Abstract
Recently, apoptosis has been considered to be an important regulator for allograft survival. The serine/threonine kinase Pim2 has been implicated in many apoptotic pathways. In a previous study, we found that pim2 was highly expressed in CD4+ T cells in an allograft model. Here, we further investigated the effects of Pim2 on allograft survival and the underlying mechanisms associated with apoptosis. The results showed that pim2 was overexpressed in grafts and spleens, particularly in spleen CD4+ T cells when acute allorejection occurred, and correlated positively with the extent of rejection. In T cells from the spleens of naive BALB/c mice treated with 5 µM 4a (a specific inhibitor of Pim2) for 24 h, the apoptosis rate increased and the phosphorylation of BAD was decreased. Furthermore, adoptive transfer of CD4+ T cells treated with 4a in vitro to allografted severe combined immunodeficiency (SCID) mice effectively prolonged allograft survival from 19.5±1.7 days to 31±2.3 days. Moreover, the results demonstrated that the CD4+CD25− effector T-cell subset was the predominate expresser of the pim2 gene as compared with the CD4+CD25+ regulatory T (Treg) cell subset. Alloantigen-induced CD4+CD25+ T cells displayed less Foxp3 expression and a low suppression of apoptosis compared with effector CD4+CD25− T cells treated with 4a. Collectively, these data revealed that Pim2 facilitated allograft rejection primarily by modulating the apoptosis of effector T cells and the function of Treg cells. These data suggested that Pim2 may be an important target for in vivo anti-rejection therapies and for the ex vivo expansion of CD4+CD25+ T cells.
Keywords: apoptosis, CD4+ T cell, Pim2, regulatory, transplantation
Introduction
Organ transplantation is the most effective therapy for patients with end-stage organ failure. However, acute rejection, which occurs within the first few weeks or months, remains a major cause of treatment failure. The exact mechanism of organ rejection is not fully understood, even though much research has been conducted on this topic. Current research is now focused on the mechanisms and relationships among different pathways during allograft rejection. Recently, the ‘immunologic constant of rejection' phenomenon has been suggested, which states that different immune-mediated tissue destruction processes (i.e., cancer, infection and autoimmunity) share common convergent final mechanisms.1 Therefore, finding a specific molecule that is associated with allograft rejection will provide new ways to downregulate allorejection and other immune-mediated diseases.
Recent data have stressed the importance of apoptosis-associated intercellular communication networks in the regulation of allograft remodeling and immune responses in transplantation. Transplanted organs must cope with diverse immunologic and metabolic stressors that augment the percentage of dying cells, where apoptosis is tightly orchestrated and culminates in the activation of caspases during rejection.2
The pim family is a small family of proto-oncogenes encoding for serine/threonine kinases that consists of three members, pim1, pim2 and pim3, which play important roles in tumorigenesis.3,4,5 The Pim kinases induce a variety of effects in different types of tumors.6 It has been reported that 5-(3-trifluoromethylbenzylidene)thiazolidine-2,4-dione (4a) is a specific inhibitor of the Pim kinases,7 which could reduce the ability of Pim to phosphorylate the BAD BH3 protein and induce G1 phase cell cycle arrest and apoptosis.8
CD4+ T cells play critical roles in alloimmune responses.9 These cells can be activated by recognizing allogeneic antigen directly or indirectly and can mediate delayed-type hypersensitivity responses to damage the allograft.10 In addition, CD4+ T cells can be differentiated into various subsets. CD4+CD25+Foxp3+ T cells, an important subset of CD4+ T cells, can suppress allograft rejection by potently suppressing the activation or function of conventional CD4+CD25− T effector cells.11 Moreover, the expression of CD25 (the α-chain of the IL-2 receptor) in mice correlates with the intracellular expression of the transcriptional factor Foxp3, which inhibits IL-2 gene transcription.12 It has been reported that donor antigen-specific CD4+CD25+Foxp3+ Regulatory T (Treg) cells can regulate alloresponses and promote donor-specific tolerance in a skin transplantation model.13 Interestingly, Foxp3 induces pim2 expression in natural Tregs, which caused Foxp3-expressing Treg cells to be selected in the presence of rapamycin.14 These studies suggest that pim2 may be associated with the function of Treg cells in allograft rejection.
The growth and survival of T cells is partly mediated through the activation of the effector enzymes AKT and target of rapamycin (TOR) in the phosphatidylinositol 3-kinase pathway.15 TOR is one key mediator of Akt-dependent signal transduction.16 Rapamycin-mediated Mammalian target of rapamycin (mTOR) inhibition blocks critical T-cell effector functions such as migration and cytokine production and limits T-cell expansion.17,18,19 Akt and Pim2 share many common downstream targets, including Bad and 4E-binding protein-1.20 Pim2 expression has been shown to be required to compensate for TOR inhibition by rapamycin in wild-type T cells, suggesting that Pim2 kinase might provide an alternative pathway for T-cell survival.21 The pim1 and pim2 mRNA expression in human cord blood-derived CD4+ T cells could be upregulated by Th1-specific cytokines (IL-12 and IFN-α) and downregulated by Th2-specific cytokine (IL-4), which suggests that Pim kinase is involved in the differentiation process of CD4+ T cells.22 Furthermore, in our previous study, an approximately fivefold overexpression of the pim2 gene, as determined by sequencing-based serial analysis of gene expression, was detected in the allograft-activated CD4+ T cells, which suggests a potential role of the Pim2 kinase in allograft rejection.23
Considering all of the aforementioned results, we hypothesize that the Pim2 kinase may participate in allograft rejection through targeting the apoptosis of CD4+ T cells and modulating Treg-suppressing activities. In this study, we investigated the role and the underlying mechanism of Pim2 during allograft rejection.
Materials and methods
Mice
Inbred strains of female BALB/c mice (H-2d) were purchased from the Animal Center of Shandong University, China. Inbred strains of female C57BL/6 (H-2b, B6) were purchased from the Laboratory Animal Center of Shanghai, China. Inbred strains of female severe combined immunodeficiency (SCID) mice were purchased from the Vitalriveri Co. Ltd (Beijing, China). The experimental female mice were 18–22 g in weight, 6–8 weeks old and bred in a pathogen-free facility. The mice were housed in microisolator cages containing sterilized feed, autoclaved bedding and water, according to the principles of laboratory animal care. The mice study was approved by the Institutional Animal Experimental Committee, and animal care and surgical procedures were performed in compliance with the standard animal experimental protocols of Shandong University School of Medicine.
