Abstract
Suberoylanilide hydroxamic acid (SAHA), as a histone deacetylase (HDAC) inhibitor (HDACi), was recently found to exhibit an immunosuppressive effect. However, whether SAHA can synergize with calcineurin inhibitors (CNIs) to inhibit allograft rejection and its underlying mechanism remain elusive. In this study, we demonstrated the synergistic effects of SAHA and non-therapeutic dose of tacrolimus (FK506) in prolonging the allograft survival in a murine cardiac transplant model. Concomitant intragraft examination revealed that allografts from SAHA-treated recipients showed significantly lower levels of IL-17 expression, and no discernable difference for IL-17 expressions was detected between SAHA- and SAHA/FK506-treated allograft as compared with allografts from FK506-treated animals. In contrast, administration of FK506 significantly suppressed interferon (IFN)-γ but increased IL-10 expression as compared with that of SAHA-treated animals, and this effect was independent of SAHA. Interestingly, SAHA synergizes with FK506 to promote Foxp3 and CTLA4 expression. In vitro, SAHA reduced the proportion of Th17 cells in isolated CD4+ T-cell population and decreased expressions of IL-17A, IL-17F, STAT3 and RORγt in these cells. Moreover, SAHA enhances suppressive function of regulatory T (Treg) cells by upregulating the expression of CTLA-4 without affecting T effector cell proliferation, and increased the proportion of Treg by selectively promoting apoptosis of T effector cells. Therefore, SAHA, a HDACi, may be a promising immunosuppressive agent with potential benefit in conjunction with CNI drugs.
Keywords: allograft rejection, HDAC inhibitor, Th17, Treg
Introduction
There is compelling evidence that the application of immunosuppressive drugs has significantly improved short-term solid organ graft survival in transplantation, but prolongation of long-term allograft survival is still a formidable challenge.1 Since the advent of calcineurin inhibitors (CNIs), marked reduction for the rates of acute rejection and improvement of 1-year graft survival have been achieved.2 Despite the beneficial effect of CNIs on renal allografts,3 long-term use of CNIs has been reported to be associated with many side effects such as renal toxicity, neurotoxicity and diabetegenesis, which in turn affects the survival rate of allografts.4 Some studies further revealed that CNIs alone might cause Th17/regulatory T (Treg) imbalance by increasing Th17/Treg ratio associated with renal dysfunction even rejection.5,6 Therefore, CNI agents are now believed to be necessary for prevention of allograft rejection but preferably with reduced doses.7 Indeed, the imbalance of Th17/Treg has been shown to be associated with the development of inflammatory disorders8,9,10 and allograft rejection.11 Treg cells contribute to the induction and maintenance of tolerance of recipients to allografts,12 while Th17 cells are important to mediate chronic allograft rejection.13 Therefore, therapeutic strategies aiming at manipulating Th17/Treg balance are now considered to be the most promising approach to prevent allograft rejection and to induce tolerance.
Alterations in histone acetylation are associated with changes in chromatin structure and transcriptional regulation of genes essential for fundamental biological processes such as cell cycle progression and/or apoptosis.14,15,16 The levels for histone acetylation are currently believed to be controlled by the steady state balance of two classes of epigenetic enzymes, histone deacetylases (HDACs) and histone acetyltransferases.17,18 Other than chromatin proteins, transcription factors and cell-signaling regulatory proteins could also be the substrates for HDACs.19 Particularly, HDAC superfamily has been receiving increasing attention for its role in modulating both innate and adaptive immune responses20,21 implicatd in allograft survival and transplantation outcome.22 As such, HDAC inhibitors (HDACis) are now under intensive study for their feasibility as potential anti-rejection agents.23,24 Suberoylanilide hydroxamic acid (SAHA), an FDA-approved drug for the treatment of cutaneous T-cell lymphoma,25 is an HDACi composed of hydroxamic acid compound that can be well tolerated by the cancer patients.26,27 While its role in anti-cancer therapy has been extensively studies, its impact on transplant rejection is yet to be explored.
Previously, we demonstrated that HDAC inhibitors can block cell cycle progression and induce apoptosis in bladder cancer cells.28 In the present study, we explored the impact of SAHA on cardiac allograft rejection. We noted that SAHA synergizes with tacrolimus (FK506) to prevent murine cardiac allograft rejection, in which SAHA enhances the proportion of Treg cells by inducing T effector (Teff) cell apoptosis.
Materials and methods
Mice and ethics statement
Male BALB/C and C57BL/6 mice (4–6 weeks, weight 15–20 g) were obtained from the Joint Ventures Sipper BK Experimental Animal Company (Shanghai, China). All animal experiments were performed in accordance with the guidelines of National Institute of Health Guide for the Care and Use of Laboratory Animals, and approved by the Scientific Investigation Board of Second Military Medical University (Shanghai, China).
Reagents
SAHA was purchased from Cayman (San Diego, CA, USA; disolved in 0.1% dimethylsulfoxide (DMSO) medium, and stored at −20 °C at a concentration of 10 mmol/l); FK506 was obtained from Astellas Pharma, Inc. (Tokyo, Japan; dissolved in methanol and freshly prepared by dilution with culture medium). Mouse CD4+ T cell Isolation Kit II and CD4+CD25+ Regulatory T Cell Isolation Kit were purchased from Miltenyi Biotec (Aubum, CA, USA). Antibodies against FOXP3 and CTLA-4 antibody were provided by Abcam (Cambridge, MA, USA). Horseradish peroxidase-labeled goat anti-mouse IgG and FITC fluorescence-labeled goat anti-mouse IgG were purchased from BioLegend (San Diego, CA, USA). Transforming growth factor (TGF)-β1, IL-6, IL-1β, anti-IFN-γ, anti-IL-4, and IL-23 were purchased from PeproTech (Rocky Hill, NJ, USA). PMA, Brefeldin A and ionomycin were from Sigma (St Louis, MO, USA). ELISA kits for mouse IL-2, interferon (IFN)-γ, IL-4 and IL-10 were purchased from eBioscience (San Diego, CA, USA). PCR primers were synthesized by Fudan Yueda Biotechnology Company (Shanghai, China).
