specificity of our phospho-specific antibody for GluA1 pS567 (Fig. 1C). Taken together, our results show that multiple kinases can target the loop1 region of AMPARs, and that a single residue can be targeted by more than one kinase.
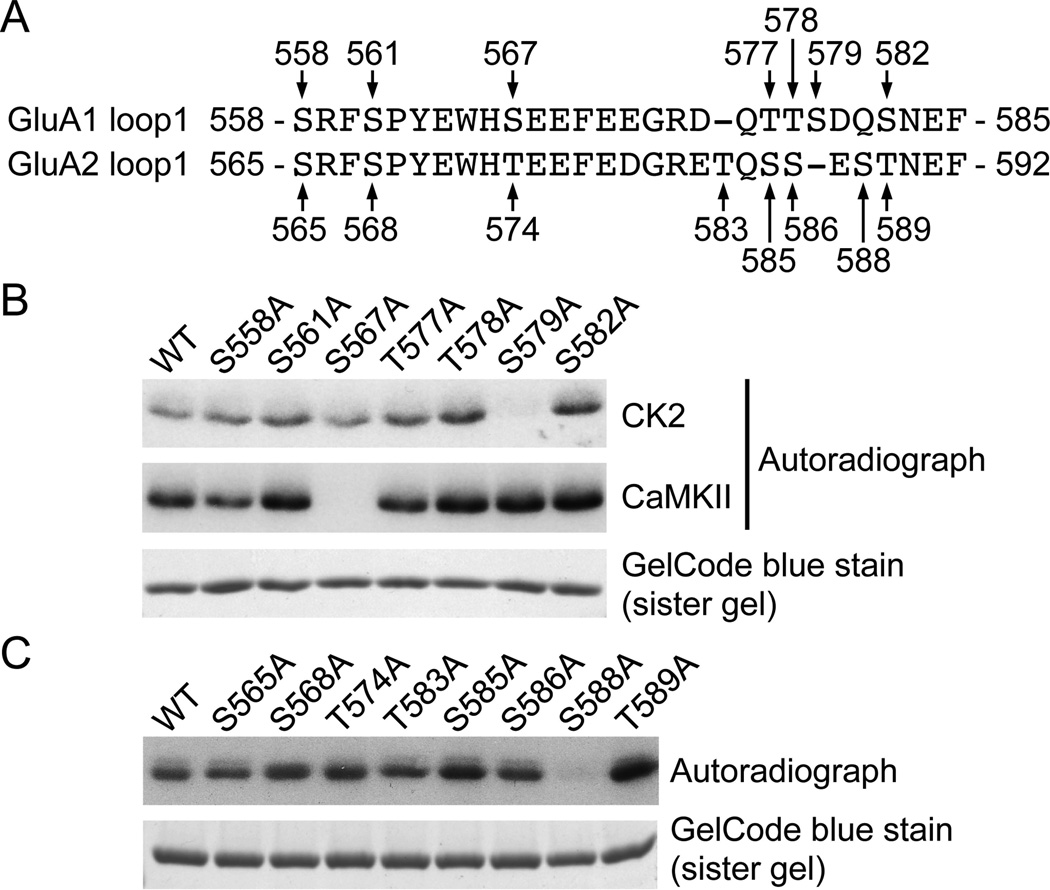
To investigate whether S567 is the only residue of the GluA1 loop1 region that is phosphorylated by CK2, we performed an in vitro kinase assay on purified GST-fusion protein where each serine or threonine was mutated to alanine (Fig. 2A). We found that the phosphorylation of GluA1 loop1 by CK2 is significantly reduced by the mutation of S579 to alanine (Fig 2B). Although we demonstrated that CK2 clearly phosphorylates GluA1 using phospho-specific immunoblotting (Fig 1C), the mutation of S567 to alanine did not dramatically change the overall phosphorylation level of GluA1 loop1 when compared to wild type (WT, Fig 2B). In addition, our results show that S567A mutation abrogated the ability of CaMKII to phosphorylate the GluA1 loop1, whereas S579A mutation was indistinguishable from WT as previously reported (Lu et al., 2010). As demonstrated in figure 1B, CK2 also robustly phosphorylates GluA2 loop1, showing a complex relationship between the sites phosphorylated by CK2. We next sought to identify the residue on GluA2 phosphorylated by CK2. We performed an in vitro kinase assay on purified GST-fusion proteins in which each serine and threonine was mutated to alanine (Fig. 2C). We found that the phosphorylation of GluA2 loop1 by CK2 is significantly reduced by the mutation of S588 into an alanine (Fig. 2C).
Figure 2. Identification of the CK2 phosphorylation sites in the loop1 region of GluA1 and GluA2.
A) Sequence alignment of the GluA1 and GluA2 intracellular loop1 residues. Arrows identify each of the serine and threonine residues in GST-GluA1 or GST-GluA2 loop1 mutated to alanine. B) GST-GluA1 loop1 fusion proteins were incubated with CK2 or CaMKII and [γ-32P]-ATP, and analyzed by autoradiography. C) GST-GluA2 loop1 fusion proteins were incubated with CK2 and [γ-32P]-ATP, and analyzed by autoradiography. GelCode blue staining shows the amount of GST-fusion protein used for an assay in (B) and (D). A typical result for each condition is shown.
