Abstract
Binding of a neurotransmitter to its ionotropic receptor opens a distantly located ion channel, a process termed allosteric activation. Here we review recent advances in the molecular mechanism by which the cys-loop receptors are activated with emphasis on the best studied nicotinic acetylcholine receptors (nAChRs). With a combination of affinity labeling, mutagenesis, electrophysiology, kinetic modeling, electron microscopy (EM), and crystal structure analysis, the allosteric activation mechanism is emerging. Specifically, the binding domain and gating domain are interconnected by an allosteric activation network. Agonist binding induces conformational changes, resulting in the rotation of a β sheet of amino-terminal domain and outward movement of loop 2, loop F, and cys-loop, which are coupled to the M2–M3 linker to pull the channel to open. However, there are still some controversies about the movement of the channel-lining domain M2. Nine angstrom resolution EM structure of a nAChR imaged in the open state suggests that channel opening is the result of rotation of the M2 domain. In contrast, recent crystal structures of bacterial homologues of the cys-loop receptor family in apparently open state have implied an M2 tilting model with pore dilation and quaternary twist of the whole pentameric receptor. An elegant study of the nAChR using protonation scanning of M2 domain supports a similar pore dilation activation mechanism with minimal rotation of M2. This remains to be validated with other approaches including high resolution structure determination of the mammalian cys-loop receptors in the open state.
Keywords: cys-loop receptors, allosteric activation, gating domain, receptor binding, coupling, ACh binding protein, nicotinic receptors, GABA receptors, glycine receptors, serotonin receptors
Introduction
The cys-loop receptor family of ligand-gated ion channels has a signature cysteine loop in the amino-terminal domain. This family includes nicotinic receptors (nAChRs), serotonin receptor type 3 (5-HT3R), γ-aminobutyric acid receptors type A and C (GABAA/C), glycine receptors, zinc-activated cation channel, and invertebrate glutamate/serotonin-activated anionic channels or GABA-gated cation channels1, 2, 3. Recently, prokaryotic proton-gated ion channels are also considered to be in the same family although they are devoid of the signature cysteine loop4. All cys-loop receptors are allosteric proteins, in which binding of agonist to the binding pocket in the subunit interface of the extracellular amino-terminal domain controls the distantly located channel domain to open the pore1. This long range coupling between binding pocket and the gating machinery requires an interconnected allosteric network, through which the binding energy can be transduced to the gating energy to open the channel5. Accumulating evidence suggests that the activation mechanisms of this receptor family are likely to be very similar. Thus, we review the activation mechanism of the cys-loop receptor family in general with emphasis on nAChRs.
Kinetic models for channel activation
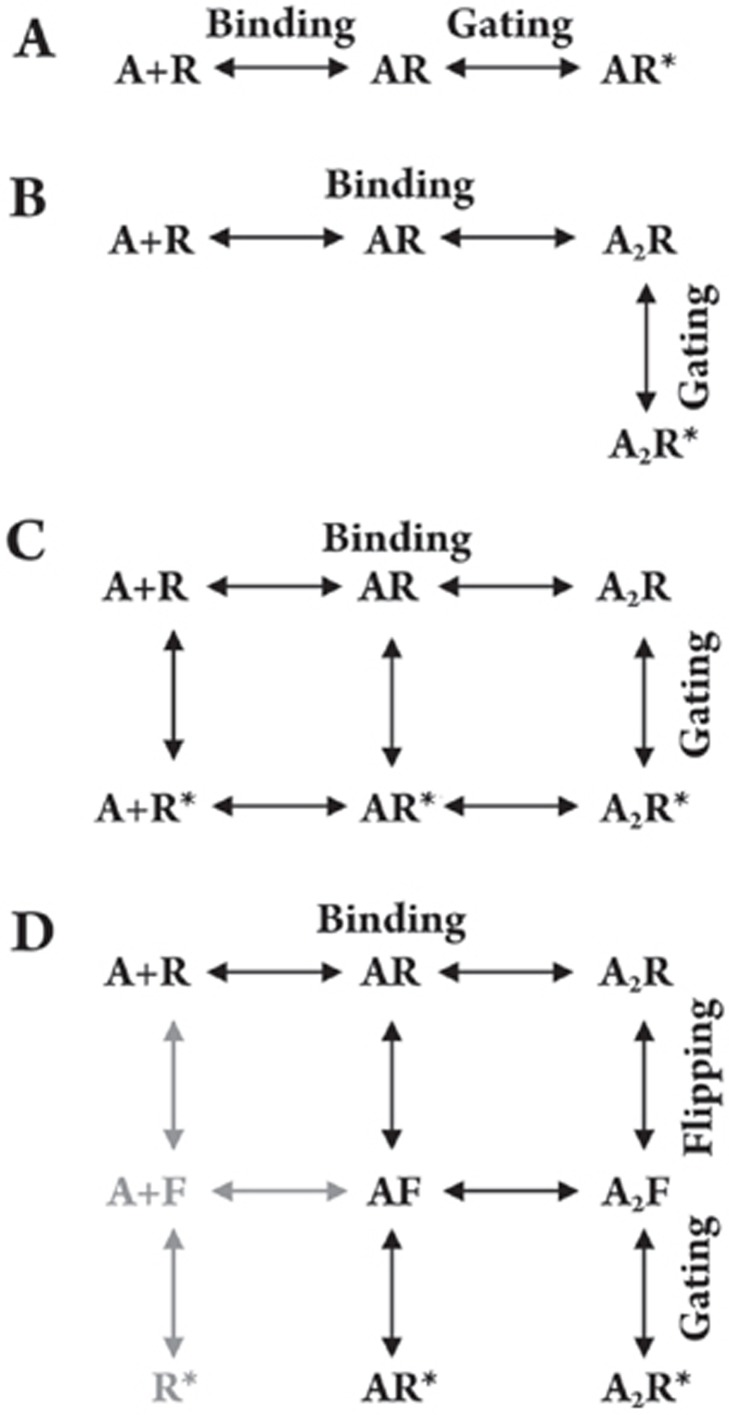
Activation of a ligand-gated ion channel includes binding and gating steps (Figure 1A)6. The Hill slope of dose-response relationships of most heteromeric cys-loop receptors is greater than one, suggesting at least two binding steps for the receptor activation (Figure 1B)7. However, radio ligand binding studies revealed that receptor binding affinities are usually in the nanomolar range, whereas current activations require agonist concentrations in the micromolar range. This long puzzled discrepancy between binding and functional studies is now known to be the result of the difference in functional states of receptors under distinct experimental conditions. Binding measures receptor affinity mainly in the high affinity desensitized state where receptor-ligand interaction has reached equilibrium, whereas electrophysiological recording measures the receptor response at the time of peak current8. In fact, binding and gating are mutually coupled9, except when receptor is in the desensitized state. The binding can influence channel gating, and gating can also alter binding affinity. In the case of a non-desensitizing GABAC receptor, channel opening can increase binding affinity to such an extent that it appears to lock the agonist in the binding pocket10. Desensitization can even increase the binding affinity further for desensitizing receptors8. In addition, the gating influence on binding is further supported by the fact that mutations of gating residues in the pore-lining domain can alter the agonist sensitivity of receptors11, 12, 13. Thus, channel gating involves global conformational changes from the binding pocket to the gating machinery. This phenomenon can be well described by Monod-Wyman-Changeux allosteric activation (MWC) model1, 14, 15 (Figure 1C). Excellent agreement of experimental data to the MWC model further strengthens this conclusion13. A recent study with comparison of the effects of partial agonists and full agonists on channel gating suggested that the increase in binding affinity and channel gating are not a single step. It can be further divided into two sequential steps: the conformational change in the binding domain and then channel opening16. That is, there is an intermediate state, termed “flip state”, with the receptor binding domain switching to a high affinity state before channel opening (Figure 1D). In summary, kinetic studies revealed that binding and gating are coupled with mutual influence with each other. However, the coupling between binding and gating is not a single step through a single rigid body. Instead, activation involves binding, sequential conformational change(s), and gating.