Skin-grafting model
The dorsal skin of C57BL/6 mice was transplanted onto the BALB/c mice under sterile conditions (allogenic transplantation). The dorsal skin of BALB/c mice was transplanted onto the BALB/c mice (isogenic transplantation) as the control. Skin grafting was performed as previously described.24 Briefly, full-thickness trunk skin from the donor mice was cut into 0.5-cm2 pieces and stored in sterile phosphate-buffered saline (PBS) until use. The recipient mice were then anesthetized with 0.6% sodium pentobarbital (50 µg/g of body weight). Then, the graft was promptly initiated in a slightly larger graft bed on the back of the recipient that was then covered with vaseline gauze and bandages. The grafts were observed daily after the removal of the bandages. When greater than 90% of the epidermal surface became necrotic, the graft was considered to be rejected.25
Histological examination
Five mice from each group (allorejected and isografted recipient mice) were killed at 14 days after transplantation. On the designated day, fragments of skin grafts and spleens were collected for histopathology. These samples were fixed in 10% formalin, dehydrated, cleared, embedded in paraffin, cut (6 µm thick) and stained with hematoxylin and eosin. For immunohistochemical staining, tissue samples were embedded in optimal cold temperature compound and snap frozen, and 6-µm sections were cut for staining. A mouse polyclonal antibody against Pim2 (sc-137049; Santa Cruz Biotechnology, Inc. 2145 Delaware Avenue, Santa Cruz, CA. 95060. U.S.A.) was diluted 1∶200 and incubated with tissue section overnight at 4 °C. The slides were washed with PBS and incubated with the horseradish peroxidase-labeled secondary antibodies at 37 °C for 30 min. After washing, the slides were then incubated with 3,3-diaminobenzidine (ZLI-9017, ZSGB-BIO) for 5 min at room temperature. Hematoxylin staining solution (C0107; Beyotime Institute of Biotechnology, Haimen, Jiangsu, China) was used to stain cell nuclei for 2 min prior to examination under a microscope.
Cell purification and culture
On days 1, 7, 10, 14 and 21 following transplantation, the spleens were taken from the recipient mice. A single cell suspension was obtained by pushing the cells through a 200-mesh copper net. Then, the erythrocytes were degraded with red blood cell lysis buffer. CD4+ T cells were purified with Mouse T cell Enrichment Columns (R&D Systems, Inc. 614 McKinley Place NE Minneapolis, MN 55413 USA), while CD4+CD25+ T cells and CD4+CD25− T cells were purified and collected with MagCellect Mouse CD4+CD25+ Regulatory T Cell Isolation Kit (R&D Systems, Inc.) All cells were cultured in RPMI-1640 medium with 10% (v/v) fetal bovine serum (FBS) at 37 °C in 5% CO2.
RNA extraction and Reverse transcription-PCR
Total RNA was extracted, reverse transcribed, amplified, and analyzed as previously described.26 Briefly, total RNA was extracted from skin grafts, spleen cells and T cells subsets using a total RNA isolation kit (TransGen) according to the manufacturer's instructions. Ultraviolet spectrophotometry was used to determine the quantity and quality of total RNA. The Gapdh gene was used to normalize cDNA quantities and was amplified with the forward primer 5′-AGGTCGGTGTGAACGGATTTG-3′ and reverse primer 5′-TGTAGACCATGTAGTTGAGGTCA-3′ (Biosune Biotechnology Co., Ltd. Shanghai, China). Pim2 cDNA was amplified with the forward primer 5′-TTCGAAACACCCGAAGGCTT-3′ and reverse primer 5′-CATAGGTCGATCAGGATGTTC-3′ (Biosune Biotechnology Co., Ltd. Shanghai, China). Foxp3 cDNA was amplified with forward primer 5′-ATCCAGCCTGCCTCTGACAAGAACC-3′ and reverse primer 5′-GGGTTGTCCAGTGGACGCACTTGGAGC-3′ (Biosune Biotechnology Co., Ltd. Shanghai, China).
Western blot analysis
Cell lysis, electrophoresis and immunoblotting were performed as described previously.27 Commercial antibodies were performed according to the manufacturer's instructions. Briefly, total protein was extracted from skin grafts, spleens and splenocytes using cell lysis buffer (p1033; Beyotime). The protein concentration was determined by the Bradford method (Beyotime). Then, 16 µl of total protein was added to 4 µl 5× loading buffer, which was then subjected to 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis. The electrophoresed proteins were transferred to a polyvinylidene difluoride membrane at 180 mA for 1 h in transfer buffer (25 mM Tris, 0.2 M glycine, 20% methanol). The membranes were blocked in 5% nonfat milk in PBS with 0.1% Tween-20 for 1 h with agitation followed by washing and then addition of primary antibodies including Pim2 (sc-137049; Santa Cruz Biotechnology), p-Bad (Ser112,AB-009; Beyotime) and β-Actin (AA128; Beyotime) (1∶500–1∶1000 dilution in 5% bovine serum albumin in PBS buffer). The membranes were incubated overnight at 4 °C. β-actin was used as the loading control. Then, horseradish peroxidase-conjugated secondary antibodies (1∶2000 dilution in 5% bovine serum albumin in PBS with 0.1% Tween-20) were added for 1 h at room temperature. The proteins were detected using the Enhanced Chemiluminescence Western Blotting Detection Reagent (Beyotime). The results are described as a ratio of the relative absorbance values of the band of the protein of interest to β-actin.
Apoptosis analysis
T cells purified from the spleens of recipient mice with graft rejection were cocultured with 5 µM 4a or DMSO as a control for 24 h, then washed with ice cold PBS and stained with annexin V-fluorescein isothiocyanate using the Annexin V-FITC Apoptosis Detection Kit (C1062; Beyotime). Cell apoptosis was detected by flow cytometry.
