Abstract
Aim:
To investigate the role of insulin receptor substrate 2 (IRS-2) in oncogenic transformation induced by v-src.
Methods:
IRS-2 gene was silenced using small interfering RNAs (siRNAs). Nuclear translocation and interaction of IRS-2 with v-src was determined using subcellular fractionation, confocal microscopy, and immunoprecipitation. The activity of the cyclin D1 promoter and r-DNA promoter was measured with a luciferase assay.
Results:
Depletion of IRS-2 inhibited R-/v-src cell growth and reverse the oncogenic transformation. IRS-2 bound to src via its two PI3-K binding sites, which are critical for activities involved in the transformation. Nuclear IRS-2 occupied the cyclin D1 and rDNA promoters. The combination of IRS-2 and v-src increased the activity of the two promoters, especially the rDNA promoter.
Conclusion:
Depletion of insulin receptor substrate 2 could reverse oncogenic transformation induced by v-src.
Keywords: insulin receptor substrate 2 (IRS-2), cellular transformation, nuclear translocation, v-src, cyclin D1 promoter, rDNA promoter, RNA interference
Introduction
The insulin-like growth factor-1 receptor (IGF-1R) mediates its functions through the activation of an intrinsic tyrosine kinase in its cytoplasmic domain. Upon activation, the receptor is autophosphorylated and recruits intracellular substrates, such as the insulin receptor substrate (IRS) proteins. IRS-1 and IRS-2 are two important adaptor molecules essential for IGF-1R intracellular signaling1. The IRS proteins have a common amino terminus that includes a pleckstrin homology (PH) domain and a phosphotyrosine-binding (PTB) domain. These two domains are comprised of roughly 300 amino acids and are followed by long tails that vary among the different IRS proteins. These domains contain a number of binding sites for different substrates (such as PI3-K, Grb2, and phosphatases)2. Despite extensive experimental studies over the past few years, the role of IRS proteins in oncogenic transformation remains elusive.
The IRS proteins are able to coordinate and amplify numerous signals that are critical during tumorigenesis. In particular, both IRS-1 and IRS-2 can similarly signal to induce proliferation and survival, two activities essential for cellular transformation3. In fibroblasts lacking IRS-1, IRS-2 can compensate for IRS-1's role in signal transduction but cannot stimulate cellular proliferation4. However, IRS-1 and IRS-2 may work differently in respect to certain phenotypes related to various oncogenes. Recently, it was shown that IRS-2 was an essential intermediate in the activation of PI3-K, promoting breast carcinoma through the α6β4-integrin receptor5. Signaling through the IRS-2 adaptor protein is important for promoting tumor cell invasion and survival (hallmarks of metastasis), and the relative IRS-2 expression level in tumors can significantly affect disease progression in patients with breast cancer5. Therefore, pathways that regulate IRS-2 expression, as well as downstream pathways activated through IRS-2, represent potential novel therapeutic targets.
The role of IGF-1R in malignant transformation is based on the observation that the targeted disruption of IGF-1R genes in mouse embryonic fibroblasts (MEFs) prevents their transformation6. MEFs that contain disrupted IGF-1R genes, referred to as R-cells, are resistant to transformation induced by numerous viral and cellular oncogenes, including the SV40 T antigen, the activated Ha-ras oncogene, the bovine papillomavirus E5 protein, the human papillomavirus E7 protein, the Ewing's sarcoma fusion protein, and activated src. To date, the only oncogenes known to transform R-cells are v-src and a mutant of Gα137. The observation that v-src is one of only two oncogenes to transform R-cells is quite remarkable because MEFs have a strong tendency to become transformed. Thus, understanding the actions of v-src in R-cells should be useful for elucidating IRS-dependent transformation.
The v-src oncogene of the Rous sarcoma virus is a 60 kDa tyrosine kinase capable of causing transformation in a variety of cell types. In v-src-transformed cells, both IGF-1R and IRS are constitutively phosphorylated. One of the most intriguing aspects of IRS signaling is that IRS proteins have noncanonical functions within the nucleus8, 9. In a similar fashion, IGF-1R and either SV40 T or v-src can cause the nuclear translocation of IRS proteins in MEFs. Once inside the nucleus, studies suggest that IRS-1 or IRS-2 binds the upstream binding factor 1 (UBF-1), playing a role in regulating RNA polymerase 1 activity and the subsequent synthesis of ribosomal RNA10, 11.
In contrast to IRS-1, few studies have addressed the role of IRS-2 in oncogenic transformation. Our previous findings indicate the potential role of IRS-2 in cellular proliferation, but the functional significance of IRS-2 in transformation needs to be clarified9, 11. In the present study, we provide evidence for the role of IRS-2 nuclear translocation in R-/v-src and BT-20 cancer cells. Additionally, we demonstrate the role of IRS-2 in the proliferation and anchorage-independent growth in combination with v-src. These data support the hypothesis that IRS-2 plays a significant role in oncogenic transformation.
Materials and methods
Cell lines and plasmids
R-cells and R-derived cells are 3T3-like cells from mouse embryos with a targeted disruption of the IGF-1R genes4. BT-20 breast cancer cells and 32D-derived cells have been previously described7. Mutant IRS-2 plasmids have been previously described6. One IRS-2 mutant is essentially inactive because of the deletion of the PTB domain between residue 155 and residue 309 (delta PTB). The second mutant has a deletion of the pleckstrin domain (delta PH). A third mutant has decreased activity because of the mutation of the PI3-K-binding-site tyrosines 608 and 939 to phenylalanines.
Gene silencing of IRS-2 was performed by RNA interference using small interfering RNAs (siRNAs). R-/v-src cells were mock-transfected with vehicle (diethylpyrocarbonate-treated water), transfected with control siRNA (scrambled), or transfected with siRNA directed against IRS-2 (siRNA IRS-2: 5′-AAUAGCUGCAAGAGCGAUGAC-3′) in each well of a 12-well plate with the Gene Porter system (Gene Therapy System, San Diego, CA, USA). The second siRNA was directed against IRS-2 (Santa Cruz Biotechnology, Inc, Santa Cruz, CA, USA) using TransIT-siTKO reagents (Mirus Bio Corporation, Madison, WI, USA) according to the manufacturer's instructions. Generation of IRS-2-stably depleted R-/v-src cells was performed by antibiotic selection with G418 after they were transfected with a pcDNA3 vector containing an antisense cDNA against IRS-2, as previously described6.