Figure 1.
Activation schemes. (A) linear model with single binding step; (B) a linear model with two binding steps6, typical for all heteromeric cys-loop receptors. However, homomeric receptors have potentially 5 binding steps. In some homomeric receptors (if not all), such as ρ1 GABAC receptor, three bindings are needed to induce significant channel openings75; (C) Monod-Wyman-Changeux allosteric activation (MWC) model13, 14; (D) adapted flip model from Lape et al16. Note that the grayed out states are rare and thus omitted in original scheme. In all models (A–D), A represents agonist; R is receptor; F is flipped state of the receptor; R* is the receptor in open state. All open states and flipped states are high affinity states, whereas resting states are low affinity states.
Functional domains of the cys-loop receptors
Binding pocket
Structure-function studies in the last two decades with combined techniques such as site-directed mutagenesis, photoaffinity labeling, and structural analysis based on electron microscopic (EM) images of tubular arrays of receptors have shaped a structural model for the cys-loop receptors. To date, the best-studied cys-loop receptor is nAChR. The agonist binding sites of the muscle type nAChR are formed in the extracellular amino-terminal domain at subunit interfaces between α and non-α subunits, whereas the binding sites for neuronal type nAChRs are formed in the subunit interface between α and β subunits for heteromeric receptors or between two α subunits in homomeric receptors17. Affinity labeling and site-directed mutagenesis have provided extensive evidence about the agonist binding site. Six loops, designated A through F, appear to participate in formation of the agonist binding pocket 17. Residues from loops A18, B19, and C20, 21, 22 of the α subunit and residues from loops D23, 24, E22, 25, and F26, 27, 28 from another subunit contribute to the formation of the binding pocket in the subunit interface.
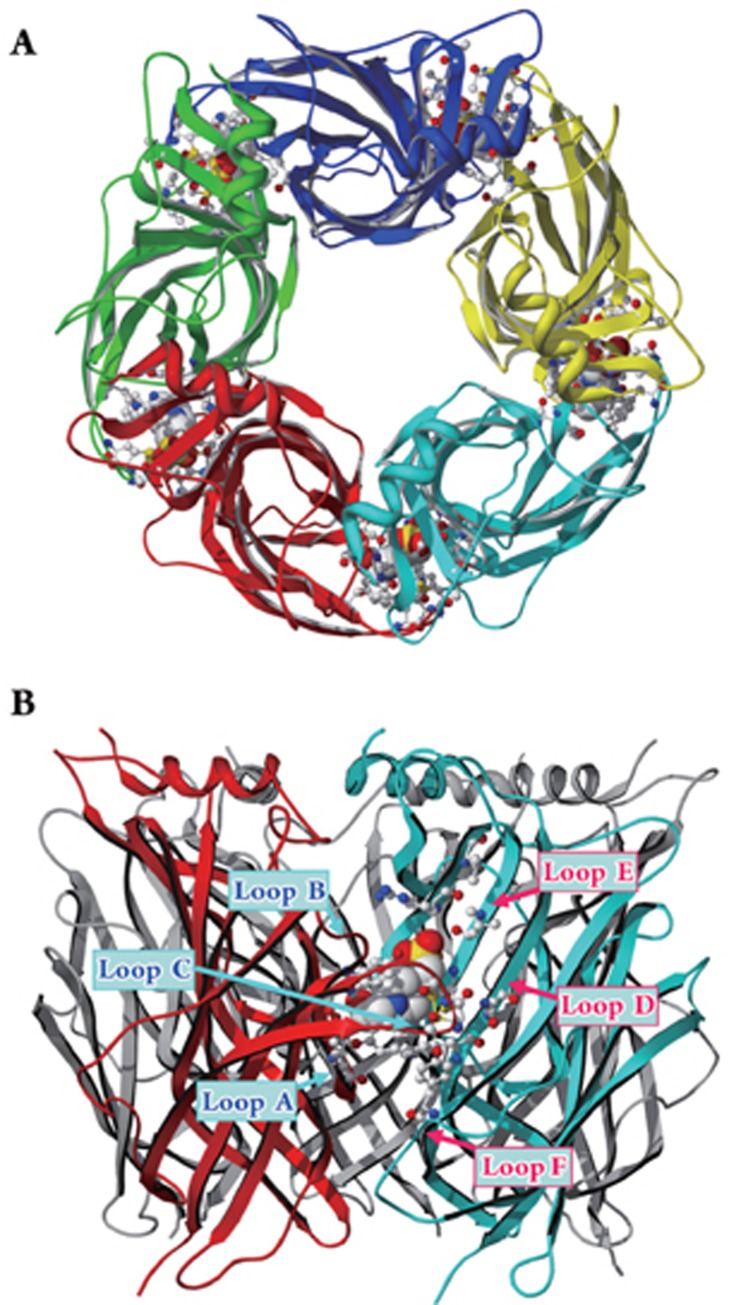
The model of the agonist binding pocket was further validated and extended by high resolution crystal structures of homologous acetylcholine binding proteins, AChBPs29, 30, 31, 32 and 4 Å resolution EM structure of the Torpedo nAChR. In this structural model, the receptor has five subunits with the agonist/antagonist-binding pocket located at the subunit interface (Figure 2A). In the heteromeric nAChRs, there are two binding pockets located in the two subunit interfaces between α and non-α subunits. For heteromeric GABAA receptors, two binding pockets are located between β and α subunits. Note that the β subunit of GABAA receptor is equivalent to the α subunit in nAChR, whereas the α subunit of GABAA receptor is in the position of the β subunit in neuronal nAChRs. For homomeric cys-loop receptors, such as α7 nAChR, ρ1 GABA receptor, α1 glycine receptor and 5-HT3Α receptor, there are 5 potential binding pockets in all five subunit interfaces. All previously identified binding loops (A through F) can be mapped onto the structural model. Loops A, B, and C from one subunit form the principal face of the binding pocket. Loops D, E, and F contributed by an adjacent subunit form the complementary face of the pocket (Figure 2B).
