Abstract
The CULLIN family of E3 ubiquitin ligases are important regulators of plant development and function. A newly identified class of CULLIN4-RING-E3 ligases (CRL4s) interacts with substrate receptors referred to as DDB1-CUL4 ASSOCIATED FACTORS (DCAFs) via a DDB1 linker protein. We have previously reported that the WD40 protein WDR55 interacts with DDB1A and is thus a putative DCAF. Mutants of WDR55 are embryo lethal, suggesting that a DDB1WDR55 complex could regulate embryo and endosperm development. Here we report that a weak allele homozygous for wdr55 display pleiotropic phenotypes in the seedling and adult stages, suggesting a novel regulatory role for WDR55 in vegetative development.
Keywords: WDR55, DDB1, vegetative development, apical dominance
Introduction
After germination of a seed, the seedling enters the vegetative phase where rosette leaves are produced by the apical meristem in a spiral arrangement separated by short internodes. The plant then enters a transient phase, marked by a reprogramming of the apical meristem into an inflorescence meristem that produces spirally patterned cauline leaves, separated by long internodes. Entering the reproductive stage, the inflorescence meristem again reorganizes and starts producing floral meristems in the same spiral pattern, where each floral meristem develops into a single flower.1 Our current understanding of vegetative development and phase transitions is mainly based on different levels of transcriptionally based regulation, incorporating various roles of environmental triggers, chromatin and distribution of hormone gradients (for review see refs. 2–6). An important level of protein regulation is, however, also mediated by UBIQUTIN-26S proteasome dependent pathways7 and an emerging perspective of both proteolytic and non-proteolytic roles of ubiquitination matches the importance of transcriptionally based regulation.8,9
We have previously demonstrated that the WD × R motif containing protein WDR55 plays a major role in reproductive development in Arabidopsis thaliana. WDR55 is essential for gametogenesis and embryogenesis and is required to break radial symmetry and establish bilateral symmetry in the apical embryo domain.10 WDR55 belongs to the WD repeat (WDR) family, which is a large family of proteins with diverse functions but with a common conserved motif; the WD repeat. WDR55 also contains a WD × R motif, which is a submotif of the DWD box [DAMAGED DNA BINDING PROTEIN1 (DDB1)-binding WD-40 box]. The DWD box and the WD × R motif are signatures for potential substrate receptors of the CULLIN4 (CUL4) RING ubiquitin ligase complexes (CRL4s). Proteins with these motifs, which interact with CUL4-DDB1 based E3 ligases, are referred to as DCAF (DDB1-CUL4 ASSOCIATED FACTOR) proteins,11 and potentially target proteins for ubiquitination after they assemble with DDB1.12,13 WDR55 interacts with DDB1A, suggesting that a putative CUL4-DDB1WDR55 E3 complex regulates embryo or endosperm development.10 The Arabidopsis CRL4s are associated with many processes such as cell cycle, DNA repair, photomorphogenesis and modulation of chromatin.14 Their mode of action appears to include both proteolytic and transcriptional mechanisms. In photomorphogenesis, CRL4SPA1-COP1 has been shown to act as a negative regulator of transcription in a CRL4 26S targeting complex.15,16 CRL4MSI4 has recently been shown to associate with the Polycomb Repressive Complex (PRC2-EMF) leading to PRC2 mediated H3K27me3 repression of FLC and MSI5 may also participate in this regulation.17 Moreover, CRL4MSI1 is required for maintaining PRC2-FIS mediated imprinting.18
We have generated a mutant that is homozygous for a weak allele of WDR55. Here we report that WDR55 is also required for proper vegetative growth and for organization of the adult plant body. wdr55 seedlings display asymmetric and often multiple cotyledons and mature plants show phenotypes often associated with auxin misregulation, suggesting that WDR55 may play a role in hormonal control of plant development.