Cardiac transplantation and histopathological examination
Cervical heterotopic heart transplantation was performed as described.29 Recipient mice were injected intraperitoneally with SAHA (50 mg/kg/day), FK506 (1 mg/kg/day) or SAHA (50 mg/kg/day) along with FK506 (1 mg/kg/day), respectively. The study end point was defined as complete cessation of cardiac beat. Survival rates of cardiac grafts were monitored by two independent observers without prior knowledge of the treatment protocols by palpation, followed by confirmation with histological analysis. Cardiac grafts were harvested at indicated time points, fixed in 10% formalin and embedded in paraffin. Sections were cut at 4 µm and counterstained for 1 min with hematoxylin–eosin. Infiltrating cells and myocardial destruction were counted under high power field (×40) by using the software Image-Pro Plus (Version 6.0.0.260).
Cell isolation and culture
Splenic CD4 T cells were isolated with anti-CD4 microbeads through positive selection by AutoMACS (Miltenyi Biotec) as instructed. CD4+CD25− and CD4+CD25+ T cells were isolated from CD4 T cells as described previously.30 Isolated T cells were cultured in RPMI1640 supplemented with 10% fetal calf serum (FCS).
Quantitative real-time PCR (qPCR)
Total RNA was extracted from allografts using the TRIzol (Invitrogen, Carlsbad, CA, U.S.) reagents according to the manufacturer's instructions. cDNA was synthesized using an oligo d(T) primer (Applied Biosystems, Foster City, California, U.S.) with a SuperScript III Reverse Transcriptase Kit (Invitrogen). A StepOne™ Real-Time PCR System (Applied Biosystems) and a SYBR RT-PCR kit (Takara, Japan) were used for quantitative real-time RT-PCR analysis. All reactions were conducted in a 20 μl reaction volume in triplicates. The relative expression levels for a target gene were normalized by GAPDH. The specificity of qPCR was verified by melting curve analysis and agarose gel electrophoresis. Primer sequences used in RT-PCR analysis are shown in Table 1.
Table 1. RT-PCR primer sequence.
| Primer | Sequence | 5′→3′ |
|---|---|---|
| TNF-α | F | AAGCCTGTAGCCCACGTCGTA |
| R | GGCACCACTAGTTGGTTGTCTTTG | |
| RORγt | F | TCACCTGACCTACCCGAGG |
| R | TCCAAGAGTAAGTTGGCCGTC | |
| GAPDH | F | AGGTCGGTGTGAACGGATTTG |
| R | TGTAGACCATGTAGTTGAGGTCA | |
| IL-17A | F | TCGCCATTCAGCAAGAAATCC |
| R | CACAGGTGCAGCCAACTTTTA | |
| IL-17F | F | TGCTACTGTTGATGTTGGGAC |
| R | AATGCCCTGGTTTTGGTTGAA | |
| STAT3 | F | CTGCTGCCGCTTTTTCCAG |
| R | GGAAGCACTCAGTAAGATGACG | |
| CD11b | F | ATGGACGCTGATGGCAATACC |
| R | TCCCCATTCACGTCTCCCA | |
| IFN-γ | F | GAACTGGCAAAAGGATGGTGA |
| R | TGTGGGTTGTTGACCTCAAAC | |
| CTLA-4 | F | AGAACCATGCCCGGATTCTG |
| R | CATCTTGCTCAAAGAAACAGCAG | |
| IL-4 | F | GGTCTCAACCCCCAGCTAGT |
| R | GCCGATGATCTCTCTCAAGTGAT | |
| FOXP3 | F | TCAAGTACCACAATATGCGACC |
| R | CCATCGGATAAGGGTGGCA |
Abbreviations: IFN, interferon; TNF, tumor-necrosis factor.
Western blot analysis
The cells were washed twice with cold phosphate-buffered saline (PBS) and then lysed in cell lysis buffer (Cell Signaling Technology, Danvers, Massachusetts, U.S.) supplemented with a protease inhibitor cocktail (Calbiochem, Darmstadt, Germany.). Protein concentrations for lysates were determined by BCA assay (Pierce, Rockford, IL, USA). Equal amount of cell lysates were loaded and subjected to SDS–PAGE. The separated proteins were next transferred onto nitrocellulose membranes, and then analyzed by Western blotting as previously described.31
Flow cytometry
Anti-CD4-PE, anti-CD25-FITC, anti-CD4-FITC, anti-IL-17A-PE, anti-Foxp3-PE, anti-CTLA4-APC, Annexin V-FITC and CFSE were purchased from either eBiosciences or BioLegend. Intracellular stainings for FOXP3, CTLA-4 and IL-17A were performed using the relevent Fix/Perm Buffer set as instructed. The intensity of intracellular fluorescence was measured using the LSR II software (BD, Franklin Lakes, New Jersey, U.S.), all data were analyzed using the Flowjo software (TreeStar, Ashland, U.S.).
Statistical analysis
Data from multiple groups were analyzed using one-way ANOVA with post hoc Bonferroni's correction (GraphPad Prism 5.0; GraphPad Software, La Jolla, CA, U.S.). Data derived from two groups were analyzed using an unpaired Student's t-test or a Mann–Whitney test (two-tailed). All data were expressed as mean±s.e.m. In all cases, P<0.05 was considered with statistical significance.
Results
SAHA prolongs the survival time of murine cardiac allografts
We first sought to address the impact of SAHA on allograft rejection, and a fully MHC-mismatched (BALB/C→C57BL/6) murine cervical heterotopic cardiac transplant model was employed for the study. Transplantation of syngeneic grafts (C57BL/6→C57BL/6) was served as controls. It was noted that cardiac arrest occurred within seven days in recipients treated with control vehicle DMSO, and in sharp contrast, adminstration of SAHA and FK506 prolonged allograft median survival time to 10 and 16 days, respectively (Figure 1a). Interestingly, adminstration of low dose of SAHA along with low dose of FK506 prolonged allograft median survival time to 27 days (Figure 1a), indicating that SAHA synergizes with FK506 to prevent allograft rejection.
