Abstract
Aim:
To study the relationship between the antihypertensive response of iptakalim and KCNJ11 polymorphisms in Chinese Han hypertensive patients.
Methods:
One hundred sixty two Chinese Han hypertensive patients were administered iptakalim (5 or 10 mg/d, po) for 8 weeks. Before the treatment and 24 h after completing the treatment blood pressure (BP) was measured. Genotyping was performed using direct sequencing.
Results:
Four common A190A, E23K, I337V and 3′UTR +62 G/A polymorphisms were found in KCNJ11. The E23K, I337V and 3′UTR +62 G/A polymorphisms were in complete linkage disequilibrium, and I337V was used as a representative. There were no significant differences in age, body mass index, sex, baseline systolic BP (SBP) and diastolic BP (DBP) among the 3 genotypes for the four polymorphisms. Significant association was found between SBP response and the polymorphisms (adjusted regression coefficient: 3.5 [1.2] mmHg; P=0.003 for the A190A polymorphism; adjusted regression coefficient: 3.1 [1.2] mmHg; P=0.012 for the I337V polymorphism). The patients with TT genotype for A190A polymorphism had higher clinical efficacy than those with CC genotype.
Conclusion:
The results suggest the KCNJ11 polymorphisms are associated with the SBP-lowering response of short-term iptakalim therapy in Chinese Han hypertensive patients.
Keywords: KCNJ11, Kir6.2, genotype, hypertension, ATP-sensitive potassium channel opener, iptakalim, polymorphisms, systolic blood pressure, diastolic blood pressure
Introduction
Essential hypertension is the leading cause of cardiovascular morbidity and mortality worldwide, affecting about 20% of the adult population and only 23%–41% of hypertensive patients receiving antihypertensive drugs achieve adequate blood pressure control1, 2. Many drugs are effective in treating hypertension, although individuals can respond differently to the same drug. Interindividual variation in the efficacy of medications may be influenced by genetic variations2, 3, 4. Hypertension pharmacogenetics seeks to find genetic predictors of response to drugs that lower blood pressure. More and more studies have investigated associations between genetic polymorphisms and response to antihypertensive drugs5, 6, 7.
Iptakalim is a novel KATP opener with antihypertensive properties targeting small arteries of hypertensive status in different models of hypertension in rats and dogs. It can produce long-lasting hypotensive effects without tolerance. At the same time it exertes a protective effect against hypertensive damage to target organs8, 9, 10, 11, 12, 13, 14, 15. Iptakalim is an effective and well tolerated antihypertensive drug. Individual variations for its antihypertensive effects have been observed16. Genetic variations that alter the structure, configuration, activity, or quantity of the drug target receptors or target-related regulation factors may contribute to individual variations in drug response17. Among the 3 different subtypes of KATP channels heterologously expressed in human embryonic kidney cells and Xenopus oocytes, iptakalim exhibits significant selectivity for SUR2B/Kir6.1 channels, mild effects on SUR2A/Kir6.2 channels, and fails to open SUR1/Kir6.2 channels. The KCNJ11 gene, which encodes the Kir6.2 subunit of the KATP channel, is among the candidate target genes for antihypertensive drug iptakalim8.
In this study, we aimed to investigate the polymorphisms of KCNJ11 in 162 Chinese patients with essential hypertension and their possible association with the antihypertensive response of iptakalim.
Materials and methods
Subjects
The material studied comes from all subjects who finished the trials in 5 centers of the phase-three clinical trials of iptakalim which is a randomized and double-blind trial performed in 14 centers in 11 cities in China. We studied 162 non-related Chinese Han patients with essential hypertension (81 men, 81 women; mean age±SD: 55±9 years; range: 26–74 years). Patients met World Health Organization-International Society of Hypertension criteria for hypertension (systolic BP [SBP]≥140 mmHg or diastolic BP [DBP]≥90 mmHg). Trained investigators assessed patents' BP on at least three different occasions. All patients underwent a complete physical examination. Secondary causes of hypertension and heart, liver, and kidney diseases were excluded by history and by physical and laboratory examinations. The diabetic patients (the fasting plasma glucose concentration>11 mmol/L) were excluded in this study. After a 2-weeks single-blind placebo run-in period, all patients were treated orally with iptakalim (Thadweik Academy of Medicine, Beijing, China) at a single daily fixed dosage of 5 mg (one tablet per day) for 4 consecutive weeks. After that, patients whose BP was less than 140/90 mmHg continued the same dose regimen for another 4 weeks. In patients whose BP was not adequately controlled (BP≥140/90 mmHg), the dose was doubled for the following 4 weeks. Patients were required to take their iptakalim around 9 am and carefully record the time they took the tablet. Blood pressure was measured before taking medicine on the first day and 24 h after 8 weeks of treatment. Three consecutive measurements were taken on the right arm of quietly seated participants with 1 min interval between replicates. If the difference between the measurements was more than 4 mmHg, the patient was asked to rest for 5 min, and then repeated the measurements. In all of our analysis, the average of three consecutive blood pressure readings was used. Written informed consent was obtained from all patients enrolled in the study in accordance with principles of the Declaration of Helsinki. The protocol of the study was approved by the institutional Ethics Committee.