Figure 2.
Crystal structure of the AChBP as a structural model of the amino-terminal domain of cys-loop receptors (created from 1I9B30). (A) Top view of the model showing a pentameric structure with the binding pocket located in the subunit interface; (B) Side view of the model with six binding loops (A through F) labeled. Loops A, B, and C from the left subunit form the principal face of the binding pocket, whereas loops D, E, and F from the right subunit form the complementary face. A HEPES buffer molecule is located in the binding pocket.
Gating machinery
The channel domain is formed by transmembrane domains (M1–M4). Studies using site-directed mutagenesis and ultrastructural analysis have identified the second transmembrane domain (M2) as the pore-lining domain in the cys-loop receptors. Hydrophilic substitutions of the conserved leucine in the mid-point of the M2 domain dramatically influence channel gating kinetics, increase agonist sensitivity, and create spontaneous opening channels in several members of the cys-loop receptors11, 12, 13, 33, 34, 35, 36, 37, 38, 39. Although earlier studies using cysteine accessibility test suggested that the gate is in the intracellular end of M239, 40, it is now clear that the accessibility of these residues in the intracellular end of M2 in the absence of agonist is due to spontaneous opening of the M2 mutant channel41. The EM structure of the Torpedo nAChR at 4Å resolution finally confirmed that the M2 domain is lining the pore, and that the gate is formed by the hydrophobic interactions between amino acid residues in the middle of the M2 domains42 (Figure 3). Mutagenesis studies also revealed that structural elements to control ionic selectivity and single channel conductance are located in the intracellular end (the beginning) of the M2 domain.
Figure 3.
Transmembrane domain and channel gate (created from PDB file of 1OED). (A) top view of transmembrane domains of all five subunits, with the M2 domains lining the pore and form the channel gate (with gate-forming residues highlighted; (B) side view of transmembrane domains with the front subunit removed.
Coupling region
Using correlated mutational analysis, we have identified an allosteric network connecting the binding pocket to the gating machinery in the cys-loop receptor family (Figure 4A, 4B)5. Through this network, binding energy can be transduced to the gating energy to open the channel. The key coupling region in this allosteric network is in the interface between amino-terminal binding domain and transmembrane gating domain for each subunit. A study using a chimeric receptor with AChBP and channel domains of 5HT3R revealed that the coupling interface requires matching of three loops (loop 2, loop 7, and loop 9) from the amino-terminal domain and one loop (M2-M3 linker) from the transmembrane domain for the receptor to be functional43. Additionally, a region in pre-M1 and the beginning of M1 that covalently links the amino-terminal domain to the transmembrane domain, is also important in channel gating (Figure 4C)44.
Figure 4.
The allosteric network and coupling loops linking binding domain to gating domain. (A) principal face of binding pocket with binding residues (red and orange) is linked to the gating machinery (L251 is shown) by the evolutionarily conserved allosteric network (yellow) identified by correlated mutations5. (created from 2BG9 chain A). (B) complementary face of binding pocket (cyan and green) and their relationship with the allosteric network (created from 2BG9 chain A). (C) the region coupling amino-terminal domain and transmembrane domains includes loop 2, loop 7 (cys-loop) and loop 9 (loop F) pre-M1/M1 and M2-M3 linker (2BG9 chain A).
Activation mechanism
As stated in the beginning, activation of the cys-loop receptor family includes binding, conformational changes, and gating steps. Briefly, agonist binding in the binding pocket of the amino-terminal domain initiates a conformational change, which then propagates to the gating machinery through the coupling region to open the channel. Propagation of “conformational wave” from the binding to channel gate is not a single step. It has been studied with single channel analysis and linear free energy relationship of gating rate constants by different mutations at each position. The results have suggested that there is a gradient change in the Φ slope factor of the linear free energy relationship, derived from the channel gating constants (opening and closing rates) of the mutations at each position, along the activation pathway45. Further analysis revealed that the gradient change in allosteric activation network is not continuous. Instead, it can be divided into several clusters based on their values of Φ slope factor. All positions in each cluster have similar Φ values, suggesting that the residues in each cluster influence channel gating similarly. In other words, each cluster probably moves as a rigid body, with synchronous movement of all residues in the cluster. It is also suggested that all residues in each cluster are coupled tightly, and conformational changes between clusters are coupled less tightly. In this scenario, the conformational change induced by agonist binding would stepwisely propagate toward the channel through discrete modules in the amino-terminal domain and finally to the gating machinery to open the channel46. This mechanism is further supported by a recent single channel analysis of partial agonist activation, which further revealed that there is a conformational change, termed flip conformation, preceding the channel opening16. The flip conformation clearly changes binding affinity before channel opening. Partial agonists have less ability to convert the receptor to the flipped high affinity state than full agonists. However, once flip conformation occurs, partial agonists and full agonists gate the channel very similarly. In the following section, we will present the detailed mechanism for this allosteric activation process.