Figure 1.
SAHA prolongs the cardiac allograft survival in murine model. (a) Fully MHC-mismatched cardiac allograft recipients (BALB/C→C57BL/6) were treated with DMSO, SAHA (50 mg/kg/day), FK506 (1 mg/kg/day) or SAHA–FK506 combination by intraperitoneal injection since the day receiving transplantation until when cardiac arrest occurred. Graft survival was assessed every day. (b, c) Pathological examination of allografts harvested on day 7 post-transplantation. Shown are section graphs from the low power field (b) and high power field (c). NC, negative control represented cardiac graft from isotransplantation. (d) Statistical analysis of the pathological graphs in (c). Infiltrating cells (left panel) and myocardial destruction (right panel) per high field were counted from four mice per group. *P<0.05, **P<0.01, compared with DMSO group. DMSO, dimethylsulfoxide; FK506, tacrolimus; SAHA, suberoylanilide hydroxamic acid.
To confirm the above results, three allografts from each study group were harvested on day 7 post-transplantion and subjected to histological analysis. In line with the above results, severe inflammatory infiltration and myocardial destruction were present in DMSO-treated allografts, while grafts from syngeneic transplantation remained intact myocardial structure (Figure 1b). Importantly, although administration of SAHA significantly suppressed inflammatory infiltration, we still observed impaired myocardial structure in the allografts. On the contrary, adminstration of SAHA along with FK506 has almost completely prevented inflammatory infiltration without perceptible changes for myocardial structure (Figure 1b–c). Together, these data support that SAHA synergizes with non-therapeutic dose of FK506 to prmote the survival of MHC fully mismatched cardiac allografts.
SAHA regulates the balance of Th17/Treg
Next, we examined the impact of SAHA on the expression of inflammatory cytokines and regulatory molecules. To this end, allografts were harvested on day 7 after transplantation for qPCR analysis. Compared with allografts derived from DMSO or FK506 treated animals, allografts from SAHA-treated recipients showed significantly lower levels of IL-17 expression, and in consistent with this result, no discernable difference for IL-17 expressions was detected between SAHA- and SAHA/FK506treated allografts (Figure 2a), indicating that the reduced IL-17 expression was a direct effect caused by SAHA. In contrast, administration of FK506 significantly suppressed IFN-γ (Figure 2b) but increased IL-10 (Figure 2c) expression as compared with that of DMSO or SAHA treated animals, and this effect was independent of SAHA, as we failed to detect a significantly difference between allografts-treated with FK506 alone and that treated with SAHA/FK506. Interestingly, a synergizing effect was noted between SAHA and FK506 on the expression of regulatory molecules. For example, SAHA synergizes with FK506 to promote Foxp3 (Figure 2d) and CTLA4 (Figure 2e) expression, while to suppress CD11b expression (Figure 2f).
Figure 2.
The effect of SAHA on the expressions of inflammatory cytokines and immunomolecules. (a–f) The allografts were harvested 7 days after transplantation and were homogenized. Foxp3, CTLA-4, IL-17, CD11b, IL-10 and IFN-γ mRNA were measured by qPCR. The data are expressed as mean±s.d. (n=3). *P<0.05, **P<0.01, compared with DMSO group. NC, negative control represented cardiac graft from isotransplantation. (g–h) The recipient thymus, draining lymph nodes and spleens were harvested 7 days after transplantation. CD4+ T cells were isolated and CD25+ and Foxp3+ T cells were measured by flow cytometry. Shown are representative of three separate experiments. DMSO, dimethylsulfoxide; FK, tacrolimus; IFN, interferon; qPCR, quantitative PCR; SAHA, suberoylanilide hydroxamic acid.
Given the role of Foxp3 and CTLA4 played in Treg cells, the above results prompted us to examine the number of Treg cells in animals of each study group. The recipient mice were sacrificed on day 7 after transplantation to harvest thymus, spleen and draining lymph nodes for analysis of Foxp3+ Treg cells by flow cytometry. Indeed, SAHA treatment significantly increased the proportion of Foxp3+ Treg cells among total CD4+ T cells in the thymus, lymph nodes and spleen (Figure 2g) along with enhanced Foxp3 expression as determined by mean fluorescence intensity (Figure 2h).
To address whether the prolonged allograft survival was solely caused by the upregulation of Treg cells, we transplanted BALB/C-derived cardiac grafts into Foxp3 deficient B6 recipient mice. To our surprise, although loss of Foxp3 attenuated allograft survival, but failed to completely eradicate the impact of SAHA on allograft survival (Figure 3a), suggesting that Treg cells only partly involved in SAHA-mediated protection against allograft rejection.
Figure 3.
SAHA regulates Th17/Treg balance. (a) Cardiac transplant was performed with Foxp3−/− B6 mice as recipients (BALB/C→Foxp3−/− C57BL/6) as described in Figure 1a. Shown was the graft survival curve. (b) Naive CD4+ T cells were isolated from mice spleen, and polarized for Th17 differentiation in the presence of different concentration of SAHA. IL-17A expressing CD4+ T cells were measured by flow cytometry. Shown are representative of three separate experiments. (c) The effect of SAHA on the expressions of STAT3, RORγt and Foxp3 measured by western blots. Shown are representative of three separate experiments. (d) Gene expressions of IL-17A, IL-17F, STAT3, RORγt, Foxp3 and CTLA-4 in CD4+ T cells measured by qPCR. The data are expressed as mean±s.d. (n=3). *P<0.05, **P<0.01, compared with Th17 group. DMSO, dimethylsulfoxide; qPCR, quantitative PCR; SAHA, suberoylanilide hydroxamic acid; Treg, regulatory T.