CD4+CD25+ T cells and CD4+CD25− T cells were purified from the spleens of recipient mice with allograft rejection as described in ‘cell purification and culture'. CD4+CD25+ T cells (5×105) treated with 5 µM 4a or DMSO as a control for 24 h were then cocultured with CD4+CD25− T cells (3×106) for another 12 h, followed by washing with ice cold PBS and staining with annexin V-fluorescein isothiocyanate and propidium iodide. Cell apoptosis was detected by flow cytometry; additionally, these cell extracts were also subjected to Western blotting with antibodies to p-BAD. All flow cytometry was performed using a FACS Calibur (BD Bioscience).
Adoptive cell transfer
The dorsal skin of C57BL/6 mice was transplanted onto the dorsal of SCID mice under sterile conditions as described above; grafts will not be rejected in these mice due to the severe immune deficiency. Twenty-one days later, after the wound had healed, 2×107 T cells (isolated from the spleens of naive BALB/c mice and cultured with 5 µM 4a or DMSO as a control for 24 h) were injected into the grafted SCID mice via intraperitoneal injection, and the condition of the allografts was observed daily.
Statistical analysis
Statistical analysis was performed with SPSS11.0 software. Parametric data were expressed as the mean±standard deviation (s.d.). The means were compared using the one-way analysis of variance (ANOVA) test. For all analyses, statistical significance was set at P<0.05.
Results
Pim2 was highly expressed in allografted mice and positively correlated with the severity of allograft rejection
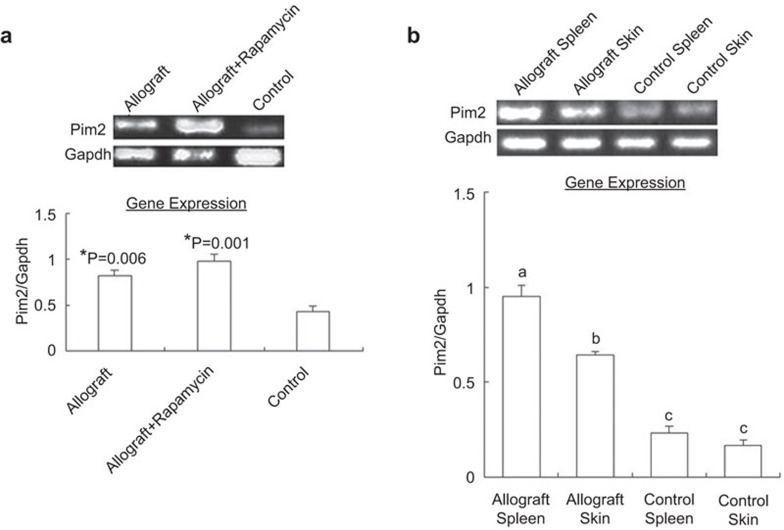
The model mice were divided into three groups, with five mice in each group: allografted mice, rapamycin-treated allografted mice (rapamycin at 1.5 mg/kg/day, 1–14 days post-transplantation) and isografted mice. At day 14 post-transplantation (allografts rejected completely, while isografts remained normal), pim2 mRNA from splenic CD4+ T cells of different recipient mice was examined. The results showed that the expression of pim2 was significantly higher both in the allografted mice and rapamycin-treated allograft mice, approximately 1.88-fold and 2.28-fold relative to isografted mice, respectively (Figure 1a). However, rapamycin treatment in vivo did not significantly change the expression of pim2 in allograft mice (P>0.05). The results also showed that pim2 mRNA from spleens and skin grafts was significantly elevated in allograft mice compared with that from isografted mice. Moreover, in allograft mice, pim2 mRNA was markedly higher in the spleens than in the skin grafts, while in isografted mice, there was no significant difference between these two tissues (Figure 1b).
Figure 1.
Pim2 gene expression was significantly increased during acute allograft rejection. (a) Pim2 mRNA in CD4+T cells. (b) Pim2 mRNA in spleens and skin grafts. The data are shown as the mean±s.e. (n=3). *indicates significance (P<0.05) vs. control. The values with different letters show significant differences (P<0.05).
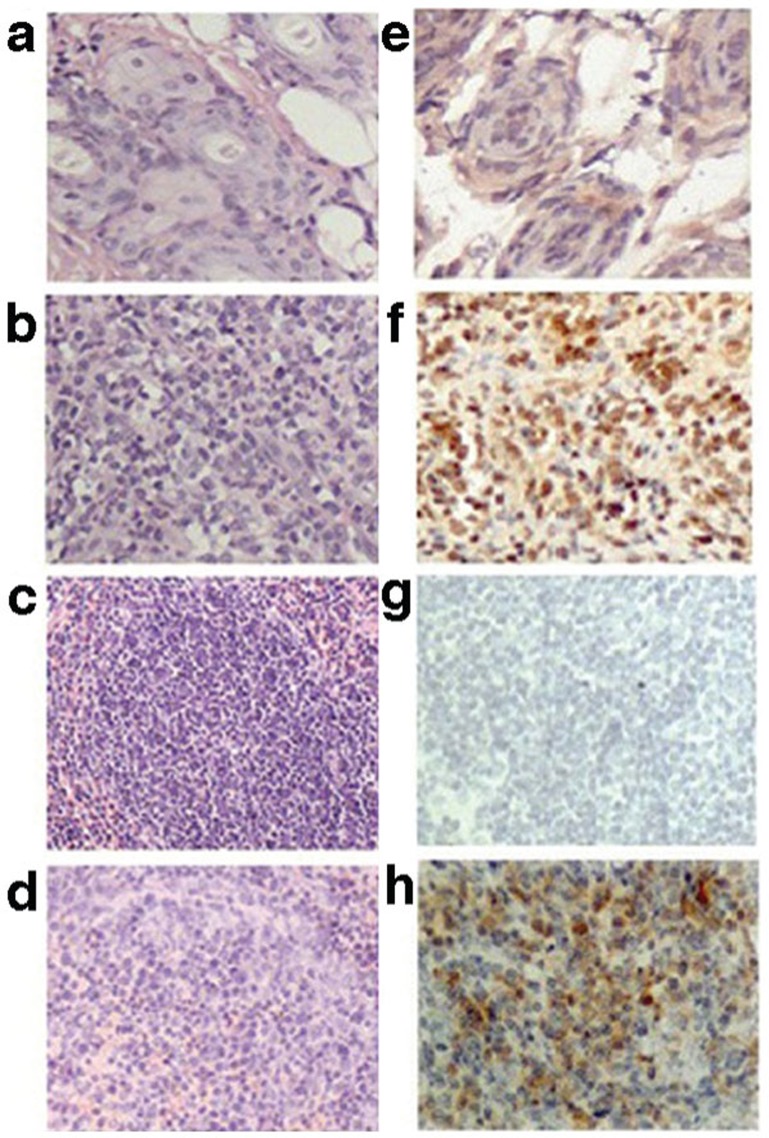
The histology of spleens and skin grafts from allorejected and isografted mice are shown in Figure 2. In the panels of the left row, there was an obvious difference in the pathology between the two groups. The allograft skins showed obvious foci of infiltrated mononuclear cells (Figure 2a), while the isograft skins appeared normal (Figure 2b). There was a minor repopulation of lymphoid areas in the white pulp of the spleens of allograft mice (Figure 2c), compared with the control spleens (Figure 2d). The panels of the right row show the in situ expression of Pim2 protein by immunohistochemical staining. Pim2 protein was markedly expressed in the cytoplasm of allografted skins (Figure 2f) and spleens (Figure 2h), but only weakly or negatively expressed in isografted skins (Figure 2e) and spleens (Figure 2g). The results suggested that Pim2 protein was involved in the pathogenesis of allograft rejection.