Colony formation in soft agar
Briefly, to compare the anchorage-independent growth of different cell lines, cells were plated in essential modified Eagle's medium containing 10% fetal bovine serum (with or without IGF-1) and 0.2% agarose (with a 0.4% agarose underlay). The numbers of colonies larger than 125 μm in diameter were determined 3 weeks after cells were plated.
Confocal microscopy
After fixation with 3% paraformaldehyde for 25 min, cells on coverslips were washed three times with PBS at room temperature, permeabilized with 0.2% Triton X-100 in PBS for 5 min, blocked with 10% normal donkey serum (sc-2044, Santa Cruz Biotechnology, Inc, Santa Cruz, CA, USA) for 20 min, and incubated for 1 h with appropriate primary and secondary antibodies. Confocal analysis was carried out according to the manufacturer's protocol.
Subcellular fractionation, Western blotting, and immunoprecipitation
For immunoprecipitation, 200 μg of nuclear or cytoplasmic lysate was incubated for 2 h at 4 °C with corresponding antibodies coupled to 20 μL of packed protein G-sepharose beads (Oncogene Science, Inc). Immunocomplexes were electrophoresed on a 4%–15% SDS-PAGE gel. Proteins were transferred onto a nitrocellulose membrane and were probed with the indicated antibodies4, 6. The blots were detected using enhanced chemiluminescence system (Amersham Biosciences, Piscataway, NJ, USA). The protein size was confirmed by molecular weight standards (Invitrogen, Carlsbad, CA, USA). The intensity of the bands on western blots was analyzed by Image J (software from NCBI).
GST pull-down analysis
All GST-fusion proteins with various regions of IRS-2 were expressed in BL-21 bacterial cells (Invitrogen, Carlsbad, CA, USA) and purified with glutathione-agarose beads using standard techniques. Cells were lysed with 50 mmol/L HEPES (pH 7.5), 1% Nonidet P-40, 1 mmol/L EGTA, 10 mmol/L NaF, 20 mmol/L sodium pyrophosphate, 10 g/mL aproptinin, and 10 μg/mL leupeptin on ice for 30 min. The resulting supernatants were incubated with the immuno-immobilized GST proteins overnight. After extensive washing with 50 mmol/L HEPES (pH 7.5), 150 mmol/L NaCl, and 0.1% Triton X-100, the proteins bound to either IRS-2 or GST (control) were analyzed by SDS-PAGE, followed by immunoblotting with appropriate antibodies.
Chromatin immunoprecipitation assay (ChIP)
Chromatin immunoprecipitation assays were performed according to the manufacturer's instructions (Upstate, Lake Placid, NY, USA) and previously described methods10. An Epicentre Fail Safe PCR System (Epicentre, Madison, WI, USA) was used for PCR. The amplification products were analyzed in a 2% agarose gel and visualized by SYBR Gold (Molecular Probes, Eugene, OR, USA) staining. Gels stained with SYBR Gold were scanned with a Typhoon Laser scanner (Typhoon 9400 Variable Mode Imager, Amersham Biosciences, Piscataway, NJ, USA).
Reporter genes
The cyclin D1 promoter and r-DNA promoter were used to transiently express luciferase. Determination of luciferase activity was performed with standard procedures. Transfection efficiency was monitored using previously described methods10. Each cell line was transfected in a single batch before being subdivided into aliquots that were measured at different time intervals after transfection and insulin stimulation. Mock-transfected cells were utilized to determine background.
Antibodies
The following antibodies were used: phospho-IRS-2 antibody against phosphorylated Tyr residues in IRS-2 (catalog No sc-17195-R, Santa Cruz Biotechnology, Inc, USA), mouse monoclonal anti-UBF (Santa Cruz Biotechnology, Inc, USA), anti-IRS-2 (Upstate, Inc, USA), v-src and mouse monoclonal anti-Grb2 (both from Transduction Laboratories, USA), goat anti-rabbit IgG-FITC (Santa Cruz Biotechnology, Inc, USA) and goat anti-mouse IgG2a-FITC (Santa Cruz Biotechnology, Inc, USA). The siRNA was from Santa Cruz Biotechnology, Inc or UBI Biotechnology, Inc.
Statistical analysis
Experiments were performed in triplicate and repeated at least three times. Results are expressed as mean±SD. All statistical analyses were performed with SigmaStat for Windows version 3.10 (Systat Software, Inc, Port Richmond, CA, USA). Differences were considered statistically significant when P<0.05.
Results
Depletion of IRS-2 inhibits growth and reverses transformation of R-/v-src cells
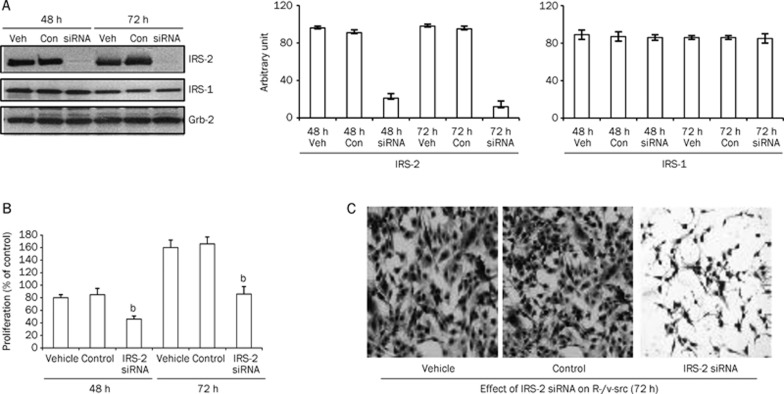
One of the distinctive features of transformed cell lines such as R-/v-src cells is the ability to proliferate in the absence of serum. Using siRNA, we effectively depleted endogenous IRS-2 when compared with vehicle-treated or control-treated cells. Additionally, this depletion had no effect on IRS-1 or Grb2, an unrelated internal control (Figure 1A). IRS-2 depletion considerably retarded the growth of R-/v-src cells in the absence of serum (Figure 1B), suggesting that endogenous IRS-2 contributes to the growth of R-/v-src cells in these conditions. To confirm the growth of identical cells that underwent treatment, we compared microscopic images of vehicle, control, and siRNA/IRS-2-transfected cells after 72 h (Figure 1C). Importantly, preliminary tests indicated that these siRNAs at this concentration had no toxic effect on R-/v-src cells with serum present (data not shown).