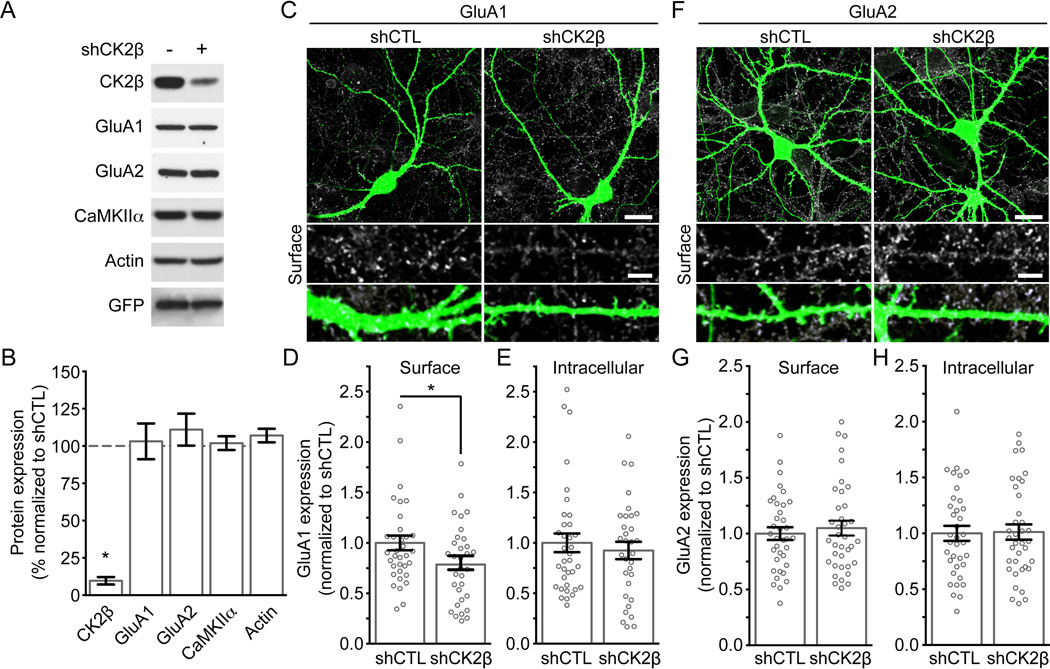
Thus far, our results show that both GluA1 and GluA2 loop1 regions are in vitro substrates for CK2. Since we know that CK2 plays a crucial role in regulating the subunit composition and synaptic content of NMDARs (Chung et al., 2004, Sanz-Clemente et al., 2010, 2013), we speculated that CK2 might also regulate surface expression of GluA1 and GluA2. To investigate the function of endogenous CK2 expression, we generated lentiviral shRNA constructs to knockdown CK2. First, we tested the efficiency of our shCK2β in cultured neurons. After infecting cultured cortical neurons with lentiviruses delivering a control (shCTL) or CK2β specific shRNA (shCK2β), we found that the expression of endogenous CK2β is greatly reduced by our shCK2β but not with the shCTL (Fig 3A and B (shCK2β to shCTL); CK2β: 0.09 ± 0.02, P < 0.0001, n = 11]. Also, the total expression of GluA1, GluA2, CaMKIIα or actin was unaffected when decreasing CK2β expression in cultured neurons [Fig. 3A and B (shCK2β to shCTL); GluA1: 1.03 ± 0.12, P = 0.80, n = 8; GluA2: 1.11 ± 0.11, P = 0.34, n = 8; CaMKIIα: 1.02 ± 0.046, P = 0.69, n = 9; actin: 1.07 ± 0.045, P = 0.18, n = 6]. We then evaluated the consequences of reducing CK2 expression on the surface expression of AMPARs. Cultured hippocampal neurons were transfected with a construct expressing EGFP and control or CK2β specific shRNAs (same shRNA-vectors used to generate lentiviruses in figure 3A) and subjected to an antibody-based confocal imaging assay (Lussier et al., 2012). We found that endogenous surface expression of GluA1 was significantly decreased in cells expressing shCK2β compared with shCTL [Fig 3F and 3G; shCTL: 1.00 ± 0.073, n = 35; shCK2β: 0.79 ± 0.069, n = 32, P = 0.0044]. In contrast, although we found that GluA2 was a better in vitro substrate for CK2 than GluA1 (Fig. 1B), our results show that GluA2 surface expression was unaffected by CK2β knockdown [Fig. 3D and 3E; shCTL: 1.00 ± 0.057, n = 36; shCK2β: 1.05 ± 0.067, n = 37, P = 0.468]. Consistent with our biochemical analysis of total levels of proteins (Fig 3A and B), confocal microscopy analysis revealed that total expression of GluA1 and GluA2 was not affected in cells expressing shCK2β when compared with their respective shCTL [Fig. 3E, GluA1; shCTL: 1.00 ± 0.093, n = 35; shCK2β: 0.92 ± 0.086, n = 32, P = 0.39; Fig. 3H, GluA2; shCTL: 1.00 ± 0.067, n = 36; shCK2β: 1.01 ± 0.069, n = 37, P = 0.87].
Figure 3. A reduction of CK2 expression in cultured hippocampal neurons decreases surface expression of GluA1, but not GluA2.
A) Lysates from cultured neurons transduced with shCTL or shCK2β lentiviruses were prepared and immunoblotted with specific antibodies as indicated. B) Bar graph of protein expression in (A) presented as mean ± SEM and * P < 0.0001 using unpaired t-test. Data obtained from at least 4 experiments. C–H) Hippocampal neurons transfected with shCTL or shCK2β were stained live for endogenous surface GluA1 (C–D) or GluA2 (F–G) under non-permeabilized conditions before being examined by confocal microscopy. Representative images show AMPAR surface expression and EGFP from a transfected neuron. Scale bar represents 20 µm. At higher magnification, the scale bar represents 5 µm. C–H) Quantitation of AMPAR expression. Bar graph is presented as mean ± SEM. The scatter plots represent the distribution of each cells analyzed and show that shCK2β, but not shCTL, reduces surface expression of GluA1 (C) but not GluA2 (E) without affecting total expression of GluA1 (E) or GluA2 (H). Images not shown for (E) and (H) Each group contain 32–37 cells from 4 independent experiments (* P < 0.05 using unpaired t-test).