Genotyping analysis
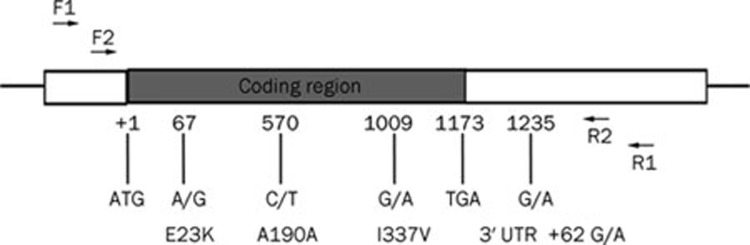
Genomic DNA was isolated from peripheral leukocytes. KCNJ11 gene was amplified by PCR and sequenced. The multiple sequence alignment program ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) and Chromas program were used for genotyping. The primers of KCNJ11 were designed using Primer3 (http://frodo.wi.mit.edu/primer3/). For specific amplification of DNA samples, two rounds of PCR amplification were carried out for KCNJ11. The primers and the amplification conditions for the first PCR were as follows: forward F1, 5′-ACTGGGATTACAGGCGTGAG-3′ and reverse R1, 5′-CCTGACCACAGGCACTTCTT-3′, initial denaturation at 95 °C for 5 min followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s, and extension at 72 °C for 2 min, and final extension at 72 °C for 5 min. The PCR product is a 1824-bp fragment which was diluted (1:50) with water and used as templates to the second PCR. Amplification conditions for the second PCR were the same as that given above but the primers used were different (forward F2, 5′-GACTCTGCAGTGAGGCCCTA-3′ and reverse R2, 5′-CCTGCTGAGGCCAGAAATAG-3′). The product of the second PCR was a 1421-bp fragment which was sequenced (ABI 3730XL sequencer, PE Applied Biosystem, USA). cDNA and protein sequences were numbered according to NCBI ORF finder (http://www.ncbi.nlm.nih.gov/gorf/orfig.cgi; GenBank NM_000525.3) with nucleotide numbering beginning with the first Met.
Statistical analysis
Values are expressed as the mean±SD. The means for continuous variables in the 2 groups were compared using t-test and the differences among three groups were analyzed by one-way analysis of variance (ANOVA). The prevalence of categorical variables was compared using χ2-test. The blood pressure (BP) response was defined as the blood pressure before treatment minus the blood pressure at the end of 8 weeks of treatment. Multiple linear regression and logistic regression analyses were used to assess the influence of KCNJ11 polymorphisms on baseline BP and antihypertensive efficacy, with adjustment for potential confounding factors, including baseline BP, age, sex, and BMI. The SPSS 13.0 statistical package (SPSS, Chicago, IL, USA) was used for analysis. All tests were two-tailed and P<0.05 was considered statistically significant. Linkage disequilibrium and haplotypes analysis were done with a SHEsis software18, 19.
Results
Clinical characteristics
For the 162 patients in this study, there were no significant differences in age, sex, BMI, smoking status, alcohol consumption, baseline BP, heart rate and biochemical parameters among the three genotypes of the four polymorphisms before iptakalim treatment. Significant decreases in SBP and DBP occurred after treatment in all genotype groups (P<0.01). The posttreatment DBP was not different among the three groups, while the posttreatment SBP was both lower in patients with TT genotype of the A190A polymorphism and AA genotype of the I337V polymorphism (Table 1).
Table 1. Clinical characteristics and biochemical parameters by KCNJ11 gene polymorphisms.