Results and Discussion
Two mutant alleles of WDR55 have previously been identified to affect gametophyte development and function, as well as the establishment of apical symmetry in the embryo. The weaker allele, wdr55-2, initially displayed a close to mendelian ratio of mutant seeds (22.8%, n = 548), but no homozygous plants were observed among the germinating progeny.10 In order to search for a small fraction of homozygous plants (~2%) anticipated from the genetic data, a large number of seeds from heterozygous wdr55-2 plants were germinated on MS-2 plates and observed over a longer period. Upon closer inspection, we identified a minor class of seedlings (3.6%, n = 1,035) that were late germinating, slow growing and considerably smaller than wild type seedlings. Using PCR genotyping and sequencing, we verified that this class was homozygous for wdr55-2.
Using the same screening setup as above, we also inspected the stronger wdr55-1 allele, but no small homozygous plants could be observed, much in agreement with the wdr55-1 early embryo abort phenotype described previously.10 Transgenic plants with a full length genomic WDR55 rescue construct restored wild type phenotype in both wdr55-1 and wdr55-2 alleles. Furthermore, in a wdr55-1/ wdr55-2 complementation cross, the wdr55-1 phenotype is partially rescued to phenocopy the wdr55-2 embryo phenotype. The same partial rescue is also found in transgenic plants with rescue constructs containing the splice form WDR55.1 controlled by its endogenous promoter. We therefore also inspected the progeny of wdr55-1 crossed with wdr55-2 on MS-2 media selecting for both mutant alleles, but were not able to identify any putative homozygous seedlings (n = 319).10
The identified homozygous mutant class appeared to be unique to the wdr55-2 allele, which has a T-DNA insert in exon 13 (out of 14) and the protein thus may be able to create the proposed propeller structure characteristic for WDR proteins, but will lack the features of the C-terminal unstructured region.10,19,20 To investigate a putative role of WDR55 in the vegetative phase and whether the main requirement of WDR55 is restricted to gametophyte and seed development, we transferred the mutants to fresh S-2 agar plates to allow further growth. Generally, wdr55-2−/−seedlings continued to develop, but compared with a wild type pattern their morphology and development largely diverged. Development of mature mutant plants was marked by a plethora of developmental defects in both roots and aerial structures, indicating that WDR55 is not only required in the reproductive phase, but represents a more general mechanism required throughout plant development. We therefore analyzed the wdr55-2 vegetative phenotype.
wdr55 Seedlings have Variable Cotyledon Numbers
wdr55-2 homozygous seedlings were substantially smaller than their heterozygous siblings and germinated later than wild type. Mutant seedlings required 24 d of growth to reach a size similar to that of 5-d-old wild type seedlings (Fig. 1A and B). The normally highly symmetrical cotyledons were severely affected and present as a single, strongly reduced cotyledon (Fig. 1C), while others often had two asymmetric cotyledons (Fig. 1D–F), cotyledons fused together (Fig. 1F) or appeared in threes (Fig. 1B and G) or in fours (Fig. 1H). The seedlings were kept on MS-2 medium to allow generation of rosette leaves. In this respect the mutant plants were highly variable but most plants produced miniature rosettes with small, serrated leaves (Fig. 1H), while others developed into unorganized multi-leaf structures (Fig. 1J). The asymmetric and radially developing seedlings may be seen in connection with a failure of establishing embryo bilateral symmetry, as observed earlier.10 The cotyledon formation defects observed resemble auxin response mutants, such as mp (MONOPTEROS).21 We have previously shown that the direct MP target DORNROESCHEN (DRN), is ectopically expressed in wdr55-2 embryos and DRN has also been reported to act in cotyledon formation.10,22
Figure 1.wdr55−/−seedling and early vegetative phase defects. (A) Light micrographs of heterozygous (A) and (B–J) homozygous wdr55-2 seedlings grown on MS-2 plates. (A) is a 5-d-old wdr55-2+/− wild type seedling, while (B) is a 24 d old wdr55-2−/− mutant seedling grown under same conditions. (C) wdr55-2−/− mutant seedling with reduced cotyledons. This phenotype class often dies or develops into seedlings reminiscent of the two-cotyledons phenotype class (E and F). The two-cotyledons phenotypes are often very small (D) with asymmetric cotyledons (E) or appear to have two fused cotyledons (F, right cotyledon). The three-cotyledons phenotype (G) is mainly symmetric but also asymmetrical seedlings were found (data not shown). The four-cotyledons phenotype (H) have either asymmetric cotyledons (two small and two large) or are symmetric (data not shown). Entering the vegetative stage, various phenotypic effects can be observed (I–J); some appear dwarfed (but flower), while others fail to flower and develop into unorganized leafy balls. (I) wdr55-2−/− dwarf plant in the vegetative phase with serrated true leaves (rosette leaves). (J) Late vegetative phase wdr55-2−/− with multiple layers of leaves. Scale bar 1 mm.