Since SAHA suppressed IL-17 expression in the allografts independent of FK506, we therefore examined the impact of SAHA on Th17 development. Naïve CD4+ T cells were isolated and then polarized to Th17 lineage in the presence of SAHA or DMSO as previously described.32 Flow cytometry analysis of polarized T cells revealed that SAHA significantly suppressed the production of Th17 cells (Figure 3b), and western blot analysis indicated that SAHA reduced the expression of RORγt and STAT3, and by which it promoted Foxp3 expression (Figure 3c). In line with these results, qPCR analysis further confirmed that SAHA downregulated the expression of IL-17A, IL-17F, STAT3 and RORγt, but upregulated Foxp3 and CTLA-4 expressions (Figure 3d). Taken together, our data suggest that SAHA regulates the balance between Th17 and Treg cells, through which it prolongs allograft survival.
SAHA enhances Treg function without affecting Teff responsiveness
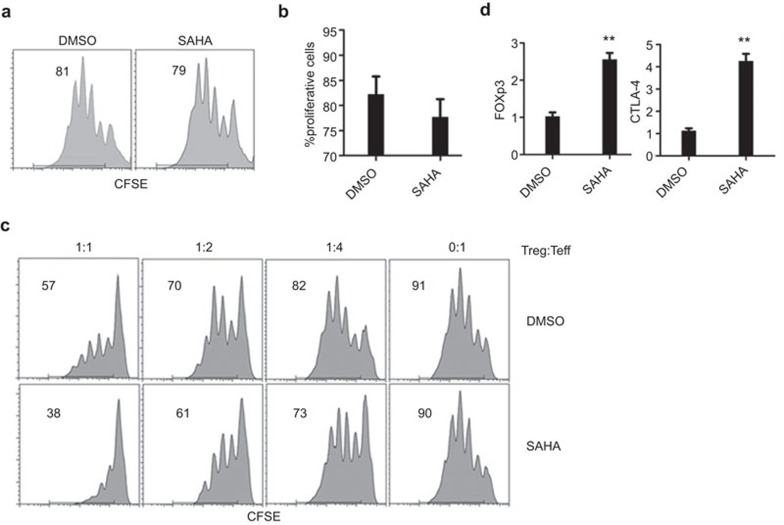
To further dissect the mechanisms underlying SAHA protection of allograft rejection, we further examined its impact on Teff cells. We isolated CD4+CD25− T cells from the spleens of recipient mice on day 7 after transplantation, and examined their proliferation in the presence of anti-CD3/CD28 antibodies. Flow cytometry analysis demonstrated a similar proliferation potency between SAHA and DMSO treated CD4+CD25− T cells (Figure 4a and b), suggesting that SAHA does not affect Teff cell responsiveness. Next, we examined the suppressive function of CD4+CD25+ Treg cells isolated from SAHA- and DMSO-treated recipient mice, respectively. It was found that CD4+CD25+ T cells derived from SAHA-treated mice showed significantly higher potency to suppress CD4+CD25− T cell proliferation as compared with their DMSO-treated counterparts (Figure 4c). Consistently, significantly higher expressions for Foxp3 and CTLA-4 were observed for Treg cells derived from SAHA treated mice as compared with that derived from DMSO-treated mice (Figure 4D). Collectively, these data suggest that SAHA specifically enhances the suppressive function of Treg cells without affecting the responsiveness of Teff cells.
Figure 4.
SAHA enhances Treg cell function without affecting T effector responsiveness. On the seventh post-transplant day, CD4+CD25+ and CD4+CD25− T cells were isolated from the recipient spleen with different treatment. (a) CD4+CD25− T cells were stimulated with anti-CD3/CD28 antibodies; the cell proliferation was examined by flow cytometry. (b) Statistical analysis for CD4+CD25− T cells proliferation in (a). The data are expressed as mean±s.d. (n=4). *P<0.05, **P<0.01. (c) CD4+CD25+ T cells were mixed with CFSE labeled CD4+CD25− T cells at different ratio. Cells division was measured by flow cytometric analysis. Shown are representative of three separate experiments. (d) Treg cells were collected from SAHA-treated and DMSO-treated mice, and analyzed the expressions of Foxp3 and CTLA-4 by qPCR. The data are expressed as mean±s.d (n=3). *P<0.05, **P<0.01, compared with DMSO group. DMSO, dimethylsulfoxide; qPCR, quantitative PCR; Teff, T effector; Treg, regulatory T; SAHA, suberoylanilide hydroxamic acid.
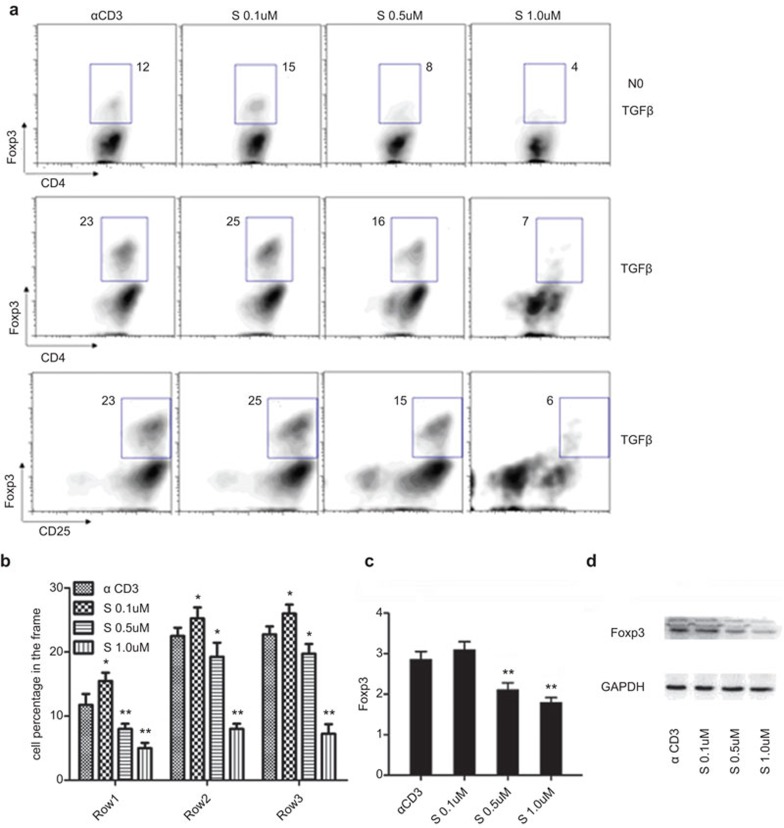
Low dose of SAHA enhances Treg cell proportion without promoting Foxp3 expression
We next intend to determine the impact of SAHA doses on Treg cells production. Unexpectedly, disparate results were obtained when various doses of SAHA were employed. It was consistently found that low dose of SAHA (0.1 μM) enhanced the proportion of CD4+Foxp3+ T cells, while high dose of SAHA (0.5 and 1 μM) significantly suppressed the generation of Foxp3+ T cells (Figure 5a and b). Furthermore, qPCR analysis disclosed that low dose of SAHA did not increase the expression of Foxp3 mRNA, whereas high dose of SAHA markedly inhibited Foxp3 expression (Figure 5c). Western blot analysis further demonstrated a significant downregulation Foxp3 protein levels along with the increase of SAHA doses (Figure 5d).