Conformational changes in the amino-terminal domain
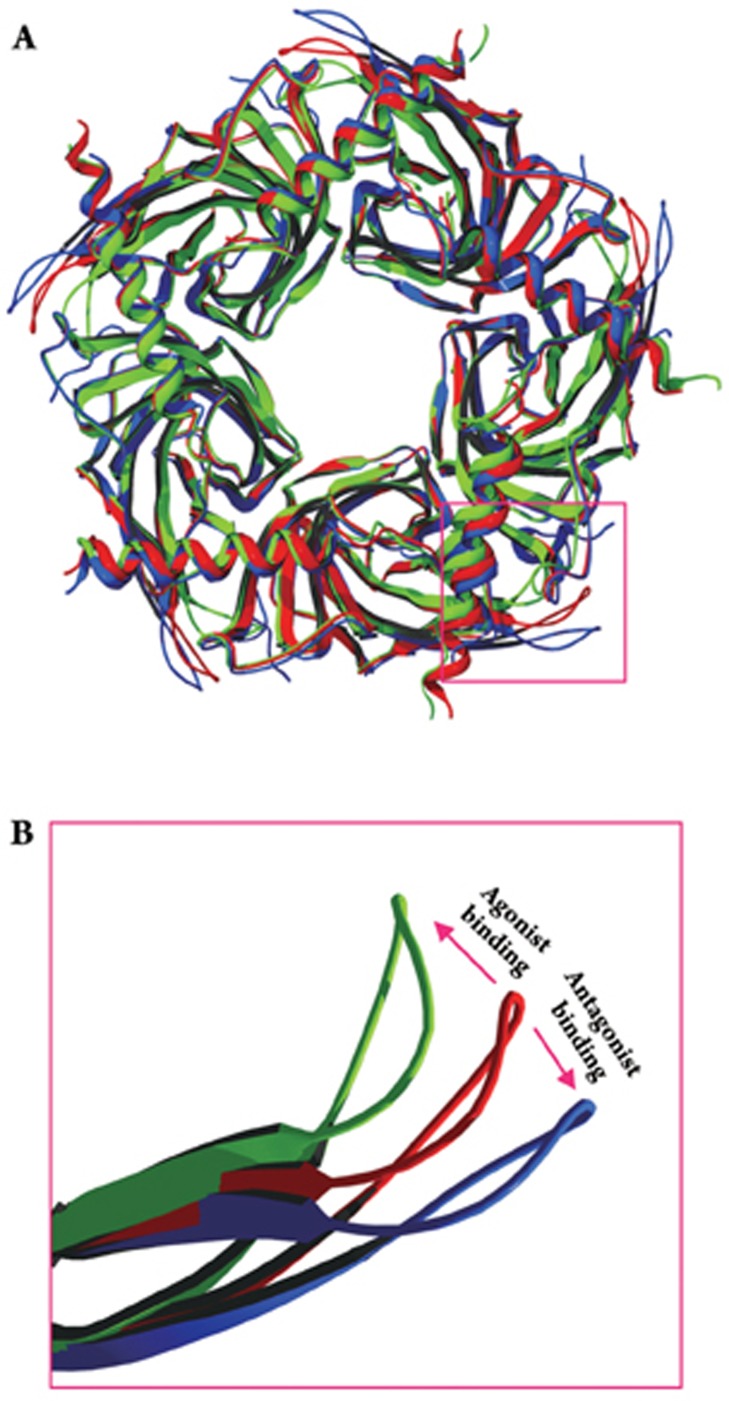
Based on the 4Å EM structure of Torpedo nAChR and comparison between α and non-α subunits, Unwin and colleagues proposed that the activation mechanism of the receptor involves agonist-induced clockwise rotation (viewed from the extracellular end) of the inner sheets in the amino-terminal domains of two α subunits. This rotation of the amino-terminal domain is then translated into the rotation of the M2 domain by direct coupling between the bottom of the inner sheet (loop 2) and top of the M2 domain (or beginning of M2–M3 liker) (Figure 5). However, this proposed mechanism is not based on the agonist-induced structural change. The agonist-induced structural change in the amino-terminal domain is best demonstrated in the crystal structures of AChBPs. When an agonist co-crystallized with the receptor, it induces an inward movement of loop C (also called loop C capping) to tighten the binding pocket (Figure 6)31, 32. This could be related to the increased binding affinity during channel activation10. New hydrogen bond formation between Y185 in loop C and K139 in β7 strand (connecting to cys-loop) in the nicotine bound state of an AChBP may suggest initial coupling. In the case of muscle type nAChR, single channel analysis demonstrated that mutations of αY19047 or αD20048 can influence channel gating. Mutant cycle analysis further revealed that αY190 (homologous to Y185 in AChBP) is coupled to αK145 (homologous to K139 in AChBP) when an agonist binds to the receptor after disrupting the salt-bridge between αD200 in loop B and αK145 in β7 strand in the resting state49. Interestingly, in a GABAA receptor, similar charge interaction between homologous residues (βE153 is at homologous position as αK145, and βK196 is at homologous position of αY190) is critical for channel activation, although with charges reversed50. Thus, while there are some variations in detailed interactions, the general mechanism of activation is conserved in the cys-loop receptor family. In addition, in GABAA receptors, another negatively charged residue in loop B (βE155) is also an important determinant for channel gating. Mutation of this residue created spontaneously opening channels, suggesting it may also serve as a trigger for channel activation51. Since the conformational change of the receptor can be divided into blocks, it is likely that E155 and E153 are in the same rigid body. Loop C also interacts with loop B for the allosteric channel gating through a backbone hydrogen bonding52.
Figure 5.
M2 rotation hypothesis42. Agonist binding induces a rotation of the inner sheet of the amino-terminal domain, which is coupled to the M2 transmembrane domain through the interaction between loop 2 in amino-terminal domain and M2-M3 linker from the transmembrane domain (created from 2BG9 chain A).
Figure 6.
Agonist- and antagonist-induced structural changes in AChBP. (A) AChBPs crystallized in apo state (red, from 2BYN), Epibatidine (agonist)-bound state (green, from 2BYQ) and ImI (antagonist)-bound state (blue, from 2BYP). Three crystal structures were loaded to Swiss PDB viewer 3.7 and fitted with magic fit function. The red box is the location of loop C from one subunit for all three structures. (B) close look of agonist and antagonist-induced movement in loop C32.
Another significant change in the crystal structure of AChBP upon agonist binding is in the binding loop F31. The conformational change of loop F during channel activation is further supported by increased photolabeling of loop F in the α1 subunit of nAChR in the open state, although the direction of the movement is not completely clear53. Mutation of a loop F residue (εD175N) of nAChR clearly influences channel gating, suggesting the importance of the loop F in channel activation54. In the ρ1 GABAC receptor, the outward movement of the lower part of loop F is supported by cysteine accessibility test and fluorescence detection, which is partially coupled to the channel gating as assessed by sensitivity of agonist-induced fluorescence change to a non-competitive antagonist55. Since one arm of loop C is linked to the bottom of loop F, it is possible that loop C inward movement would pry the bottom part of loop F in the same subunit and create an outward movement of it. In addition, upon agonist binding, the backbone of αS191 in loop C can form a hydrogen bond with an aspartate residue (γD174/δD180) in loop F of the complementary face of the muscle type nAChR56. This dynamic hydrogen bonding between loop C and loop F upon agonist binding could pull loop F outward toward loop C. The outward movement of loop F is then potentially coupled to loop 2 and M2–M3 linker to pull the channel open.