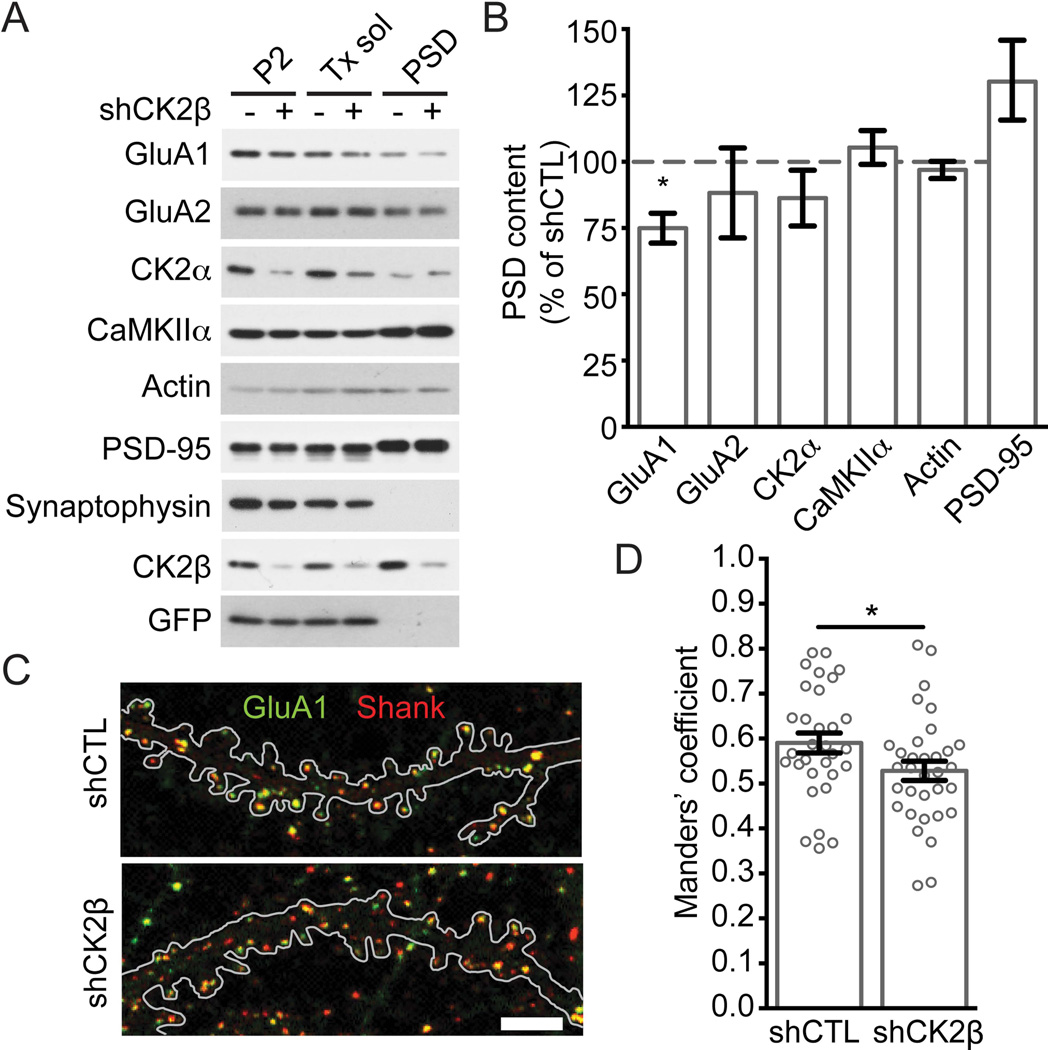
To investigate the importance of endogenous CK2 expression in regulating synaptic expression of glutamate receptors, we used lentiviruses harboring shRNAs for CK2β to knockdown CK2 and used a biochemical assay to examine the synaptic protein expression in cultured neurons. Cultured cortical neurons expressing control or CK2β specific shRNA were collected, a subcellular fractionation was performed, and fractions were probed with the indicated antibodies (Fig. 4A). Our results show that the pellet fraction, which is defined as the Triton X-100 insoluble fraction of P2 / crude synaptosomes, contained PSD-95 and was depleted in synaptophysin. Our procedure efficiently isolated synaptic proteins and can be used to examine the synaptic content of glutamate receptors in cultured neurons. We examined the expression of synaptic proteins by specific immunoblotting and found that reduced CK2β expression decreases synaptic expression of GluA1, but not GluA2, PSD-95, CK2α, CaMKIIα or actin [Fig. 5B, shCK2β to shCTL; GluA1: 0.75 ± 0.056, P = 0.0068; GluA2: 0.88 ± 0.17, P = 0.52; CK2α: 0.86 ± 0.11, P = 0.25; CaMKIIα: 1.05 ± 0.064, P = 0.44; actin: 0.97 ± 0.032, P = 0.39; PSD-95: 1.30 ± 0.15, P = 0.10; N= 6 experiments]. To confirm the reduction in GluA1 synaptic content, we performed a colocalization assay between GluA1 and the post-synaptic protein Shank; we used Shank because it is a cleaner synaptic marker (punctate pattern) than PSD-95 for cultured hippocampal neurons. Our results indicate a slight but significant reduction in GluA1 synaptic expression when knocking down CK2β, as determined by the Manders’ overlap coefficient of colocalization between Shank and GluA1 (Fig 4C and 4D: shCTL: 0.59 ± 0.022, n = 31; shCK2β: 0.53 ± 0.021, n = 33, P = 0.043, N = 5 experiments).
Figure 4. Reduced CK2 expression in cultured neurons decreases GluA1 in the synaptic fraction.
A) Subcellular fractionation was performed on cultured cortical neurons transduced with shCTL or shCK2β lentiviruses. Samples were prepared as described under Materials and Methods, and immunoblotted with specific antibodies as indicated. B) Quantitation of proteins found in the PSD / pellet fraction in (A). Bar graph shows that shCK2β but not shCTL decreases the expression of GluA1 and is presented as mean ± SEM. Quantitation performed from 6 experiments (*P < 0.05 for GluA1 using unpaired t-test). C–D) Hippocampal neurons transfected with shCTL or shCK2β were stained for endogenous total GluA1 (green) and Shank (red) before being examined by confocal microscopy. C) Representative images are shown, and the outline of the EGFP signal from a transfected neuron is depicted. Scale bar represents 5 µm. D) Quantitation of overlap between Shank and GluA1. Bar graph is presented as mean ± SEM while the scatter plot represents the distribution of each cells analyzed. Each group contains 31–33 cells from 5 independent experiments (* P < 0.05 using unpaired t-test).
Figure 5. Disruption of CK2 phosphorylation of GluA1 decreases surface expression.
A) Amino acid sequence of GluA1 intracellular loop1. Arrowheads identify residues S567 and S579 that are phosphorylated by CK2 (Fig.1 B, C and 2B) and mutated to alanine in (C), while arrows indicate acidic residues in GluA1 loop1 mutated to glutamine (B–C). B–C) GST-GluA1 loop1 was incubated with CK2 or CaMKII and [γ-32P]-ATP, and analyzed by autoradiography. A typical assay is shown. The arrowhead in (C) represents a non-specific band, and a similar pattern was also observed in Fig. 1B. D) Lysates from HEK293T cells expressing Flag-GluA1 WT, S579A or D580Q molecular replacement constructs in pRK5 vector were immunoblotted with specific antibodies as indicated. Typical result is shown. E) Hippocampal neurons were transduced with GluA1 specific shRNA lentiviruses, transfected with Flag-GluA1 WT, S579A or D580Q molecular replacement constructs, stained under non-permeabilized (surface) and permeabilized (intracellular) conditions. The expression of Flag-GluA1 was assessed by confocal microscopy. Representative images show AMPAR surface and intracellular / total expression from transfected neurons. Scale bar represents 10 µm. F) Quantitation of Flag-GluA1 WT, S579A and D580Q in (E) and performed as described in Material and Methods. Bar graph is presented as mean ± SEM while the scatter plot represents the distribution of each cells analyzed (each group contains 38–39 cells from 3 independent experiments, * P < 0.05 and ** P < 0.01 using Kruskal-Wallis one-way ANOVA with Dunn’s multiple comparison test).
