Abstract
Early-stage clinical trials of oncolytic virotherapy have reported the safety of several virus platforms, and viruses from three families have progressed to advanced efficacy trials. In addition, preclinical studies have established proof-of-principle for many new genetic engineering strategies. Thus, the virotherapy field now has available a diverse collection of viruses that are equipped to address unmet clinical needs owing to improved systemic administration, greater tumour specificity and enhanced oncolytic efficacy. The current key challenge for the field is to develop viruses that replicate with greater efficiency within tumours while achieving therapeutic synergy with currently available treatments.
The concept of oncolytic virotherapy originates from clinical reports of cancer regression that coincides with natural viral infections1. Virotherapy is currently being developed by genetically modifying viruses for the selective infection and destruction of cancer cells2,3. Many viruses have specific tissue tropisms that can be exploited as a starting point for preferential infection and replication within the tumour microenvironment, killing cancer cells while replicating and spreading within disease foci. Clinical trials of oncolysis have been performed for decades4, but virus engineering strategies have only recently been developed to closely monitor virus replication and to address clinically relevant challenges, such as efficient systemic delivery, tight tumour specificity and improved efficacy in combination with current cancer therapies. By exploiting our ever greater understanding of tumour biology (BOX 1), these advances support the clinical translation of many new and diverse viruses that have been rationally designed to have greater safety and efficacy in the clinic3,5.
Box 1 | Targeted oncolysis: exploiting improved understanding of tumour biology.
Genetically modified viruses can exploit different classes of tumour-specific abnormalities for efficient and specific oncolysis. First, tumour targeting can take advantage of the preferential expression of certain proteins on the cell surface; these proteins can be repurposed as receptors for virus attachment and cell entry. Second, promoters and enhancers that are particularly active in tumour cells can be used to drive the expression of certain viral genes, thereby governing viral replication. Third, viral gene expression can be made more tumour-specific by inserting sequences that are complementary to endogenous microRNAs (miRNAs) into viral genomes. Fourth, tumour-associated antigens can become immunogenic if they are exposed to the immune system during viral infection and in the context of immunostimulatory transgene expression.
Most first-generation oncolytic viruses targeted only one of these tumour-specific characteristics, but most viruses that are currently in preclinical trials target two or more simultaneously. These developments are made possible by our improved understanding of tumour biology, which is reflected by the availability of databases that profile different tumour characteristics; for example, databases of microarray expression data, such as the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) can be used to identify both the transcription levels of surface proteins and the promoter activity abnormalities that are specific to diseases of interest. Transcriptome profiling tools, such as RNA-sequencing133 and the Encylcopedia of DNA Elements (ENCODE) database134, can be used to put these data in the context of transcription and protein expression in normal tissues. Tumour-specific miRNA sequences can be queried in databases, such as the miRNA database (miRBase) and microRNA.org, and Cancer Genome Atlas researchers are mapping the genetic changes in 18 different types of cancer.
As the molecular pathophysiology of different diseases is characterized135, quantitative insights will emerge about the frequency with which different tumour-associated antigens are detected in patient populations. These insights will enable recombinant viruses that have broad oncolytic activities to be used for the treatment of specific diseases. Moreover, gene expression profiles from patients will help to identify those individuals that have the highest probability of responding well to therapy with a specific virus.
Excellent reviews have thoroughly covered the results of current clinical trials of oncolytic virotherapy3,6. In this Review, as an introduction to the field, we summarize the most advanced clinical trials for viruses from nine different families that are currently being tested as anticancer therapies3. TABLE 1 lists these selected examples and the modifications of the engineered viruses, the routes of administration and the use of combination therapies for each trial. We focus on three points: first, the increasing diversity of viral families that are being developed for oncolysis; second, the notable safety of currently used viruses, which has, in many cases, been shown at the highest doses achievable by today's manufacturing processes; and third, the successes that have been achieved using oncolytic viruses that express immunostimulatory transgenes. As a transition to next-generation preclinical viruses, we also highlight the continued clinical need for improved delivery to and replication within systemic tumours, as well as therapeutic synergy with both the immune system and currently available cancer therapeutics.
Table 1.
Selected examples of current clinical trials with viruses from nine families
| Virus family | Phase and type of clinical trial |
Virus name | Genetic changes* | Route | Combination therapy |
NCT number |
|---|---|---|---|---|---|---|
|
| ||||||
| DNA viruses | ||||||
|
| ||||||
Herpesviridae
|
Phase III complete; melanoma, primary end point met | Talimogene laherparepvec | • Armed with GM-CSF | Intratumoural | None | NCT00769704 |
| • Tumour-specific replication: ICP34.5 deleted, US11 altered | ||||||
| • Enabled antigen presentation: ICP47 deleted | ||||||
|
| ||||||
Adenoviridae
|
Phase II and III; bladder cancer | CG0070 | • Armed with GM-CSF | Intratumoural | None | NCT01438112 |
| • Preferential replication in RB-deficient tumours | ||||||
|
| ||||||
| Approved therapeutic (China); SCCHN | Oncorine (H101) | Preferential replication in tumours: E1B-55k deleted | Intravesicular | Cisplatin | PMID15601557 | |
|
| ||||||
Poxviridae
|
Phase IIB; hepatocellular carcinoma | JX-594 (Pexa-Vec) | • Armed with GM-CSF | Intratumoural | None | NCT01387555; PMID23396206 |
| • Preferential replication in tumours: thymidine kinase deleted | ||||||
|
| ||||||
Parvoviridae

|
Phase I and II; glioma | ParvOryx | None | Intravenous; intratumoural | None | NCT01301430 |
|
| ||||||
| RNA viruses | ||||||
Paramyxoviridae
|
Phase I; myeloma | MV-NIS | • Armed with human NIS | Intravenous | Cyclophosphamide | NCT00450814 |
| • Preferential replication in tumours (incompetent to block STAT1 and MDA5) | ||||||
|
| ||||||
Picornaviridae
|
Phase I; glioma | PVS-RIPO | Translation controlled by the internal ribosome entry site of another virus | Intratumoural | None | NCT01491893 |
|
| ||||||
Rhabdoviridae
|
Phase I; hepatocellular carcinoma | VSV-hlFNβ | Armed with human IFNβ | Intratumoural | None | NCT01628640 |
|
| ||||||
Retroviridae

|
Phase I and II; glioma | Toca 511 | Armed with cytosine deaminase | Intratumoural; intravenous; into resection cavity | 5-fluorocytosine | NCT01156584; NCT01470794; NCT01985256 |
|
| ||||||
Reoviridae
|
Phase III; SCCHN | Reolysin | None | Intravenous | Carboplatin paclitaxel | NCT01166542 |
GM-CSF, granulocyte–macrophage colony-stimulating factor; hlFNβ, human interferon-β; ICP, infected cell protein; MDA5, melanoma differentiation-associated protein 5; MV, measles virus; NCT, national clinical trial; N IS, sodium–iodide symporter; PVS, poliovirusSabin; RB, retinoblastoma protein; RIPO, Rhinovirus–poliovirus hybrid; SCCHN, squamous cell carcinoma of the head and neck; STAT1, signal transducer and activator of transcription 1; US, unique sequence; VSV, vesicular stomatitis virus.
Changes can be in addition to intrinsic virus properties that favour oncolysis.
Virus families have evolved specificities for different cell types, and this natural diversity is being used for therapeutic development. Currently, there are viruses from nine different families in clinical trials: Adenoviridae7, Picornaviridae8, Herpesviridae9,10, Paramyxoviridae11,12, Parvoviridae13, Reoviridae14,15, Poxviridae16–18, Retroviridae19 and Rhabdoviridae20,21 (TABLE 1). Viruses from all of these families (except Reoviridae and Parvoviridae) have been engineered to have greater tumour specificity and/or efficacy than their parental strains. A reverse genetics system has become available for reovirus22, which will enable further development of this virus family in the future. Broadened virus availability is exemplified by the newly discovered picornavirus Seneca Valley virus, which was found to preferentially infect neuroendocrine tumours, such as small cell lung cancer23. Indeed, robust replication was specifically observed in patients who had small cell lung cancer8, and this has led to the initiation of a Phase II clinical trial.
Generally, oncolytic virotherapy has been a well-tolerated experimental clinical therapy after both localized and systemic administration5. The most common adverse effects are fever and general flu-like symptoms, but more serious toxicities have been documented in rare cases5,6. In addition, no transmission of an oncolytic virus from treated patients to carers or other contacts has been noted, although shedding of virus has been documented in the urinary and respiratory tracts, especially after systemic administration. Continuing technological advances in virus production should enable more aggressive dosing in future trials3, placing greater onus on the tumour specificity of future viruses to maintain current safety profiles.
An important result from clinical trials is the success of therapeutic protocols that are based on the expression of the immunostimulatory cytokine granulocyte–macrophage colony-stimulating factor (GM-CSF). Replicating viruses from three different families (TABLE 1), talimogene laherparepvec (Amgen; Herpesviridae), JX-594 (Jennerex Biotherapeutics; Poxviridae) and CG0070 (Cell Genesys; Adenoviridae), combine replicative onco lysis with GM-CSF-mediated stimulation of granulocytes and monocytes to induce inflammation and adaptive immunity against tumour antigens. All three viruses have shown efficacy in advanced clinical trials: the results of a Phase II trial for talimogene laherparepvec in melanoma have been reported9 and a Phase III trial in melanoma has recently been completed; JX-594 has been tested in a randomized Phase II trial in hepatocellular carcinoma18; and CG0070 is currently being tested in a Phase II trial for bladder cancer24.
Although Phase I and II clinical trials are not designed to directly measure efficacy, most current-generation viruses in clinical trials have fallen short of the efficacy expectations that were set by preclinical models2,3. Improving efficacy is a multifactorial challenge, and in this Review, we summarize the preclinical engineering strategies for the virus families that have been most extensively studied so far. We describe engineering strategies that focus on overcoming continued clinical challenges: resisting antibody neutralization using genetic and chemical virus shielding; improving tumour specificity by targeting tumour-associated receptors and controlling post-entry viral replication; and improving therapeutic synergy with the immune system, chemotherapy and radiotherapy. By carefully pairing diverse virus families with engineering strategies and combination therapies, next-generation viruses can be rationally designed for greater efficacy against diseases that have unmet clinical needs.