The wdr55-2−/− roots were generally shorter than in wild type and appeared to have more root hairs (Fig. 1D and G, data not shown). To check root cell identity we crossed wdr55-02 to the GLABRA2 (GL2) reporter line pGL2:GUS, which is expressed in developing hairless epidermal cells of the root shortly after these cells are generated by the meristematic region.23 We found that the pGL2::GUS expression pattern in wdr55-2−/− background was similar to wild type, however the signal intensity appeared to be much weaker (Fig. 2). Both gl2 homozygous and heterozygous mutants have ectopic root hairs and GL2 is thought to be dosage dependent.23 The observed weaker expression in the mutant thus supports the notion that wdr55-2 roots have more or ectopic root hairs. Analysis of the GL2 promoter reporter activity also suggested that the meristematic zone in wdr55-2−/−is reduced compared with wild type, consistent with the reduced growth of the mutant roots (Fig. 2A and D).
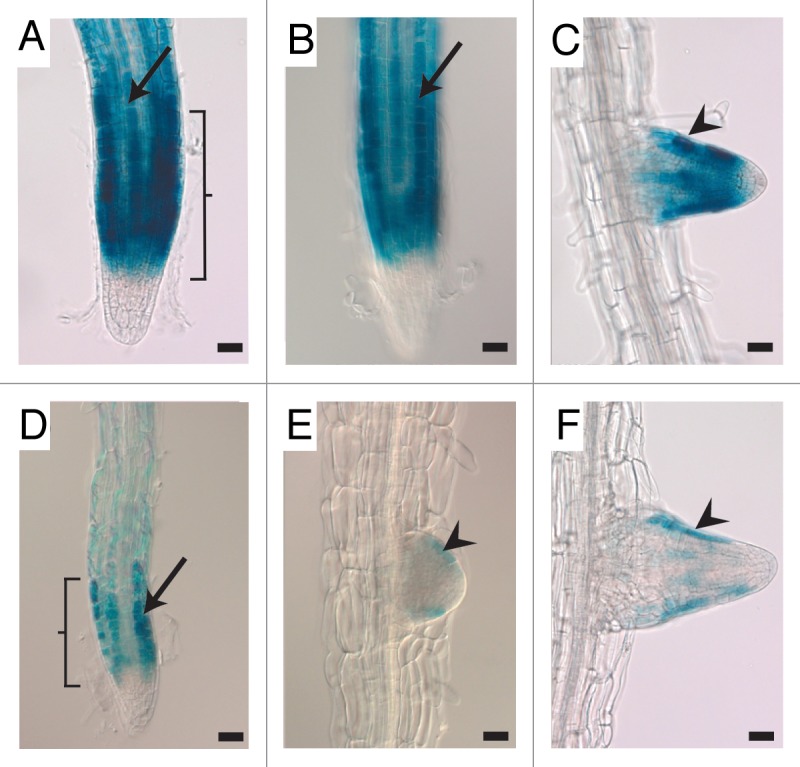
Figure 2.pGL2::GUS expression is reduced in wdr55-2−/−seedling roots. (A–F) Light micrographs of pGL2:GUS expression in wdr55-2+/− seedling roots (A–C) and in wdr55-2−/− seedling roots (D–F). The expression of pGL2:GUS in wild type wdr55-2+/− roots is found in differentiating epidermal cells located over tangential cortical cell walls which are hairless positions in both the main root (A and B, arrow) and in lateral roots (C, arrowhead). The pGL2:GUS expression in mutant wdr55-2−/− seedling roots is similarly located, but the expression level is much weaker in both the main root (D, arrow) and in lateral roots (E and F, arrowheads). Brackets in (A) and (D) marks the meristematic zone. GL2, GLABRA2. Scale bar 20 µm.