Based on our findings, the endogenous expression of CK2 regulates the surface and synaptic expression of the AMPAR subunit GluA1, but not GluA2 (Fig. 3 and 4). Because CK2 phosphorylates many proteins and the use of an shRNA may generate off target effects, we also examined wild type (WT) versus mutated GluA1. Our in vitro data showed that S579 in the GluA1 loop was the major residue phosphorylated by CK2 (Fig. 2B). As we mentioned previously, CK2 is a kinase that phosphorylates residues in close proximity to acidic amino acids. Therefore, we also examined if the mutagenesis of any acidic amino acids surrounding S567 or S579 might disrupt the ability of CK2 to phosphorylate GluA1 loop1. Using an in vitro kinase assay with purified GST-fusion protein of the GluA1 loop1, we found that mutating D580 to glutamine (Q) (D580Q) abolished the phosphorylation of GluA1 loop1 by CK2, whereas the other point mutations did not (Fig. 5B). Because a mutant designed to inhibit CK2 phosphorylation might also prevent CaMKII phosphorylation of S567 in the loop1 region of GluA1, we performed an in vitro kinase assay to compare the ability of CK2 and CaMKII to phosphorylate the GluA1 loop1 region containing various mutations. As predicted, the CK2 phospho-defective mutants S579A and D580Q are still phosphorylated by CaMKII, whereas the S567A and the double mutant S567, S579/AA are not (Fig. 5C). Furthermore, the single mutants GluA1 loop1 S579A, GluA1 loop1 D580Q as well as the double mutant GluA1 loop1 S567, S579/AA block the phosphorylation of GluA1 by CK2 (Fig. 5C).
Up until now, our results demonstrate that CK2 is required for specifically regulating surface expression of GluA1, and CK2 phosphorylates predominantly S579 of GluA1. In addition, mutating D580 to glutamine (Q) at position +1 from S579 eliminates CK2 phosphorylation while keeping intact the ability of CaMKII to phosphorylate S567. Consequently, in addition to the phospho-deficient S579A mutant, the D580Q GluA1 mutant might be a powerful molecular tool to investigate the function of CK2 phosphorylation of the GluA1 loop1 region. Specific immunoblotting of HEK293T transfected cells shows that the expression of GluA1 S579A and D580Q mutants is indistinguishable from GluA1 WT (Fig. 5D). To determine if eliminating the ability of CK2 to phosphorylate GluA1 will modify its surface expression, we used a molecular replacement approach in which cultured hippocampal neurons were infected at DIV5 with lentiviruses expressing EGFP and a specific GluA1 shRNA before being transfected at DIV14 with an shRNA resistant Flag-GluA1 WT, S579A or D580Q construct. At DIV17, cells were subjected to an antibody-based confocal imaging assay. We found that the surface expression of GluA1 S579A and GluA1 D580Q were significantly decreased when compared to GluA1 WT (Fig. 5E and 5F; GluA1 WT: 1.00 ± 0.060, n = 38 cells; GluA1 S579A: 0.74 ± 0.052, n = 38 cells, *P < 0.05 (S579A to WT), GluA1 D580Q: 0.71 ± 0.065, n = 39 cells, **P < 0.01 (D580Q to WT), 3 experiments]. Therefore, CK2 phosphorylation of the loop region of GluA1 is required for maintaining a constant level of GluA1-containing AMPARs at the cell surface.
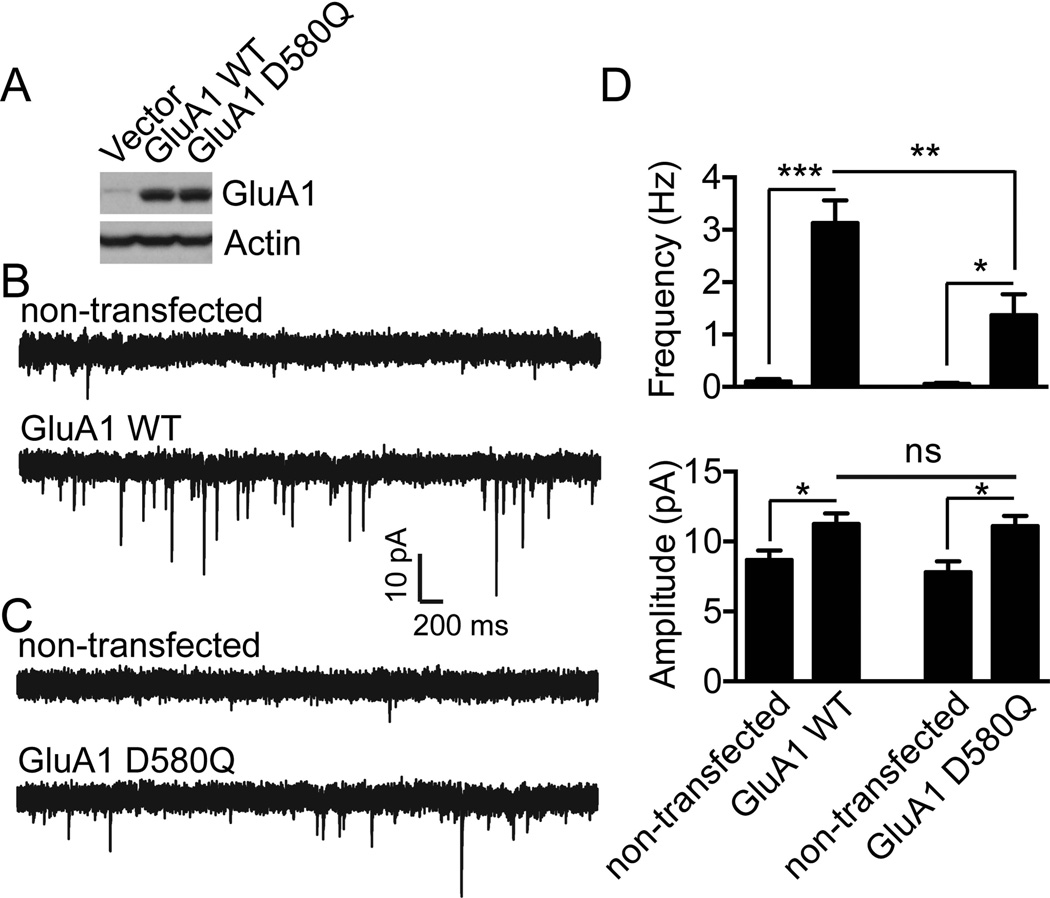
Finally we sought to determine the role of CK2 phosphorylation of GluA1 Loop1 in synaptic transmission. Once again, an immunoblot confirmed that the expression of GluA1 D580Q was similar to GluA1 WT (Fig. 6A). Hippocampal neurons cultured from GluA1 knockout mice were transfected with GluA1 WT or GluA1 D580Q. After transfection, we performed whole-cell voltage clamp recording to measure AMPAR-mediated miniature excitatory postsynaptic currents (mEPSCs). Compared to non-transfected cells, expression of GluA1 WT significantly increased mEPSC frequency (non-transfected, 0.099 ± 0.044 Hz; GluA1 WT, 3.13 ± 0.43 Hz; n = 9 for both conditions; ***P = 0.0001) and amplitude (non-transfected, 8.68 ± 0.68 pA; GluA1 WT, 11.26 ± 0.76 pA; n = 9 for both conditions; *P = 0.023) (Fig. 6B and 6D). Similar to GluA1 WT expression, transfection of the GluA1 D580Q mutant into GluA1 knockout cells also significantly enhanced mEPSC frequency (non-transfected, 0.055 ± 0.025 Hz, n = 6; GluA1 D580Q 1.37 ± 0.40 Hz, n = 10; * P = 0.009) and amplitude (non-transfected, 7.80 ± 0.79 pA, n = 6; GluA1 D580Q, 11.11 ± 0.74, n = 10; *P = 0.009) (Fig. 6C and 6D). However the enhancement of mEPSC frequency, but not mEPSC amplitude, in cells expressing GluA1 D580Q was significantly smaller than in cells expressing GluA1 WT (**P = 0.008) (Fig. 6D), suggesting that trafficking of GluA1 D580Q containing AMPA receptors to a subset of synapses was suppressed. Taken together, these data support that CK2 activity is critical for the regulation of synaptic delivery of AMPA receptors and demonstrate that inhibition of GluA1 Loop1 phosphorylation by CK2 inhibits AMPA receptor trafficking to a subpopulation of synapses.