Avoiding virus neutralization
Systemic administration of oncolytic viruses via the vasculature gives the virus access to all perfused regions of primary and metastatic tumours, making it the preferred administration route for the treatment of metastatic disease. However, systemic delivery also makes the virus susceptible to inactivation by pre-existing antibodies in the blood, which can arise in patient populations via natural contagion, scheduled immunization or prior administration of a therapeutic virus. Pre-existing and induced antiviral immune responses can be pharmacologically tempered to limit the neutralization of therapeutic viruses25–29. In this section, we discuss virus engineering strategies that can shield therapeutic viruses from pre-existing neutralizing antibodies by changing or physically masking the epitopes that are recognized by the antibodies. Multiple engineering strategies that differ in the type and magnitude of modification have been applied to different virus families, but the end result is always a chimeric virus with an engineered serotype that is not recognized by the pre-existing antibodies present in the target patient population (FIG. 1). We use vesicular stomatitis virus (VSV), adenovirus and measles virus to illustrate these different strategies, highlighting both the type and the extent of each modification, as well as discussing the applicability of these strategies to other virus families and combination regimens with pharmacological immunosuppression.
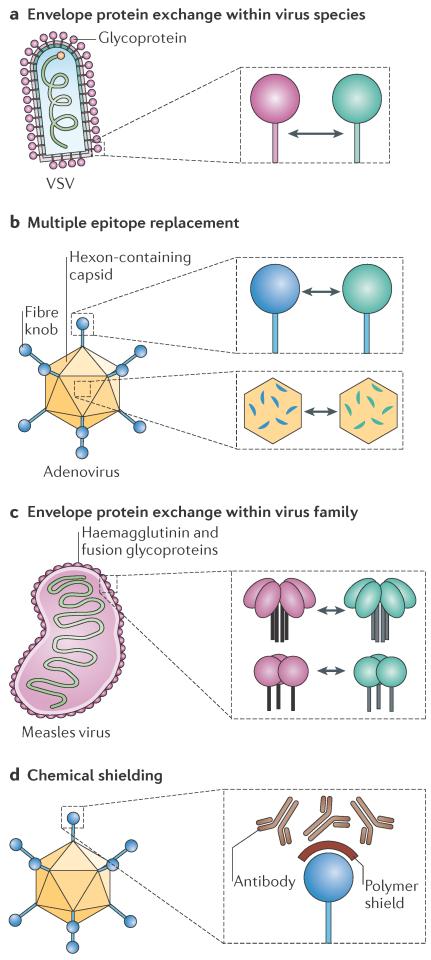
Figure 1. Vector-shielding strategies.
a | Interchange of different serotypes from the same virus species is shown for vesicular stomatitis virus (VSV), which has only one envelope glycoprotein. b | Replacement of multiple immunogenic epitopes in different proteins is shown for adenovirus; exchange can be achieved using full domains (such as the fibre knob) or individual motifs (such as hexon hypervariable loops). c | Generation of a new serotype is shown for measles virus. The two glycoproteins of this monotypic virus are substituted by the glycoproteins of an animal virus of the same genus. d | Chemical shielding of viral epitopes is shown for adenovirus; small polymers can be added to purified viral particles.
Serotype exchange
The most basic method of genetic shielding is serotype exchange, whereby a different sero-type of the same virus species is engineered onto the core of an established virus, generating a chimeric virus that has the donor serotype but the original core (FIG. 1a). This strategy requires the availability, for a given virus, of multiple serotypes, which typically have less than 60% identity between their surface-exposed glycoproteins or capsid proteins30. For example, all neutralizing epitopes that are present on VSV particles are found in the VSV-G glycoprotein, therefore serotype switching can be accomplished by simply replacing the entire VSV-G glycoprotein gene sequence with that of another serotype (FIG. 1a). Serotype switching for adenovirus is more complicated because multiple surface-exposed capsid proteins, such as the hexon or fibre knob proteins, contain neutralizing epitopes, as recently reviewed in REF. 31. The hexon capsid protein encodes seven distinct hypervariable regions that all contribute to determining the serotype of the virus32; by modifying all seven hypervariable regions, the human adenovirus 5 (HAdV-5) serotype, against which most of the population has neutralizing immunity, can be substituted for the much rarer HAdV-48 serotype33 (FIG. 1b). Replacing the HAdV-5 fibre knob with that from HAdV-3 also generated a less immunogenic chimeric virus that was resistant to neutralization by serum from HAdV-5-immunized mice during gene transfer in vivo and ex vivo34 (FIG. 1b). However, in addition to changing serotypes, knob modifications can change receptor specificity: replacing the fibre of HAdV-5 with the fibre from HAdV-16 or HAdV-50 changes receptor specificity from coxsackie–adenovirus receptor (CAR) to the ubiquitous CD46 molecule35. Notwithstanding tropism modifications, which are discussed below, these genetic exchanges can create viruses that have unique serotypes built around a common viral backbone.
Novel serotypes
A variation of the serotype exchange strategy has been implemented for monotypic viruses, such as measles virus, which have only one serotype. The measles virus glycoproteins were replaced with those of canine distemper virus (CDV), which is also a Morbillivirus, to generate an infectious virus that is not neutralized by anti-measles virus antibodies36 (FIG. 1c). Importantly, CDV and measles virus, and possibly all members of the Morbillivirus genus, enter cells using the same primary receptors — signalling lymphocytic activation molecule (SLAM) and nectin 4 — although there are differences in cross-species receptor recognition37,38. The applicability of glycoprotein exchange is limited by the availability of suitable envelope donors that are compatible with measles virus; the glycoproteins from a closely related virus of another genus did not sustain efficient assembly of chimeric particles39. Much like serotype exchange, glycoprotein or capsid exchange between viruses requires a balance between protein diversity that is sufficient for avoiding cross-neutralization and sequence and structural similarities that are necessary to support particle formation.
Chemical modifications
Shielding strategies can be used to generate a repertoire of viruses that are built around a common core but that have unique serotypes. Such a repertoire can then be used for sequential rounds of therapy that maintain efficacy even as patients develop immunity to previously used viruses. This sequential administration strategy using engineered serotypes can enhance efficacy by expanding the therapeutic window for virotherapy and, at the same time, maintaining common targeting and arming strategies.
Chemical modifications of virus particles help to overcome some of the challenges that are associated with genetic shielding. Polymer deposition on particles can physically shield epitopes from antibody neutralization (FIG. 1d); polyethylene glycol (PEG) and poly-N-(2-hydroxypropyl) methacrylamide (poly-HPMA) polymers are two examples of polymers that are used to shield oncolytic viruses40,41. Chemical shielding has been most extensively applied to Adenoviridae to protect particles from inactivation in the blood and decrease off-target liver transduction42–45, but other virus families, such as Rhabdoviridae46 and Poxviridae47, have also been chemically modified. The utility of chemical shielding depends on viruses retaining their ability to enter cells after polymer deposition, which has been successful with the icosahedral adenovirus and enveloped viruses, such as VSV and vaccinia virus. Alternatively, polymer shields that are linked to specific ligands can be used to restore and target virus entry41,48, as discussed below. Importantly, chemical shielding strategies simplify virus production, but in vivo virus replication will leave progeny particles unprotected, potentially limiting efficacy in applications that depend on virus spread.
Tumour targeting
In this Review, we use the term targeting to describe virus modifications that confer greater specificity for tumour cells by improving infection of diseased tissues and decreasing infection of healthy tissues. This specificity can be enhanced either at the stage of virus entry into target cells or post-entry during replication. Entry targeting can be achieved by fusing or conjugating specificity domains that modify receptor usage of virus particles. Post-entry, tumour-specific replication targeting, using promoters and engineered microRNA (miRNA) target sequences, can restrict virus replication in off-target tissues. These targeting strategies differ fundamentally from those that were used in the first-generation oncolytic viruses, which were often based on the removal of virulence factors that are redundant for replication in tumours49. Although this enhances safety in normal tissues, it often limits replicative fitness in target tissues50. Instead, these novel retargeting strategies can be applied to viruses that have more wild-type characteristics to minimize off-target toxicity without compromising virus replication within disease foci.
All viruses require interactions with surface molecules on host cells to start infection. Viruses can be specifically retargeted to recognize molecules that are preferentially or exclusively expressed on tumour cells (BOX 1). This strategy requires modifications to be made to the receptor-binding proteins that are present on viral particles. These modifications are either genetic and generate chimeric proteins, or they use chemical adaptors to link specificity domains to virus particles51. Targeting of enveloped viruses from the Paramyxoviridae and Herpesviridae families has rapidly progressed owing to the plasticity of their glycoproteins and the separation of receptor-binding and membrane-fusion functions, which are mediated by different proteins52. By contrast, non-enveloped icosahedral viruses, such as the Adenoviridae, have very stringent structural constraints on particle assembly, which limit viable modifications to short peptides or the chemical retargeting of assembled particles44,53 (FIG. 2).
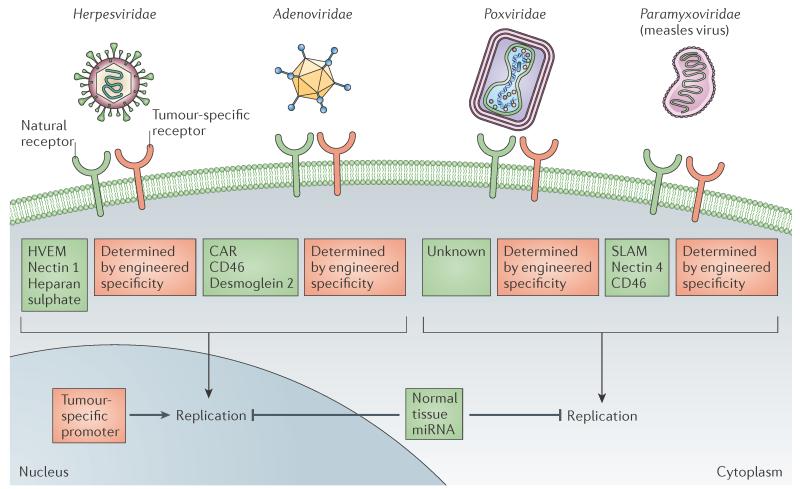
Figure 2. Principles of tumour targeting — illustrated for four virus families.