Figure 5.
Low concentration of SAHA enhances Treg cell proportion without promoting Foxp3 expression in vitro. (a) Naive CD4+ T cells were isolated from mice spleen and activated with anti-CD3/CD28 in the presence or absence of TGF-β, with different concentrations of SAHA. The effect of SAHA on the generation of CD4+Foxp3+ T cells was assessed by flow cytometric analysis. (b) Statistical analysis for the effect of SAHA on the generation of CD4+Foxp3+ T cells in (a). The data are expressed as mean±s.d. (n=4). *P<0.05, **P<0.01, compared with α CD3 group. (c) Foxp3 expression assessed by qPCR. The data are expressed as mean±s.d. (n=3). *P<0.05, **P<0.01, compared with α CD3 group. (d) Foxp3 expression was assessed by western blots. Data are representative of three separate experiments. qPCR, quantitative PCR; TGF, transforming growth factor; Treg, regulatory T; SAHA, suberoylanilide hydroxamic acid.
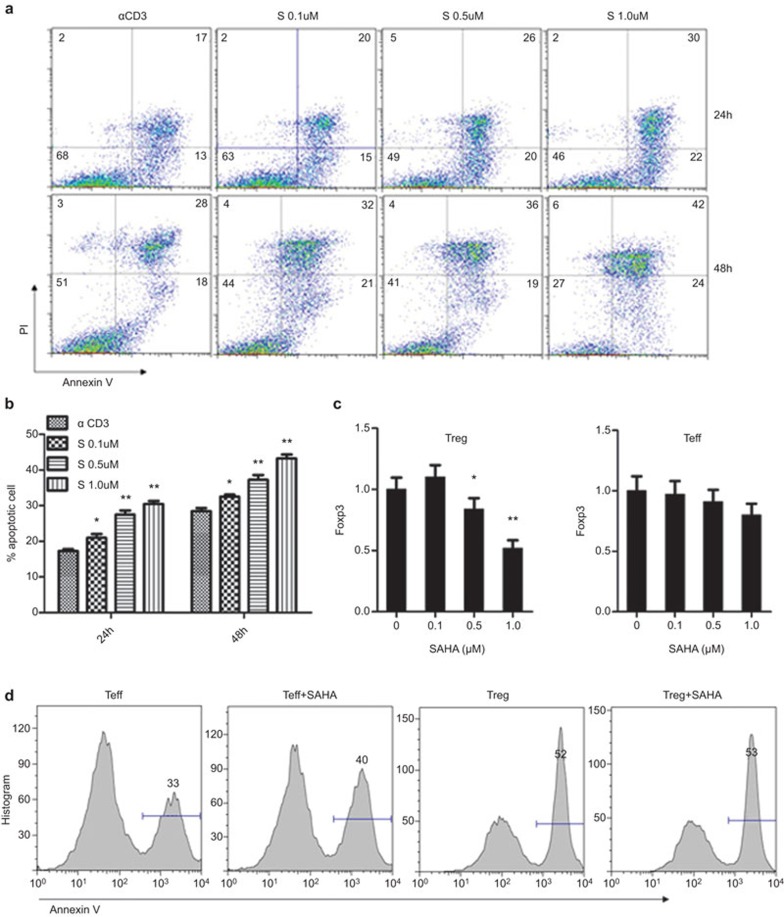
Low dose of SAHA selectively induces Teff cell apoptosis
To address why low dose of SAHA did not alter Foxp3 expressions but increased the proportion of Treg cells, we investigated the impact of SAHA on Teff cell apoptosis. Upon anti-CD3/CD28-stimulated activation, around 17% of activated Teff cells underwent apoptosis. Surprisingly, SAHA dose-dependently promoted Teff cell apoptosis (Figure 6a and b). Given that low dose of SAHA failed to enhance Foxp3 mRNA, we assume that the increase for the proportion of Treg cells upon low dose of SAHA treatment was caused by enhanced Teff cell apoptosis or Teff cell to Treg cell conversion. To test this notion, we isolated CD4+CD25+ Treg and CD4+CD25− non-Treg cells using magnetic beads, and activated them by anti-CD3 treatment in the presence of SAHA (0.1 μM). qPCR analysis revealed that low dose of SAHA neither upregulated Foxp3 expression nor promoted the conversion of CD4+CD25− T cells to Treg cells (Figure 6c). This result prompted us to examine whether SAHA induced Teff cell apoptosis. Indeed, flow cytometry analysis confirmed that low dose of SAHA selectively promoted the apoptosis of Teff cells, but without a perceptible impact on Treg cells (Figure 6d). Together, these data suggest that low dose of SAHA enhances the proportion of Foxp3+ T cells by promoting Teff cell apoptosis.
Figure 6.