Although the conformational changes in loops A, D, and E are not observed in the crystal structures of AChBPs in the presence of agonists, mutagenesis and functional studies in intact channels have suggested that loops A, D, and E are also involved in channel gating. For example, mutation(s) of ɛW55/δW57 in binding loop D of muscle type nAChR dramatically reduces channel opening rate57. Mutations of α7W55 in homomeric α7 nAChR alter channel gating kinetics, with slowed desensitization58. In the case of the ρ1 GABAC receptor, a mutation of the homologous residue (Y102S) in loop D created spontaneously opening channels59, further suggesting the importance of this aromatic residue in loop D in initial conformational changes induced by an agonist. Similarly, in the ρ1 GABAC receptor, a mutation (F146C) in loop A and a mutation in loop E (Q160C) also create spontaneously opening channels60. Unlike AChBP, the amino-terminal domain of a cys-loop receptor is coupled to transmembrane domain. It is likely that in the resting state, the conformation of the amino-terminal domain is different from the resting state of the soluble AChBP protein. Thus, it is understandable that in an intact cys-loop receptor, these three binding loops also undergo conformational rearrangement during channel function. This possibility is further supported by the site-specific fluorescence monitoring during channel activation. For example, in the ρ1 GABAC receptor, GABA-induced fluorescence change was detected in loop E (L166C) and in the top of the receptor (S66C), which can be partially (in L166C) or completely (in S66C) blocked by non-competitive antagonist picrotoxin61. In summary, it appears that all six loops have some contributions to the channel gating, which involves global conformational change in the receptor. It is likely that the coordinated movement of all six binding loops cause inner sheet rotation.
Conformational changes in the gating machinery
EM imaging of the Torpedo nAChR in the open state with 9 Å resolution showed that channel opening involves a rotation of the pore-lining kinked rod structures62. These pore lining rod structure are further confirmed to be second transmembrane domain (M2) by the EM image at 4Å resolution. As mentioned above, kinked M2 domains line the pore and form the channel gate by hydrophobic interaction in the middle of the transmembrane domains. The M2 rotation presumably disrupts the hydrophobic interactions of the gate forming residues and thus widens the pore to allow ions to flow through (Figure 7A).
Figure 7.
M2 rotation model vs M2 tilting and pore dilation model. (A) M2 rotation hypothesis[62] for channel opening (created from 1OED); (B) top view of M2 tilting (created by magic fitting 2BL0 AE chains (closed state with M2 colored with cyan and M3 with blue) to 3EHZ (open state with M2 colored with pink and M3 with red). (C) side view of M2–M3 tilting and pore dilation with front two subunits removed (same crystal structures, fit, and colors as in B). Arrows indicate the direction of M2–M3 tilting.
However, two recent studies using the crystal structure of the bacterial proton-gated ion channels, which are bacterial counterparts of the mammalian cys-loop receptor family, have suggested a novel mechanism: pore dilation caused by the tilting of the M2 and M3 domain as a rigid body along the axis parallel to the membrane63, 64. The bacterial pentameric ligand-gated ion channel homologue from Erwinia chrysanthemi (ELIC) was apparently crystallized in the resting closed state. The outer segments of the M2 domains of this receptor interact with each other to form a hydrophobic barrier, the channel gate, to prevent ion flux. The bacterial Gloeobacter violaceus pentameric ligand-gated ion channel homologue (GLIC) was crystallized with high proton concentration (low pH) and was apparently in the open state. The major difference in channel domain of the two structures is that the upper part of M2–M3 domains tilts out in GLIC. As a result, the pore diameter in outer half of M2 becomes larger for ion conduction, and the intracellular end of the pore becomes smaller for ionic selectivity and single channel conductance (Figure 7B, 7C). Thus, the activation for bacterial ligand-gated ion channel involves mainly tilting of M2–M3 as a rigid body in the channel domain. Now, the question is whether this activation mechanism is also applicable to mammalian cys-loop receptors. In the nAChR, although high resolution structural model in the open state is still not available, single channel analysis of protonation scanning of pore lining domain suggests that M2 rotation in the open state is minimal, supporting the pore dilation mechanism65. Although this mechanism in the cys-loop receptors needs to be further validated with other approaches, the gating mechanism of this receptor family is likely to be conserved across species.
Coupling between amino-terminal domain and the gating machinery
As mentioned above, coupling between binding and gating domains requires matching of three loops (loop 2, loop 7/cys-loop, and loop 9/loop F) from the amino-terminal domain and one loop (M2–M3 linker) from the transmembrane domain43 and pre-M1 and the beginning of M144. Since M2-M3 linker is not conserved across the entire cys-loop receptor family, detailed coupling residues could vary depending on subfamilies, although the general mechanism is likely to be conserved. In muscle type nAChR, αV46 in loop 2 is coupled to S269 and P272 in the M2-M3 linker, whereas αE45 in loop 2 is coupled to R209 in pre-M166. Since the pre-M1 domain is also directly linked to loop C, the authors believe that loop C capping can directly result in the rotation of the pre-M1, which is coupled to loop 2 and then in turn to M2–M3 linker to open the channel. They proposed that this coupling between pre-M1 and loop 2 serves as the principal activation pathway. However, another study, also using single channel analysis, suggested that the coupling between pre-M1 and loop 2 is relatively weak and thus plays a less important role in channel gating67. αP272 in M2-M3 linker is coupled not only to V46 in loop 2 but also to V135 in the cys-loop68. The homologous proline in M2-M3 linker of 5-HT3R controls channel opening and closing through its backbone cis-trans isomerization69. However, this proline is only conserved in nAChRs and 5-HT3R. In GABAA receptor, the couplings between loop2/cys-loop to M2–M3 linker in both α and β subunits are through a charge interaction70, 71. However, based on the relative tolerance of charge reversal, neutralization, or introduction in several members of the cys-loop receptor family, Xiu et al concluded that it is the overall charge pattern, but not specific charge interaction, in the coupling interface that controls channel gating in the cys-loop receptor family72. The conserved arginine in pre-M1 of GABAA receptor β subunit (R216) also plays a pivotal role in channel activation by both GABA and pentobarbital, suggesting similar coupling mechanism at this level73. In the ρ1 GABAC receptor, the same arginine is coupled to E92 in loop 274.
Concluding remarks
In summary, the amino-terminal binding domain is coupled to the channel gate through an interconnected allosteric network. Both binding pocket and gating machinery have a tendency to close. Thus, they are coupled with a tension, so that the closures of binding pocket and gate are mutually exclusive, unless their coupling is disrupted as in the case of desensitization. In the resting state, the gating machinery has stronger force than the binding pocket to close, so that the equilibrium shifts toward closing of the channel gate and opening of the binding pocket. However, when the gating machinery is loosened, as in the case of hydrophilic mutation in the gate-forming residues, the channel opens spontaneously with simultaneous closure of binding pocket as reflected by increased binding affinity. In the wild type receptor, the closure of the binding pocket, mainly induced by agonist-binding, would alter the energy landscape of the receptor to open the channel. The conformational change in the binding pocket rearranges the interaction between loop C and loop F/cys-loop, which potentially causes an outward movement of both loop F and cys-loop. At the same time, the conformational change of the loop C may also pry the pre-M1 and M1 domain to move outward. This outward motion pulls M2–M3 linker directly (through M1) and indirectly (through pre-M1 and loop 2 coupling). Conformational change also involves rotation of inner β-sheet in amino-terminal domain, probably through coordinated movement of all six binding loops, making loop 2 to move toward periphery. The outward movement of these three loops and pre-M1 and M1 is then coupled to the M2–M3 linker, pulling channel lining M2 to open (more likely causing pore dilation than M2 rotation). The above summary also includes some speculations of the authors. The detailed coupling and gating mechanism still awaits future investigation with functional analysis (especially mutant cycle analysis) combined with real time monitoring of conformational changes during channel activation by fluorescence technique guided with structural models when high resolution crystal structures of mammalian cys-loop receptor are available.