From top to bottom: targeting cell entry (detargeting from natural receptors and retargeting to tumour surface markers) and post-entry targeting (targeting of transcription, replication or microRNAs (miRNAs)). CAR, coxsackie–adenovirus receptor; HVEM, herpesvirus entry mediator; SLAM, signalling lymphocytic activation molecule.
Receptor targeting: Paramyxoviridae
Among enveloped viruses, Paramyxoviridae remain the preferred platform for the development of new targeting strategies because receptor binding and membrane fusion are mediated by two different proteins, the attachment protein (haemagglutinin, haemagglutinin-neuraminidase or glycoprotein) and the fusion protein, respectively. Adding specificity domains, such as single-chain antibodies, to the carboxyl terminus of the attachment protein sustains binding to designated receptors and subsequent membrane fusion is achieved by the unmodified fusion protein (FIG. 1). Recombinant measles virus particles that express a retargeted haemagglutinin in place of the standard haemagglutinin can be reliably generated and stably passaged54. In addition, entry via the natural receptors can be ablated by mutating specific haemagglutinin protein residues that are necessary for binding and/or entry55. This targeting principle has recently been extended to designed ankyrin repeat proteins (DARPins), which are engineered repeat-motif proteins that are smaller than single-chain antibodies and can be combined to achieve multiple specificities56; for example, it was shown that DARPins can simultaneously retarget measles virus to two different tumour markers57. This dual retargeting strategy could theoretically be used to target both tumour parenchyma (for debulking) and CD133+ cancer-initiating cells (for prolonged growth inhibition58) using a single virus, as well as to safeguard against tumour resistance due to heterogeneous or downregulated receptor expression.
Receptor targeting: Herpesviridae
The herpes simplex virus (HSV) entry mechanism is more complex than that of the Paramyxoviridae. HSV relies on five proteins, glycoprotein C, glycoprotein B, glycoprotein D and the glycoprotein H–L dimer, for receptor binding and membrane fusion, which can occur at the plasma membrane or in endocytic vesicles59,60. Glycoprotein D is responsible for binding three different receptors: the herpesvirus entry mediator (HVEM); the cell adhesion molecule nectin 1; and 3-o-sulphotransferase-modified heparan sulphate59,60 (FIG. 2). Despite this complexity, HSV retargeting shares common themes with measles virus: glycoprotein D can be engineered to express ligands, such as interleukin-13 (IL-13)61 or urokinase plasminogen activator62,63, or single-chain antibodies against human epithelial growth factor receptor 2 (HER2; also known as ERBB2)64 near its amino terminus, which retargets HSV to antigens expressed on gliomas and breast tumours, respectively. Furthermore, the natural receptor tropism can be ablated by sterically blocking the receptor-binding interfaces of glycoprotein D with a single-chain antibody64 or by using a single chain antibody to replace the entire imunoglobulin core of the glycoprotein65, which results in simultaneous retargeting of the virus to HER2 and detargeting from its natural receptors. In addition, glycoprotein C functions with glycoprotein B during initial cell binding and can be used to retarget entry using small deletions and appended single-chain antibodies66. Modifications to glycoprotein B and glycoprotein D have also been combined: entry-accelerating mutants of glycoprotein B have been paired with single-chain variable fragment (sc-Fv)-engineered envelope glycoprotein D to improve the efficiency of HSV retargeting to epidermal growth factor receptor, which is expressed on glioblastomas67. Although the entry mechanism of HSV is more complex than that of measles virus, both viruses can be genetically engineered for targeted entry using similar strategies.
Receptor retargeting: Adenoviridae
Adenovirus entry targeting is more demanding than that of Paramyxoviridae or Herpesviridae owing to the constraints of the icosahedral particle structure. Nevertheless, short heterologous peptides and specificity domains that recognize tumour-associated antigens have been inserted into the HI loop and the C terminus of the receptor-binding trimeric fibre protein, which is located at the vertices of the capsid53,68–71. The HI loop, in particular, is flexible and the inserted short peptides have minimal negative effects on virus fitness, even in combination with mutations that detarget viruses from natural adenovirus receptors68. In addition, an adenoviral minor capsid protein, the hexon-interlacing protein IX (also known as `cement' protein IX), can be used to successfully retarget entry using specificity domains, such as single-domain antibodies that do not require oxidation for folding, but not domains that are folded in the endoplasmic reticulum, such as single-chain antibodies72. A second strategy for adenovirus retargeting uses adaptors to non-covalently link particles to larger specificity domains44,53 (FIG. 1): X-ray crystallography structural data were recently used to develop universal adaptors that have high affinity for adenovirus fibre. These adaptors were then linked to DARPin specificity domains to retarget the cell entry of coated particles73. Such a universal adaptor makes it possible to target many different tumour markers using a single starting virus but, as discussed above for shielding, chemical virus retargeting is limited to a single round of replication.
Post-entry targeting
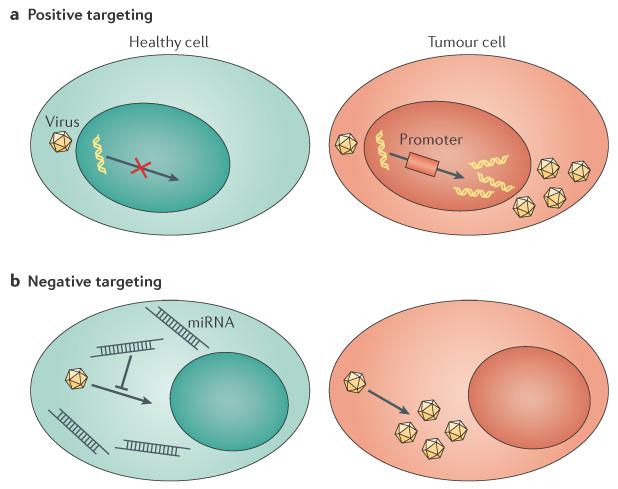
In this section we consider two post-entry targeting principles: positive targeting — which selectively promotes the expression of viral genes or engineered transgenes in target cells using tumour-specific transcriptional control74,75 — and negative targeting — which restricts infection in non-target cells using tissue-specific miRNAs that recognize target sequences that have been engineered into oncolytic virus genomes (FIG. 3). These strategies are complementary and are being applied, sometimes in combination, to multiple virus families (FIG. 2).
Figure 3. Post-entry targeting.
a | Positive transcription targeting relies on promoters that are highly expressed in cancer cells to stimulate the preferential expression of viral genes or transgenes in tumours. b | Negative targeting depends on microRNA (miRNA) expression in normal cells to restrict the replication of vectors that express miRNA-recognition sequences within their genomes. Tumours have decreased expression of certain miRNAs, which renders them unable to restrict vector replication.
Tumorigenesis is, in part, driven by aberrantly high transcription levels of genes that are not expressed in normal tissue (BOX 1). Positive transcriptional targeting can control the expression of virus genes as they have been determined to be dependent on overexpressed, tumour-specific promoters76 (FIG. 3a). Positive transcriptional targeting has been most extensively applied to adenovirus using differentially expressed tumour-associated promoters, such as telomerase (reviewed in REFS 77–79). Novel tumour-associated promoters can be identified for individual diseases using gene expression profiling, for example, by comparing transcriptional profiles between hepatocellular carcinoma (HCC) and normal liver76. This strategy has been used to identify and validate HCC-specific promoter expression in vitro and in HCC xenografts in mice76, and then to exploit those promoters to activate the expression of wild-type virulence factors, such as HSV-infected cell protein 27 (ICP27) and ICP34.5, specifically in diseased tissues80,81. Positive replication targeting can be applied to any virus family that relies on the cellular machinery for transcription but not to viruses that use virally encoded polymerases for replication in the cytoplasm, such as measles virus and vaccinia virus (FIG. 2).
An alternative strategy for regulating replication is the insertion of miRNA target sequences within the untranslated regions (UTRs) of virus transcripts to provide negative post-transcriptional regulation in non-target tissues (FIG. 3 b). Proof-of-principle for this targeting strategy was first shown using an oncolytic picornavirus that had miRNA target sequences for muscle-specific miRNA in its genome. Indeed, virus replication in muscles was minimal owing to the cellular miRNA machinery recognizing and degrading viral transcripts, thereby eliminating toxic myositis without negatively affecting viral oncolysis82. Negative replication targeting is versatile and has been applied to Adenoviridae83–88, Herpesviridae89,90, Paramyxoviridae91, Poxviridae92 and Rhabdoviridae93. The common principle is to express perfect-match miRNA target sequences, often multiple sequences in tandem, in the UTRs of essential viral genes. These target sequences are chosen on the basis of abnormally low expression of specific miRNAs in tumours. With increasing knowledge about miRNA expression in tumours94,95 (BOX 1) and the relative ease with which these target sequences can be incorporated into virus genomes without negatively affecting replication, this tumour-targeting strategy has broad applicability (FIG. 2).
Combined post-entry targeting
Positive and negative replication targeting have recently been combined to create an HSV virus that depends on the liver-specific apolipoprotein E–α-1-antitrypsin (AAT) promoter for the expression of envelope glycoprotein H and is restricted in normal liver (but not in tumours) by three differentially expressed miRNAs90. This combination of positive and negative targeting effectively blocks replication of this virus in tissues other than hepatic tumours without the need to remove the virulence factors that are necessary for optimal oncolytic efficacy. Oncolytic adenovirus has also been dually targeted using positive and negative replication engineering to restrict virus infection in normal liver but not in multiple tumour types85. When combined, these targeting strategies effectively limit virus replication to tumour cells, even if promoter and miRNA expression are insufficient for tight restriction alone. As next-generation viruses that have more wild-type characteristics are developed, it will be possible to counteract their potential for greater toxicity using combinations of entry and post-entry retargeting strategies that are applicable to many different virus families.