Figure 1.
Depletion of endogenous IRS-2 proteins effectively reduces the proliferation of R-/v-src cells in serum-free conditions. (A) Gene knockdown of IRS-2 was achieved by siRNA. Densitometric analysis of endogenous IRS-2 and IRS-1 were quantified by Image J (software from NCBI) and normalized to the internal control Grb-2. Values in arbitrary units from three independent experiments are shown. (B) Cells at 48 and 72 h were processed and analyzed for growth. Values are expressed as percent increase over serum free medium (SFM). bP<0.05 compared with vehicle-treated control cells by ANOVA. (C) Microscopic images of R-/v-src cells treated with siRNA designed against IRS-2. Untreated cells and cells treated with siRNA were compared at 72 h.
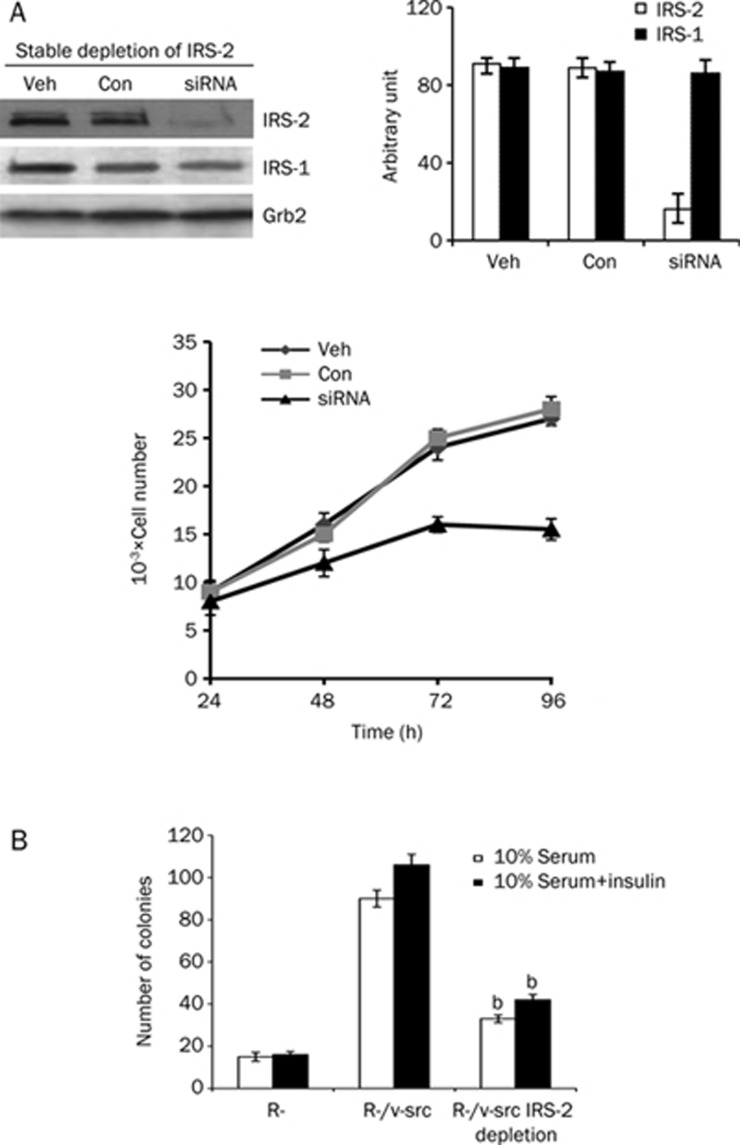
Based on these data, we sought to confirm the role of IRS-2 in the transformation of R-/v-src cells by stably depleting endogenous IRS-2. Antisense IRS-2 siRNAs dramatically reduced IRS-2 protein expression in R-/v-src cells (Figure 2A, top). Furthermore, the stable depletion of IRS-2 strongly decreased the growth of R-/v-src cells under conditions of serum starvation (Figure 2A, bottom), confirming our transient siRNA experimental findings.
Figure 2.
Depletion of IRS-2 inhibits growth and reverses the transformation of R-/v-src cells. (A) R-/v-src cells were transfected with a specific siRNA for IRS-2 and either DMSO (Veh) or control siRNA (Con) as described in the Materials and methods. IRS-2 expression in cell lysates was detected by immunoblotting at 48 h after transfection. Cells were washed, transferred to SFM, and counted at the indicated times. The level of endogenous IRS-2 and IRS-1 was detected by immunoblotting, and protein expression was quantified by Image J (software from NCBI) and normalized to the internal control Grb-2. Mean±SD. bP<0.05 compared to control oligo-treated cells by ANOVA. (B) Colony formation of R-/v-src cells in soft agar after IRS-2 siRNA transfection. Cells were counted after 4 weeks. Colonies >125 μm were scored as positive. Mean±SD.
Another critical measurement of cellular transformation is the ability of cancer cells to grow in an anchorage-independent manner. Therefore, we wanted to determine whether or not the stable depletion of IRS-2 in R-/v-src cells would affect their ability to form colonies in soft agar. We determined that only R-cells expressing wild-type IRS-2 and v-src formed colonies in soft agar (Figure 2B). Adding insulin increased the number of colonies on the plates. As expected, the number of colonies formed by R-/v-src cells lacking IRS-2 was significantly decreased in soft agar, confirming the fact that cellular transformation requires the activation of IRS-2 and is v-src independent.
Our findings indicate that IRS-2 is essential for the role of v-src to encourage serum-independent and anchorage-independent cellular growth, two key properties of aggressive tumorigenesis.