Figure 2.
Expression of Pim2 within grafted skin and spleen. The left panels show sections with hematoxylin and eosinstaining, and the right panels show immunohistochemical staining using anti-Pim2. (a, e) Isograft skin. (b, f) Allograft skin. (c, g) Splenic areas of isografted mice. (d, h) Splenic areas of allografted mice. The results have been repeated in three independent experiments.
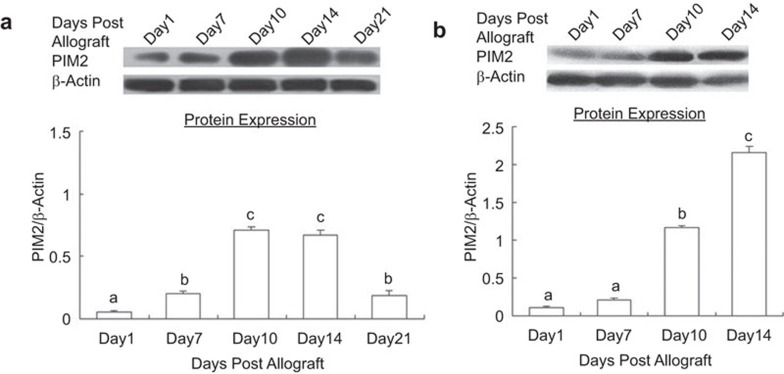
The expression of Pim2 in spleens and grafted skins at various times post-allotransplantation was also confirmed using immunoblotting. The results showed that Pim2 protein was upregulated at day 7 in the spleens of recipient mice, which is 3.82-fold of those at day 1 (P<0.05). The highest levels were observed at day 10, which is 13.51-fold compared with those at day 1 (P<0.05). There were no significant differences between day 10 and day 14 or between day 7 and day 21 (P>0.05) (Figure 3a). Correspondingly, the accumulation of Pim2 in allograft skins showed the same tendency as in the spleens, except that upregulation occurred at day 10 (10.79-fold) and that the highest level observed was at day 14 (19.81-fold), compared with that at day 1 (P<0.05) (Figure 3b). This pattern of Pim2 expression overlapped with the progression of graft rejection in response to the allogeneic antigen. These data indicated that the kinetic expression of Pim2 protein positively correlated with the severity of graft rejection, suggesting that Pim2 played important roles in the alloresponse and graft survival.
Figure 3.
Pim2 protein expression varied at different times post-allotransplantation. (a) Spleens of allograft mice. (b) Allograft skins. The data are shown as mean±s.e. (n=3). The values with different letters show significant differences (P<0.05).
Blockade of Pim2 kinase by 4a prevented acute rejection in vivo
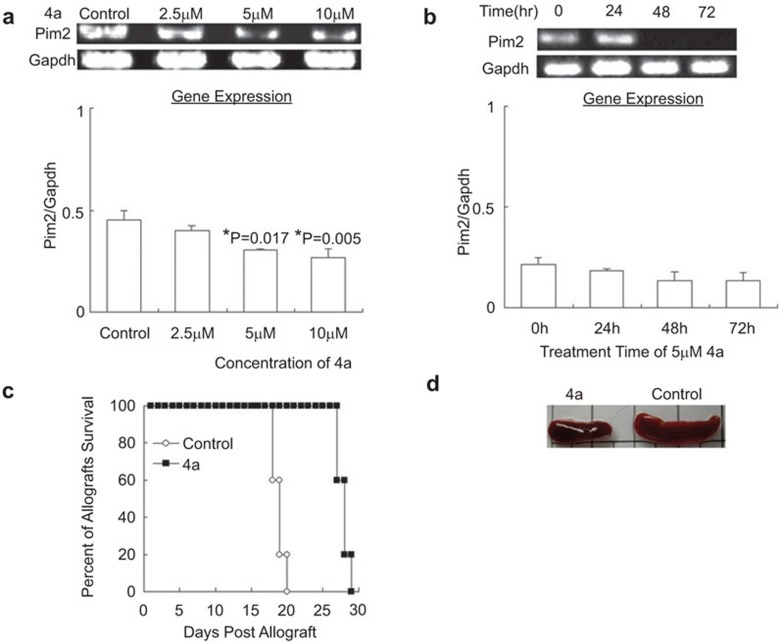
Next, we investigated the effect of blocking Pim2 kinase on allograft survival. After determination of a suitable concentration of 4a, naive BALB/c mice were treated with different concentrations of 4a for 24, 48 and 72 h, followed by analysis of pim2 mRNA from spleen T cells. The results showed that pim2 mRNA was decreased in a dose-dependent manner, with a significant effect shown in the group treated with 5 µM 4a (Figure 4a), but that little change was observed among the different treatment timepoints (P>0.05) (Figure 4b).
Figure 4.