Note
Please note that a correction has been made to the doi of this article since initial online publication on 11 May 2009. The doi published here is the correct doi. Acta Pharmacologica Sinica would like to apologise for any inconvenience caused.
Acknowledgments
We thank Arizona Biomedical Research Commission (grant ABRC0702) and Barrow Neurological Foundation for financial support (to Yong-chang CHANG) for structure function relationship studies of a cys-loop receptor. We also thank Dr Alan GIBSON in the Barrow Neurological Institute for his help in proofreading the manuscript.
References
- Changeux JP, Edelstein SJ. Allosteric receptors after 30 years. Neuron. 1998;21:959–80. doi: 10.1016/s0896-6273(00)80616-9. [DOI] [PubMed] [Google Scholar]
- Lester HA, Dibas MI, Dahan DS, Leite JF, Dougherty DA. Cys-loop receptors: new twists and turns. Trends Neurosci. 2004;27:329–36. doi: 10.1016/j.tins.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Ortells MO, Lunt GG. Evolutionary history of the ligand-gated ion-channel superfamily of receptors. Trends Neurosci. 1995;18:121–7. doi: 10.1016/0166-2236(95)93887-4. [DOI] [PubMed] [Google Scholar]
- Tasneem A, Iyer LM, Jakobsson E, Aravind L. Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels. Genome Biol. 2004;6:R4. doi: 10.1186/gb-2004-6-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Reilly K, Chang Y. Evolutionarily conserved allosteric network in the cys-loop family of ligand-gated ion channels revealed by statistical covariance analyses. J Biol Chem. 2006;281:18184–92. doi: 10.1074/jbc.M600349200. [DOI] [PubMed] [Google Scholar]
- del Castillo J, Katz B. Interaction at end-plate receptors between different choline derivatives. Proc R Soc Lond B Biol Sci. 1957;146:369–81. doi: 10.1098/rspb.1957.0018. [DOI] [PubMed] [Google Scholar]
- Amin J, Weiss DS. GABAA receptor needs two homologous domains of the β subunit for activation by GABA, but not by pentobarbital. Nature. 1993;366:565–9. doi: 10.1038/366565a0. [DOI] [PubMed] [Google Scholar]
- Chang Y, Gansah E, Chen Y, Ye J, Weiss DS. Desensitization mechanism of GABA receptor revealed by single oocyte binding and receptor function. J Neurosci. 2002;22:7982–90. doi: 10.1523/JNEUROSCI.22-18-07982.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun D. Binding, gating, affinity, and efficacy: the interpretation of structure-activity relationships for agonists and of the effects of mutating receptors. Br J Pharmacol. 1998;125:923–48. doi: 10.1038/sj.bjp.0702164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Weiss DS. Channel opening locks agonist onto the GABAC receptor. Nat Neurosci. 1999;2:219–25. doi: 10.1038/6313. [DOI] [PubMed] [Google Scholar]
- Chang Y, Wang R, Barot S, Weiss DS. Stoichiometry of a recombinant GABAA receptor. J Neurosci. 1996;16:5415–24. doi: 10.1523/JNEUROSCI.16-17-05415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarca C, Nowak MW, Zhang H, Tang L, Deshpande P, Lester HA. Channel gating governed symmetrically by conserved leucine residues in the M2 domain of nicotinic receptors. Nature. 1995;376:514–6. doi: 10.1038/376514a0. [DOI] [PubMed] [Google Scholar]
- Chang Y, Weiss DS. Allosteric activation mechanism of the α1β2γ2 γ-aminobutyric acid type A receptor revealed by mutation of the conserved M2 leucine. Biophys J. 1999;77:2542–51. doi: 10.1016/s0006-3495(99)77089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein SJ, Changeux JP. Allosteric proteins after thirty years: the binding and state functions of the neuronal α7 nicotinic acetylcholine receptors. Experientia. 1996;52:1083–90. doi: 10.1007/BF01952106. [DOI] [PubMed] [Google Scholar]
- Monod J, Wyman J, Changeux JP. On the nature of allosteric proteins: a plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Lape R, Colquhoun D, Sivilotti LG. On the nature of partial agonism in the nicotinic receptor superfamily. Nature. 2008;454:722–7. doi: 10.1038/nature07139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corringer PJ, Le Novère N, Changeux JP. Nicotinic receptors at the amino-acid level. Annu Rev Pharmacol Toxicol. 2000;40:431–58. doi: 10.1146/annurev.pharmtox.40.1.431. [DOI] [PubMed] [Google Scholar]
- Galzi JL, Revah F, Black D, Goeldner M, Hirth C, Changeux JP. Identification of a novel amino acid alpha-tyrosine 93 within the cholinergic ligands-binding sites of the acetylcholine receptor by photoaffinity labeling. Additional evidence for a three-loop model of the cholinergic ligands-binding sites. J Biol Chem. 1990;265:10430–7. [PubMed] [Google Scholar]
- Dennis M, Giraudat J, Kotzyba-Hibert F, Goeldner M, Hirth C, Chang JY, et al. Amino acids of the Torpedo marmorata acetylcholine receptor alpha subunit labeled by a photoaffinity ligand for the acetylcholine binding site. Biochemistry. 1988;27:2346–57. doi: 10.1021/bi00407a016. [DOI] [PubMed] [Google Scholar]
- Kao PN, Karlin A. Acetylcholine receptor binding site contains a disulfide cross-link between adjacent half-cystinyl residues. J Biol Chem. 1986;261:8085–8. [PubMed] [Google Scholar]
- Middleton RE, Cohen JB. Mapping of the acetylcholine binding site of the nicotinic acetylcholine receptor: [3H]nicotine as an agonist photoaffinity label. Biochemistry. 1991;30:6987–97. doi: 10.1021/bi00242a026. [DOI] [PubMed] [Google Scholar]
- Fu DX, Sine SM. Competitive antagonists bridge the alpha-gamma subunit interface of the acetylcholine receptor through quaternary ammonium-aromatic interactions. J Biol Chem. 1994;269:26152–7. [PubMed] [Google Scholar]
- O'Leary ME, Filatov GN, White MM. Characterization of d-tubocurarine binding site of Torpedo acetylcholine receptor. Am J Physiol. 1994;266:C648–53. doi: 10.1152/ajpcell.1994.266.3.C648. [DOI] [PubMed] [Google Scholar]
- Corringer PJ, Galzi JL, Eisele JL, Bertrand S, Changeux JP, Bertrand D. Identification of a new component of the agonist binding site of the nicotinic alpha 7 homooligomeric receptor. J Biol Chem. 1995;270:11749–52. doi: 10.1074/jbc.270.20.11749. [DOI] [PubMed] [Google Scholar]