IRS-2 domains are required for interaction with v-src and PI3-K-binding tyrosine residues
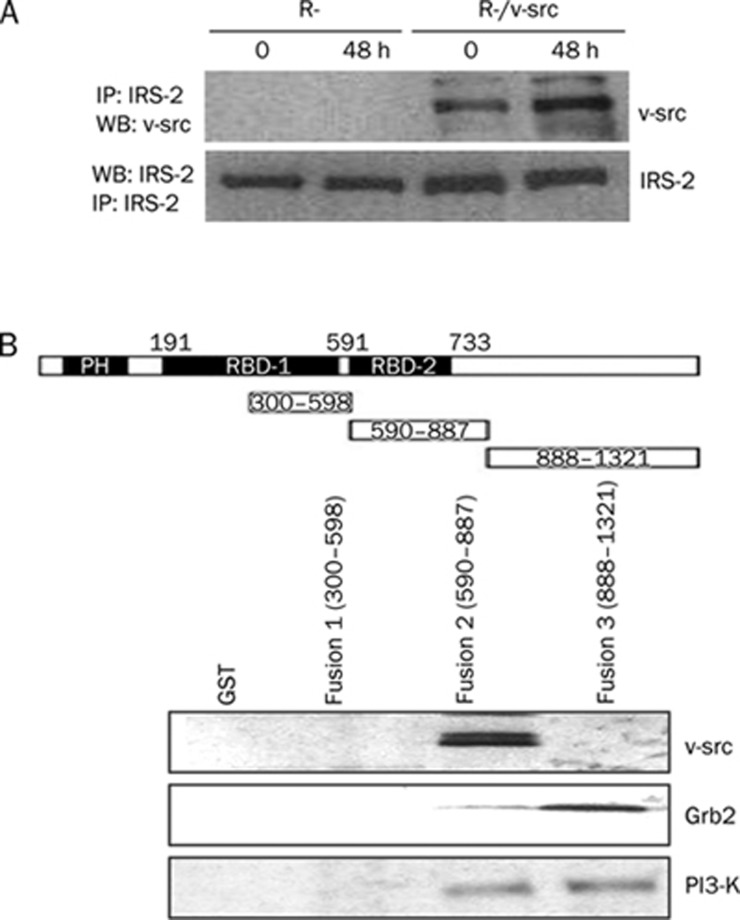
To elucidate the role of IRS-2 in the growth inhibition of R-/v-src cells, we investigated the interaction between IRS-2 and v-src by immunoprecipitating (IP) lysates from R- and R-/v-src cells. When lysates were immunoprecipitated with IRS-2 antibodies, and Western blots were performed with antibodies against v-src, we detected an interaction between v-src and IRS-2 (Figure 3A). To further elucidate the domain required for this interaction, we performed a GST pull-down assay to determine the presence of proteins known to interact with IRS proteins and v-src, such as PI3-K and Grb2. The results shown in Figure 3B indicated that the p85 subunit of PI3-K could be pulled down with amino acids between residues 590 and 1321 of IRS-2. Additionally, the stretch of amino acids from residues 590 to 887 also pulled down v-src.
Figure 3.
Interaction between IRS-2 and v-src. (A) IPs performed with an antibody against IRS-2, and Western blots performed with an anti-v-src antibody. (B) Diagram of IRS-2 GST constructs (RBD, receptor binding domain) and pull-down experiments for PI3-K, Grb2, and v-src.
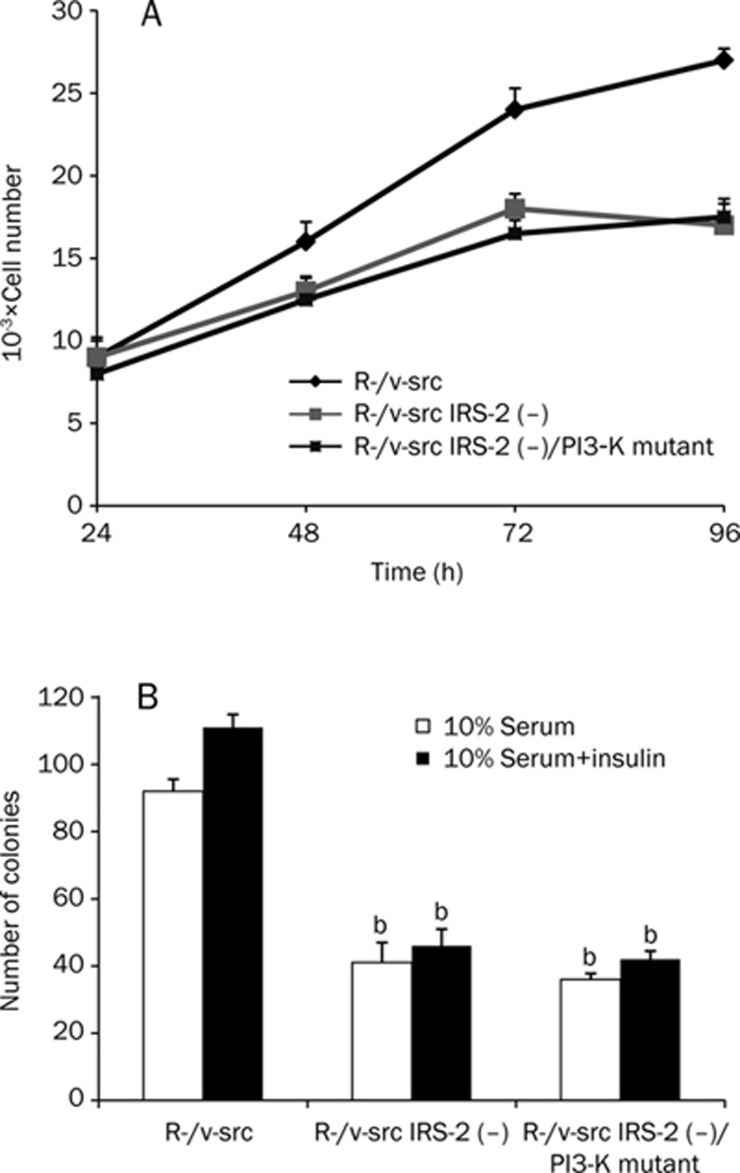
These data suggest that the sequences responsible for the binding of IRS-2 to v-src are required for IRS-2's involvement in transformation. To identify the importance of this PI3-K binding site, we stably expressed IRS-2 with a PI3-K binding site mutation in IRS-2 depleted R-/v-src cells. As shown in Figure 4, although IRS-2 still bound v-src (at one or several other sites other than the PI3-K site), the deletion of this essential PI3-K binding site abolished transformation, indicating that PI3-K is required in this process.
Figure 4.
V-src co-precipitates Tyr-phosphorylated IRS-2. Cell lysates were immunoprecipitated with an antibody to v-src. Western blots with antibodies directed against anti-phospho-IRS-2 and v-src were performed.