Adoptive transfer of 4a-treated T cell-prolonged allograft survival. (a) Effect of 4a at various concentrations for 24 h on pim2 mRNA expression in spleen cells of naive BALB/c mice. Pim2 mRNA was significantly decreased in 5 or 10 µM 4a-treated cells. *indicates significance (P<0.05) vs. control. (b) Spleen cells of naive BALB/c mice were incubated with 5 µM 4a for different times. No significant differences were found among the different time points (P>0.05). (c) Survival time of skin allograft. T cells from normal BABL/c mice were treated by 5 µM 4a or DMSO (control) for 24 h in vitro and then transferred to allograft-SCID mice, followed by daily monitoring of the grafts (4a vs. control, P<0.05). (d) Spleen sizes from allograft-SCID mice when the skin allograft in the control group was rejected completely. The data are shown as mean±s.e. (n=3). SCID, severe combined immunodeficiency.
The SCID mice were grafted with the skin from B6 mice, and the wounds healed 3 weeks later; the alloimmune response was then reconstituted with the adoptive transfer of 2×107 T cells from naive splenic T cells treated with 5 µM 4a for 24 h or with cells treated with DMSO as a control. All of the mice in the control group had severe rejection at day 19.5±1.7 postcell transfer, while the allograft rejection in the experimental group was observed at day 31±2.3 (Figure 4c). Therefore, blockade of Pim2 significantly postponed acute rejection mediated by T cells. Moreover, we found that there was a twofold decrease in spleen weight compared with the control group (Figure 4d) at same time. As 4a was able to prolong skin allograft survival, abrogating Pim kinase activity could possibly be utilized as a novel approach to prevent graft rejection.
Pim2 prolonged allograft survival by decreasing apoptosis of effector CD4+CD25− T cells
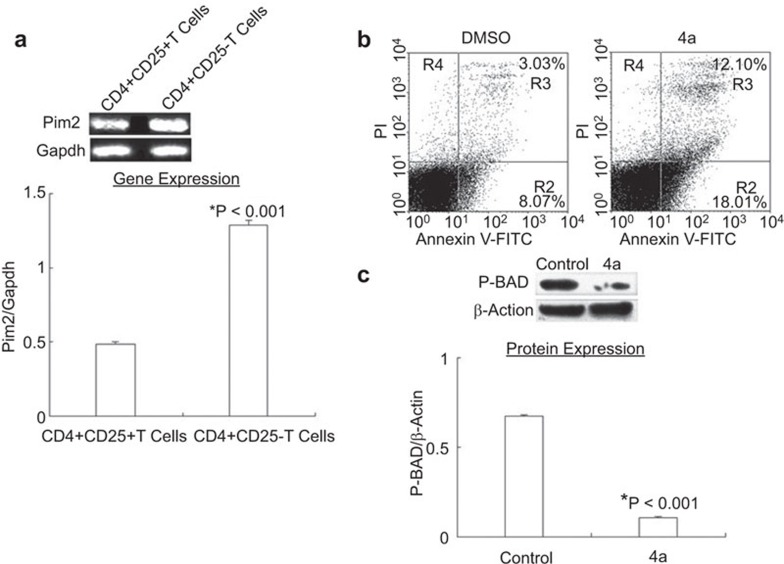
Based on the finding that pim2 was overexpressed in unfractionated CD4+ T cells during acute allograft rejection, we analyzed the expression of Pim2 in different CD4+ T-cell subsets. CD4+ T cells could be divided into effector CD4+CD25− T cells and suppressive CD4+CD25+ T cells.18 It has been reported that pim2 mRNA and protein were readily detected in human CD4+CD25+ T cells14 with a twofold higher expression of pim2 than in CD4+CD25− T-cells,6 whereas other reports have not found any differences between these two subsets.24,25 However, all of the data shown in our experiments demonstrated that pim2 facilitates the allograft rejection, suggesting that this gene may be expressed in effector CD4+CD25− T cells. To prove this hypothesis, CD4+CD25−T cells and CD4+CD25+ T cells were purified from recipient spleens at the height of allorejection, and pim2 expression was detected separately. The results showed that expression of pim2 mRNA in CD4+CD25− T cells was 2.66-fold higher than that of CD4+CD25+ T cells (Figure 5a), which may indicate that Pim2 kinase more strongly promoted CD4+CD25− T-cell survival.
Figure 5.
Pim2 preferably mediated the survival of alloantigen-induced CD4+CD25− T cells. (a) At day 14 postallografting, CD4+CD25− T cells and CD4+CD25+ T cells were purified from allograft mice and pim2 mRNA was measured. CD4+CD25− T cells expressed more pim2 mRNA than the CD4+CD25+ T cells. The data are shown as the mean±s.e. (n=3). *indicates significance (P<0.001) vs. CD4+CD25+ T cells. (b) T cells from allorejected mice was incubated with 10 µM 4a or DMSO for 24 h, stained with annexin V and PI and analyzed by flow cytometry. The data are representative of three independent experiments. (c) P-BAD protein was examined in these T cells by western blotting with β-actin levels as a loading control. The data are shown as mean±s.e. (n=3). p-BAD protein was significantly decreased in 4a-treated cells. (*indicates P<0.05 vs. DMSO).
To further explore the potential effect of Pim2 on the apoptosis of alloreactive T cells, T cells that were purified from recipient spleens at the height of allorejection were subjected to apoptosis analysis after treatment with 5 µM 4a for 24 h. The result demonstrated a notable increase in apoptotic cells in the 4a-treated group from 8.07% to 18.01% (Figure 5b). In addition, the phosphorylation of Bad (Ser112), a Pim2 kinase substrate and apoptotic regulator, was significantly reduced in the 4a-treated group (Figure 5c). These results suggested that the absence of Pim2 activity increased the level of T-cell apoptosis by reducing the phosphorylation of apoptotic regulator Bad (Ser112) during allograft rejection, which indicates that pim2 may promote the allograft rejection by inhibiting T-cell apoptosis.