Low concentration of SAHA selectively induces apoptosis of Teff cells in vitro. (a) Naive CD4+ T cells were isolated from mice spleen, Treg (CD4+CD25+) and Teff (CD4+CD25−) cells were then purified using magnetic beads and stimulated with anti-CD3/CD28 in the presence of different concentrations of SAHA for 24 or 48 h, cells apoptosis were analyzed by flow cytometry with PI and Annexin V staining. (b) Statistical analysis for the effect of SAHA on cell apoptosis in (a). The data are expressed as mean±s.d. (n=3). *P<0.05, **P<0.01, compared with α CD3 group. (c) Foxp3 expression in Treg cells (left) and Teff cells (right). The data are expressed as mean±s.d. (n=3). *P<0.05, **P<0.01, compared with SAHA (0 μM) group. (d) Apoptosis of Teff cells and Treg cells in low concentration of SAHA. Data are representatives of three separate experiments. qPCR, quantitative PCR; SAHA, suberoylanilide hydroxamic acid; Teff, T effector; Treg, regulatory T.
Discussion
In clinics, immunosuppressive agents are usually combinedly used for post transplant recipients to enhance suppressive effect, and at the same time, to reduce side effects caused by high-dose drug administration. As a cornerstone of immunosuppression in transplantation, the administration of CNI agents markedly reduced the incidence of acute rejection by blocking the mainstream of T-cell signal transduction,33 making the management of chronic rejection the main task for transplant physicians. Unfortunately, the precise mechanism of chronic rejection remains elusive. We therefore put forward the hypothesis that the chronic rejection is possibly mediated by the activation of some bypasses under the circumstances of long-term CNI use and therapeutic inhibition of these bypasses by some mild immunosuppressants might be beneficial to prolong graft survival.
HDACis, which are initially used in clinic as anticancer agents, have a broad effect on the biology of a variety of functional cells. Previous studies have demonstrated that HDAC is also actively involved in some inflammatory diseases,34,35 making these agents hold promise as immunomodulatory drugs. HDACis, such as FR276457 and TSA, have been reported to prolong the median survival time of the transplanted grafts in a rodent animal model as a monotherapy, and even more effective in combination with FK506 or rapamycin,23,36 which is consistent with our observation in the current study. Obviously, a combination of SAHA and FK506 is more effective than either of them alone in the prevention of allograft rejection, which lead us to explore the pharmacological mechanism of SAHA.
Over the past years, the roles of Treg cells in prevention of transplant rejection and induction of immune tolerance has drawn an increasing attention from many investigators.37 Theraputic manipulation of Foxp3 acetylation using HDACis was demonstrated to promote the development and suppressive functions of Treg cells, with beneficial results obtained in models of colitis,38 arthritis39 and transplant rejection.40 However, the understanding about the effect of HDACis on Treg cells are still controversy. Some studies revealed that HDACis could upregulate the expression of Foxp3 in CD4+ T cells, thus promoting the conversion of naive T cells to Treg cells in vitro,41,42,43 while Liu et al.44 argued that trichostatin A (TSA) reduces the expression of Foxp3 and the number of Treg cells both in vitro and in vivo. Our results are not in agreement with any of the published studies. In the current study, we demonstrated disparate effects of SAHA on Foxp3 expression under varying SAHA concentrations. High dose of SAHA (0.5 and 1 μM) significantly suppressed the generation of Foxp3+ T cells, while low dose (0.1 μM) of SAHA enhances the proportion of Treg in CD4+ T-cell population by selectively inducing apoptosis of Teff without affecting Treg cells under in vitro circumstances. These data not only perfectly explain the conflicting results from previous studies of HDACis' effect on Tregs, but also indicate that attention should be paid to the dose of SAHA in its potential clinical application in the future.
The involvement of CD4+ cells in immune responses of transplant rejection has long been established,45 of which Th2 polarization is believed to protect allograft from rejection due to the ability of IL-4 for inhibition of Th1 differentiation.46 Besides, as a newly described subset of CD4+ T cells, Th17 has been proved to play a key role in the pathogenesis of inflammatory and immune diseases.47 Bosisio et al.48 has revealed that HDACis could inhibit the Th1- and Th17-inducing potential of dentritic cells by decreasing the production of relevant cytokines. However, there is still rare report concerning the regulation of HDACis on Th17 differentiation. In this study, the intragraft examination revealed that SAHA upregulated the expressions of Foxp3 and CTLA-4, but downregulated CD11b and IL-17. As CD11b is associated with the activation of monocytes and neutrophilic granulocytes,49 the downregulation of CD11b revealed an improvement on intragraft infiltration. As Th17 are not the only IL-17 producing cell,50 we isolated CD4+ T cells and confirmed the downregulation of IL-17 in CD4+ T cell subset by FCM analysis in vitro. Although IL-17 has already been shown to participate in the pathogenesis of chronic rejection in a murine cardiac transplant model, yet our results in this study seem to be a challenge to Itoh's report,51 in which gamma delta (γδ) T cells were believed to be the predominant source of IL-17 production. Moreover, SAHA treatment also down-regulates the expression of STAT3, which is believed to be a master regulator of Th17, including cell differentiation and cytokine production.52 All these results may serve as evidence for the hypothesis that SAHA probably prevents allograft rejection through modulating the balance between Treg and Th17 and drives the balance towards Treg deviation.
Acknowledgments
This work was supported by grants from the National Key Basic Research Program of China (2009CB522402) and the National Natural Science Foundation of China (31170856, U0832009 and 30772046).