Arming
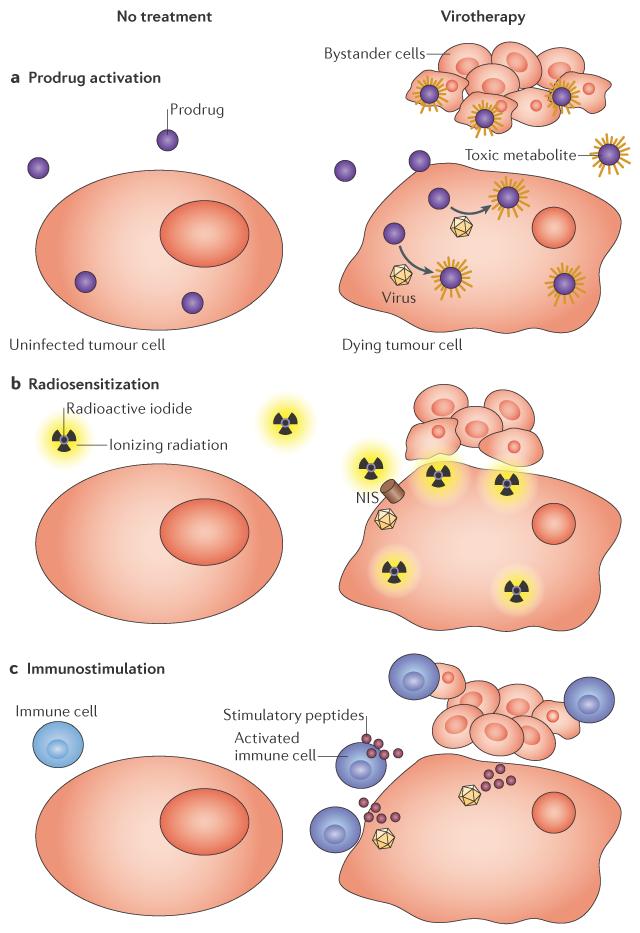
Oncolytic viruses must infect and kill tumour cells to achieve efficacy, and despite intratumoural replication, accessing and infecting 100% of tumour cells remains a major clinical challenge for therapeutic viruses. Therefore, the therapeutic efficacy of oncolytic viruses can be enhanced using strategies that induce `bystander cell killing', whereby a protein that is expressed by the oncolytic virus sensitizes both the infected cell and surrounding uninfected cells to subsequent combination therapies or immune destruction. Prodrug convertases that are expressed from oncolytic viruses can enhance the efficacy of chemotherapy by activating prodrugs, ion transport proteins can promote radiation poisoning of tumours owing to the concentration of radioisotopes, and immunostimulatory factors can induce innate and adaptive immune responses to tumour-associated antigens (FIG. 4). We discuss these three types of virus arming below.
Figure 4. Arming strategies that induce bystander cell killing.
a | Convertase enzymes that are expressed in infected cells metabolize prodrugs into toxic metabolites that diffuse and kill uninfected tumour cells. b | The sodium–iodide symporter (NIS) concentrates radioactive ions in infected cells, which induces radiation poisoning of uninfected bystander tumour cells. c | Immunostimulatory transgenes that are expressed in infected cells prime responses against tumour antigens, which causes the systemic destruction of tumour cells.
Prodrug convertases
Prodrug convertases include the thymidine kinase96,97, the cytosine deaminase97 and the purine nucleoside phosphorylase (PNP)98 systems. The corresponding genes have all been expressed by oncolytic viruses to activate non-toxic precursors, which generate highly toxic metabolites in the tumour microenvironment99. The HSV thymidine kinase phosphorylates ganciclovir to generate ganciclovir triphosphate, cytosine deaminase converts chemotherapeutic 5-fluorocytosine (5-FC) to 5-fluorouracil (5-FU), and PNP converts fludarabine phosphate into 2-fluoroadenine99. These nucleoside analogues are incorporated into the DNA of replicating cells, which halts replication and ultimately results in cell death99. The common goal of these strategies is to reduce systemic toxicity by giving lower doses of minimally toxic chemotherapeutic drugs that are only converted to highly toxic metabolites within the tumour microenvironment (FIG. 4a). Ganciclovir triphosphate cannot easily diffuse between cells after activation100, making it a good option for the selective elimination of the toxicity that is induced by retrovirally transduced cells101,102. However, the activated metabolites 5-FU and 2-fluoroadenine can both diffuse out of the infected cell and into surrounding cells98,103 to induce chemotherapeutic bystander killing (FIG. 4a), making them clinically relevant arming strategies for virotherapy. We focus in this section on the most clinically advanced viruses that are armed with the cytosine deaminase and PNP transgenes, as well as on promising preclinical viruses that are ready to enter future clinical trials.
Cytosine deaminase and PNP have both been incorporated into several virus classes that have been preclinically tested, including viruses that are based on Herpesviridae104, Adenoviridae105, Poxviridae106,107, Paramyxoviridae108,109 and Rhabdoviridae110. The most advanced cytosine deaminase virus is a replication-competent retrovirus known as Toca 511, which integrates the cytosine deaminase transgene into the genome of infected cells to establish permanent reservoirs of tumour cells that are sensitive to subsequent rounds of chemotherapy using 5-FC19. Toca 511 is currently being tested in combination with 5-FC in Phase I and II clinical trials using intratumoural administration in patients with grade 4 glioblastoma multiforme (TABLE 1). HSV and VSV viruses that express cytosine deaminase are also being preclinically developed for combination therapies using 5-FC104,110.
Intratumoural administration of an adenovirus with the PNP transgene in combination with intravenous fludarabine has been used to treat patients with head and neck tumours111 (clinicaltrials.gov identifier: NCT01310179). In general, cytosine deaminase and PNP transgenes can be applied to many virus families because their small sizes and low cellular toxicities incur minimal negative effects on in vitro virus fitness or production. However, the timing of prodrug dosing in vivo must be optimized to ensure that virus replication and spread is sufficient for maximal synergistic effects with the chemotherapeutic prodrug104,109,112. In this respect, a PNP-expressing measles virus that has been retargeted to CD20 has been extensively tested in combination with fludarabine in preclinical models of lymphoma108,109.
Radiosensitization
The normal physiological function of the human sodium–iodide symporter (NIS) is to transport iodide ions into cells, which occurs predominantly in the thyroid but also in the stomach, salivary glands and mammary glands113,114. When NIS is expressed from the genome of an oncolytic virus, infected cells concentrate iodide or similar isotopes intracellularly. During virotherapy, γ-emitting isotopes, such as 123I and pertechnetate, can be administered to visualize virus replication using single-photon emission computed tomography (SPECT; BOX 2), whereas β-emitting isotopes, such as 131I and 188Re, can be administered to specifically induce radiation poisoning within the tumour microenvironment (FIG. 4b), in analogy to the clinically well-established radiotherapy that is used for metastatic thyroid cancer.
Box 2 | In vivo imaging.
The optimization of virotherapy, in particular the arming strategies that are outlined in this Review, generally requires the characterization of virus replication in target tissues and an understanding of its determinants. Single-photon emission computed tomography (SPECT) and positron emission tomography (PET) are clinically relevant imaging approaches that are capable of achieving impressive resolution in small animal models and in humans. These imaging modalities rely on transgenes, such as the sodium–iodide symporter (NIS), that cause infected cells to accumulate radioactive tracers that have detectable emissions. Some tracers, including those that emit γ-radiation, have more effective tissue penetration than fluorescence or bioluminescence. Only effective tissue penetration enables whole-body analyses to be performed in the clinic.
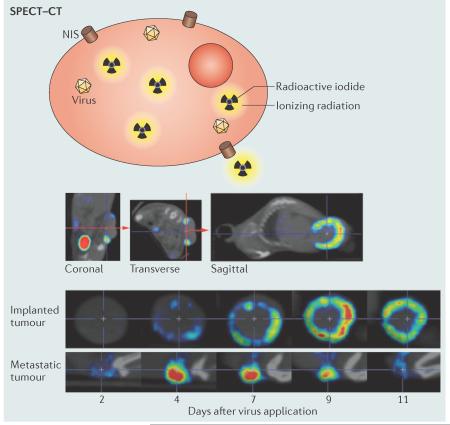
Dedicated imaging systems have been developed for small animals, and recent advances in SPECT imaging have achieved submillimetre resolutions for whole-animal tomographic analyses. The figure shows an in vivo analysis of viral replication by SPECT–computed tomography (SPECT–CT) imaging as an example. NIS, expressed from an oncolytic measles virus, induces isotope accumulation within infected cells. The top row of images illustrates how the base of the implanted tumours is aligned in the coronal and transverse planes to obtain three-dimensional tumour images with equivalent orientation at different times. The central and bottom rows of images document virus replication and show a ring being formed in the outer region of the implanted tumour by day 9 after virotherapy (middle row), whereas infection and replication in a metastatic tumour are more intense and peak earlier (bottom row). Thus, SPECT–CT imaging allows four-dimensional (three spatial dimensions plus time) analyses of virus spread in tumours in living hosts. SPECT–CT, as well as advanced PET and magnetic resonance imaging (MRI) strategies that are excellently reviewed in REFS 136–138, are broadly applicable to many oncolytic virus families and can inform sequential rounds of therapy, correlate virus replication and distribution with measures of clinical efficacy and, in worst-case scenarios, can identify off-target virus replication.
SPECT–CT images are reproduced, with permission, from ref. 118 © (2013) American Society for Gene and Cell Therapy (ASGCT). High-resolution videos of SPECT–CT data can be viewed in ref. 118.
The NIS transgene system has undergone extensive preclinical development in multiple virus families, and radiovirotherapy has consistently achieved synergistic tumour destruction in several radiosensitive preclinical disease models115–117. Measles virus is an especially efficacious virus for NIS-mediated imaging (BOX 2) and radiovirotherapy. Preclinical studies of lymphoma118, ovarian cancer119, myeloma120 and mesothelioma121, in addition to many other disease models117, have used NIS expression and SPECT–computed tomography (SPECT–CT) imaging to visualize and quantify virus replication and enhance disease regression using combination radio virotherapy. Phase I clinical trials using NIS-expressing measles virus have been initiated for ovarian cancer, myeloma, mesothelioma and head and neck cancer (clinicaltrials.gov identifiers: NCT00408590, NCT00450814, NCT01503177 and NCT01846091, respectively), with all four studies using SPECT–CT intervention to image virus replication in patients. The results of these clinical trials and the continued translation of diverse NIS-expressing viruses in different tumour types will inform the applicability of SPECT–CT imaging and radiovirotherapy interventions to future trials.