- Sine SM, Kreienkamp HJ, Bren N, Maeda R, Taylor P. Molecular dissection of subunit interfaces in the acetylcholine receptor: identification of determinants of alpha-conotoxin M1 selectivity. Neuron. 1995;15:205–11. doi: 10.1016/0896-6273(95)90077-2. [DOI] [PubMed] [Google Scholar]
- Czajkowski C, Kaufmann C, Karlin A. Negatively charged amino-acid residues in the nicotinic receptor delta subunit that contribute to the binding of acetylcholine. Proc Natl Acad Sci USA. 1993;90:6285–9. doi: 10.1073/pnas.90.13.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Czajkowski C, Karlin A. The contributions of aspartyl residues in the acetylcholine receptor gamma and delta subunits to the binding of agonists and competitive antagonists. J Biol Chem. 1996;271:13497–503. doi: 10.1074/jbc.271.23.13497. [DOI] [PubMed] [Google Scholar]
- Prince RJ, Sine SM. Molecular dissection of subunit interfaces in the acetylcholine receptor. Identification of residues that determine agonist selectivity. J Biol Chem. 1996;271:25770–7. doi: 10.1074/jbc.271.42.25770. [DOI] [PubMed] [Google Scholar]
- Celie PHN, Klaassen RV, van Rossum-Fikkert SE, van Elk R, van Nierop P, Smit AB, et al. Crystal structure of acetylcholine-binding protein from Bulinus truncatus reveals the conserved structural scaffold and sites of variation in nicotinic acetylcholine receptors. J Biol Chem. 2005;280:26457–66. doi: 10.1074/jbc.M414476200. [DOI] [PubMed] [Google Scholar]
- Brejc K, Dijk WJV, Klaassen RV, Schuurmans M, Oost JVD, Smit AB, et al. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–76. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- Celie PHN, Rossum-Fikkert SEv, Dijk WJV, Brejc K, Smit AB, Sixma TK. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–14. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- Hansen SB, Sulzenbacher G, Huxford T, Marchot P, Taylor P, Bourne Y. Structures of Aplysia AChBP complexes with nicotinic agonists and antagonists reveal distinctive binding interfaces and conformations. EMBO J. 2005;24:3635–46. doi: 10.1038/sj.emboj.7600828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revah F, Bertrand D, Gaizi JL, Devillers-Thiery A, Mulle C, Hussy N, et al. Mutations in the channel domain alter desensitization of a neuronal nicotinic receptor. Nature. 1991;353:846–9. doi: 10.1038/353846a0. [DOI] [PubMed] [Google Scholar]
- Yakel JL, Lagrutta A, Adelman JP, North RA. Single amino acid substitution affects desensitization of the 5-hydroxytryptamine type 3 receptor expressed in Xenopus oocytes. Proc Natl Acad Sci USA. 1993;90:5030–3. doi: 10.1073/pnas.90.11.5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov GN, White MM. The role of conserved leucines in the M2 domain of the acetylcholine receptor in channel gating. Mol Pharmacol. 1995;48:379–84. [PubMed] [Google Scholar]
- Chang Y, Weiss DS. Substitutions of the highly conserved M2 leucine create spontaneously opening ρ1 γ-aminobutyric acid receptors. Mol Pharmacol. 1998;53:511–23. doi: 10.1124/mol.53.3.511. [DOI] [PubMed] [Google Scholar]
- Pan ZH, Zhang D, Zhang X, Lipton SA. Agonist-induced closure of constitutively open γ-aminobutyric acid channels with mutated M2 domains. Proc Natl Acad Sci USA. 1997;94:6490–5. doi: 10.1073/pnas.94.12.6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin A, Akabas MH. Toward a structural basis for the function of nicotinic acetylcholine receptors and their cousins. Neuron. 1995;15:1231–44. doi: 10.1016/0896-6273(95)90004-7. [DOI] [PubMed] [Google Scholar]
- Xu M, Akabas MH. Identification of channel-lining residues in the M2 membrane-spanning segment of the GABAA receptor α1 subunit. J Gen Physiol. 1996;107:195–205. doi: 10.1085/jgp.107.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akabas MH, Kaufmann C, Archdeacon P, Karlin A. Identification of acetylcholine receptor channel-lining residues in the entire M2 segment of the α subunit. Neuron. 1994;13:919–27. doi: 10.1016/0896-6273(94)90257-7. [DOI] [PubMed] [Google Scholar]
- Bali M, Akabas MH. The location of a closed channel gate in the GABAA receptor channel. J Gen Physiol. 2007;129:145–59. doi: 10.1085/jgp.200609639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–55. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- Bouzat C, Gumilar F, Spitzmaul G, Wang HL, Rayes D, Hansen SB, et al. Coupling of agonist binding to channel gating in an ACh-binding protein linked to an ion channel. Nature. 2004;430:896–900. doi: 10.1038/nature02753. [DOI] [PubMed] [Google Scholar]
- Bouzat C, Bartos M, Corradi J, Sine SM. The interface between extracellular and transmembrane domains of homomeric cys-loop receptors governs open-channel lifetime and rate of desensitization. J Neurosci. 2008;28:7808–19. doi: 10.1523/JNEUROSCI.0448-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosman C, Zhou M, Auerbach A. Mapping the conformational wave of acetylcholine receptor channel gating. Nature. 2000;403:773–6. doi: 10.1038/35001586. [DOI] [PubMed] [Google Scholar]
- Purohit P, Mitra A, Auerbach A. A stepwise mechanism for acetylcholine receptor channel gating. Nature. 2007;446:930–3. doi: 10.1038/nature05721. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhang Y, Akk G, Sine SM, Auerbach A. Activation kinetics of recombinant mouse nicotinic acetylcholine receptors: mutations of alphasubunit tyrosine 190 affect both binding and gating. Biophys J. 1995;69:849–59. doi: 10.1016/S0006-3495(95)79959-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akk G, Sine SM, Auerbach A. Binding sites contribute unequally to the gating of mouse nicotinic αD200N acetylcholine receptors. J Physiol. 1996;496:185–96. doi: 10.1113/jphysiol.1996.sp021676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtasimova N, Free C, Sine SM. Initial coupling of binding to gating mediated by conserved residues in the muscle nicotinic receptor. J Gen Physiol. 