Figure 6. Disruption of GluA1 phosphorylation by CK2 impairs GluA1 synaptic targeting.
A) Lysates from HEK293T cells expressing Flag-GluA1 WT or D580Q constructs in pIRES2-EGFP vector were prepared and immunoblotted with specific antibodies as indicated. Typical result is shown. B–C) Dissociated hippocampal cultures from GluA1 knockout mice were transfected with GluA1 WT (B) or with CK2 phospho-deficient mutant (GluA1 D580Q) (C). Whole-cell voltage clamp recordings to measure miniature EPSCs (mEPSCs) were performed after transfection. As shown in (D), although both GluA1 WT and GluA1 D580Q expression enhanced mEPSC frequency and amplitude, mEPSC frequency in neurons transfected with GluA1 WT was significantly higher than that in neurons transfected with GluA1 D580Q (for GluA1 WT, n = 9 for non-transfected and GluA1 WT transfected neurons; for GluA1 D580Q, n = 6 for non-transfected control and n = 10 for GluA1 D580Q). Error bars represent S.E.M. *P < 0.05, **P < 0.01, *** P < 0.001.
DISCUSSION
The processes controlling synaptic neurotransmission and the synaptic content of glutamate receptors are fundamental for learning and memory. In addition to the expression of postsynaptic receptors and signaling molecules, posttranslational modifications of these proteins clearly provide another level of complexity in synaptic function (Lussier et al., 2011, 2012; Lu & Roche, 2012; Nasu-Nishimura et al., 2010). Posttranslational modifications regulate protein activity, localization and interaction with other cellular components such as proteins or lipids. It has become increasingly clear that kinase activity and the phosphorylation of AMPARs are key mechanisms regulating their surface expression and intracellular routing. All four AMPAR subunits are substrates for kinases, and the function of a particular kinase depends on the residue phosphorylated within a specific subunit. For example, the phosphorylation state of S831 and S845 in the C-terminus of GluA1 regulates its intracellular trafficking in an activity-dependent manner (Barria et al., 1997; Ehlers, 2000; Lee et al., 2000, 2003; Mammen et al., 1997; Roche et al., 1996; Serulle et al. 2007), and the phosphorylation of S818 promotes the interaction of GluA1 with protein 4.1N resulting in increased receptor insertion at extrasynaptic membranes (Lin et al., 2009). Additionally, the phosphorylation of GluA1 S831 regulates channel conductance, revealing that biophysical properties of AMPAR may be modified by phosphorylation (Banke et al., 2000; Derkach et al., 1999; Kristensen et al., 2011). Surprisingly, molecular and genetic approaches show that none of those sites in the GluA1 C-terminus is essential for regulating basal synaptic transmission, whereas a CaMKII phosphorylation site in the loop1 region of GluA1 regulates the synaptic expression of GluA1 (Lu et al., 2010). In the current study we show that CK2 activity specifically regulates the surface expression of the GluA1 subunit by targeting the intracellular loop1 region.
We previously demonstrated that S567 of GluA1 loop1 region was a CaMKII site, and that mimicking the phosphorylation of S567 with an aspartate reduced the synaptic delivery of GluA1 as well as its colocalization with the synaptic marker PSD-95 (Lu et al., 2010). CaMKII, which is abundant at synapses (Kelly et al., 1984; Kennedy et al., 1983), is rapidly activated following synaptic stimulation and calcium influx and is recruited to spines. When using various pharmacological approaches to modify neuronal activity or CaMKII activity, we have been unable to modulate the phosphorylation of GluA1 S567 compared to the robust modulation of the phosphorylation of GluA1 S831 (unpublished observations). We now clearly demonstrate that GluA1 S567 is phosphorylated by CK2 as well as CaMKII in vitro. CK2 is a constitutively active serine/threonine kinase with important functions as a regulator of excitatory synapses and in processes of learning and memory. The present study shows that regulation of S567 is almost certainly not specific to CaMKII as other kinases can target this residue. Furthermore, CK2 can phosphorylate multiple residues on both GluA1 and GluA2 adding to the potential complexity of the loop1 regulation of AMPARs.
The high number of S/T/Y residues in the loop1 region of AMPARs is quite intriguing as they may serve as a hotspot for kinases. Within a short segment of approximately 30 amino acids, the number of S/T/Y residues ranges from 4 in GluA3 to 9 in GluA2 (Fig. 1A). However, it is the presence of a group of acidic residues downstream of GluA1 S567 (Fig. 1A) that led us to consider that CK2 might also target S567. We next extended our investigation to all AMPAR subunits since the residues surrounding GluA1 S567 are similar to those around the corresponding S/T in GluA2 and GluA4 (Fig. 1A). Surprisingly, we found that the GluA2 subunit is a better in vitro substrate than GluA1 for CK2, whereas GluA4 was barely phosphorylated (Fig. 1B). Interestingly, GluA1 surface expression and synaptic localization was specifically affected when knocking down CK2β (Fig. 3A–D and Fig. 4) and we therefore focused our attention on trying to understand the mechanism of GluA1 phosphorylation by CK2.
In addition to CaMKII and CK2, we also found that casein kinase 1 (CK1) can phosphorylate GluA1 and GluA2 in vitro (Fig. 1B). CK1, one of the first serine/threonine kinases to be isolated and characterized (Gross & Anderson, 1998), is basally active but the activity of the CK1δ and CK1ε isoforms can be enhanced by dephosphorylation in response to the activation of metabotropic glutamate receptors (Liu et al., 2001; 2002). To our knowledge, only one study has evaluated the possible function for CK1 on glutamatergic synaptic transmission. Although the direct phosphorylation of glutamate receptors by CK1 was not evaluated, N-methyl-D-aspartate receptor (NMDAR) miniature excitatory postsynaptic currents (mEPSCs) were increased while AMPAR mEPSCs remained unchanged by the inhibition of CK1 in coronal brain slices from three-week old mice (Chergui et al., 2005). Since the basal synaptic activity of AMPARs was not affected by CK1 inhibition, perhaps CK1 acts on the regulation of intracellular trafficking, membrane or synaptic insertion.