| Variable | KCNJ11 A190A genotype | KCNJ11 I337V genotype | ||||||
|---|---|---|---|---|---|---|---|---|
| CC (n=58) | CT (n=64) | TT (n=40) | P# | GG (n=35) | GA (n=70) | AA (n=57) | P# | |
| Dose (no increase/increase) | 24/34 | 39/25 | 24/16 | 0.063 | 17/18 | 35/35 | 35/22 | 0.352 |
| Sex (male/female) | 30/28 | 34/30 | 17/23 | 0.544 | 18/17 | 35/35 | 28/29 | 0.977 |
| Age (years) | 55.6 (8.5) | 55.3 (9.0) | 55.5 (9.7) | 0.981 | 54.7 (8.6) | 55.1 (8.2) | 56.4 (10.0) | 0.631 |
| Smoking status (never/current) | 43/15 | 46/18 | 33/7 | 0.463 | 27/8 | 48/22 | 47/10 | 0.191 |
| Alcohol consumption (never/current) | 43/15 | 49/15 | 32/8 | 0.800 | 24/11 | 55/15 | 45/12 | 0.458 |
| BMI (kg/m2) | 25.2 (2.4) | 25.3 (2.7) | 25.5 (2.9) | 0.845 | 25.1 (2.4) | 25.0 (2.7) | 25.8 (2.7) | 0.197 |
| Sodium (mmol/L) | 141.5 (3.2) | 141.9 (3.0) | 141.4 (2.6) | 0.558 | 141.1 (3.1) | 141.9 (2.9) | 141.7 (3.1) | 0.459 |
| Potassium (mmol/L) | 4.1 (0.3) | 4.1 (0.4) | 4.2 (0.4) | 0.058 | 4.1 (0.3) | 4.1 (0.4) | 4.2 (0.4) | 0.235 |
| Chloride (mmol/L) | 104.0 (3.1) | 103.4 (2.9) | 103.6 (3.0) | 0.568 | 103.8 (3.0) | 103.7 (3.1) | 103.5 (2.8) | 0.833 |
| Glucose (mmol/L) | 5.7 (1.4) | 5.5 (1.3) | 5.3 (1.3) | 0.338 | 5.7 (1.5) | 5.6 (1.5) | 5.4 (1.1) | 0.512 |
| Uric acid (μmol/L) | 307.4 (79.3) | 314.8 (96.8) | 324.2 (75.9) | 0.638 | 308.8 (74.7) | 313.4 (98.3) | 319.3 (75.7) | 0.841 |
| Total cholesterol (mmol/L) | 5.3 (1.1) | 5.1 (0.9) | 5.2 (0.8) | 0.446 | 5.3 (1.3) | 5.2 (0.9) | 5.1 (0.8) | 0.441 |
| Triglyceride (mmol/L) | 2.3 (1.8) | 2.0 (1.3) | 1.8 (1.0) | 0.271 | 2.2 (1.8) | 2.2 (1.6) | 1.8 (1.0) | 0.264 |
| HDL cholesterol (mmol/L) | 1.3 (0.3) | 1.4 (0.4) | 1.4 (0.5) | 0.365 | 1.4 (0.3) | 1.3 (0.4) | 1.4 (0.4) | 0.548 |
| GPT (U/L) | 25.9 (13.3) | 24.7 (13.9) | 25.6 (13.0) | 0.875 | 28.1 (15.1) | 24.1 (12.8) | 25.2 (12.9) | 0.353 |
| Pretreatment SBP (mmHg) | 153.6 (14.0) | 152.6 (12.1) | 151.4 (11.6) | 0.710 | 151.1 (12.1) | 153.8 (13.3) | 152.2 (12.2) | 0.564 |
| Posttreatment SBP (mmHg) | 145.0 (14.3)c | 141.3 (12.0)c | 137.3 (11.4)c | 0.015 | 142.8 (13.9)c | 144.1 (13.3)c | 137.8 (11.4)c | 0.019 |
| Pretreatment DBP (mmHg) | 100.4 (4.3) | 99.7 (3.8) | 99.8 (4.1) | 0.622 | 99.7 (3.9) | 100.2 (4.3) | 99.8 (3.8) | 0.791 |
| Posttreatment DBP (mmHg) | 90.4 (10.9)c | 88.2 (7.1)c | 87.5 (8.0)c | 0.208 | 89.0 (10.2)c | 90.0 (8.9)c | 87.3 (8.0)c | 0.247 |
| Pretreatment HR (beats/min) | 72.0 (10.2) | 70.1 (9.3) | 71.0 (11.1) | 0.582 | 72.0 (9.4) | 70.1 (9.3) | 71.4 (11.3) | 0.628 |
| Posttreatment HR (beats/min) | 70.1 (8.5) | 70.1 (8.4) | 71.0 (9.6) | 0.523 | 70.7 (7.8) | 69.8 (8.5) | 67.5 (9.4) | 0.186 |
| Change in HR at 8 weeks (beats/min) | 1.9 (8.4) | 0.9 (10.2) | 3.0 (9.4) | 0.544 | 1.3 (8.4) | 0.3 (9.2) | 3.8 (9.8) | 0.103 |
Data are presented as the mean (SD).
#ANOVA and Pearson's χ2 tests were applied to continuous and categorical variables, respectively. The E23K, I337V and 3′UTR +62 G/A polymorphisms are in complete linkage disequilibrium (LD).
cP<0.01 between pretreatment and posttreatment BP. Change in HR at 8 weeks is pretreatment HR minus posttreatment HR.