References
- Nankivell BJ, Kuypers DR. Diagnosis and prevention of chronic kidney allograft loss. Lancet. 2011;378:1428–1437. doi: 10.1016/S0140-6736(11)60699-5. [DOI] [PubMed] [Google Scholar]
- Hariharan S, Johnson CP, Bresnahan BA, Taranto SE, McIntosh MJ, Stablein D. Improved graft survival after renal transplantation in the United States, 1988 to 1996. N Engl J Med. 2000;342:605–612. doi: 10.1056/NEJM200003023420901. [DOI] [PubMed] [Google Scholar]
- Pascual M, Theruvath T, Kawai T, Tolkoff-Rubin N, Cosimi AB. Strategies to improve long-term outcomes after renal transplantation. N Engl J Med. 2002;346:580–590. doi: 10.1056/NEJMra011295. [DOI] [PubMed] [Google Scholar]
- Casey MJ, Meier-Kriesche HU. Calcineurin inhibitors in kidney transplantation: friend or foe. Curr Opin Nephrol Hypertens. 2011;20:610–615. doi: 10.1097/MNH.0b013e32834b4343. [DOI] [PubMed] [Google Scholar]
- van de Wetering J, Koumoutsakos P, Peeters A, van der Mast BJ, de Kuiper P, Ijzermans JN, et al. Discontinuation of calcineurin inhibitors treatment allows the development of FOXP3+ regulatory T-cells in patients after kidney transplantation. Clin Transplant. 2011;25:40–46. doi: 10.1111/j.1399-0012.2010.01311.x. [DOI] [PubMed] [Google Scholar]
- Li Y, Shi Y, Huang Z, Bai Y, Niu Q, Cai B, et al. CNI induced Th17/Treg imbalance and susceptibility to renal dysfunction in renal transplantation. Int Immunopharmacol. 2011;11:2033–2038. doi: 10.1016/j.intimp.2011.08.015. [DOI] [PubMed] [Google Scholar]
- Ponticelli C, Scolari MP. Calcineurin inhibitors in renal transplantation still needed but in reduced doses: a review. Transplant Proc. 2010;42:2205–2208. doi: 10.1016/j.transproceed.2010.05.036. [DOI] [PubMed] [Google Scholar]
- Zeng C, Shi X, Zhang B, Liu H, Zhang L, Ding W, et al. The imbalance of Th17/Th1/Tregs in patients with type 2 diabetes: relationship with metabolic factors and complications. J Mol Med (Berl) 2011;90:175–186. doi: 10.1007/s00109-011-0816-5. [DOI] [PubMed] [Google Scholar]
- Shen Y, Tang XY, Yang YC, Ke X, Kou W, Pan CK, et al. Impaired balance of Th17/Treg in patients with nasal polyposis. Scand J Immunol. 2011;74:176–185. doi: 10.1111/j.1365-3083.2011.02546.x. [DOI] [PubMed] [Google Scholar]
- Niu Q, Cai B, Huang ZC, Shi YY, Wang LL.Disturbed Th17/Treg balance in patients with rheumatoid arthritis. Rheumatol Int 2011. in press. [DOI] [PubMed]
- Hanidziar D, Koulmanda M. Inflammation and the balance of Treg and Th17 cells in transplant rejection and tolerance. Curr Opin Organ Transplant. 2010;15:411–415. doi: 10.1097/MOT.0b013e32833b7929. [DOI] [PubMed] [Google Scholar]
- Wood KJ. Regulatory T cells in transplantation. Transplant Proc. 2011;43:2135–2136. doi: 10.1016/j.transproceed.2011.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abadja F, Sarraj B, Ansari MJ. Significance of T helper 17 immunity in transplantation. Curr Opin Organ Transplant. 2012;17:8–14. doi: 10.1097/MOT.0b013e32834ef4e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Mol Oncol. 2007;1:19–25. doi: 10.1016/j.molonc.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- Hake SB, Xiao A, Allis CD. Linking the epigenetic ‘language' of covalent histone modifications to cancer. Br J Cancer. 2007;96 Suppl:R31–R39. [PubMed] [Google Scholar]
- Singh BN, Zhang G, Hwa YL, Li J, Dowdy SC, Jiang SW. Nonhistone protein acetylation as cancer therapy targets. Expert Rev Anticancer Ther. 2010;10:935–954. doi: 10.1586/era.10.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger T, Lugrin J, Le Roy D, Goy G, Mombelli M, Koessler T, et al. Histone deacetylase inhibitors impair innate immune responses to Toll-like receptor agonists and to infection. Blood. 2011;117:1205–1217. doi: 10.1182/blood-2010-05-284711. [DOI] [PubMed] [Google Scholar]
- Grabiec AM, Tak PP, Reedquist KA. Function of histone deacetylase inhibitors in inflammation. Crit Rev Immunol. 2011;31:233–263. doi: 10.1615/critrevimmunol.v31.i3.40. [DOI] [PubMed] [Google Scholar]
- Schildberg FA, Hagmann CA, Bohnert V, Tolba RH. Improved transplantation outcome by epigenetic changes. Transpl Immunol. 2010;23:104–110. doi: 10.1016/j.trim.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- Moreira JM, Scheipers P, Sorensen P. The histone deacetylase inhibitor Trichostatin A modulates CD4+ T cell responses. BMC Cancer. 2003;3:30. doi: 10.1186/1471-2407-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codd R, Braich N, Liu J, Soe CZ, Pakchung AA. Zn(II)-dependent histone deacetylase inhibitors: suberoylanilide hydroxamic acid and trichostatin A. Int J Biochem Cell Biol. 2009;41:736–739. doi: 10.1016/j.biocel.2008.05.026. [DOI] [PubMed] [Google Scholar]
- Marks PA, Jiang X. Histone deacetylase inhibitors in programmed cell death and cancer therapy. Cell Cycle. 2005;4:549–551. doi: 10.4161/cc.4.4.1564. [DOI] [PubMed] [Google Scholar]
- Marks PA, Richon VM, Miller T, Kelly WK. Histone deacetylase inhibitors. Adv Cancer Res. 2004;91:137–168. doi: 10.1016/S0065-230X(04)91004-4. [DOI] [PubMed] [Google Scholar]
- Qu W, Kang YD, Zhou MS, Fu LL, Hua ZH, Wang LM. Experimental study on inhibitory effects of histone deacetylase inhibitor MS-275 and TSA on bladder cancer cells. Urol Oncol. 2010;28:648–654. doi: 10.1016/j.urolonc.2008.11.018. [DOI] [PubMed] [Google Scholar]