wdr55-2 Plants Display Reduced Apical Dominance
Developing and mature wdr55-2−/− plants were dwarfed and displayed various pleiotropic defects (Fig. 3). We noted that phyllotaxis appeared to diverge from a wild type pattern, but this was not examined in detail. Furthermore, the first rosette leaves were serrated, but lost this appearance in subsequent leaves. The internodes, the stems between two adjacent buds or leaf forming nodes in the wdr55-2 plants appeared to change route after each lateral shoot (Fig. 4A–C). In wild type, internodes grow in a straight path (Fig. 4B, left). wdr55-2 internodes, however, alternate directions after each node, giving rise to a Y-shaped appearance (Fig. 4D and E).

Figure 3. Comparison of wild type and wdr55-2−/− adult plants. (A–C) Adult wild type (A and C, right) and homozygous wdr55-2−/− plants (B and C, left). The wild type plant in (A) is 5-wk-old and the wdr55-2−/− plant is 11-wk-old (B). (C) Direct comparison of a homozygous wdr55-2−/− plant (left) and wild type (right) of the same age. The wdr55-2−/− plant remains dwarfed and bushy, due to many secondary shoots.

Figure 4. Internodes growth and lack of apical dominance in wdr55-2−/− adult plants. (A–E) Images of wdr55-2−/− plants, with comparison to wild type (B, right). (A) The adult wdr55-2−/− plant has a bushy appearance indicating reduced apical dominance and the growth path of the internodes change direction after each lateral shoot [compare wdr55-2−/− (B, left and C–E) and wild type (B, right)]. (F) Graph showing the average number of primary shoots, secondary shoots and number of inflorescences in 62–66-d-old wdr55-2−/− plants (still growing) compared with wild type 66-d-old Columbia wild type (senescent). wdr55-2−/− have an increased frequency of secondary shoots and inflorescences. The frequency of primary shoots is reduced (43%, see text for details).
The adult wdr55-2−/− plant was found to have an increased frequency of secondary shoots (Fig. 4A and F). Less than half of the plants display a clear primary shoot (43%; n, 14) and there was an increase in total number of inflorescences (Fig. 4F). The homozygous wdr55-2 plants and wild type Col plants used for this experiment were 66-d-old and while wdr55 plants still produced new flowers and secondary shoots, the wild type control was senescent. The frequency of secondary shoots and inflorescences is therefore rather an underestimate and considerably higher if plants representing the same developmental stage are compared (compare Figs. 3A and B).
Plant hormones are key players in setting up the plant body plan and mediate positional information and patterning in processes ranging from female gametogenesis and embryogenesis to floral organ development by the establishment of gradients.24,25 In the adult plant, the leading shoot dominates the growth of the auxiliary buds in the axialla of the petioles of the leaves below, a process known as apical dominance26. Notably, formation of secondary inflorescences in wild type plants is inhibited by auxin which is produced in the shoot apex of the primary inflorescence and transported downwards. Cytokinin is an activator of bud growth and might serve as an upward (basal-apical) mobile signal in the primary stem.3 Auxin and cytokinin act antagonistically and it is therefore hypothesized that auxin inhibits bud growth by restricting the amount of cytokinin transported to the buds. Auxin gradients specify apical-basal patterning and dynamic auxin fluxes pattern organ initiation at the shoot apex. Mutations that disrupt auxin transport or signaling and organ formation also disrupt axillary meristem formation, showing that auxin or its transport is required for axillary meristem formation, at least during floral growth.2