Immunostimulation
Advanced clinical trials of oncolytic viruses from three different virus families have used the combination of lytic infection and immunostimulatory transgene expression to induce anti tumour immunity (FIG. 4c). The most successful strategy that has been used so far is the expression of GM-CSF to stimulate the production of granulocytes and monocytes, which in turn stimulate adaptive immunity against tumour-associated antigens122. HSV, adenoviruses and vaccinia viruses that express GM-CSF are currently used in clinical trials and have repeatedly been shown to have clinical efficacy 122,123, especially in diseases that are amenable to immunotherapy, such as melanoma (BOX 1). These advanced clinical studies have shown that these viruses work via immunotherapy rather than via virus-mediated tumour lysis: only moderate replication of vaccinia JX-594 was documented in tumour biopsies or measured by the detection of GM-CSF in the blood, but lytic replication or recovery of infectious virus from harvested tumours were not reported18,124. The HSV virus talimogene laherparepvec9,125 is currently being tested against melanoma in a Phase III clinical trial that will directly compare the virus expressing GM-CSF with GM-CSF administration alone. This study will greatly improve our understanding of the relative contributions of virus replication and transgene expression in stimulating antitumour immunity. The successes of current oncolytic viruses that express immunostimulatory transgenes, even in the absence of robust intratumoural replication, highlights the potential for new viruses that have greater replication and transgene expression to stimulate improved, and potentially curative, immune responses against the tumour microenvironment.
Future directions
Our increasing knowledge of the determinants of virus tropism, and of the proteins and gene expression pathways that are altered in tumour tissues, are important resources for developing next-generation viruses. With few exceptions, such as vaccinia virus and VSV, virus families have adapted to tissue niches, which are often defined by specific receptors; for example, measles virus initially uses SLAM to establish systemic infection in lymphatic organs, and then nectin 4 to infect epithelia126–129. Oncolysis, similarly to wild-type virus spread, depends not only on efficient cell entry via specific receptors, but also on efficient replication; in particular, viruses often activate or exploit certain gene expression pathways that facilitate their replication, and similar events may be required for efficient oncolysis. Consequently, efficient cell entry might not always result in efficient oncolysis; for example, measles virus-based oncolytic viruses are likely to replicate most efficiently in SLAM-positive haematological malignancies, such as lymphoma, and in nectin 4-positive epithelial malignancies, such as breast130, ovarian131 and lung132 tumours. As there is an increasingly diverse pool of oncolytic viruses and our knowledge of tumour biology is improving, choosing virus classes to target specific tumour types, and then stratifying individual patients on the basis of disease susceptibility to virotherapy, should soon become standard practice.
Oncolytic virotherapy has, to date, proven to be very safe in humans. By contrast, the therapeutic efficacy of oncolytic viruses in humans has been less than expected from preclinical studies. Early protocols were based on highly attenuated viruses, and generalized attenuation interfered with clinical efficacy. Next-generation viruses will benefit from retaining wild-type replicative potential in disease tissue, combined with engineered entry and post-entry restriction mechanisms that maintain current safety profiles in off-target tissues. In addition, arming strategies that combine chemo-, radio- and immuno-therapies will be potentiated by greater virus replication, and advances in in vivo imaging will enable real-time tracking of virus spread (BOX 2).
New knowledge and new viruses can now be used to address existing clinical needs. Although many challenges remain for oncolytic virotherapy, such as the production and validation of an increasing number of therapeutic viruses, there are strategies to overcome them. There is no single best virotherapy approach for all tumours, but creative engineering strategies that use viruses from diverse families are yielding new viruses that are likely to be highly efficacious in specific applications. Understanding the susceptibility and resistance of different tumour types and progress in stratifying individual patients will facilitate the recruitment of those patients who are most likely to benefit from a new virus. In synergy with approved cancer therapies and antitumour immunity, these new viruses are expected to achieve the clinical promise of oncolytic virotherapy.
Acknowledgements
This work was supported by the US National Cancer Institute Grant R01 CA 139398. T.S.M's salary was supported in part by grant T32 GM065841 from the US National Institute of Health and US National Institute of General Medical Sciences.
Glossary
- Granulocyte–macrophage colony-stimulating factor
(GM-CSF). An immunostimulatory cytokine that functions as a growth factor for granulocytes and monocytes
- Serotype
A collection of viruses grouped according to common antigenic epitopes that are recognized by the same immune serum
- Coxsackie–adenovirus receptor
(CAR). An immunoglobulin-like transmembrane cell adhesion protein that is used by some coxsackievirus and adenovirus species as a receptor
- CD46
A ubiquitously expressed type-1 transmembrane protein that functions to regulate complement. It functions as a receptor for vaccine strains of measles virus and some adenovirus species
- Parenchyma
The bulk of tumours; consists of clonally transformed cells
- CD133+ cancer-initiating cells
A subset of tumour cells that have stem cell-like characteristics. They are capable of initiating new tumour growth and may drive tumour recurrence after therapy
- Prodrug
A chemotherapeutic drug that is dependent on enzymatic activation to achieve full activity
- Radioisotopes
Elements with unstable nuclei that emit γ-rays or α- or β-ionizing radiation
- Nucleoside analogues
Chemotherapeutic drugs that incorporate into replicating DNA as nucleosides but halt DNA replication, which leads to cell death
- γ-emitting isotopes
Radioactive isotopes that emit ionizing radiation in the form of high-energy electromagnetic γ rays
- β-emitting isotopes
Radioactive isotopes that emit ionizing radiation in the form of high-energy, high-speed electrons or protons
Footnotes
Competing interests statement The authors declare no competing interests.
References
- 1.Dock G. The influence of complicating diseases upon leukemia. Am. J. Med. Sci. 1904;127:563–592. [Google Scholar]
- 2.Cattaneo R, Miest T, Shashkova EV, Barry MA. Reprogrammed viruses as cancer therapeutics: targeted, armed and shielded. Nature Rev. Microbiol. 2008;6:529–540. doi: 10.1038/nrmicro1927. [DOI] [PMC free article] [PubMed] [Google Scholar]; This Review provides a thorough introduction to virus engineering strategies.
- 3.Russell SJ, Peng K-W, Bell JC. Oncolytic virotherapy. Nature Biotech. 2012;30:658–670. doi: 10.1038/nbt.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]; This Review gives a comprehensive overview of current virotherapy clinical trials.
- 4.Kelly E, Russell SJ. History of oncolytic viruses: genesis to genetic engineering. Mol. Ther. 2007;15:651–659. doi: 10.1038/sj.mt.6300108. [DOI] [PubMed] [Google Scholar]; This paper provides a narrative introduction to the history of oncolytic virotherapy.
- 5.Liu T-C, Galanis E, Kirn D. Clinical trial results with oncolytic virotherapy: a century of promise, a decade of progress. Nature Rev. Clin. Oncol. 2007;4:101–117. doi: 10.1038/ncponc0736. [DOI] [PubMed] [Google Scholar]
- 6.Eager RM, Nemunaitis J. Clinical development directions in oncolytic viral therapy. Cancer Gene Ther. 2011;18:305–317. doi: 10.1038/cgt.2011.7. [DOI] [PubMed] [Google Scholar]
- 7.Xia ZJ, et al. Phase III randomized clinical trial of intratumoral injection of E1B gene-deleted adenovirus (H101) combined with cisplatin-based chemotherapy in treating squamous cell cancer of head and neck or esophagus. Ai Zheng. 2004;23:1666–1670. [PubMed] [Google Scholar]
- 8.Rudin CM, et al. Phase I clinical study of Seneca Valley Virus (SVV-001), a replication-competent picornavirus, in advanced solid tumors with neuroendocrine features. Clin. Cancer Res. 2011;17:888–895. doi: 10.1158/1078-0432.CCR-10-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senzer NN, et al. Phase II clinical trial of a granulocyte–macrophage colony-stimulating factor-encoding, second-generation oncolytic herpesvirus in patients with unresectable metastatic melanoma. J. Clin. Oncol. 2009;27:5763–5771. doi: 10.1200/JCO.2009.24.3675. [DOI] [PubMed] [Google Scholar]
- 10.Kaufman HL, et al. Local and distant immunity induced by intralesional vaccination with an oncolytic herpes virus encoding GM-CSF in patients with stage IIIc and IV melanoma. Ann. Surg. Oncol. 2010;17:718–730. doi: 10.1245/s10434-009-0809-6. [DOI] [PubMed] [Google Scholar]
- 11.Freeman AI, et al. Phase I/II trial of intravenous NDV-HUJ oncolytic virus in recurrent glioblastoma multiforme. Mol. Ther. 2006;13:221–228. doi: 10.1016/j.ymthe.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 12.Galanis E, et al. Phase I trial of intraperitoneal administration of an oncolytic measles virus strain engineered to express carcinoembryonic antigen for recurrent ovarian cancer. Cancer Res. 2010;70:875–882. doi: 10.1158/0008-5472.CAN-09-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper is an example of monitoring virus replication in patients using a soluble protein that is expressed by the virus.
- 13.Nüesch JPF, Lacroix J, Marchini A, Rommelaere J. Molecular pathways: rodent parvoviruses — mechanisms of oncolysis and prospects for clinical cancer treatment. Clin. Cancer Res. 2012;18:3516–3523. doi: 10.1158/1078-0432.CCR-11-2325. [DOI] [PubMed] [Google Scholar]
- 14.Vidal L, et al. A Phase I study of intravenous oncolytic reovirus type 3 Dearing in patients with advanced cancer. Clin. Cancer Res. 2008;14:7127–7137. doi: 10.1158/1078-0432.CCR-08-0524. [DOI] [PubMed] [Google Scholar]
- 15.Forsyth P, et al. A phase I trial of intratumoral administration of reovirus in patients with histologically confirmed recurrent malignant gliomas. Mol. Ther. 2008;16:627–632. doi: 10.1038/sj.mt.6300403. [DOI] [PubMed] [Google Scholar]
- 16.Park B-H, et al. Use of a targeted oncolytic poxvirus, JX-594, in patients with refractory primary or metastatic liver cancer: a phase I trial. Lancet Oncol. 2008;9:533–542. doi: 10.1016/S1470-2045(08)70107-4. [DOI] [PubMed] [Google Scholar]
- 17.Hwang T-H, et al. A mechanistic proof-of-concept clinical trial with JX-594, a targeted multi-mechanistic oncolytic poxvirus, in patients with metastatic melanoma. Mol. Ther. 2011;19:1913–1922. doi: 10.1038/mt.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heo J, et al. Randomized dose-finding clinical trial of oncolytic immunotherapeutic vaccinia JX-594 in liver cancer. Nature Med. 2013;19:329–336. doi: 10.1038/nm.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes the planning of a Phase III randomized trial with a placebo control arm on the basis of the results of a Phase II clinical trial.