Interestingly, GluA2 seems to be a better substrate in vitro than GluA1 for both CK2 and CK1 (constitutively active CK1δ isoform in our assay). However, the substrate recognition motifs for CK1 and CK2 are quite different. For phosphorylation by CK1, the most efficient motif requires the presence of a phosphorylated S/T at position n−3 (where n represents the target S/T of interest). However, the enzyme can also phosphorylate a S/T at position n+3 from a cluster of acidic residues (Graves & Roach, 1995). In the case of CK2, multiple acidic residues located between the position n−2 to n+5 from the target S/T determine the substrate specificity and recognition site for phosphorylation, with the acidic residues downstream of the target S/T being the most important (Meggio & Pinna, 2003). This is the case at least for the GluA1 loop1 region where we were able to abrogate CK2 phosphorylation of GluA1 by changing the acidic residue D580 to Q at position n+1 from S579 (Fig. 5B and 5C). The expression of the GluA1 D580Q mutant in hippocampal neurons behaves exactly as the GluA1 S579A mutant (Fig. 5E and F) supporting our conclusion that CK2 regulates the surface expression of GluA1-containing AMPARs.
GluA1 forms GluA1A2 heteromers that generate about 80% of AMPARs synaptic content in hippocampus (Lu et al., 2009). Therefore, it was quite surprising to find that knocking down CK2 specifically reduced the surface and synaptic expression of GluA1, while GluA2 remained unchanged (Fig. 3 and 4). However we cannot exclude the possibility that neurons from acute hippocampal slices and hippocampal dissociated neuronal cultures may contain AMPARs with different compositions. Additionally, our recording experiments using a molecular replacement approach in GluA1 knockout hippocampal neurons confirmed that the phosphorylation of GluA1 by CK2 regulates GluA1 trafficking to synapses (Fig. 6). We may now speculate that CK2 regulates a population of homomeric GluA1 that is generally targeted to synapses or a specific subset of synapses. Also, our results suggest that the phosphorylation of GluA1 loop1 by CK2 is not the only determinant leading to its synaptic targeting, since we observed a reduction but not the complete exclusion from surface and synapses (Fig. 3A and D, Fig.4, Fig. 5E and F, Fig. 6). At this moment, the mechanism leading to GluA1 loop1 phosphorylation by CK2 remains unclear, and only future studies will expose the precise mechanism, as well as the possibility of interference and regulation between residues phosphorylated by several kinases such as CaMKII and CK1 identified in the current study (Fig. 1B).
CK2 is an important regulator of excitatory synapses, phosphorylating many important components of the PSD such as the GluN2B subunit of NMDARs, (Chung et al., 2004; Sanz-Clemente et al., 2010; 2013). Specifically, CK2 phosphorylation of S1480 on GluN2B within its PDZ domain is critical in removing the receptor from synapses, leading to its internalization and its replacement by GluN2A-containing NMDARs. Importantly, the GluN2B for GluN2A switch can be blocked by using a CK2 inhibitor (Sanz-Clemente et al, 2010). Interestingly, the activity of CK2 increases after the induction of LTP in the rat hippocampus (Charriaut-Marlangue et al., 1991) and leads to the rapid replacement of synaptic GluN2B with synaptic GluN2A in young animals (Bellone & Nicoll, 2007). The pharmacological inhibition of CK2 can block LTP (Belmeguenai et al., 2010; Kimura & Matsuki, 2008). In addition to its implication in normal brain function, CK2 has been associated with Parkinson’s disease where it was found in Lewy bodies (Ryu et al., 2008), and described to phosphorylate synuclein and synphilin-1 (Lee et al., 2004; Okochi et al., 2000). Interestingly, CK2 expression decreases in aging brain (Loerch et al., 2008) and in patients suffering from Alzheimer’s disease (Aksenova et al., 1991), Furthermore, the CK1 isoforms CK1α, CK1δ and CK1ε have been associated with Alzheimer’s disease (Li et al., 2004; Schwab et al., 2000). The possibility that CK1 or CK2 activity may also regulate AMPAR membrane expression or intracellular trafficking during synaptic plasticity or neurological disorders is intriguing but remains to be investigated. However, our present study demonstrates a direct and important role for the constitutive activity of CK2 in regulating surface expression and synaptic transmission of AMPARs through the phosphorylation of the loop1 region of GluA1. Importantly, our research illustrates the need for a deeper examination of the function of kinases such as CK2 on the intracellular loop1 region of AMPARs in order to understand the basis of normal brain function and synaptic basis of diseases.
ACKNOWLEDGMENTS
We thank John D. Badger II for technical assistance, the NINDS DNA sequencing facility, and Rolf Sprengel for providing us with the GluA1 knockout mouse line. The NINDS Intramural Research Program (to W.L. and K.W.R.) supported this research
ABBREVIATIONS
- ACSF
artificial cerebrospinal fluid
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- AMPAR
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor
- CaMKII
Ca2+/calmodulin-dependent protein kinase II
- CK1
casein kinase 1
- CK2
casein kinase 2
- DIV
days in vitro
- GluA
glutamate receptor AMPA-type
- GluN
glutamate receptor NMDA-type
- mEPSC
miniature excitatory postsynaptic current
- NMDA
N-methyl-D-aspartate
- NMDAR
N-methyl-D-aspartate receptor
- PKA
protein kinase A
- PKC
protein kinase C.
Footnotes
All the authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
All the authors contributed to the experimental design, interpretation and analysis of data. M.P.L. and X.G. performed experiments. M.P.L. and K.W.R. wrote the manuscript.