Pim kinase is required for the activation and function of alloantigen-induced CD4+CD25+ T cells
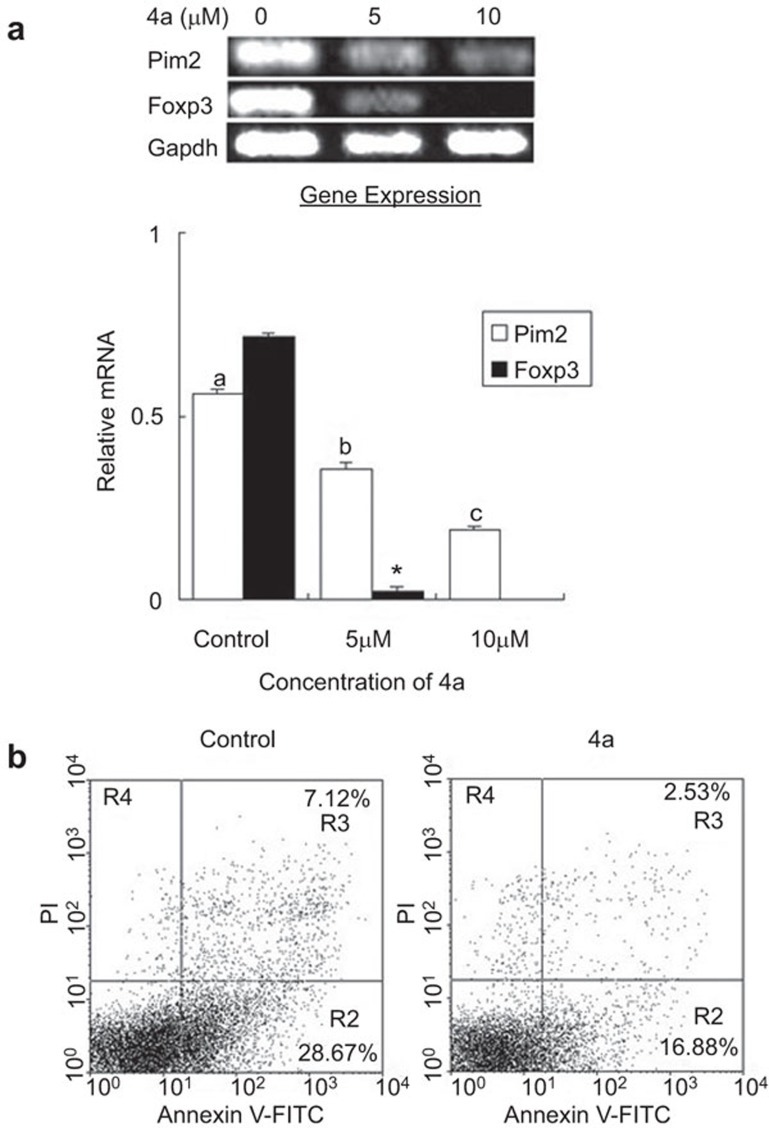
Considering the negative regulation of CD4+CD25+ T cells observed during allorejection, we further investigated the effect of Pim2 on this subset. Expression of Foxp3 and pim2 in splenic T cells isolated from the allograft mice and treated with 5 or 10 µM 4a for 24 h was analyzed. The results showed that 5 or 10 µM 4a treatment resulted in the significant decrease of Foxp3 and pim2 expression (Figure 6a). Because Foxp3 is the key transcriptional factor of CD4+CD25+ T cells, these data may reflect that Pim2 enforced an alloantigen-induced CD4+CD25+FOXP3+ T-cell phenotype and promoted the ex vivo expansion of CD4+CD25+FOXP3+ T cells.
Figure 6.
4a impaired the activation and function of alloantigen-induced CD4+CD25+ T cells. (a) 4a treatment significantly reduced pim2 and Foxp3 mRNA expression in vitro in spleen cells from acute allorejected mice. Control: treated with DMSO for 24 h; 5 µM: treated with 5 µM 4a for 24 h; 10 µM: treated with 10 µM 4a for 24 h. The data are shown as the mean±s.e. (n=3). The values with different letters show significant differences (P<0.001); *indicates significance (P<0.001) vs. control. (b) 4a treatment reduced the ability of CD4+CD25+ T cells to induce CD4+CD25− T-cell apoptosis. CD4+CD25+ T cells were added to CD4+CD25− T cells at a ratio of 1∶6. The data are shown as mean±s.e. (n=3). The data are representative of three independent experiments.
Next, we explored whether Pim2 was required for alloantigen-induced CD4+CD25+ T-cell suppression activity. CD4+CD25−T cells and CD4+CD25+ T cells were purified from the freshly isolated spleens of allograft mice with rejection. CD4+CD25+ T cells treated with or without 5 µM 4a for 24 h were cocultured with CD4+CD25− T cells for 12 h at a ratio of 1∶6 separately, and then examined for cell apoptosis by flow cytometric analysis. The results showed that treatment with 4a led to an obvious decrease in the percentage of apoptosis (from 28.67% to 16.88%) (Figure 6b), thus suggesting that the blocking of Pim2 impaired the function of CD4+CD25+ T cells to induce CD4+CD25− T-cell apoptosis. In other words, the alloantigen-induced CD4+CD25+ T cells appear to display high suppressive activity in the presence of Pim2. This result is consistent with the report that Foxp3 induces pim2 expression, allowing CD4+CD25+ T cells to evade many rapamycin-imposed signaling blocks and to expand preferentially.14 Collectively, Pim2 is indispensable for the ex vivo activation and function of alloantigen-induced CD4+CD25+ T cells, which has opened a new method for the ex vivo expansion of CD4+CD25+ T cells.
Discussion
Transplanted organs are exposed to various types of cellular stresses including ischemia-reperfusion injury and allograft rejection. These conditions can significantly increase the apoptotic load of allografts and may foster maladaptive healing and scarring. Apoptotic cells can transfer bioactive molecules through the release of apoptotic bodies that in turn shape the phenotype and functions of the recipient cells. Apoptosis may also affect the innate and adaptive immune responses, shifting the balance between tolerance and rejection of allogenic tissue. Increasing evidence16,28 shows that the Pim2 target is common to many anti-apoptotic pathways. The pim1, pim2, and pim3 oncogenes belong to a serine/threonine kinase family. It has been shown that Pim2 phosphorylates BAD on serine 112, thus reversing BAD-induced cell death.16,29 The prosurvival mechanism of Pim2 is of great biological and clinical significance as it may be associated with the growth, invasion, and progression of a variety of tumors.