2005;126:23–39. doi: 10.1085/jgp.200509283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalan SP, Czajkowski C. A conserved salt bridge critical for GABAA receptor function and loop C dynamics. Proc Natl Acad Sci USA. 2008;105:13604–9. doi: 10.1073/pnas.0801854105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell JG, McDevitt RA, Czajkowski C. Mutation of glutamate 155 of the GABAA receptor β2 subunit produces a spontaneously open channel: a trigger for channel activation. J Neurosci. 2004;24:11226–35. doi: 10.1523/JNEUROSCI.3746-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grutter T, de Carvalho LP, Novere NL, Corringer PJ, Edelstein SJ, Changeux JP. An H-bond between two residues from different loops of the acetylcholine binding site contributes to the activation mechanism of nicotinic receptors. EMBO J. 2003;22:1990–2003. doi: 10.1093/emboj/cdg197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite JF, Blanton MP, Shahgholi M, Dougherty DA, Lester HA. Conformation-dependent hydrophobic photolabeling of the nicotinic receptor: electrophysiology-coordinated photochemistry and mass spectrometry. Proc Natl Acad Sci USA. 2003;100:13054–9. doi: 10.1073/pnas.2133028100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akk G, Zhou M, Auerbach A. A mutational analysis of the acetylcholine receptor channel transmitter binding site. Biophys J. 1999;76:207–18. doi: 10.1016/S0006-3495(99)77190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Xue F, Chang Y. Agonist- and antagonist-induced conformational changes of loop F and their contributions to the ρ1 GABA receptor function. J Physiol. 2009;587:139–53. doi: 10.1113/jphysiol.2008.160093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleitsman KR, Kedrowski SMA, Lester HA, Dougherty DA. An intersubunit hydrogen bond in the nicotinic acetylcholine receptor that contributes to channel gating. J Biol Chem. 2008;283:35638–43. doi: 10.1074/jbc.M807226200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akk G. Contributions of the non-α subunit residues (loop D) to agonist binding and channel gating in the muscle nicotinic acetylcholine receptor. J Physiol. 2002;544:695–705. doi: 10.1113/jphysiol.2002.029413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay EA, Giniatullin R, Skorinkin A, Yakel JL. Aromatic residues at position 55 of rat α7 nicotinic acetylcholine receptors are critical for maintaining rapid desensitization. J Physiol. 2008;586:1105–15. doi: 10.1113/jphysiol.2007.149492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres V, Weiss DS. Identification of a tyrosine in the agonist binding site of the homomeric ρ1 GABA receptor that, when mutated, produces spontaneous opening. J Biol Chem. 2002;277:43741–8. doi: 10.1074/jbc.M202007200. [DOI] [PubMed] [Google Scholar]
- Sedelnikova A, Smith CD, Zakharkin SO, Davis D, Weiss DS, Chang Y. Mapping ρ1 GABAC receptor agonist binding pocket: constructing a complete model. J Biol Chem. 2005;280:1535–42. doi: 10.1074/jbc.M409908200. [DOI] [PubMed] [Google Scholar]
- Chang Y, Weiss DS. Site-specific fluorescence reveals distinct structural changes with GABA receptor activation and antagonism. Nat Neurosci. 2002;5:1163–8. doi: 10.1038/nn926. [DOI] [PubMed] [Google Scholar]
- Unwin N. Acetylcholine receptor channel imaged in the open state. Nature. 1995;373:37–43. doi: 10.1038/373037a0. [DOI] [PubMed] [Google Scholar]
- Bocquet N, Nury H, Baaden M, Poupon CL, Changeux JP, Delarue M, et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–4. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- Hilf RJC, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457:115–9. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- Cymes GD, Ni Y, Grosman C. Probing ion-channel pores one proton at a time. Nature. 2005;438:975–80. doi: 10.1038/nature04293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WY, Sine SM. Principal pathway coupling agonist binding to channel gating in nicotinic receptors. Nature. 2005;438:243–7. doi: 10.1038/nature04156. [DOI] [PubMed] [Google Scholar]
- Purohit P, Auerbach A. Acetylcholine receptor gating at extracellular transmembrane domain interface: the “Pre-M1” linker. J Gen Physiol. 2007;130:559–68. doi: 10.1085/jgp.200709857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WY, Free CR, Sine SM. Nicotinic receptor interloop proline anchors β1–β2 and Cys loops in coupling agonist binding to channel gating. J Gen Physiol. 2008;132:265–78. doi: 10.1085/jgp.200810014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lummis SC, Beene DL, Lee LW, Lester HA, Broadhurst RW, Dougherty DA. Cis-trans isomerization at a proline opens the pore of a neurotransmitter-gated ion channel. Nature. 2005;438:248–52. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- Kash TL, Jenkins A, Kelley JC, Trudell JR, Harrison NL. Coupling of agonist binding to channel gating in the GABAA receptor. Nature. 2003;421:272–75. doi: 10.1038/nature01280. [DOI] [PubMed] [Google Scholar]
- Kash TL, Dizon MJF, Trudell JR, Harrison NL. Charged residues in the β2 subunit involved in GABAA receptor activation. J Biol Chem. 2004;279:4887–93. doi: 10.1074/jbc.M311441200. [DOI] [PubMed] [Google Scholar]
- Xiu X, Hanek AP, Wang J, Lester HA, Dougherty DA. A unified view of the role of electrostatic interactions in modulating the gating of Cys loop receptors. J Biol Chem. 2005;280:41655–66. doi: 10.1074/jbc.M508635200. [DOI] [PubMed] [Google Scholar]
- Mercado J, Czajkowski C. Charged residues in the α1 and β2 pre-M1 regions involved in GABAA receptor activation. J Neurosci. 2006;26:2031–40. doi: 10.1523/JNEUROSCI.4555-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lester HA, Dougherty DA. Establishing an ion pair interaction in the homomeric ρ1 γ-aminobutyric acid type A receptor that contributes to the gating pathway. J Biol Chem. 2007;282:26210–6. doi: 10.1074/jbc.M702314200. [DOI] [PubMed] [Google Scholar]
- Amin J, Weiss DS. Insight into the activation mechanism of ρ1 GABA receptors obtained by coexpression of wild type and activation impaired subunits. Proc Biol Sci. 1996;263:273–82. doi: 10.1098/rspb.1996.0042. [DOI] [PubMed] [Google Scholar]