REFERENCES
- Aksenova MV, Burbaeva GS, Kandror KV, Kapkov DV, Stepanov AS. The decreased level of casein kinase 2 in brain cortex of schizophrenic and Alzheimer’s disease patient. FEBS Lett. 1991;279:55–57. doi: 10.1016/0014-5793(91)80249-3. [DOI] [PubMed] [Google Scholar]
- Banke TG, Bowie D, Lee H, Huganir RL, Schousboe A, Traynelis SF. Control of GluR1 AMPA receptor function by cAMP-dependent protein kinase. J. Neurosci. 2000;20:89–102. doi: 10.1523/JNEUROSCI.20-01-00089.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barria A, Derkach V, Soderling T. Identification of the Ca2+/calmodulin-dependent protein kinase II regulatory phosphorylation site in the alpha-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate-type glutamate receptor. J. Biol. Chem. 1997;272:32727–32730. doi: 10.1074/jbc.272.52.32727. [DOI] [PubMed] [Google Scholar]
- Bellone C, Nicoll RA. Rapid bidirectional switching of synaptic NMDA receptors. Neuron. 2007;55:779–785. doi: 10.1016/j.neuron.2007.07.035. [DOI] [PubMed] [Google Scholar]
- Belmeguenai A, Hosy E, Bengtsson F, Pedroarena CM, Piochon C, Teuling E, He Q, Ohtsuki G, De Jeu MT, Elgersma Y, De Zeeuw CI, Jörntell H, Hansel C. Intrinsic plasticity complements long-term potentiation in parallel fiber input gain control in cerebellar Purkinje cells. J. Neurosci. 2010;30:13630–13643. doi: 10.1523/JNEUROSCI.3226-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm J, Kang MG, Johnson RC, Esteban J, Huganir RL, Malinow R. Synaptic incorporation of AMPA receptors during LTP is controlled by a PKC phosphorylation site on GluR1. Neuron. 2006;51:213–225. doi: 10.1016/j.neuron.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 8.Carroll RC, Beattie EC, von Zastrow M, Malenka RC. Role of AMPA receptor endocytosis in synaptic plasticity. Nat. Rev. Neurosci. 2001;2:315–324. doi: 10.1038/35072500. [DOI] [PubMed] [Google Scholar]
- Charriault-Marlangue C, Otani S, Creuzet C, Ben-Ari Y, Loeb T. Rapid activation of hippocampal casein kinase II during long-term potentiation. Proc. Natl Acad. Sci. USA. 1991;88:10232–10236. doi: 10.1073/pnas.88.22.10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chergui K, Svenningson P, Greengard P. Physiological role for casein kinase 1 in glutamatergic synaptic transmission. J. Neurosci. 2005;25:6601–6609. doi: 10.1523/JNEUROSCI.1082-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HJ, Huang HY, Lau HF, Huganir RL. Regulation of the NMDA receptor complex and trafficking by activity-dependent regulation of the NR2B subunit PDZ domain ligand. J. Neurosci. 2004;24:10248–10259. doi: 10.1523/JNEUROSCI.0546-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkach V, Barria A, Soderling TR. Ca2+/calmodulin-kinase II enhances channel conductance of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionate type glutamate receptors. Proc. Natl Acad. Sci. USA. 1999;96:3269–3274. doi: 10.1073/pnas.96.6.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD. Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron. 2000;28:511–525. doi: 10.1016/s0896-6273(00)00129-x. [DOI] [PubMed] [Google Scholar]
- Esteban JA, Shi SH, Wilson C, Nuriya M, Huganir RL, Malinow R. PKA phosphorylation of AMPA subunits controls synaptic trafficking underlying plasticity. Nat. Neurosci. 2003;6:136–143. doi: 10.1038/nn997. [DOI] [PubMed] [Google Scholar]
- 15.Graves PR, Roach PJ. Role of COOH-terminal phosphorylation in the regulation of the casein kinase I delta. J. Biol. Chem. 2005;270:21689–21694. doi: 10.1074/jbc.270.37.21689. [DOI] [PubMed] [Google Scholar]
- Groc L, Choquet D. AMPA and NMDA glutamate receptor trafficking: Multiple roads for reaching and leaving the synapse. Cell Tissue Res. 2006;326:423–438. doi: 10.1007/s00441-006-0254-9. [DOI] [PubMed] [Google Scholar]
- Gross SD, Anderson RA. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell Signal. 1998;10:699–711. doi: 10.1016/s0898-6568(98)00042-4. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- Hanley JG. Endosomal sorting of AMPA receptors in hippocampal neurons. Biochem. Soc. Trans. 2010;38:460–465. doi: 10.1042/BST0380460. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Shi SH, Esteban JA, Piccini A, Poncer JC, Malinow R. Driving AMPA receptors into synapses by LTP and CaMKII: Requirement for GluR1 and PDZ domain interaction. Science. 2000;287:2262–2267. doi: 10.1126/science.287.5461.2262. [DOI] [PubMed] [Google Scholar]
- 21.Hirling H. Endosomal trafficking of AMPA-type glutamate receptors. Neuroscience. 2009;158:36–44. doi: 10.1016/j.neuroscience.2008.02.057. [DOI] [PubMed] [Google Scholar]
- Kelly PT, McGuinness TL, Greengard P. Evidence of that the major postsynaptic density protein is a component of a Ca2+/calmodulin-dependent protein kinase. Proc. Natl Acad. Sci. USA. 1984;81:945–949. doi: 10.1073/pnas.81.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MB, Bennett MK, Erondu NE. Biochemical and immunochemical evidence that the “major postsynaptic density protein” is a subunit of a calmodulin-dependent protein kinase. Proc. Natl Acad. Sci. USA. 1983;80:7357–7361. doi: 10.1073/pnas.80.23.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura R, Matzuki N. Protein kinase CK2 modulates synaptic plasticity by modification of synaptic NMDA receptors in the hippocampus. J. Physiol. 2008;586:3195–3206. doi: 10.1113/jphysiol.2008.151894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen AS, Jenkins MA, Banke TG, Schousboe A, Makino Y, Johnson RC, Huganir R, Traynelis SF. Mechanism of Ca2+/calmodulin-dependent kinase II regulation of AMPA receptor gating. Nat. Neurosci. 2011;14:727–735. doi: 10.1038/nn.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Tanaka M, Park K, Lee SS, Kim YM, Junn E, Lee SH, Mouridian MM. Casein kinase II-mediated phosphorylation regulates alpha synuclein/synphilin-1 interaction and inclusion body formation. J. Biol. Chem. 2004;279:6834–6839. doi: 10.1074/jbc.M312760200. [DOI] [PubMed] [Google Scholar]
- Lee HK, Barbarosie M, Kameyama K, Bear MF, Huganir RL. Regulation of distinct AMPA receptor phosphorylation sites during bidirectional synaptic plasticity. Nature. 2000;405:955–959. doi: 10.1038/35016089. [DOI] [PubMed] [Google Scholar]