- 19.Perez OD, et al. Design and selection of Toca 511 for clinical use: modified retroviral replicating vector with improved stability and gene expression. Mol. Ther. 2012;20:1689–1698. doi: 10.1038/mt.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willmon CL, et al. Expression of IFN-β enhances both efficacy and safety of oncolytic vesicular stomatitis virus for therapy of mesothelioma. Cancer Res. 2009;69:7713–7720. doi: 10.1158/0008-5472.CAN-09-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenks N, et al. Safety studies on intrahepatic or intratumoral injection of oncolytic vesicular stomatitis virus expressing interferon-β in rodents and nonhuman primates. Hum. Gene Ther. 2010;21:451–462. doi: 10.1089/hum.2009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi T, et al. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe. 2007;1:147–157. doi: 10.1016/j.chom.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy PS, et al. Seneca Valley virus, a systemically deliverable oncolytic picornavirus, and the treatment of neuroendocrine cancers. J. Natl Cancer Inst. 2007;99:1623–1633. doi: 10.1093/jnci/djm198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burke JM, et al. A first in human phase 1 study of CG0070, a GM-CSF expressing oncolytic adenovirus, for the treatment of nonmuscle invasive bladder cancer. J. Urol. 2012;188:2391–2397. doi: 10.1016/j.juro.2012.07.097. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda K, et al. Oncolytic virus therapy of multiple tumors in the brain requires suppression of innate and elicited antiviral responses. Nature Med. 1999;5:881–887. doi: 10.1038/11320. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda K, et al. Complement depletion facilitates the infection of multiple brain tumors by an intravascular, replication-conditional herpes simplex virus mutant. J. Virol. 2000;74:4765–4775. doi: 10.1128/jvi.74.10.4765-4775.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakimoto H, et al. The complement response against an oncolytic virus is species-specific in its activation pathways. Mol. Ther. 2002;5:275–282. doi: 10.1006/mthe.2002.0547. [DOI] [PubMed] [Google Scholar]
- 28.Kambara H, Saeki Y, Chiocca EA. Cyclophosphamide allows for in vivo dose reduction of a potent oncolytic virus. Cancer Res. 2005;65:11255–11258. doi: 10.1158/0008-5472.CAN-05-2278. [DOI] [PubMed] [Google Scholar]
- 29.Peng K-W, et al. Using clinically approved cyclophosphamide regimens to control the humoral immune response to oncolytic viruses. Gene Ther. 2013;20:255–261. doi: 10.1038/gt.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallione CJ, Rose JK. Nucleotide sequence of a cDNA clone encoding the entire glycoprotein from the New Jersey serotype of vesicular stomatitis virus. J. Virol. 1983;46:162–169. doi: 10.1128/jvi.46.1.162-169.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaufmann JK, Nettelbeck DM. Virus chimeras for gene therapy, vaccination, and oncolysis: adenoviruses and beyond. Trends Mol. Med. 2012;18:365–376. doi: 10.1016/j.molmed.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Bradley RR, et al. Adenovirus serotype 5-specific neutralizing antibodies target multiple hexon hypervariable regions. J. Virol. 2012;86:1267–1272. doi: 10.1128/JVI.06165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts DM, et al. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature. 2006;441:239–243. doi: 10.1038/nature04721. [DOI] [PubMed] [Google Scholar]; This paper is a `tour-de-force' that shows how neutralizing epitopes can be substituted by other sequences in an icosahedral capsid.
- 34.Särkioja M, et al. Changing the adenovirus fiber for retaining gene delivery efficacy in the presence of neutralizing antibodies. Gene Ther. 2008;15:921–929. doi: 10.1038/gt.2008.56. [DOI] [PubMed] [Google Scholar]
- 35.Kuhlmann KFD, et al. Fiber-chimeric adenoviruses expressing fibers from serotype 16 and 50 improve gene transfer to human pancreatic adenocarcinoma. Cancer Gene Ther. 2009;16:585–597. doi: 10.1038/cgt.2009.4. [DOI] [PubMed] [Google Scholar]
- 36.Miest TS, et al. Envelope-chimeric entry-targeted measles virus escapes neutralization and achieves oncolysis. Mol. Ther. 2011;19:1813–1820. doi: 10.1038/mt.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study is an example of glycoprotein exchange between different virus species that avoids neutralization.
- 37.Tatsuo H, Ono N, Yanagi Y. Morbilliviruses use signaling lymphocyte activation molecules (CD150) as cellular receptors. J. Virol. 2001;75:5842–5850. doi: 10.1128/JVI.75.13.5842-5850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tatsuo H, Yanagi Y. The morbillivirus receptor SLAM (CD150) Microbiol. Immunol. 2002;46:135–142. doi: 10.1111/j.1348-0421.2002.tb02678.x. [DOI] [PubMed] [Google Scholar]
- 39.Hudacek AW, Navaratnarajah CK, Cattaneo R. Development of measles virus-based shielded oncolytic vectors: suitability of other paramyxovirus glycoproteins. Cancer Gene Ther. 2013;20:109–116. doi: 10.1038/cgt.2012.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kreppel F, Kochanek S. Modification of adenovirus gene transfer vectors with synthetic polymers: a scientific review and technical guide. Mol. Ther. 2008;16:16–29. doi: 10.1038/sj.mt.6300321. [DOI] [PubMed] [Google Scholar]
- 41.Fisher KD, Seymour LW. HPMA copolymers for masking and retargeting of therapeutic viruses. Adv. Drug Deliv. Rev. 2010;62:240–245. doi: 10.1016/j.addr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Kim P-H, et al. Bioreducible polymer-conjugated oncolytic adenovirus for hepatoma-specific therapy via systemic administration. Biomaterials. 2011;32:9328–9342. doi: 10.1016/j.biomaterials.2011.08.066. [DOI] [PubMed] [Google Scholar]
- 43.Doronin K, Shashkova EV, May SM, Hofherr SE, Barry MA. Chemical modification with high molecular weight polyethylene glycol reduces transduction of hepatocytes and increases efficacy of intravenously delivered oncolytic adenovirus. Hum. Gene Ther. 2009;20:975–988. doi: 10.1089/hum.2009.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report shows that chemical modification of the viral surface can have a positive effect on targeting.
- 44.Khare R, Chen CY, Weaver EA, Barry MA. Advances and future challenges in adenoviral vector pharmacology and targeting. Curr. Gene Ther. 2011;11:241–258. doi: 10.2174/156652311796150363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi J-W, Lee J-S, Kim SW, Yun C-O. Evolution of oncolytic adenovirus for cancer treatment. Adv. Drug Deliv. Rev. 2012;64:720–729. doi: 10.1016/j.addr.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 46.Tesfay MZ, et al. PEGylation of vesicular stomatitis virus extends virus persistence in blood circulation of passively immunized mice. J. Virol. 2013;87:3752–3759. doi: 10.1128/JVI.02832-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naito T, Kaneko Y, Kozbor D. Oral vaccination with modified vaccinia virus Ankara attached covalently to TMPEG-modified cationic liposomes overcomes pre-existing poxvirus immunity from recombinant vaccinia immunization. J. Gen. Virol. 2007;88:61–70. doi: 10.1099/vir.0.82216-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim P-H, et al. Active targeting and safety profile of PEG-modified adenovirus conjugated with herceptin. Biomaterials. 2011;32:2314–2326. doi: 10.1016/j.biomaterials.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 49.Naik S, Russell SJ. Engineering oncolytic viruses to exploit tumor specific defects in innate immune signaling pathways. Expert Opin. Biol. Ther. 2009;9:1163–1176. doi: 10.1517/14712590903170653. [DOI] [PubMed] [Google Scholar]
- 50.Haralambieva I, et al. Engineering oncolytic measles virus to circumvent the intracellular innate immune response. Mol. Ther. 2007;15:588–597. doi: 10.1038/sj.mt.6300076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verheije MH, Rottier PJM. Retargeting of viruses to generate oncolytic agents. Adv. Virol. 2012;2012:798526. doi: 10.1155/2012/798526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Navaratnarajah CK, Miest TS, Carfi A, Cattaneo R. Targeted entry of enveloped viruses: measles and herpes simplex virus I. Curr. Opin. Virol. 2012;2:43–49. doi: 10.1016/j.coviro.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coughlan L, et al. Tropism-modification strategies for targeted gene delivery using adenoviral vectors. Viruses. 2010;2:2290–2355. doi: 10.3390/v2102290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider U, Bullough F, Vongpunsawad S, Russell SJ, Cattaneo R. Recombinant measles viruses efficiently entering cells through targeted receptors. J. Virol. 2000;74:9928–9936. doi: 10.1128/jvi.74.21.9928-9936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vongpunsawad S, Oezgun N, Braun W, Cattaneo R. Selectively receptor-blind measles viruses: identification of residues necessary for SLAM-or CD46-induced fusion and their localization on a new hemagglutinin structural model. J. Virol. 2004;78:302–313. doi: 10.1128/JVI.78.1.302-313.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boersma YL, Plückthun A. DARPins and other repeat protein scaffolds: advances in engineering and applications. Curr. Opin. Biotechnol. 2011;22:849–857. doi: 10.1016/j.copbio.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 57.Friedrich K, et al. DARPin-targeting of measles virus: unique bispecificity, effective oncolysis, and enhanced safety. Mol. Ther. 2013;21:849–859. doi: 10.1038/mt.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first example of targeting viral entry to two different tumour markers using new specificity domains that can be easily combined.