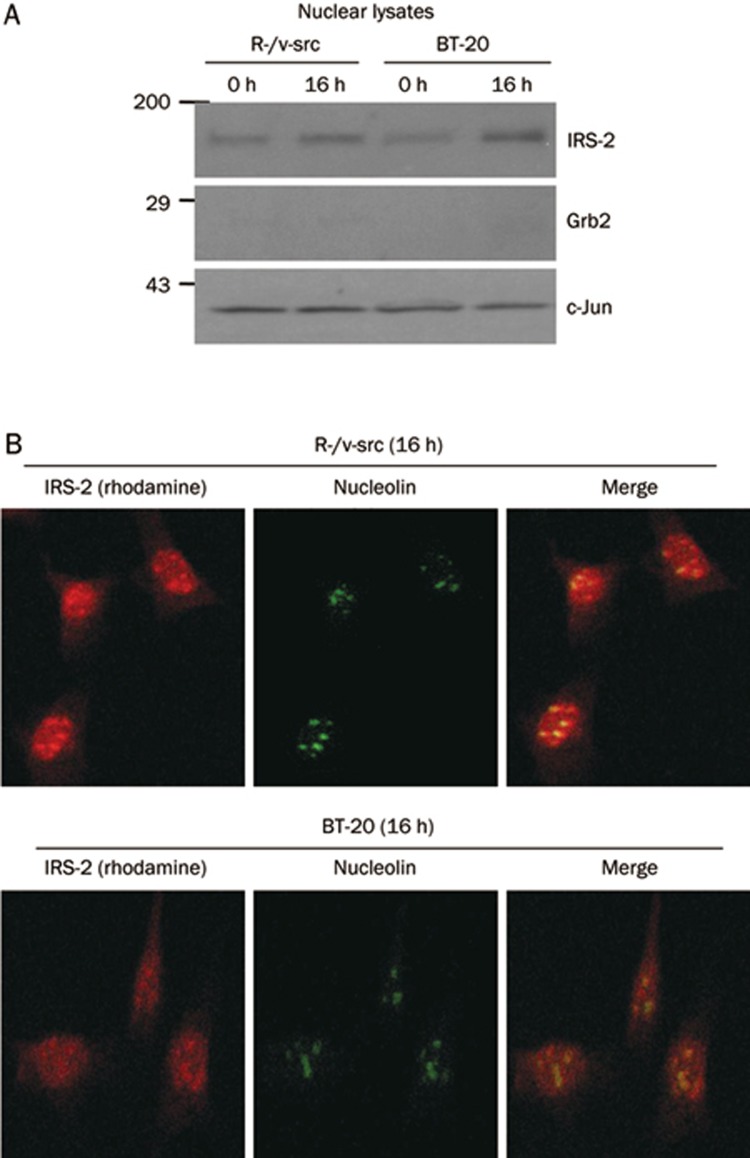
Nuclear translocation of IRS-2 in R-/v-src and BT-20 cells
Nuclear translocation is considered to be one of the most important steps in the process of cellular transformation7. Following subcellular fractionation, IRS-2 was present in the nuclear fraction of R-/v-src and BT-20 cells (Figure 5A). Grb2 and c-Jun antibodies were used to monitor the purity of these subcellular fractions8. Nuclear translocation of IRS-2 in R-/v-src and BT-20 cells was then confirmed by confocal microscopy. After stimulation with insulin for 16 h, merged images of R-/v-src and BT-20 cells stained with antibodies directed against IRS-2 (rhodamine, red) and nucleolin (FITC, green) (Figure 5B, top) showed that a substantial fraction of IRS-2 was detectable in the nuclei of these cells, indicated by the co-expression of IRS-2 and nucleolin (greenish-yellow nucleolin).
Figure 5.
Nuclear translocation of IRS-2 in R-/v-src and BT-20 cells. (A) Subcellular localization of IRS-2 in R-/v-src and BT-20 cells. Nuclear fractions were prepared from cells in serum-free medium after insulin stimulation (0 and 16 h). To control for the purity of the subcellular fractions, we used antibodies to c-Jun (for nuclear purity) and antibodies to Grb2 (for cytosolic purity). (B) Confocal microscopy of BT-20 cells stained with antibodies directed against IRS-2 and nucleolin. IRS-2 is found in the nucleus as well as in the cytoplasm at the indicated times with insulin stimulation indicated by the arrow.
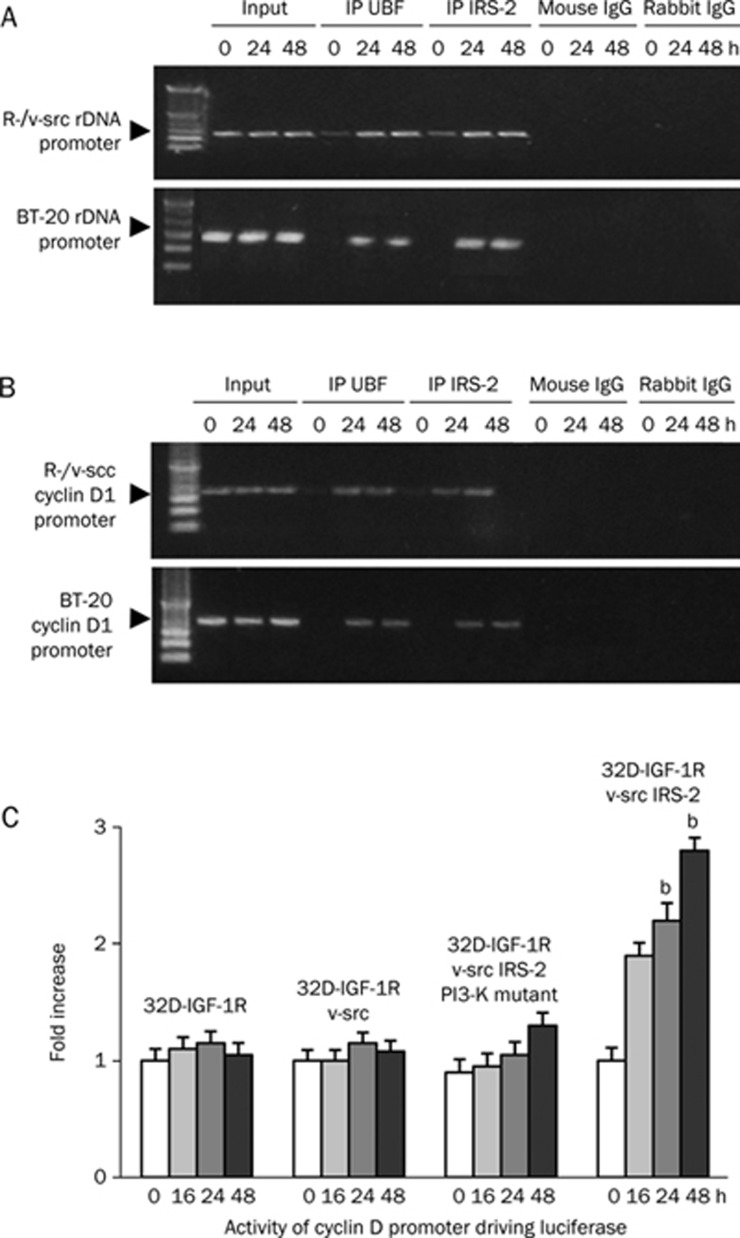
Recruitment of nuclear IRS-2 to the rDNA and cyclin D1 promoters
The presence of IRS-2 in the nuclei of transformed cells suggested that IRS-2 might be associated with the regulatory sequences of target genes such as rDNA and cyclin D1, which are involved in cellular transformation and cell-cycle regulation. To determine whether IRS-2 was associated with the regulatory sequences of target genes, we investigated the recruitment of IRS-2 to these promoters by ChIP. IRS-2 was detected with ChIP at the rDNA and cyclin D1 promoters. Because upstream binding factor (UBF) is always present at the rDNA and cyclin D1 promoters, this protein served as a positive control10, 11. Mouse and rabbit IgG served as the negative controls. These data are presented in Figure 6A and 6B, which shows ChIP results from untreated R-/v-src cells in serum-free medium (SFM) and cells stimulated with insulin for 24 and 48 h. In the R-/v-src cells, IRS-2 was detectable at both the cyclin D1 and rDNA promoters 24 and 48 h after insulin stimulation. A weak signal was already detectable at both promoters in unstimulated cells, but this signal became more clear after 24 h of stimulation.
Figure 6.
Recruitment of IRS-2 to the rDNA and cyclin D1 promoter. The main panel shows the results of experiments in which sonicated chromatin from R-/v-src and BT-20 cells was immunoprecipitated with antibodies against IRS-2 and UBF (positive control). (A and B): the first lane is time 0 (no stimulation with insulin), followed by insulin stimulation for 24 and 48 h, respectively. (C) Activity of the cyclin D1 promoters in 32D-derived cell lines.
To further validate this result, we used ChIP assays to investigate whether or not IRS-2 was present at the rDNA and cyclin D1 promoters of BT-20 cells. IRS-2 was still detectable at both promoters. It seems that the mechanism by which IRS-2 promotes transformation is, at least in part, due to its participation in a complex of the rDNA promoter and UBF1, a protein that upregulates RNA polymerase I activity12, 13.
Activity of the cyclin D1 and rDNA promoters