- Lee HK, Takamiya K, Han JS, Man H, Kim CH, Rumbaugh G, Yu s, Ding L, He C, Petralia RS, Wenthold RJ, Gallagher M, Huganir RL. Phosphorylation of the AMPA receptor GluR1 subunit is required for synaptic plasticity and retention of spatial memory. Cell. 2003;112:631–643. doi: 10.1016/s0092-8674(03)00122-3. [DOI] [PubMed] [Google Scholar]
- Li G, Yin H, Kuret J. Casein kinase 1 delta phosphorylates Tau and disrupts its binding to microtubules. J. Biol. Chem. 2004;279:15938–15945. doi: 10.1074/jbc.M314116200. [DOI] [PubMed] [Google Scholar]
- Lin DT, Makino Y, Sharma K, Hayashi T, Neve R, Takamiya K, Huganir RL. Regulation of AMPA receptor extrasynaptic insertion by 4.1N, phosphorylation and palmitoylation. Nat. Neurosci. 2009;12:879–887. doi: 10.1038/nn.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Ma XH, Ule J, Bibb BA, Nishi A, DeMaggio AJ, Yan Z, Nairn AC, Greengard P. Regulation of cyclin-dependent kinase 5 and casein kinase 1 by metabotropic glutamate receptors. Proc. Natl Acad. Sci. USA. 2001;98:11062–11068. doi: 10.1073/pnas.191353898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Virshup DM, Nairn AC, Greengard P. Mechanism of regulation of casein kinase I activity by group I metabotropic glutamate receptors. J. Biol. Chem. 2002;277:45393–45399. doi: 10.1074/jbc.M204499200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerch PM, Lu T, Dakin KA, Vann JM, Isaacs A, Geula C, Wang J, Pan Y, Gabuzda DH, Li C, Prolla TA, Yankner BA. Evolution of the aging brain transcriptome and synaptic regulation. PLoS ONE. 2008;3:e3329. doi: 10.1371/journal.pone.0003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Roche KW. Posttranslational regulation of AMPA receptor trafficking and function. Curr. Opin. Neurobiol. 2012;22:470–479. doi: 10.1016/j.conb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Isozaki K, Roche KW, Nicoll RA. Synaptic targeting of AMPA receptors is regulated by a CaMKII site in the first intracellular loop of GluA1. Proc. Natl Acad. Sci. USA. 2010;107:22266–22271. doi: 10.1073/pnas.1016289107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Shi Y, Jackson AC, Bjorgan K, During MJ, Sprengel R, Seeburg PH, Nicoll RA. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron. 2009;62:254–268. doi: 10.1016/j.neuron.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier MP, Herring BE, Nasu-Nishimura Y, Neutzner A, Karbowski M, Youle RJ, Nicoll RA, Roche KW. Ubiquitin ligase RNF167 regulates AMPA receptor-mediated synaptic transmission. Proc. Natl Acad. Sci. USA. 2012;109:19426–19431. doi: 10.1073/pnas.1217477109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier MP, Nasu-Nishimura Y, Roche KW. Activity-dependent ubiquitination of the AMPA receptor subunit GluA2. J. Neurosci. 2011;31:3077–3081. doi: 10.1523/JNEUROSCI.5944-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammen AL, Kameyama K, Roche KW, Huganir RL. Phosphorylation of the alpha-amino-3-hydroxy-5-methylisoxazole4-propionic acid receptor GluR1 subunit by calcium/calmodulin-dependent kinase II. J. Biol. Chem. 1997;272:32528–32533. doi: 10.1074/jbc.272.51.32528. [DOI] [PubMed] [Google Scholar]
- Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- Nasu-Nishimura Y, Jaffe H, Isaac JT, Roche KW. Differential regulation of kainate receptor trafficking by phosphorylation of distinct site on GluR6. J. Biol. Chem. 2010;285:2847–2856. doi: 10.1074/jbc.M109.081141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okochi M, Walter J, Koyama A, Nakajo S, Baba M, Iwatsubo T, Meijer L, Kahle PJ, Haass C. Constitutive phosphorylation of the Parkinson’s disease associated alpha-synuclein. J. Biol. Chem. 2000;275:390–397. doi: 10.1074/jbc.275.1.390. [DOI] [PubMed] [Google Scholar]
- Pallares J, Llobet D, Santacana M, Eritja N, Velasco A, Cuevas D, Lopez S, Palomar-Asenjo V, Yeramian A, Dolcet X, Matias-Guiu X. CK2beta is expressed in endometrial carcinoma and has a role in apoptosis resistance and cell proliferation. Am. J. Patho. 2009;174:287–296. doi: 10.2353/ajpath.2009.080552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinna LA, Ruzzene M. How do protein kinases recognize their substrate? Biochim. Biophys. Acta. 1996;1314:191–225. doi: 10.1016/s0167-4889(96)00083-3. [DOI] [PubMed] [Google Scholar]
- Roche KW, O'Brien RJ, Mammen AL, Bernhardt J, Huganir RL. Characterization of multiple phosphorylation sites on the AMPA receptor GluR1 subunit. Neuron. 1996;16:1179–1188. doi: 10.1016/s0896-6273(00)80144-0. [DOI] [PubMed] [Google Scholar]
- Ryu MY, Kim DW, Arima K, Mouridian MM, Kim SU, Lee G. Localization of CKII beta subunits in Lewy bodies of Parkinson’s disease. J. Neurol. Sci. 2008;266:9–12. doi: 10.1016/j.jns.2007.08.027. [DOI] [PubMed] [Google Scholar]
- Sanz-Clemente A, Gray JA, Ogilvie KA, Nicoll RA, Roche KW. Activated CaMKII couples GluN2B and casein kinase 2 to control synaptic NMDA receptors. Cell Rep. 2013;3:607–614. doi: 10.1016/j.celrep.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Clemente A, Matta JA, Isaac JT, Roche KW. Casein kinase 2 regulates the NR2 subunit composition of synaptic NMDA receptor. Neuron. 2010;67:984–996. doi: 10.1016/j.neuron.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab C, DeMaggio AJ, Ghoshal N, Binder LI, Kuret J, McGeer PL. Casein kinase 1 delta is associated with pathological accumulation of tau in several neurodegenerative diseases. Neurobiol. Aging. 2000;21:503–510. doi: 10.1016/s0197-4580(00)00110-x. [DOI] [PubMed] [Google Scholar]
- Serulle Y, Zhang S, Ninan I, Puzzo D, McCarthy M, Khatri L, Arancio O, Ziff EB. A GluR1-cGKII interaction regulates AMPA receptor trafficking. Neuron. 2007;56:670–688. doi: 10.1016/j.neuron.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wenthold RJ, Petralia RS, Blahos J, II, Niedzielski AS. Evidence for multiple AMPA receptor complexes in hippocampal CA1/CA2 neurons. J. Neurosci. 1996;16:1982–1989. doi: 10.1523/JNEUROSCI.16-06-01982.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zamanillo D, Sprengel R, Hvalby O, Jensen V, Burnashev N, Rozov A, Kaiser KM, Köster HJ, Borchardt T, Worley P, Lübke J, Frotscher M, Kelly PH, Sommer B, Andersen P, Seeburg PH, Sakmann B. Importance of AMPA receptors for hippocampal synaptic plasticity but not spatial learning. Science. 1999;284:1805–1811. doi: 10.1126/science.284.5421.1805. [DOI] [PubMed] [Google Scholar]