Allele and genotype frequencies
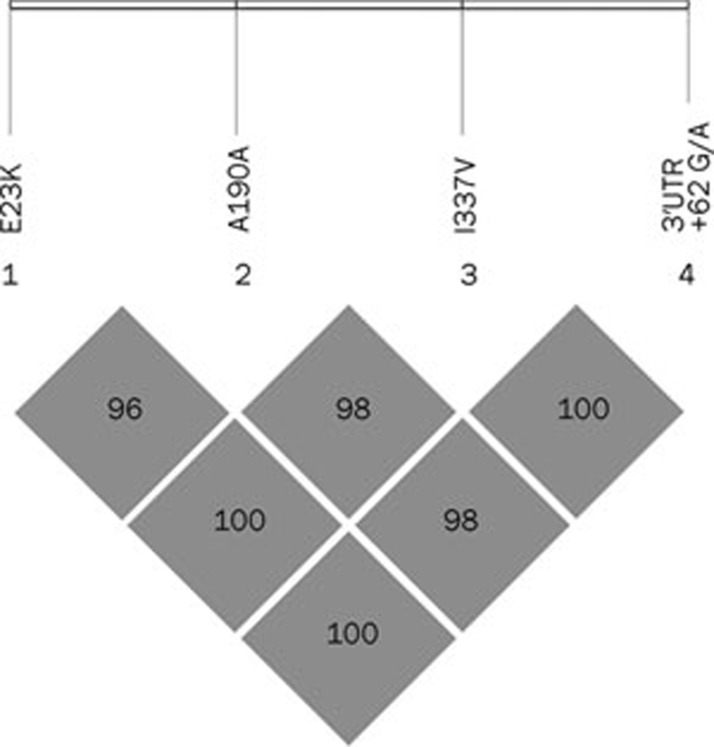
Five polymorphisms were identified in 162 Chinese hypertensive patients. Four known polymorphisms were E23K, A190A, I337V and 3′UTR +62 G/A (Figure 1). Complete information on genotypes and phenotypes was available for 162 patients. For A190A polymorphism, the frequencies of the C and T alleles were 55.6% and 44.4%, respectively, and the frequencies of the CC, CT, and TT genotypes were 58/162 (35.8%), 64/162 (39.5%), and 40/162 (24.7%), respectively. E23K, I337V, and 3′UTR +62 G/A were found to be in complete LD (D'=1, Figure 2). We will use I337V polymorphism as a representative polymorphism for the next analysis. The frequencies of the G and A alleles were 43.2% and 56.8%, respectively, and the frequencies of the GG, GA, and AA genotypes were 35/162 (21.6%), 70/162 (43.2%), and 57/162 (35.2%), respectively. Patients with TT genotype of the A190A polymorphism are all AA genotype of the I337V polymorphism. The study population showed no deviation in genotype frequencies from those predicted by the Hardy-Weinberg equilibrium. In addition, we identified two synonymous mutations (C480T, N160N; C579T, H193H) and one missense mutation (C1105T, R369C) in three patients, and they are all heterozygous. Blood pressure of the patient with the novel R369C missense mutation which is yet unknown functional consequences can be well controlled by iptakalim. The fifth polymorphism found in this population is I284I, and the frequencies of genotypes were 157 CC, 3 CA, 1 AA, and 1 CT. Haplotype frequencies were calculated with the SHEsis program. There are 16 possible haplotypes of these four loci (E23K, A190A, L267L, and I337V), but only three haplotypes were observed due to complete linkage disequilibrium among E23K, L267L, and I337V. They are H1 (A-C-G-G, 43%), H2 (G-C-A-A, 13%), and H3 (G-T-A-A, 44%). Two kinds of Kir6.2 pore-forming proteins may be seen in this population: one is 23K/337V, and the other is 23E/337I. The patients with H1H1 genotype (A-C-G-G/A-C-G-G, n=35) may have 23K/337V pore-forming proteins. Those with H1H2 or H1H3 genotypes (A-C-G-G/G-C-A-A or A-C-G-G/G-T-A-A, n=70) may have 23K/337V and 23E/337I pore-forming proteins. Those with H2H2, H2H3 or H3H3 genotypes (G-C-A-A/G-C-A-A, G-C-A-A/G-T-A-A, or G-T-A-A/G-T-A-A, n=57) may have 23E/337I pore-forming proteins. Due to complete LD among E23K, I337V, and 3′UTR +62 G/A, these genotypes can be represented by the GG (H1H1, n=35), GA (H1H2 or H1H3, n=70) and AA (H2H2, H2H3, or H3H3, n=57) genotypes of the I337V polymorphism, respectively. Therefore I337V can be used as a representative polymorphism for the next analysis.
Figure 1.
The most common polymorphisms in KCNJ11 were E23K, A190A, I337V, 3′ UTR +62 G/A.
Figure 2.
Linkage disequilibrium plot of computed pairwise LD statistics for four polymorphisms.
Association of antihypertensive response with the four polymorphisms
The reduction in SBP and DBP was both largest in the TT (AA) group and smallest in the CC (GG) group. Reductions in SBP were statistically significant between the TT (AA) group and CC (GG) group for the A190A and I337V polymorphisms (−14.8 [10.3] and −9.0 [14.7] mmHg, respectively for the A190A polymorphism, P=0.042; −14.4 [10.7] and −8.3 [12.8] mmHg, respectively for the I337V polymorphism, P=0.037). The relationship between antihypertensive response and the A190A and I337V polymorphisms was further examined by multiple linear regression analysis, with adjustment for age, sex, BMI and baseline BP (Table 2). Significant association was found between SBP response and the A190A and I337V polymorphisms (SBP response-crude β±SE: 2.8 [1.4], P=0.042; adjusted β±SE: 3.5 [1.2], P=0.003 for the A190A polymorphism; crude β±SE: 3.2 [1.4], P=0.026; adjusted β±SE: 3.1 [1.2], P=0.012 for the I337V polymorphism).