Most of the general visual appearance of homozygous wdr55-2 plants is due to reduced apical dominance. The loss of apical dominance is typically attributed to defect in auxin responseand was first thought to result from defects in groups of SCF (CUL1-based E3 ubiquitin ligases).27 The SCF is involved in auxin signaling by degrading Aux/IAAs in response to auxin, which releases the inhibition of ARFs.8 Reduced levels of the COP9 signalosome which regulates the E3 ligase activity, exhibits a decreased auxin response based on the observation of an increase in secondary inflorescences as well as a reduced plant size and a decrease in internode length.28 Similar apical dominance defects were observed in the cul4cs (CUL4 cosuppression) lines even though the CUL1 protein level was not affected. Consequently, CUL4 could also be involved in auxin response in a similar manner to CUL1.27 The loss of apical dominance in wdr55-2 may therefore be attributed to a function of WDR55 in a CUL4- DDB1 complex. In line with this, mutants of DCAF1 (which also has the WD × R motif and interacts with DDB1 and CUL4)11 also have pleiotropic defects and loss of apical dominance. However, in lack of direct evidence, this remains speculation. WDR55 could alternatively have a non-proteolytic function and be involved in chromatin regulation as suggested for the CUL4-DDB1 substrate receptor and PRC2 protein MSI1.18
In our previous work we have identified a requirement for WDR55 in seed development and plant reproduction.10 Here we report that WDR55 also is required for vegetative growth and development. The use of a weak allele to generate homozygous wdr55 plants opens new avenues for studying the role of WDR55 using high-throughput genomics methodology and may allow more straightforward identification of WDR55 interactors and substrates. WDR55 interacts with DDB1A10 and is thus likely to form a complex with CUL4. Given the involvement of WDR55 in both vegetative and reproductive development, WDR55-DDB1A is an interesting biological system for studying the function of the CUL4 complex. Finding potential targets of WDR55 and placing WDR55 in a genetic and biological network will hopefully further our insight into the compound role of CUL4 and therefore aid in our understanding of this form of regulation in both plants and mammals.
Materials and Methods
Plant material and general procedures
The wdr55-2 (WiscDsLox430F06) line is a T-DNA insertional mutant previously characterized in Bjerkan et. al.10 Seeds were surface sterilized with ethanol, bleach and Tween20 before transfer to MS media supplemented with 2% sucrose (MS-2). Seeds were stratified on MS-2 plates at 4°C overnight before incubation at 18°C under long day conditions (16 h.) until germination. Seedlings were then transferred to soil and grown under the same conditions as above.
DNA work and genotyping
Genomic DNA isolation was made using the SP Plant DNA kit (OMEGA bio-tek) according to the manufacturer’s instruction. Genotyping was performed using genomic and T-DNA insertion specific primers; ASP Wiscdslox 430F06 (5′-TTCCTCTTGCTGCCTGTAAAGC-3ʹ), SP Wiscdslox 430F06 (5ʹ-CAACAGAATCATACAGCCGATTGG-3ʹ) and p745 (5ʹ-AACGTCCGCAATGTGTTATTAAGTTGTC-3ʹ), as described previously.10
Histology and imaging
A Zeiss Axioplan Imaging2 microscope system equipped with Nomarski optics was used when examining tissue by light microscopy. Some sporophytic tissues were examined using a Leica WILD MZ8 stereo light microscope and images were captured using a Nikon Coolpix 995 mounted on the microscope. Histochemical GUS assays were performed by incubating sampled tissue in a substrate staining solution (0.05 M NaPO4, 0.1% Triton X-100, 2 mM K4Fe(CN)63H6O, 2 mM K3Fe(CN)6 and 2mM X-Gluc) overnight in the dark at 37°C. The substrate was then removed and the tissue was stored in 50% ethanol at 4°C. Slides were prepared by mounting the material directly in clearing solution (8:2:1, Chloral Hydrate:H2O:Glycerol) and inspected after at least 1 h incubation at 4°C.