The presence of IRS-2 at the rDNA and cyclin D1 promoters indicated that IRS-2 had a functional effect on rDNA and cyclin D1 activity during proliferation. The influence of this promoter occupancy was confirmed by determining the activities of the cyclin D1 and rDNA promoters in 32D-derived cell lines that were expressing v-src in addition to wild type or mutant IRS-2. For these experiments, the 32D-derived cell lines were transiently transfected with luciferase reporter genes. Then, luciferase activity was determined after insulin stimulation. Cyclin D1 promoter activity was slightly increased in parental 32D-IGF-1R cells and in 32D-IGF-1R cells expressing only v-src (Figure 6C). When IRS-2 and v-src were added combinatorially, the promoter activity was dramatically increased. However, when v-src was expressed with a PI3-K binding site deletion mutant of IRS-2, the promoter activity of cyclin D1 was comparable to the promoter's activity in parental 32D-IGF-1R cells. In conclusion, it seems as though IRS-2 contains distinct receptor binding domains that are different from IRS-1. Combined with the noncanonical functions of IRS-2 protein within the nucleus, IRS-2 may also be the principal cause of cellular transformation.
Discussion
In this study, we provide several lines of evidence that indicate the functional involvement of IRS-2 in oncogenic transformation. First, we demonstrated that the down-regulation of IRS-2 could inhibit growth and reverse transformation of R-/v-src cells. In addition, we determined that IRS-2 bound to v-src through its two PI3-K-binding tyrosine residues. Furthermore, it appears that these two residues are critical for cellular transformation. Moreover, we demonstrated that IRS-2 could translocate to the nuclei of R-/v-src and BT-20 breast cancer cells. Finally, IRS-2 occupied the cyclin D1 and rDNA promoters, and when combined with v-src, increased the activity of these two promoters (especially the rDNA promoter). These results indicate that IRS-2 plays a significant role in the activation of cell cycle progression and transformation by v-src.
The role of IGF-1R in cancer was previously established in MEFs from mice carrying a targeted disruption of the IGF-1R genes (R-cells), which are usually refractory to transformation by viral and cellular oncogenes (including the SV40 T antigen)1. However, the 60 kDa tyrosine kinase v-src oncogene is one of only two oncogenes able to bypass the requirement of IGF-1R to promote both proliferation and anchorage-independent growth in R-cells. This exception indicated that IGF-1R and its downstream signaling pathways might be required for transformation2.
We have recently reported the nuclear translocation of IRS-2 in R+ cells, but the functional consequence of this translocation remains unclear6. In this study, we confirmed that the nuclear translocation of IRS-2 in R-/v-src and BT-20 cells was involved in cell growth and transformation. Furthermore, we also demonstrated that the presence of IRS-2 at the rDNA and cyclin D1 promoters had a functional effect on their activity, and actually caused a marked activation of both promoters14. These findings are in agreement with the observation that IRS-2 interacts with src and that IGF-1R activates STAT proteins that are src-responsive15.
Most transformation studies have focused on IRS-1, whereas the investigation of IRS-2's function in transformation has been limited4. However, recent studies indicate that the distinct biological actions of IRS-2 during cellular transformation have been underestimated. One such study indicated that IRS-2 exhibited the ability to initiate and promote mammary tumorigenesis5. The lengthy time required to form mammary tumors in IRS-2 transgenic animals suggests that other oncogenes act in concert with IRS-2, presumably by activating IRS-2. In addition, we have previously shown that unlike IRS-1, both wild-type IRS-2 and a truncated IRS-2 protein lacking the PH and PTB domains inhibit differentiation and sustain the IGF-1-dependent growth of 32D-IGF-1R cells6.