Table 2. Multiple linear regression analysis of the association between blood pressure (BP) response and the KCNJ11 A190A and I337V polymorphisms.
| Fall in BP mean (SD), mmHg | Difference in BP response | ||||||
|---|---|---|---|---|---|---|---|
| CC (GG) | CT (GA) | TT (AA) | Alone β (SE) | P | Adjust β (SE) | P | |
| KCNJ11 A190A | |||||||
| Systolic | 8.6 (14.5) | 11.2 (14.4) | 14.3 (10.4) | 2.8 (1.4) | 0.042 | 3.5 (1.2) | 0.003 |
| Diastolic | 10.0 (9.5) | 11.6 (7.1) | 12.3 (6.9) | 1.2 (0.8) | 0.144 | 1.2 (0.8) | 0.114 |
| KCNJ11 I337V | |||||||
| Systolic | 8.3 (12.8) | 9.8 (15.7) | 14.4 (10.7) | 3.2 (1.4) | 0.026 | 3.1 (1.2) | 0.012 |
| Diastolic | 10.7 (8.8) | 10.3 (8.2) | 12.5 (7.2) | 1.0 (0.8) | 0.221 | 1.0 (0.8) | 0.210 |
Each polymorphism was first considered by itself (ie, alone) as a predictor of BP response, then after adjustment for the concomitant variables (ie, after covariates) which included pretreatment (baseline) BP, gender, age, and body mass index. β (SE) is the regression coefficient (standard error). Genotype in the bracket is for the I337V polymorphism.
These findings were confirmed in the logistic regression analysis, in which the continuous BP values were transformed to dichotomous variables based on clinical antihypertensive efficacy (Table 3). Antihypertensive efficacy was based on achievement of the normal clinical standard, defined as a SBP<140 mmHg (SBP analysis), a DBP<90 mmHg (DBP analysis), and a SBP<140 mmHg and a DBP<90 mmHg (SBP and DBP analysis); failure to achieve this standard constituted lack of antihypertensive efficacy. After adjustment for age, sex, BMI and baseline BP, the proportion of patients achieving antihypertensive efficacy was significantly larger in those with the TT genotype than those with the CC genotype for the A190A polymorphism (SBP analysis – 60.0% vs 34.5%; adjusted odds ratio [OR]: 3.33; 95% CI: 1.30–8.55; P=0.012; DBP analysis – 67.5% vs 44.8%; adjusted odds ratio [OR]: 2.47; 95% CI: 0.97–6.3; P=0.057; SBP and DBP analysis – 50.0% vs 25.9%; adjusted odds ratio [OR]: 3.65; 95% CI: 1.36–9.83; P=0.010). Though the proportion of patients achieving antihypertensive efficacy in those with the AA genotype was all highest for the I337V polymorphism, it was only significantly larger compared with those with the GG genotype in DBP analysis (DBP analysis – 68.4% vs 48.6%; adjusted odds ratio [OR]: 2.66; 95% CI: 0.96–6.0; P=0.049).
Table 3. Logistic regression analysis of the association between antihypertensive efficacy and KCNJ11 A190A and I337V polymorphisms.
| KCNJ11 A190A polymorphism | |||||||
|---|---|---|---|---|---|---|---|
| Clinical efficacya | Crude | Adjustedb | |||||
| No (%) | OR | 95% CI | P | OR | 95% CI | P | |
| SBP | |||||||
| CC (GG) | 20 (34.5) | 1.00 | – | – | 1.00 | -- | – |
| CT (GA) | 27 (42.2) | 1.386 | 0.665–2.889 | 0.383 | 1.403 | 0.609–3.232 | 0.426 |
| TT(AA) | 24 (60.0) | 2.850 | 1.239–6.553 | 0.014 | 3.332 | 1.298–8.551 | 0.012 |
| DBP | |||||||
| CC (GG) | 26 (44.8) | 1.00 | – | – | 1.00 | – | – |
| CT (GA) | 40 (62.5) | 2.051 | 0.995–4.230 | 0.052 | 2.218 | 0.987–4.986 | 0.054 |
| TT (AA) | 27 (67.5) | 2.556 | 1.103–5.920 | 0.029 | 2.474 | 0.972–6.297 | 0.057 |
| SBP and DBP | |||||||
| CC (GG) | 15 (25.9) | 1.00 | – | – | 1.00 | – | – |
| CT (GA) | 25 (39.1) | 1.838 | 0.848–3.981 | 0.123 | 2.058 | 0.849–4.987 | 0.110 |
| TT (AA) | 20 (50.0) | 2.867 | 1.220–6.735 | 0.016 | 3.651 | 1.356–9.830 | 0.010 |
| KCNJ11 I337V polymorphism | |||||||
|---|---|---|---|---|---|---|---|
| Clinical efficacya | Crude | Adjustedb | |||||
| No (%) | OR | 95% CI | P | OR | 95% CI | P | |
| SBP | |||||||
| CC (GG) | 15 (42.9) | 1.00 | – | – | 1.00 | – | – |
| CT (GA) | 25 (35.7) | 0.741 | 0.323–1.697 | 0.478 | 0.806 | 0.316–2.055 | 0.651 |
| TT(AA) | 31 (54.4) | 1.590 | 0.681–3.714 | 0.284 | 2.186 | 0.826–5.785 | 0.115 |
| DBP | |||||||
| CC (GG) | 17 (48.6) | 1.00 | – | – | 1.00 | – | – |
| CT (GA) | 37 (52.9) | 1.187 | 0.527–2.674 | 0.679 | 1.376 | 0.497–2.735 | 0.494 |
| TT (AA) | 39 (68.4) | 2.294 | 0.964–5.457 | 0.061 | 2.658 | 0.957–5.983 | 0.049 |
| SBP and DBP | |||||||
| CC (GG) | 11 (31.4) | 1.00 | – | – | 1.00 | – | – |
| CT (GA) | 23 (32.9) | 1.068 | 0.447–2.550 | 0.883 | 1.220 | 0.453–3.289 | 0.694 |
| TT (AA) | 26 (45.6) | 1.830 | 0.756–4.428 | 0.180 | 2.715 | 0.971–7.594 | 0.057 |
BP, blood pressure; SBP, systolic BP; DBP, diastolic BP. Genotype in the bracket is for the I337V polymorphism.