Acknowledgments
We would like to thank Roy Falleth and Solveig Hauge Engebretsen for assistance. We thank our colleagues Reza Shirzadi, Ellen Dehnes Andersen and Veronica Gregis for careful reading and comments. We are grateful to Jonathan Bramsiepe for providing the pGL2:GUS line. This work was supported by a Norwegian Research Council FUGE Starting Grant (183190/S10) and a FRIPRO Grant (214052/F20) to PEG.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/25347
References
- 1.Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–7. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- 2.McSteen P, Leyser O. Shoot branching. Annu Rev Plant Biol. 2005;56:353–74. doi: 10.1146/annurev.arplant.56.032604.144122. [DOI] [PubMed] [Google Scholar]
- 3.Leyser O. The fall and rise of apical dominance. Curr Opin Genet Dev. 2005;15:468–71. doi: 10.1016/j.gde.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Saini S, Sharma I, Kaur N, Pati PK. Auxin: a master regulator in plant root development. Plant Cell Rep. 2013;32:741–57. doi: 10.1007/s00299-013-1430-5. [DOI] [PubMed] [Google Scholar]
- 5.Lau S, Slane D, Herud O, Kong J, Jürgens G. Early embryogenesis in flowering plants: setting up the basic body pattern. Annu Rev Plant Biol. 2012;63:483–506. doi: 10.1146/annurev-arplant-042811-105507. [DOI] [PubMed] [Google Scholar]
- 6.Song J, Angel A, Howard M, Dean C. Vernalization - a cold-induced epigenetic switch. J Cell Sci. 2012;125:3723–31. doi: 10.1242/jcs.084764. [DOI] [PubMed] [Google Scholar]
- 7.Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10:385–97. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 8.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–90. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 9.Dreher K, Callis J. Ubiquitin, hormones and biotic stress in plants. Ann Bot. 2007;99:787–822. doi: 10.1093/aob/mcl255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjerkan KN, Jung-Roméo S, Jürgens G, Genschik P, Grini PE. Arabidopsis WD repeat domain55 Interacts with DNA damaged binding protein1 and is required for apical patterning in the embryo. Plant Cell. 2012;24:1013–33. doi: 10.1105/tpc.111.089425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Feng S, Chen F, Chen H, Wang J, McCall C, et al. Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell. 2008;20:1437–55. doi: 10.1105/tpc.108.058891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–3. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 13.He YJ, McCall CM, Hu J, Zeng Y, Xiong Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 2006;20:2949–54. doi: 10.1101/gad.1483206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernhardt A, Lechner E, Hano P, Schade V, Dieterle M, Anders M, et al. CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. Plant J. 2006;47:591–603. doi: 10.1111/j.1365-313X.2006.02810.x. [DOI] [PubMed] [Google Scholar]
- 15.Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, et al. Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell. 2010;22:108–23. doi: 10.1105/tpc.109.065490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012;17:584–93. doi: 10.1016/j.tplants.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Pazhouhandeh M, Molinier J, Berr A, Genschik P. MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:3430–5. doi: 10.1073/pnas.1018242108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dumbliauskas E, Lechner E, Jaciubek M, Berr A, Pazhouhandeh M, Alioua M, et al. The Arabidopsis CUL4-DDB1 complex interacts with MSI1 and is required to maintain MEDEA parental imprinting. EMBO J. 2011;30:731–43. doi: 10.1038/emboj.2010.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/S1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 20.Ishida T, Kinoshita K. Prediction of disordered regions in proteins based on the meta approach. Bioinformatics. 2008;24:1344–8. doi: 10.1093/bioinformatics/btn195. [DOI] [PubMed] [Google Scholar]
- 21.Berleth T, Jurgens G. The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development. 1993;118:575–87. [Google Scholar]
- 22.Cole M, Chandler J, Weijers D, Jacobs B, Comelli P, Werr W. DORNROSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development. 2009;136:1643–51. doi: 10.1242/dev.032177. [DOI] [PubMed] [Google Scholar]
- 23.Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, et al. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development. 1996;122:1253–60. doi: 10.1242/dev.122.4.1253. [DOI] [PubMed] [Google Scholar]
- 24.Alabadí D, Blázquez MA, Carbonell J, Ferrándiz C, Pérez-Amador MA. Instructive roles for hormones in plant development. Int J Dev Biol. 2009;53:1597–608. doi: 10.1387/ijdb.072423da. [DOI] [PubMed] [Google Scholar]
- 25.Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136:1005–16. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Napoli CA, Beveridge CA, Snowden KC. Reevaluating concepts of apical dominance and the control of axillary bud outgrowth. Curr Top Dev Biol. 1999;44:127–69. doi: 10.1016/S0070-2153(08)60469-X. [DOI] [PubMed] [Google Scholar]
- 27.Chen H, Shen Y, Tang X, Yu L, Wang J, Guo L, et al. Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell. 2006;18:1991–2004. doi: 10.1105/tpc.106.043224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, et al. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science. 2001;292:1379–82. doi: 10.1126/science.1059776. [DOI] [PubMed] [Google Scholar]