- 58.Bach P, et al. Specific elimination of CD133+ tumor cells with targeted oncolytic measles virus. Cancer Res. 2013;73:865–874. doi: 10.1158/0008-5472.CAN-12-2221. [DOI] [PubMed] [Google Scholar]
- 59.Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nature Rev. Microbiol. 2011;9:369–381. doi: 10.1038/nrmicro2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Campadelli-Fiume G, Menotti L, Avitabile E, Gianni T. Viral and cellular contributions to herpes simplex virus entry into the cell. Curr. Opin. Virol. 2012;2:28–36. doi: 10.1016/j.coviro.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 61.Zhou G, Ye G-J, Debinski W, Roizman B. Engineered herpes simplex virus 1 is dependent on IL 13Rα2 receptor for cell entry and independent of glycoprotein D receptor interaction. Proc. Natl Acad. Sci. USA. 2002;99:15124–15129. doi: 10.1073/pnas.232588699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kamiyama H, Zhou G, Roizman B. Herpes simplex virus 1 recombinant virions exhibiting the amino terminal fragment of urokinase-type plasminogen activator can enter cells via the cognate receptor. Gene Ther. 2006;13:621–629. doi: 10.1038/sj.gt.3302685. [DOI] [PubMed] [Google Scholar]
- 63.Zhou G, Roizman B. Separation of receptor-binding and profusogenic domains of glycoprotein D of herpes simplex virus 1 into distinct interacting proteins. Proc. Natl Acad. Sci. USA. 2007;104:4142–4146. doi: 10.1073/pnas.0611565104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Menotti L, Cerretani A, Hengel H, Campadelli-Fiume G. Construction of a fully retargeted herpes simplex virus 1 recombinant capable of entering cells solely via human epidermal growth factor receptor 2. J. Virol. 2008;82:10153–10161. doi: 10.1128/JVI.01133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Menotti L, et al. Inhibition of human tumor growth in mice by an oncolytic herpes simplex virus designed to target solely HER-2-positive cells. Proc. Natl Acad. Sci. USA. 2009;106:9039–9044. doi: 10.1073/pnas.0812268106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grandi P, et al. Targeting HSV-1 virions for specific binding to epidermal growth factor receptor-vIII-bearing tumor cells. Cancer Gene Ther. 2010;17:655–663. doi: 10.1038/cgt.2010.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Uchida H, et al. Effective treatment of an orthotopic xenograft model of human glioblastoma using an EGFR-retargeted oncolytic herpes simplex virus. Mol. Ther. 2013;21:561–569. doi: 10.1038/mt.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coughlan L, et al. Combined fiber modifications both to target αvβ6 and detarget the coxsackievirus–adenovirus receptor improve virus toxicity profiles in vivo but fail to improve antitumoral efficacy relative to adenovirus serotype 5. Hum. Gene Ther. 2012;23:960–979. doi: 10.1089/hum.2011.218. [DOI] [PubMed] [Google Scholar]
- 69.Ballard EN, Trinh VT, Hogg RT, Gerard RD. Peptide targeting of adenoviral vectors to augment tumor gene transfer. Cancer Gene Ther. 2012;19:476–488. doi: 10.1038/cgt.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Magnusson MK, et al. A transductionally retargeted adenoviral vector for virotherapy of Her2/neu-expressing prostate cancer. Hum. Gene Ther. 2012;23:70–82. doi: 10.1089/hum.2011.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.MacLeod SH, Elgadi MM, Bossi G, Sankar U. HER3 targeting of adenovirus by fiber modification increases infection of breast cancer cells in vitro, but not following intratumoral injection in mice. Cancer Gene Ther. 2012;19:888–898. doi: 10.1038/cgt.2012.79. [DOI] [PubMed] [Google Scholar]
- 72.Poulin KL, et al. Retargeting of adenovirus vectors through genetic fusion of a single-chain or single-domain antibody to capsid protein IX. J. Virol. 2010;84:10074–10086. doi: 10.1128/JVI.02665-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dreier B, et al. Development of a generic adenovirus delivery system based on structure-guided design of bispecific trimeric DARPin adapters. Proc. Natl Acad. Sci. USA. 2013;110:E869–E877. doi: 10.1073/pnas.1213653110. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows the structure-guided design of a clamp-like adaptor that has been engineered on the top of the adenovirus knob domain.
- 74.Chung RY, Saeki Y, Chiocca EA. B-myb promoter retargeting of herpes simplex virus γ34.5 gene-mediated virulence toward tumor and cycling cells. J. Virol. 1999;73:7556–7564. doi: 10.1128/jvi.73.9.7556-7564.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kambara H, Okano H, Chiocca EA, Saeki Y. An oncolytic HSV-1 mutant expressing ICP34.5 under control of a nestin promoter increases survival of animals even when symptomatic from a brain tumor. Cancer Res. 2005;65:2832–2839. doi: 10.1158/0008-5472.CAN-04-3227. [DOI] [PubMed] [Google Scholar]
- 76.Foka P, et al. Novel tumour-specific promoters for transcriptional targeting of hepatocellular carcinoma by herpes simplex virus vectors. J. Gene Med. 2010;12:956–967. doi: 10.1002/jgm.1519. [DOI] [PubMed] [Google Scholar]
- 77.Dorer DE, Nettelbeck DM. Targeting cancer by transcriptional control in cancer gene therapy and viral oncolysis. Adv. Drug Deliv. Rev. 2009;61:554–571. doi: 10.1016/j.addr.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 78.Toth K, Dhar D, Wold WSM. Oncolytic (replication-competent) adenoviruses as anticancer agents. Expert Opin. Biol. Ther. 2010;10:353–368. doi: 10.1517/14712590903559822. [DOI] [PubMed] [Google Scholar]
- 79.Harley CB. Telomerase and cancer therapeutics. Nature Rev. Cancer. 2008;8:167–179. doi: 10.1038/nrc2275. [DOI] [PubMed] [Google Scholar]
- 80.Lee CYF, et al. Transcriptional and translational dual-regulated oncolytic herpes simplex virus type 1 for targeting prostate tumors. Mol. Ther. 2010;18:929–935. doi: 10.1038/mt.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maldonado AR, et al. Molecular engineering and validation of an oncolytic herpes simplex virus type 1 transcriptionally targeted to midkine-positive tumors. J. Gene Med. 2010;12:613–623. doi: 10.1002/jgm.1479. [DOI] [PubMed] [Google Scholar]
- 82.Kelly EJ, Hadac EM, Greiner S, Russell SJ. Engineering microRNA responsiveness to decrease virus pathogenicity. Nature Med. 2008;14:1278–1283. doi: 10.1038/nm.1776. [DOI] [PubMed] [Google Scholar]; This paper is an example of controlling virus toxicity using engineered miRNA recognition sequences within the viral genome.
- 83.Ylösmäki E, et al. Generation of a conditionally replicating adenovirus based on targeted destruction of E1A mRNA by a cell type-specific microRNA. J. Virol. 2008;82:11009–11015. doi: 10.1128/JVI.01608-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cawood R, et al. Use of tissue-specific microRNA to control pathology of wild-type adenovirus without attenuation of its ability to kill cancer cells. PLoS Pathog. 2009;5:e1000440. doi: 10.1371/journal.ppat.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sugio K, et al. Enhanced safety profiles of the telomerase-specific replication-competent adenovirus by incorporation of normal cell-specific microRNA-targeted sequences. Clin. Cancer Res. 2011;17:2807–2818. doi: 10.1158/1078-0432.CCR-10-2008. [DOI] [PubMed] [Google Scholar]
- 86.Jin H, et al. Use of microRNA Let-7 to control the replication specificity of oncolytic adenovirus in hepatocellular carcinoma cells. PLoS ONE. 2011;6:e21307. doi: 10.1371/journal.pone.0021307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang Z, Zhang X, Newman K, Liu X. MicroRNA regulation of oncolytic adenovirus 6 for selective treatment of castration-resistant prostate cancer. Mol. Cancer Ther. 2012;11:2410–2418. doi: 10.1158/1535-7163.MCT-12-0157. [DOI] [PubMed] [Google Scholar]
- 88.Ylösmäki E, et al. MicroRNA-mediated suppression of oncolytic adenovirus replication in human liver. PLoS ONE. 2013;8:e54506. doi: 10.1371/journal.pone.0054506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee CYF, Rennie PS, Jia WWG. MicroRNA regulation of oncolytic herpes simplex virus-1 for selective killing of prostate cancer cells. Clin. Cancer Res. 2009;15:5126–5135. doi: 10.1158/1078-0432.CCR-09-0051. [DOI] [PubMed] [Google Scholar]
- 90.Fu X, Rivera A, Tao L, De Geest B, Zhang X. Construction of an oncolytic herpes simplex virus that precisely targets hepatocellular carcinoma cells. Mol. Ther. 2012;20:339–346. doi: 10.1038/mt.2011.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leber MF, et al. MicroRNA-sensitive oncolytic measles viruses for cancer-specific vector tropism. Mol. Ther. 2011;19:1097–1106. doi: 10.1038/mt.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hikichi M, et al. MicroRNA regulation of glycoprotein B5R in oncolytic vaccinia virus reduces viral pathogenicity without impairing its antitumor efficacy. Mol. Ther. 2011;19:1107–1115. doi: 10.1038/mt.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kelly EJ, Nace R, Barber GN, Russell SJ. Attenuation of vesicular stomatitis virus encephalitis through microRNA targeting. J. Virol. 2010;84:1550–1562. doi: 10.1128/JVI.01788-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baer C, Claus R, Plass C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013;73:473–477. doi: 10.1158/0008-5472.CAN-12-3731. [DOI] [PubMed] [Google Scholar]
- 95.Wang Z, et al. Transcriptional and epigenetic regulation of human microRNAs. Cancer Lett. 2013;331:1–10. doi: 10.1016/j.canlet.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 96.Boviatsis EJ, et al. Long-term survival of rats harboring brain neoplasms treated with ganciclovir and a herpes simplex virus vector that retains an intact thymidine kinase gene. Cancer Res. 1994;54:5745–5751. [PubMed] [Google Scholar]
- 97.Aghi M, Kramm CM, Chou TC, Breakefield XO, Chiocca EA. Synergistic anticancer effects of ganciclovir/thymidine kinase and 5-fluorocytosine/ cytosine deaminase gene therapies. J. Natl Cancer Inst. 1998;90:370–380. doi: 10.1093/jnci/90.5.370. [DOI] [PubMed] [Google Scholar]