aClinical efficacy was defined as a SBP<140 mmHg (SBP analysis), a DBP<90 mmHg (DBP analysis), and a SBP<140 mmHg and DBP<0 mmHg (SBP and DBP analysis).
bThe generalized estimating equation model was adjusted for age, sex, body mass index, and baseline BP.
Discussion
Kir6.2 is one of the pore-forming subunits of the KATP channels which provide a unique link between cellular energetics and electrical excitability20. The KCNJ11 gene encoding human KATP subunit Kir6.2 is located on chromosome 11p15.1 with only one 1173-bp exon. This work investigated the polymorphisms of KCNJ11 gene in 162 Chinese hypertensive population treated by iptakalim. Our data revealed that five polymorphisms were found in this population. Four known polymorphisms were E23K, A190A, I337V, and 3′UTR +62 G/A. E23K/I337V/3′UTR +62 G/A were found to be in complete LD in this population. The fifth polymorphism found in this population is I284I, and the frequencies of genotypes were 157 CC, 3 CA, 1 AA, and 1 CT. In addition, we identified two synonymous mutations (C480T, N160N; C579T, H193H) and one nonsynonymous mutation (C1105T, R369C) in three patients, respectively. The other polymorphisms reported in Germans were not found in this population21. This might somewhat reflect the racial difference.
The association of the four common polymorphisms of KCNJ11 gene and the BP response to iptakalim were further investigated. The four polymorphisms were significantly associated with SBP response to iptakalim (adjusted regression coefficient: 3.5 [1.2] mmHg; P=0.003 for the A190A polymorphism; adjusted regression coefficient: 3.1 [1.2] mmHg; P=0.012 for the I337V polymorphism). Patients with the TT (for the A190A polymorphism) and AA genotype (for the I337V polymorphism) exhibited greater systolic BP reduction and higher clinical efficacy than those with the CC (for the A190A polymorphism) and GG genotype (for the I337V polymorphism), respectively. Therefore the four polymorphisms may be used as genetic markers to predict SBP response and antihypertensive efficacy to iptakalim. To our knowledge, this study appears to be the first study to investigate the polymorphisms of KCNJ11 gene and the association of these polymorphisms with BP and BP response to antihypertensive drugs in Chinese Han hypertensive patients.
E23K and I337V are missense mutations which have functional consequences, however, A190A is synonymous mutation which has not very likely important functional consequences. The reason for A190A as a better marker for predicting SBP response of iptakalim may be explained by its linkage disequilibrium with other putative effective polymorphisms/mutations or the transcription level of the different genotypes. Only three haplotypes were observed due to complete LD among E23K, I337V, and 3′UTR +62 G/A. They are H1 (A-C-G-G, 43%), H2 (G-C-A-A, 13%), and H3 (G-T-A-A, 44%). Therefore there are two kinds of Kir6.2 pore-forming proteins in this population, one is 23K/337V, and the other is 23E/337I. The SBP response to iptakalim in the patients with the 23E/337I homozygotes (GG genotype for E23K polymorphism; AA genotype for I337V polymorphism) is greater than those with the 23K/337V homozygotes (AA genotype for E23K polymorphism; GG genotype for I337V polymorphism). The difference of Kir6.2 subunit may affect the function of KATP channels. Kir6.2 (23K/337V) polymorphic KATP channel has been reported that it decreases ATP sensitivity of KATP channels at physiological ATP concentrations and it has increased acyl CoA sensitivity which resides primarily in the Kir6.2 subunit22. The E23K polymorphism in KCNJ11 gene appeared to be related to high susceptibility to coronary heart disease23. This polymorphism is also associated with left ventricular size in hypertensive individuals24. The results of Kir6.2-knockout mice experiment indicate that the Kir6.2 subunit mediates the depression of cardiac excitability and contractility induced by KCOs and it plays no discernible role in the arterial tree25. KCNJ11 E23K polymorphisms may also affect therapeutic efficacy of repaglinide26, 27. Iptakalim exhibits mild effects on SUR2A/Kir6.2 channels8. It is possible that the different composition of Kir6.2 pore-forming proteins might in part account for the different SBP response to iptakalim in Chinese Han hypertensive patients.