In this study, we investigated the role of Pim2 in acute allograft rejection and the effect of a Pim2 kinase inhibitor on allograft survival. We found that pim2 was increasingly expressed as the degree of the allograft rejection intensified. Specifically, CD4+CD25− T cells expressed more pim2 mRNA than CD4+CD25+ T cells during allograft rejection. The expression difference of pim2 in CD4+ T cells subsets could be attributed to differences in the upstream activation and regulation of Pim2 kinase. Generally, Pim2 is undetectable in naive CD4+CD25− T cells, while resting Foxp3-expressing Tregs constitutively express Pim2, conferring a replicative advantage in cultures containing rapamycin.14 However, CD4+ T cells were activated by the alloantigens when acute allograft rejection occurred. Alloresponse CD4+ T cells secrete a large amount of IL-2 and express IL-2 receptor. The triggering of the IL-2 receptor stimulates the Janus family kinases Jak1 and Jak3 and induces the tyrosine phosphorylation and DNA binding of the STAT (signal transducer and activator of transcription) family transcription factors STAT5 and STAT3.27 The STAT proteins serve as transcription factors for the pim genes and upregulate pim expression by binding to the pim promoter.16 Thus, our finding of the high expression of pim2 during allorejection conforms with other reports. In contrast, transcriptional factor Foxp3 that maintains the function of Tregs inhibits IL-2 gene transcription.12 Therefore, we presumed that pim2 expression is tightly regulated by cytokine-induced JAK/STAT pathways in CD4+CD25− T cells, while pim2 expression is induced by Foxp3 in CD4+CD25+ T cells during acute allograft rejection. This report identified a critical role of Pim2 as a specific serine-threonine kinase that is associated with transplantation rejection.
Here, we reported that 4a prolonged allograft survival for approximately 12 days rather than inducing long-term tolerance. Multiple factors may contribute to this outcome, including the efficacy and specificity of 4a for Pim2 kinase, the microenvironment factors, the adoptive cell number and viability, and, most importantly, the alternative survival signaling pathways that exist in activated T cells. Pim2 and Akt-1 are critical components of overlapping but independent pathways, either of which is sufficient to promote the growth and survival of hematopoietic cells.20 This has led to the speculation that inhibitors of Pim2 would be more effective in inducing allograft tolerance in combination with mTOR-specific inhibitors. Despite the established clinical efficacy of rapamycin in delaying allotransplant rejection, it stays only partially immunosuppressive for allograft rejection. The complete immunosuppression by rapamycin in the allograft rejection model may require the concomitant inhibition of Pim2 activity. Thus, further investigations should aim to evaluate the therapeutic utility of a combination of a Pim2 inhibitor and rapamycin, which may represent a new strategy to augment the therapeutic efficacy of rapamycin in the induction of allograft tolerance.
In addition, it was found that Pim2 was required for the proliferation and function of alloantigen-induced CD4+CD25+ T cells in this study, suggesting a considerable role for Pim2 in the therapeutic efficacy of in vitro expanded Tregs. Although the therapeutic potential of Tregs in preventing allograft rejection has been well documented, the technical issues of Treg purification and in vitro expansion for in vivo use to induce transplantation tolerance remain to be optimized.30 In many studies, only an inhibitor and not an activator of Pim2 has been applied. It has been inferred that an activator of Pim2 might significantly restore the survival and function of Treg cells in vitro. Therefore, the use of Pim2 in expanding Treg cultures is a promising method to enable adoptive Treg cell therapy.
Current research is focused on the identification and functional characterization of pim2 pathways during acute allograft rejection. Given the known Pim2 kinase overexpression profiles, PIM inhibitors will continue to emerge as novel anti-rejection therapeutics and may have potential utility for many other illnesses such as cancer and inflammatory diseases, as described in the ‘immunologic constant of rejection'.
In conclusion, in this study, we first defined the role of Pim2 in acute allograft rejection and evaluated the therapeutic potential of Pim2 inhibition for graft survival. These data suggested that pim2-related T-cell apoptosis played an important role in allograft rejection and that pim2 may be a new target for the diagnosis and therapy of allorejection. Moreover, we found that pim2 was highly expressed on CD4+CD25− T cells and that the inhibition of Pim2 could impair the function of CD4+CD25+ T cells in inducing CD4+CD25− T-cell apoptosis. It is necessary to further understand the effects and the interactions of the different isoforms of the Pim kinases during acute allograft rejection. It will also be important to analyze the direct effect of pim2 on CD4+CD25+ T cells and this gene's relationship with mTOR during allorejection, which may be very useful for the expansion of CD4+CD25+ T cells and for the safe application of rapamycin, an mTOR inhibitor agent being used in clinical organ transplantation.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 81071172, G. Hou) and the Natural Science Foundation of Shandong Province (No. ZR2010CM025, G. Hou).