- Wang Q, Liu Y, Li XK. Simplified technique for heterotopic vascularized cervical heart transplantation in mice. Microsurgery. 2005;25:76–79. doi: 10.1002/micr.20082. [DOI] [PubMed] [Google Scholar]
- Xiao L, Fu ZR, Liu F, Zhang LD, Shi XM, Shen XY, et al. Suppression of allograft rejection by Tim-1-Fc through cross-linking with a novel Tim-1 binding partner on T cells. PLoS ONE. 2011;6:e21697. doi: 10.1371/journal.pone.0021697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Hou J, Zhou J, Zhao W, Xu H, Zheng Y, et al. Phosphatase SHP-1 promotes TLR- and RIG-I-activated production of type I interferon by inhibiting the kinase IRAK1. Nat Immunol. 2008;9:542–550. doi: 10.1038/ni.1604. [DOI] [PubMed] [Google Scholar]
- Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- Clipstone NA, Crabtree GR. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- McKinsey TA. The biology and therapeutic implications of HDACs in the heart. Handb Exp Pharmacol. 2011;206:57–78. doi: 10.1007/978-3-642-21631-2_4. [DOI] [PubMed] [Google Scholar]
- Ito K, Ito M, Elliott WM, Cosio B, Caramori G, Kon OM, et al. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N Engl J Med. 2005;352:1967–1976. doi: 10.1056/NEJMoa041892. [DOI] [PubMed] [Google Scholar]
- Kinugasa F, Yamada T, Noto T, Matsuoka H, Mori H, Sudo Y, et al. Effect of a new immunosuppressant histon deacetylase (HDAC) inhibitor FR276457 in a rat cardiac transplant model. Biol Pharm Bull. 2008;31:1723–1726. doi: 10.1248/bpb.31.1723. [DOI] [PubMed] [Google Scholar]
- Wang L, de Zoeten EF, Greene MI, Hancock WW. Immunomodulatory effects of deacetylase inhibitors: therapeutic targeting of FOXP3+ regulatory T cells. Nat Rev Drug Discov. 2009;8:969–981. doi: 10.1038/nrd3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zoeten EF, Wang L, Sai H, Dillmann WH, Hancock WW. Inhibition of HDAC9 increases T regulatory cell function and prevents colitis in mice. Gastroenterology. 2010;138:583–594. doi: 10.1053/j.gastro.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saouaf SJ, Li B, Zhang G, Shen Y, Furuuchi N, Hancock WW, et al. Deacetylase inhibition increases regulatory T cell function and decreases incidence and severity of collagen-induced arthritis. Exp Mol Pathol. 2009;87:99–104. doi: 10.1016/j.yexmp.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zoeten EF, Wang L, Butler K, Beier UH, Akimova T, Sai H, et al. Histone deacetylase 6 and heat shock protein 90 control the functions of Foxp3+ T-regulatory cells. Mol Cell Biol. 2011;31:2066–2078. doi: 10.1128/MCB.05155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon C, Kim SH, Park KS, Choi BK, Lee HS, Park JB, et al. Use of epigenetic modification to induce FOXP3 expression in naive T cells. Transplant Proc. 2009;41:1848–1854. doi: 10.1016/j.transproceed.2009.02.101. [DOI] [PubMed] [Google Scholar]
- Lucas JL, Mirshahpanah P, Haas-Stapleton E, Asadullah K, Zollner TM, Numerof RP. Induction of Foxp3+ regulatory T cells with histone deacetylase inhibitors. Cell Immunol. 2009;257:97–104. doi: 10.1016/j.cellimm.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Johnson J, Pahuja A, Graham M, Hering B, Hancock WW, Bansal-Pakala P. Effects of histone deacetylase inhibitor SAHA on effector and FOXP3+ regulatory T cells in rhesus macaques. Transplant Proc. 2008;40:459–461. doi: 10.1016/j.transproceed.2008.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zhang C, Sun J. Deacetylase inhibitor trichostatin A down-regulates Foxp3 expression and reduces CD4+CD25+ regulatory T cells. Biochem Biophys Res Commun. 2010;400:409–412. doi: 10.1016/j.bbrc.2010.08.090. [DOI] [PubMed] [Google Scholar]
- Chen N, Gao Q, Field EH. Prevention of Th1 response is critical for tolerance. Transplantation. 1996;61:1076–1083. doi: 10.1097/00007890-199604150-00016. [DOI] [PubMed] [Google Scholar]
- Tay SS, Plain KM, Bishop GA. Role of IL-4 and Th2 responses in allograft rejection and tolerance. Curr Opin Organ Transplant. 2009;14:16–22. doi: 10.1097/MOT.0b013e32831ebdf5. [DOI] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- Bosisio D, Vulcano M, del Prete A, Sironi M, Salvi V, Salogni L, et al. Blocking TH17-polarizing cytokines by histone deacetylase inhibitors in vitro and in vivo. . J Leukoc Biol. 2008;84:1540–1548. doi: 10.1189/jlb.0708401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DL, Griggs KM, Bersten AD, de Pasquale CG. Systemic inflammation and cell activation reflects morbidity in chronic heart failure. Cytokine. 2011;56:593–599. doi: 10.1016/j.cyto.2011.08.029. [DOI] [PubMed] [Google Scholar]
- Cai Y, Shen X, Ding C, Qi C, Li K, Li X, et al. Pivotal role of dermal IL-17-producing gammadelta T cells in skin inflammation. Immunity. 2011;35:596–610. doi: 10.1016/j.immuni.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh S, Nakae S, Axtell RC, Velotta JB, Kimura N, Kajiwara N, et al. IL-17 contributes to the development of chronic rejection in a murine heart transplant model. J Clin Immunol. 2010;30:235–240. doi: 10.1007/s10875-009-9366-9. [DOI] [PubMed] [Google Scholar]
- Harris TJ, Grosso JF, Yen HR, Xin H, Kortylewski M, Albesiano E, et al. Cutting edge: An in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J Immunol. 2007;179:4313–4317. doi: 10.4049/jimmunol.179.7.4313. [DOI] [PubMed] [Google Scholar]