The importance of IRS-2 in cellular proliferation may be due to its nuclear translocation. There have been reports that delta-PHPTB IRS-2 may have biological activity16, 17. The delta-PHPTB IRS-2 is fully capable (as is wild type IRS-2 and IRS-1) of both stimulating growth and inhibiting differentiation of 32D-IGF-1R cells. In contrast, IRS-1 proteins lacking the same PH and PTB domains are completely inactive in blocking differentiation and stimulating IL-3-independent growth of 32D-IGF-1R cells6. Down-regulation of IRS-2 may affect the protein's nuclear functions, bypassing its conventional signaling pathway17, 18.
Our results show that in R-/v-src cells, transformation by v-src requires IRS-2, and that this mechanism seems to be based on an interaction between the two proteins. As shown in the GST pull-down assay, the sequence between the amino acid residues 590 and 887 in IRS-2 potentially interacts with v-src. Therefore, the phosphorylation status of IRS-2 and its effect on binding to v-src is of interest. To exclude interference from IRS-1, we performed the same experiments in the BT-20 cell line, which has no endogenous IRS-1, allowing an excellent model to verify the specific interaction between phospho-IRS-2 and v-src. This interaction was confirmed by Western blotting (with phospho-IRS-2 antibodies that specifically detect phosphorylated IRS-2 at Tyr612) an IP lysates of v-src (Figure S1). Results indicated that v-src co-precipitated phosphorylated IRS-2.
Wild type IRS-2 is detectable at both the rDNA and cyclin D1 promoters, and the PI3-K binding sites of IRS-2 are required for v-src binding19, 20. The importance of IRS-2 in v-src transformation was also demonstrated by the inability of IRS-2 protein with mutated PI3-K binding sites to activate the rDNA and cyclin D1 promoters. We propose that IRS-2 binds v-src, which leads to the phosphorylation of IRS-2 in the absence of IGF-1R signaling. This interaction requires the PI3-K binding sites of IRS-2, which causes the subsequent nuclear translocation of IRS-2 and its occupancy at the rDNA and cyclin D1 promoters that is observed during transformation21, 22. Oncogenes v-src not only directly associate with IRS-2, but also are dependent on IRS-2 tyrosine phosphorylation for their mitogenic and transforming activity.
Despite its nuclear localization, IRS-2 phosphorylates UBF, suggesting that its activation of cell proliferation and rRNA synthesis are regulated through conventional signal transduction pathways23, 24. The activity of UBF1 depends on its phosphorylation state and cellular levels25. It was reported that IRS-2 is more effective than IRS-1 in the InR-mediated phosphorylation of ERKs26. There is a possible contribution of ERKs to the overall activity of UBF1 because of the fact that ERKs phosphorylate UBF1 at two threonine residues (residues 117 and 201). Mutations at these residues inactivate UBF127, suggesting a possible link between these signaling pathways28, 29. Further studies are required to determine the relative roles of IRS-1 and IRS-2, as well as the upstream activators and downstream effectors of IRS-2 during oncogenic transformation.
Conclusion
We have shown that oncogenic transformation by v-src in MEFs requires the association of v-src with IRS-2. We have also shown that IRS-2 binds src through two tyrosine residues that also bind PI3-K. The interaction of v-src, coupled with the nuclear translocation of IRS-2, might be responsible for cell growth and transformation.
Abbreviations
IGF-1R, IGF-1 receptor; IRS, insulin receptor substrate; MEF, mouse embryo fibroblast; PH, pleckstrin homology; PI3-K, phosphatidylinositol 3-kinase; PTB, phosphotyrosine binding domain; UBF, upstream binding factor.
Author contribution
Hong-zhi SUN and Shu-fang WU designed and executed the experiments and drafted the manuscript. Bo ZHOU and Lin XU conducted most of the experiments and contributed to manuscript preparation. Wei-jin ZANG contributed to the overall experimental design.
Acknowledgments
We are grateful for the donation of plasmids from Dr Renato Baserga at the Kimmel Cancer Center. This work was supported by grants from the National Natural Science Foundation of China (Key Program, No 30930105; General Program, No 30971392) and the National Basic Research Program of China (973 Program, No 2007CB512005). This work was also supported by the New Century Excellent Talent in University from the Ministry of Education program (NCET-08-0435) and the “Tengfei” Supporting Program of Xi'an Jiaotong University.
Footnotes
Supplementary figure is available at Acta Pharmacologica Sinica website of NPG.
Supplementary Information
V-src favors co-precipitation of Tyr phosphorylated IRS-2.