References
- Spivey TL, Uccellini L, Ascierto ML, Zoppoli G, de Giorgi V, Delogu LG, et al. Gene expression profiling in acute allograft rejection: challenging the immunologic constant of rejection hypothesis. J Transl Med. 2011;9:174. doi: 10.1186/1479-5876-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallet N, Dieudé M, Cailhier J, Hébert M. The molecular legacy of apoptosis in transplantation. Am J Transplant. 2012;12:1378–1384. doi: 10.1111/j.1600-6143.2012.04015.x. [DOI] [PubMed] [Google Scholar]
- Bachmann M, Moroy T. The serine/threonine kinase PIM-1. Int J Biochem Cell Biol. 2005;37:726–730. doi: 10.1016/j.biocel.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Breuer ML, Cuypers HT, Berns A. Evidence for the involvement of PIM-2, a new common proviral insertion site, in progression of lymphomas. EMBO J. 1989;8:743–748. doi: 10.1002/j.1460-2075.1989.tb03434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkers H, Allen J, Knipscheer P, Romeijn L, Hart A, Vink E, et al. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Gen. 2002;32:153–159. doi: 10.1038/ng950. [DOI] [PubMed] [Google Scholar]
- Alvarado Y, Giles FJ, Swords RT. The PIM kinases in hematological cancers. Expert Rev Hematol. 2012;5:81–96. doi: 10.1586/ehm.11.69. [DOI] [PubMed] [Google Scholar]
- Xia Z, Knaak C, Ma J, Beharry ZM, McInnes C, Wang W, et al. Synthesis and evaluation of novel inhibitors of Pim-1 and Pim-2protein kinases. J Med Chem. 2009;52:74–86. doi: 10.1021/jm800937p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beharry Z, Zemskova M, Mahajan S, Zhang F, Ma J, Xia Z, et al. Novel benzylidene-thiazolidine-2,4-diones inhibit Pim protein kinase activity and induce cell cycle arrest in leukemia and prostate cancer cells. Mol Cancer Ther. 2009;8:1473–1483. doi: 10.1158/1535-7163.MCT-08-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa F, Schiopu A, Wood KJ. Role of T cells in graft rejection and transplanttation tolerance. Expert Rev Clin Immunol. 2010;6:155–169. doi: 10.1586/eci.09.64. [DOI] [PubMed] [Google Scholar]
- Rocha PN, Plumb TJ, Crowley SD, Coffman TM. Effector mechanisms in transplant rejection. Immunol Rev. 2003;196:51–64. doi: 10.1046/j.1600-065x.2003.00090.x. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic selftolerance maintained by activated T cells expressing IL-2 receptor alphachains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995;155:1151–1164. [PubMed] [Google Scholar]
- Kaser T, Gerner W, Hammer SE, Patzl M, Saalmüller A. Phenotypic and functional characterisation of porcine CD4+CD25high regulatory T cells. Vet Immunol Immunopathol. 2008;122:153–158. doi: 10.1016/j.vetimm.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Golshayan D, Jiang S, Tsang J, Garin MI, Mottet C, Lechler RI. In vitro-expanded donor alloantigen-specific CD4+CD25+ regulatory T cells promote experimental transplantation tolerance. Blood. 2007;109:827–835. doi: 10.1182/blood-2006-05-025460. [DOI] [PubMed] [Google Scholar]
- Basu S, Golovina T, Mikheeva T, June CH, Riley JL. Foxp3-mediated induction of pim2 allows human T regulatory cells to preferentially expand in rapamycin. J Immunol. 2008;180:5794–5798. doi: 10.4049/jimmunol.180.9.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyasu S. The role of PI3K in immune cells. Nat. Immunol. 2003;4:313–319. doi: 10.1038/ni0403-313. [DOI] [PubMed] [Google Scholar]
- Fox CJ, Hammerman PS, Cinalli RM, Master SR, Chodosh LA, Thompson CB. The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev. 2003;17:1841–1854. doi: 10.1101/gad.1105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Bluestone JA. The Foxp3+ regulatory T cell: a jack of all trades, master of regulation. Nat Immunol. 2008;9:239–244. doi: 10.1038/ni1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Plain KM, Verma N, Robinson C, Boyd R, Hodgkinson SJ, et al. The cellular basis of cardiac allograft rejection. IX. Ratio of naive CD4+CD25+ T cells/CD4+CD25− T cells determines rejection or tolerance. Transpl Immunol. 2006;15:311–318. doi: 10.1016/j.trim.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Golovina TN, Mikheeva T, Brusko TM, Blazar BR, Bluestone JA, Riley JL. Retinoic acid and rapamycin differentially affect and synergistically promote the ex vivo expansion of natural human T regulatory cells. PLoS One. 2011;6:e15868. doi: 10.1371/journal.pone.0015868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerman PS, Fox CJ, Birnbaum MJ, Thompson CB. Pim and Akt oncogenes are independent regulators of hematopoietic cell growth and survival. Blood. 2005;105:4477–4483. doi: 10.1182/blood-2004-09-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox CJ, Hammerman PS, Thompson CB. The Pim kinases control rapamycin-resistant T cell survival and activation. J Exp Med. 2005;201:259–266. doi: 10.1084/jem.20042020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aho TL, Lund RJ, Ylikoski EK, Matikainen S, Lahesmaa R, Koskinen PJ. Expression of human pim family genes is selectively up-regulated by cytokines promoting T helper type 1, but not T helper type 2 cell differentiation. Immunology. 2005;116:82–88. doi: 10.1111/j.1365-2567.2005.02201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wang D, Zhang C, Song J, Liang T, Jin W, et al. Alternatively expressed genes identified in the CD4+ T cells of allograft rejection mice. Cell Transpl. 2011;20:333–350. doi: 10.3727/096368910X552844. [DOI] [PubMed] [Google Scholar]
- Learn CA, Fecci PE, Schmittling RJ, Xie W, Karikari I, Mitchell DA, et al. Profiling of CD4+, CD8+, and CD4+CD25+CD45RO+FoxP3+ T cells in patients with malignant glioma reveals differential expression of the immunologic transcriptome compared with T cells from healthy volunteers. Clin Cancer Res. 2006;12:7306–7315. doi: 10.1158/1078-0432.CCR-06-1727. [DOI] [PubMed] [Google Scholar]
- Sugimoto N, Oida T, Hirota K, Nakamura K, Nomura T, Uchiyama T, et al. Foxp3-dependent and-independent molecules specific for CD25+CD4+ natural regulatory T cells revealed by DNAmicroarray analysis. Int Immunol. 2006;18:1197–1209. doi: 10.1093/intimm/dxl060. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing andmature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- Macintyre AN, Finlay D, Preston G, Sinclair LV, Waugh CM, Tamas P, et al. Protein kinase B controls transcriptional programs that directcytotoxic T cell fate but is dispensable for T cell metabolism. Immunity. 2011;34:224–236. doi: 10.1016/j.immuni.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Lugt NM, Domen J, Verhoeven E, Linders K, van der Gulden H, Allen J, et al. Proviral tagging in E mu-myc transgenic mice lacking the Pim-1 proto-oncogene leads to compensatory activation of Pim-2. Embo J. 1995;14:2536–2544. doi: 10.1002/j.1460-2075.1995.tb07251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B, Zemskova M, Holder S, Chin V, Kraft A, Koskinen PJ, et al. The PIM-2 kinase phosphorylates BAD on serine 112 andreverses BAD-induced cell death. J Biol Chem. 2003;278:45358–45367. doi: 10.1074/jbc.M307933200. [DOI] [PubMed] [Google Scholar]
- Muller YD, Golshayan D, Ehirchiou D, Wekerle T, Seebach JD, Bühler LH. T regulatory cells in xenotransplantation. Xenotransplantation. 2009;16:121–128. doi: 10.1111/j.1399-3089.2009.00531.x. [DOI] [PubMed] [Google Scholar]