The limitation of this study is the lack of normal control group. This study shows that the four KCNJ11 polymorphisms were not related to baseline BP in the hypertensive patients. KCNJ11 polymorphisms were reported to be associated with blood pressure variation and hypertension in the Japanese and Korean population28, 29, 30. Further large scale association studies compared with control subjects are needed to clarify whether these polymorphisms are associated with the pathogenesis of hypertension in Chinese Han population.
In conclusion, we found five polymorphisms in KCNJ11 and demonstrated that polymorphisms of KCNJ11 are associated with the antihypertensive response of iptakalim in Chinese Han hypertensive patients. Patients with the TT genotype (for the A190A polymorphism) exhibited a more pronounced SBP reduction and higher clinical efficacy in response to iptakalim compared to patients with the CC genotype. It may be used as marker for predicting SBP response to iptakalim. This implies the advantage of genotyping for selecting the optimal antihypertensive treatment for individual patient.
Author contribution
Hai WANG designed research; Rui-feng DUAN performed research; Wen-yu CUI recruited patients; and Rui-feng DUAN analyzed data and wrote the paper.
Acknowledgments
We thank the patients for their participation. This study was supported by the grants from National Key Technologies R&D program of China (No 2008ZXJ09004-018).
References
- Zhang S, Mao G, Zhang Y, Tang G, Wen Y, Hong X, et al. Association between human atrial natriuretic peptide Val7Met polymorphism and baseline blood pressure, plasma trough irbesartan concentrations, and the antihypertensive efficacy of irbesartan in rural Chinese patients with essential hypertension. Clin Ther. 2005;27:1774–84. doi: 10.1016/j.clinthera.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Taverne K, de Groot M, de Boer A, Klungel O. Genetic polymorphisms related to the renin-angiotensin-aldosterone system and response to antihypertensive drugs. Expert Opin Drug Metab Toxicol. 2010;6:439–60. doi: 10.1517/17425250903571670. [DOI] [PubMed] [Google Scholar]
- Trotta R, Donati MB, Iacoviello L. Trends in pharmacogenomics of drugs acting on hypertension. Pharmacol Res. 2004;49:351–6. doi: 10.1016/j.phrs.2003.07.018. [DOI] [PubMed] [Google Scholar]
- Arnett DK, Claas SA. Pharmacogenetics of antihypertensive treatment: detailing disciplinary dissonance. Pharmacogenomics. 2009;10:1295–307. doi: 10.2217/pgs.09.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherva R, Ford CE, Eckfeldt JH, Davis BR, Boerwinkle E, Arnett DK. Pharmacogenetic effect of the stromelysin (MMP3) polymorphism on stroke risk in relation to antihypertensive treatment: the genetics of hypertension associated treatment study. Stroke. 2011;42:330–5. doi: 10.1161/STROKEAHA.110.593798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Gong Y, Langaee TY, Davis HM, Elewa H, Beitelshees AL, et al. Genetic variation in the beta2 subunit of the voltage-gated calcium channel and pharmacogenetic association with adverse cardiovascular outcomes in the international verapamil SR-trandolapril study genetic substudy (INVEST-GENES) Circ Cardiovasc Genet. 2010;3:548–55. doi: 10.1161/CIRCGENETICS.110.957654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JA, Boerwinkle E, Zineh I, Chapman AB, Bailey K, Cooper-DeHoff RM, et al. Pharmacogenomics of antihypertensive drugs: rationale and design of the Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) study. Am Heart J. 2009;157:442–9. doi: 10.1016/j.ahj.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Huang J, Cui W, Long C, Zhang Y, Wang H. Targeting hypertension with a new adenosine triphosphate-sensitive potassium channel opener iptakalim. J Cardiovasc Pharmacol. 2010;56:215–28. doi: 10.1097/FJC.0b013e3181e23e2b. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang YL, Chen YP. Targeting small arteries of hypertensive status with novel ATP-sensitive potassium channel openers. Curr Vasc Pharmacol. 2005;3:119–24. doi: 10.2174/1570161053586895. [DOI] [PubMed] [Google Scholar]
- Gao S, Long CL, Wang RH, Wang H. KATP activation prevents progression of cardiac hypertrophy to failure induced by pressure overload via protecting endothelial function. Cardiovasc Res. 2009;83:444–56. doi: 10.1093/cvr/cvp099. [DOI] [PubMed] [Google Scholar]
- Wang H, Tang Y, Wang L, Long CL, Zhang YL. ATP-sensitive potassium channel openers and 2,3-dimethyl-2-butylamine derivatives. Curr Med Chem. 2007;14:133–55. doi: 10.2174/092986707779313390. [DOI] [PubMed] [Google Scholar]