References
- Nagle JA, Ma ZF, Byrne MA, White MF, Shaw LM. Involvement of insulin receptor substrate 2 in mammary tumor metastasis. Mol Cell Biol. 2004;24:9726–35. doi: 10.1128/MCB.24.22.9726-9735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baserga R, Peruzzi F, Reiss K. The IGF-I receptor in cancer biology. Int J Cancer. 2003;107:873–7. doi: 10.1002/ijc.11487. [DOI] [PubMed] [Google Scholar]
- Baserga R. The insulin-like growth factor-1 receptor as a target for cancer therapy. Expert Opin Ther Targets. 2005;9:753–68. doi: 10.1517/14728222.9.4.753. [DOI] [PubMed] [Google Scholar]
- Sun H, Tu X, Prisco M, Wu A, Casiburi I, Baserga R. Insulin-like growth factor I receptor signaling and nuclear translocation of insulin receptor substrates 1 and 2. Mol Endocrinol. 2003;17:472–86. doi: 10.1210/me.2002-0276. [DOI] [PubMed] [Google Scholar]
- Dearth RK, Cui XJ, Kim HJ, Hadsell DL, Lee AV. Oncogenic transformation by the signaling adaptor proteins insulin receptor substrate (IRS)-1 and IRS-2. Cell Cycle. 2007;6:705–13. doi: 10.4161/cc.6.6.4035. [DOI] [PubMed] [Google Scholar]
- Sun H, Baserga R. Deletion of the pleckstrin and phosphotyrosine binding domains of insulin receptor substrate-2 does not impair its ability to regulate cell proliferation in myeloid cells. Endocrinology. 2004;145:5332–43. doi: 10.1210/en.2004-0668. [DOI] [PubMed] [Google Scholar]
- Dalmizrak O, Wu A, Chen J, Sun H, Utama FE, Zambelli D, et al. Insulin receptor substrate-1 regulates the transformed phenotype of BT-20 human mammary cancer cells. Cancer Res. 2007;67:2124–30. doi: 10.1158/0008-5472.CAN-06-3954. [DOI] [PubMed] [Google Scholar]
- Reiss K, Wang JY, Romano G, Tu X, Peruzzi F, Baserga R. Mechanism of regulation of cell adhesion and motility by insulin receptor substrate-1 in prostate cancer cells. Oncogene. 2001;20:490–500. doi: 10.1038/sj.onc.1204112. [DOI] [PubMed] [Google Scholar]
- Sun H, Tu X, Baserga R. A mechanism for cell size regulation by the insulin and IGF-1 receptors. Cancer Res. 2006;66:11106–9. doi: 10.1158/0008-5472.CAN-06-2641. [DOI] [PubMed] [Google Scholar]
- Drakas R, Tu X, Baserga R. Control of cell size through phosphorylation of upstream binding factor1 by nuclear phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2004;101:9272–6. doi: 10.1073/pnas.0403328101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baserga R. Customizing the targeting of IGF-1 receptor. Future Oncol. 2009;5:43–50. doi: 10.2217/14796694.5.1.43. [DOI] [PubMed] [Google Scholar]
- Voit R, Grummt I. Phosphorylation of UBF at serine 388 is required for interaction with RNA polymerase I and activation of rDNA transcription. Proc Natl Acad Sci U S A. 2001;98:13631–6. doi: 10.1073/pnas.231071698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L, Song Y, Tan C, Bui T. Inhibition of RNA polymerase I transcription in differentiated myeloid leukemia cells by inactivation of selectivity factor-1. Cell Growth Diff. 2000;11:63–70. [PubMed] [Google Scholar]
- Sun H, Baserga R. The role of insulin receptor substrate-1 in transformation by v-src. J Cell Physiol. 2008;215:725–32. doi: 10.1002/jcp.21352. [DOI] [PubMed] [Google Scholar]
- White MF. The IRS-signalling system: a network of docking proteins that mediate insulin action. Mol Cell Biochem. 1998. 182. pp. 3–11. [PubMed]
- Klippel A, Escobedo MA, Wachowicz MS, Apell G, Brown TW, Giedlin MA, et al. Activation of phosphatidylinositol 3-kinase is sufficient for cell cycle entry and promotes cellular changes characteristic of oncogenic transformation. Mol Cell Biol. 1998;18:5699–711. doi: 10.1128/mcb.18.10.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byron SA, Horwitz KB, Richer JK, Lange CA, Zhang X, Byron SA, et al. Insulin receptor substrates mediate distinct biological responses to insulin-like growth factor receptor activation in breast cancer cells. Br J Cancer. 2006;95:120–8. doi: 10.1038/sj.bjc.6603354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis T, Chen J, Wu A, Prisco M, Baserga R. Transformation by the Simian Virus 40 T antigen is regulated by IGF-1 receptor and IRS-1 signaling. Oncogene. 2006;25:32–42. doi: 10.1038/sj.onc.1209013. [DOI] [PubMed] [Google Scholar]
- Del Rincon SV, Guo Q, Morelli C, Shiu HY, Surmacz E, Miller WH. Retinoic acid mediates degradation of IRS-1 by the ubiquitin-proteasome pathway, via a PKC-dependent mechanism. Oncogene. 2004;23:9269–79. doi: 10.1038/sj.onc.1208104. [DOI] [PubMed] [Google Scholar]
- Peruzzi F, Prisco M, Dews M, Salomoni P, Grassilli E, Romano G, et al. Multiple signaling pathways of the IGF-I receptor in protection from apoptosis. Mol Cell Biol. 1999;19:7203–15. doi: 10.1128/mcb.19.10.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenush L, Fernandez R, Myers MG, Jr, Grammer TC, Sun XJ, Blenis J, et al. The Drosophila insulin receptor activates multiple signaling pathways but requires insulin receptor substrate proteins for DNA synthesis. Mol Cell Biol. 1996;16:2509–17. doi: 10.1128/mcb.16.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L, Song Y, Tan C, Bui T. Inhibition of RNA polymerase I transcription in differentiated myeloid leukemia cells by inactivation of selectivity factor-1. Cell Growth Diff. 2000;11:63, 70. [PubMed] [Google Scholar]
- Baserga R. Customizing the targeting of IGF-1 receptor. Future Oncol. 2009;5:43–50. doi: 10.2217/14796694.5.1.43. [DOI] [PubMed] [Google Scholar]
- Sawka-Verhelle D, Tartare-Deckert S, White MF, van Obberghen E. Insulin receptor substrate-2 binds to the insulin receptor through its phosphotyrosine-binding domain and through a newly identified domain comprising amino acids 591–786. J Biol Chem. 1996;271:5980–3. doi: 10.1074/jbc.271.11.5980. [DOI] [PubMed] [Google Scholar]
- Drakas R, Tu X, Baserga R. Control of cell size through phosphorylation of upstream binding factor1 by nuclear phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2004;101:9272–6. doi: 10.1073/pnas.0403328101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Thirone ACP, Huang X, Klip A. Differential contribution of insulin receptor substrate-1 versus 2 to insulin signaling and glucose uptake in L6 myotubes. J Biol Chem. 2005;280:19426–35. doi: 10.1074/jbc.M412317200. [DOI] [PubMed] [Google Scholar]
- Stefanovsky VY, Pelletier G, Hannan R, Gagnon-Kugler T, Rothblum LI, Moss T. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol Cell. 2001;8:1063–73. doi: 10.1016/s1097-2765(01)00384-7. [DOI] [PubMed] [Google Scholar]
- Klippel A, Escobedo MA, Wachowicz MS, Apell G, Brown TW, Giedlin MA, et al. Activation of phosphatidylinositol 3-kinase is sufficient for cell cycle entry and promotes cellular changes characteristic of oncogenic transformation. Mol Cell Biol. 1998;18:5699–711. doi: 10.1128/mcb.18.10.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG, Vogt PK. An essential role for protein synthesis in oncogenic cellular transformation. Oncogene. 2004;23:3145–50. doi: 10.1038/sj.onc.1207550. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
V-src favors co-precipitation of Tyr phosphorylated IRS-2.