- 98.Parker WB, et al. Metabolism and metabolic actions of 6-methylpurine and 2-fluoroadenine in human cells. Biochem. Pharmacol. 1998;55:1673–1681. doi: 10.1016/s0006-2952(98)00034-3. [DOI] [PubMed] [Google Scholar]
- 99.Ardiani A, et al. Enzymes to die for: exploiting nucleotide metabolizing enzymes for cancer gene therapy. Curr. Gene Ther. 2012;12:77–91. doi: 10.2174/156652312800099571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mesnil M, Piccoli C, Tiraby G, Willecke K, Yamasaki H. Bystander killing of cancer cells by herpes simplex virus thymidine kinase gene is mediated by connexins. Proc. Natl Acad. Sci. USA. 1996;93:1831–1835. doi: 10.1073/pnas.93.5.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barese CN, et al. Thymidine kinase suicide gene-mediated ganciclovir ablation of autologous gene-modified rhesus hematopoiesis. Mol. Ther. 2012;20:1932–1943. doi: 10.1038/mt.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mailly L, Leboeuf C, Tiberghien P, Baumert T, Robinet E. Genetically engineered T-cells expressing a ganciclovir-sensitive HSV-tk suicide gene for the prevention of GvHD. Curr. Opin. Invest. Drugs. 2010;11:559–570. [PubMed] [Google Scholar]
- 103.Huber BE, Austin EA, Richards CA, Davis ST, Good SS. Metabolism of 5-fluorocytosine to 5-fluorouracil in human colorectal tumor cells transduced with the cytosine deaminase gene: significant antitumor effects when only a small percentage of tumor cells express cytosine deaminase. Proc. Natl Acad. Sci. USA. 1994;91:8302–8306. doi: 10.1073/pnas.91.17.8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yamada S, et al. Oncolytic herpes simplex virus expressing yeast cytosine deaminase: relationship between viral replication, transgene expression, prodrug bioactivation. Cancer Gene Ther. 2012;19:160–170. doi: 10.1038/cgt.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dias JD, et al. Targeted chemotherapy for head and neck cancer with a chimeric oncolytic adenovirus coding for bifunctional suicide protein FCU1. Clin. Cancer Res. 2010;16:2540–2549. doi: 10.1158/1078-0432.CCR-09-2974. [DOI] [PubMed] [Google Scholar]
- 106.Foloppe J, et al. Targeted delivery of a suicide gene to human colorectal tumors by a conditionally replicating vaccinia virus. Gene Ther. 2008;15:1361–1371. doi: 10.1038/gt.2008.82. [DOI] [PubMed] [Google Scholar]
- 107.Puhlmann M, Gnant M, Brown CK, Alexander HR, Bartlett DL. Thymidine kinase-deleted vaccinia virus expressing purine nucleoside phosphorylase as a vector for tumor-directed gene therapy. Hum. Gene Ther. 1999;10:649–657. doi: 10.1089/10430349950018724. [DOI] [PubMed] [Google Scholar]
- 108.Ungerechts G, et al. Lymphoma chemovirotherapy: CD20-targeted and convertase-armed measles virus can synergize with fludarabine. Cancer Res. 2007;67:10939–10947. doi: 10.1158/0008-5472.CAN-07-1252. [DOI] [PubMed] [Google Scholar]
- 109.Ungerechts G, et al. Mantle cell lymphoma salvage regimen: synergy between a reprogrammed oncolytic virus and two chemotherapeutics. Gene Ther. 2010;17:1506–1516. doi: 10.1038/gt.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows the interplay between virus replication and prodrug therapy for optimizing therapeutic outcomes.
- 110.Leveille S, Samuel S, Goulet M-L, Hiscott J. Enhancing VSV oncolytic activity with an improved cytosine deaminase suicide gene strategy. Cancer Gene Ther. 2011;18:435–443. doi: 10.1038/cgt.2011.14. [DOI] [PubMed] [Google Scholar]
- 111.Sorscher EJ, Hong JS, Allan PW, Waud WR, Parker WB. In vivo antitumor activity of intratumoral fludarabine phosphate in refractory tumors expressing E. coli purine nucleoside phosphorylase. Cancer Chemother. Pharmacol. 2012;70:321–329. doi: 10.1007/s00280-012-1908-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Seo E, et al. Effective gene therapy of biliary tract cancers by a conditionally replicative adenovirus expressing uracil phosphoribosyltransferase: significance of timing of 5-fluorouracil administration. Cancer Res. 2005;65:546–552. [PubMed] [Google Scholar]
- 113.Dai G, Levy O, Carrasco N. Cloning and characterization of the thyroid iodide transporter. Nature. 1996;379:458–460. doi: 10.1038/379458a0. [DOI] [PubMed] [Google Scholar]
- 114.Kogai T, Brent GA. The sodium iodide symporter (NIS): regulation and approaches to targeting for cancer therapeutics. Pharmacol. Ther. 2012;135:355–370. doi: 10.1016/j.pharmthera.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Baril P, Martin-Duque P, Vassaux G. Visualization of gene expression in the live subject using the Na/I symporter as a reporter gene: applications in biotherapy. Br. J. Pharmacol. 2010;159:761–771. doi: 10.1111/j.1476-5381.2009.00412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Touchefeu Y, Franken P, Harrington KJ. Radiovirotherapy: principles and prospects in oncology. Curr. Pharm. Des. 2012;18:3313–3320. doi: 10.2174/1381612811209023313. [DOI] [PubMed] [Google Scholar]
- 117.Penheiter AR, Russell SJ, Carlson SK. The sodium iodide symporter (NIS) as an imaging reporter for gene, viral, and cell-based therapies. Curr. Gene. Ther. 2012;12:33–47. doi: 10.2174/156652312799789235. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is a thorough review of the NIS reporter imaging system in multiple clinical applications.
- 118.Miest TS, Frenzke M, Cattaneo R. Measles virus entry through the signaling lymphocyte activation molecule governs efficacy of mantle cell lymphoma radiovirotherapy. Mol. Ther. 2013;21:2019–2031. doi: 10.1038/mt.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hasegawa K. Dual therapy of ovarian cancer using measles viruses expressing carcinoembryonic antigen and sodium iodide symporter. Clin. Cancer Res. 2006;12:1868–1875. doi: 10.1158/1078-0432.CCR-05-1803. [DOI] [PubMed] [Google Scholar]
- 120.Myers RM, et al. Preclinical pharmacology and toxicology of intravenous MV-NIS, an oncolytic measles virus administered with or without cyclophosphamide. Clin. Pharmacol. Ther. 2007;82:700–710. doi: 10.1038/sj.clpt.6100409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li H, Peng K-W, Dingli D, Kratzke RA, Russell SJ. Oncolytic measles viruses encoding interferon-β and the thyroidal sodium iodide symporter gene for mesothelioma virotherapy. Cancer Gene Ther. 2010;17:550–558. doi: 10.1038/cgt.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tong AW, et al. Oncolytic viruses for induction of anti-tumor immunity. Curr. Pharm. Biotechnol. 2012;13:1750–1760. doi: 10.2174/138920112800958913. [DOI] [PubMed] [Google Scholar]
- 123.Breitbach CJ, Thorne SH, Bell JC, Kirn DH. Targeted and armed oncolytic poxviruses for cancer: the lead example of JX-594. Curr. Pharm. Biotechnol. 2012;13:1768–1772. doi: 10.2174/138920112800958922. [DOI] [PubMed] [Google Scholar]
- 124.Breitbach CJ, et al. Intravenous delivery of a multi-mechanistic cancer-targeted oncolytic poxvirus in humans. Nature. 2011;477:99–102. doi: 10.1038/nature10358. [DOI] [PubMed] [Google Scholar]; This paper documents the efficient systemic delivery of replicating oncolytic virus to patient tumours.
- 125.Hu JC, et al. A phase I study of OncoVEXGM-CSF, a second-generation oncolytic herpes simplex virus expressing granulocyte macrophage colony-stimulating factor. Clin. Cancer Res. 2006;12:6737–6747. doi: 10.1158/1078-0432.CCR-06-0759. [DOI] [PubMed] [Google Scholar]
- 126.Leonard VHJ, et al. Measles virus blind to its epithelial cell receptor remains virulent in rhesus monkeys but cannot cross the airway epithelium and is not shed. J. Clin. Invest. 2008;118:2448–2458. doi: 10.1172/JCI35454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Leonard VHJ, Hodge G, Reyes-Del Valle J, McChesney MB, Cattaneo R. Measles virus selectively blind to signaling lymphocytic activation molecule (SLAM; CD150) is attenuated and induces strong adaptive immune responses in rhesus monkeys. J. Virol. 2010;84:3413–3420. doi: 10.1128/JVI.02304-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mühlebach MD, et al. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature. 2011;480:530–533. doi: 10.1038/nature10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Noyce RS, et al. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog. 2011;7:e1002240. doi: 10.1371/journal.ppat.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fabre-Lafay S, et al. Nectin-4 is a new histological and serological tumor associated marker for breast cancer. BMC Cancer. 2007;7:73. doi: 10.1186/1471-2407-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Derycke MS, et al. Nectin 4 overexpression in ovarian cancer tissues and serum: potential role as a serum biomarker. Am. J. Clin. Pathol. 2010;134:835–845. doi: 10.1309/AJCPGXK0FR4MHIHB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Takano A, et al. Identification of nectin-4 oncoprotein as a diagnostic and therapeutic target for lung cancer. Cancer Res. 2009;69:6694–6703. doi: 10.1158/0008-5472.CAN-09-0016. [DOI] [PubMed] [Google Scholar]
- 133.Wang Z, Gerstein M, Snyder M. RNA-seq: a revolutionary tool for transcriptomics. Nature Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.The ENCODE Project An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 136.Yaghoubi SS. Positron emission tomography reporter genes and reporter probes: gene and cell therapy applications. Theranostics. 2012;2:374–391. doi: 10.7150/thno.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Koba W, et al. Imaging devices for use in small animals. Semin. Nucl. Med. 2011;41:151–165. doi: 10.1053/j.semnuclmed.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 138.Ray P. Multimodality molecular imaging of disease progression in living subjects. J. Biosci. 2011;36:499–504. doi: 10.1007/s12038-011-9079-0. [DOI] [PubMed] [Google Scholar]