- Wang H, Long CL, Zhang YL. A new ATP-sensitive potassium channel opener reduces blood pressure and reverses cardiovascular remodeling in experimental hypertension. J Pharmacol Exp Ther. 2005;312:1326–33. doi: 10.1124/jpet.104.078220. [DOI] [PubMed] [Google Scholar]
- Chen YP, Qiu CR, Wang H. Cardiovascular pharmacological characterization of novel 2,3-dimethyl-2-butylamine derivatives in rats. Life Sci. 2004;75:2131–42. doi: 10.1016/j.lfs.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Chen YP, Cui WY, Wang H. Selective actions of iptakalim on the subtypes of KATP channels. Chin Pharmacol Bull. 2006;22:278–84. [Google Scholar]
- Long CL, Qin XC, Pan ZY, Chen K, Zhang YF, Cui WY, et al. Activation of ATP-sensitive potassium channels protects vascular endothelial cells from hypertension and renal injury induced by hyperuricemia. J Hypertens. 2008;26:2326–38. doi: 10.1097/HJH.0b013e328312c8c1. [DOI] [PubMed] [Google Scholar]
- Duan RF, Cui WY, Gao HY, Wang XP, Hu SK, Liu W, et al. Correlation between ACE gene I/D polymorphism and clinical efficacy of iptakalim in Chinese Han hypertensive population. Chin J Clin Pharmacol Ther. 2010;15:961–6. [Google Scholar]
- Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–91. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–8. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis ( http://analysis.bio-x.cn ). Cell Res 200919519–23. [DOI] [PubMed] [Google Scholar]
- Nichols CG. KATP channels as molecular sensors of cellular metabolism. Nature. 2006;440:470–6. doi: 10.1038/nature04711. [DOI] [PubMed] [Google Scholar]
- Jeron A, Hengstenberg C, Holmer S, Wollnik B, Riegger GA, Schunkert H, et al. KCNJ11 polymorphisms and sudden cardiac death in patients with acute myocardial infarction. J Mol Cell Cardiol. 2004;36:287–293. doi: 10.1016/j.yjmcc.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Riedel MJ, Boora P, Steckley D, de Vries G, Light PE. Kir6.2 polymorphisms sensitize beta-cell ATP-sensitive potassium channels to activation by acyl CoAs: a possible cellular mechanism for increased susceptibility to type 2 diabetes. Diabetes. 2003;52:2630–5. doi: 10.2337/diabetes.52.10.2630. [DOI] [PubMed] [Google Scholar]
- Xiong C, Zheng F, Wan J, Zhou X, Yin Z, Sun X. The E23K polymorphism in Kir6.2 gene and coronary heart disease. Clin Chim Acta. 2006;367:93–7. doi: 10.1016/j.cca.2005.11.032. [DOI] [PubMed] [Google Scholar]
- Reyes S, Terzic A, Mahoney DW, Redfield MM, Rodeheffer RJ, Olson TM. KATP channel polymorphism is associated with left ventricular size in hypertensive individuals: a large-scale community-based study. Hum Genet. 2008;123:665–7. doi: 10.1007/s00439-008-0519-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Li RA, Miki T, Uemura H, Sakamoto N, Ohmoto-Sekine Y, et al. Functional roles of cardiac and vascular ATP-sensitive potassium channels clarified by Kir6.2-knockout mice. Circ Res. 2001;88:570–7. doi: 10.1161/01.res.88.6.570. [DOI] [PubMed] [Google Scholar]
- He YY, Zhang R, Shao XY, Hu C, Wang CR, Lu JX, et al. Association of KCNJ11 and ABCC8 genetic polymorphisms with response to repaglinide in Chinese diabetic patients. Acta Pharmacol Sin. 2008;29:983–9. doi: 10.1111/j.1745-7254.2008.00840.x. [DOI] [PubMed] [Google Scholar]
- Yu M, Xu XJ, Yin JY, Wu J, Chen X, Gong ZC, et al. KCNJ11 Lys23Glu and TCF7L2 rs290487(C/T) polymorphisms affect therapeutic efficacy of repaglinide in Chinese patients with type 2 diabetes. Clin Pharmacol Ther. 2010;87:330–5. doi: 10.1038/clpt.2009.242. [DOI] [PubMed] [Google Scholar]
- Koo BK, Cho YM, Park BL, Cheong HS, Shin HD, Jang HC, et al. Polymorphisms of KCNJ11 (Kir6.2 gene) are associated with Type 2 diabetes and hypertension in the Korean population. Diabet Med. 2007;24:178–86. doi: 10.1111/j.1464-5491.2006.02050.x. [DOI] [PubMed] [Google Scholar]
- Kokubo Y, Tomoike H, Tanaka C, Banno M, Okuda T, Inamoto N, et al. Association of sixty-one non-synonymous polymorphisms in forty-one hypertension candidate genes with blood pressure variation and hypertension. Hypertens Res. 2006;29:611–9. doi: 10.1291/hypres.29.611. [DOI] [PubMed] [Google Scholar]
- Sakamoto Y, Inoue H, Keshavarz P, Miyawaki K, Yamaguchi Y, Moritani M, et al. SNPs in the KCNJ11-ABCC8 gene locus are associated with type 2 diabetes and blood pressure levels in the Japanese population. J Hum Gene. 2007;52:781–93. doi: 10.1007/s10038-007-0190-x. [DOI] [PubMed] [Google Scholar]