Abstract
Legume plants are able to establish root nodule symbioses with nitrogen-fixing bacteria, called rhizobia. Recent studies revealed that the root nodule symbiosis has co-opted the signaling pathway that mediates the ancestral mycorrhizal symbiosis that occurs in most land plants. Despite being unable to induce nodulation, rhizobia have been shown to be able to infect and colonize the roots of non-legumes such as rice. One fascinating question is whether establishment of such associations requires the common symbiosis (Sym) genes that are essential for infection of plant cells by mycorrhizal fungi and rhizobia in legumes. Here, we demonstrated that the common Sym genes are not required for endophytic colonization of rice roots by nitrogen-fixing rhizobia.
Keywords: rice, rhizobia, nitrogen fixation, legume, common symbiosis genes
The nitrogen-fixing bacteria, called rhizobia, are able to infect the roots of leguminous plants and induce the formation of root nodules. Within the root nodule, the bacteria can convert atmospheric nitrogen into ammonia, a biological form that can be directly used by the plant. Over the past decade, tremendous progress has been made in our understanding of the nodulation signaling pathway in legumes.1 It has become increasingly evident that the root nodule symbiosis has co-opted the signaling pathway that mediates the ancestral mycorrhizal symbiosis, a widespread mutualistic association between mycorrhizal fungi and the vast majority of land plants.1-5 As such, most, if not all, legume genes required for root nodule symbiosis are already present in non-legumes, and these non-legume genes have been shown to function similarly to their legume counterparts.2,6 These discoveries have reignited an old dream of plant biologists, i.e., to transfer the nitrogen-fixing symbiosis to cereals and other non-leguminous crops.4,7
Despite being unable to induce nodulation, rhizobia have been shown to be able to infect and colonize the roots of non-legumes such as rice (Oryza sativa).8-10 This so-called “endophytic” interaction also can promote plant growth, even though the exact mechanisms for such positive responses are not well understood.9,11 Characterization of the infection and colonization processes of the rice-rhizobial association revealed that the bacteria primarily enter the plant tissue through root hairs and/or crack entry located near the sites of newly emerging lateral roots.10 Similar infection strategies are also used by the rhizobia to enter the roots of the non-legume Paraponia and the water-tolerant legume Sesbania rostrata, both of which can nodulate with rhizobia.12-16 Intriguingly, similar to what occur in legumes, both root hair curling and infection thread-like structures were also observed on the rice roots inoculated with rhizobia.10 However, in contrast to the root nodule symbiosis, in which bacteria colonize intracellularly, bacteria mainly reside in the intercellular spaces within the rice roots.10
Given the seeming similarity of the infection processes, one fascinating question is whether the crack entry or root hair infection of rice by rhizobia requires the common symbiosis (Sym) genes that are essential for infection of plant cells by mycorrhizal fungi and rhizobia in legumes.2 To address this question, we examined the infection and colonization phenotypes of wild-type plants and the symbiosis-defective mutants representing three different common Sym genes. These mutants include the retrotransposon Tos17 insertion lines Os-dmi3 (NF8513) and Os-cyclops (NC2794) and the T-DNA insertion line Os-castor (C04353). All these rice mutants have been shown to be impaired in mycorrhizal symbiosis.17-20 Of the 3 common Sym genes (for review see the ref. 2), Os-CASTOR encodes a nuclear-localized potassium channel protein; Os-DMI3 codes for a Ca2+/calmodulin-dependent protein kinase (CCaMK); and Os-CYCLOPS encodes a protein that interacts with and phophosphorylated by DMI3.21
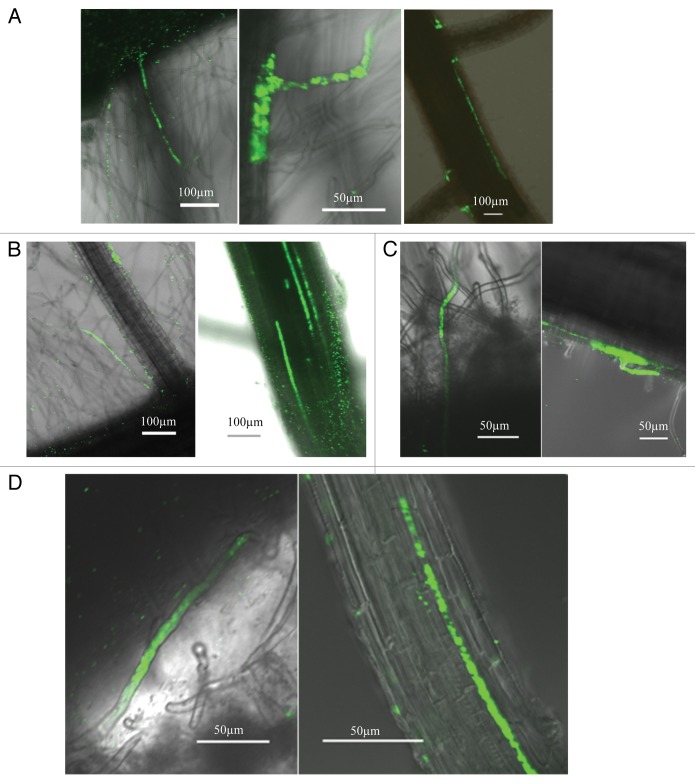
We inoculated wild-type and mutant plants with the GFP-tagged Rhizobium leguminosarum bv. trifolii strain R4 under hydrophobic conditions following the protocol described by Perrine-Walker et al.10 We examined the early infection processes 48 h post inoculation as well as the late colonization events 21 d post inoculation using an Olympus FV1000 confocal microscope. Infection of rice roots could be readily observed 48 h post inoculation for both wild-type and mutant plants, with GFP-labeled bacteria present in the root hairs or attaching to the cracks of emerging lateral roots (Fig. 1). At 21 d post inoculation, we could observe the spreading of GFP-labeled bacteria into intercellular spaces of rice root tissues forming short or long lines of GFP-tagged cells (Fig. 1). There were no distinguishable differences observed between the wild-type and mutant plants. Thus, our experiments suggest that the common Sym genes are not involved in the infection and endophytic colonization of rice roots by rhizobia.
Figure 1. Infection and colonization of rice roots of wild-type and mutant plants by GFP-tagged Rhizobium leguminosarum bv. trifolii strain R4. (A) Infection and colonization of rice roots of the wild-type genotype, Nipponbare, showing root hair infection (left), crack entry followed by intercellular colonization (right), and root hair infection followed by the colonization of the intercellular space between the epidemics and the underlying cortex (middle).( B–D) Infection and colonization of rice roots of mutants Os-castor (B), Os-dmi3 (C) and Os-cyclops (D). For each panel, root hair infection 48 h post inoculation was shown on the left and the intercellular colonization of rhizobia observed 21 d post inoculation was shown on the right. Three independent experiments were performed, and at least 25 plants per genotype were tested for each experiment.
Intracellular invasion through infection thread formation in root hairs is a common strategy that rhizobia use to enter the roots of legumes; however, many legumes also retain more primitive intercellular infection strategies to cope with adverse conditions that prevent intracellular invasion.16 The best studied example is the semi-aquatic legume Sesbania rostrata, in which both intercellular and intracellular infection strategies are adopted.15,16 Under waterlogged conditions, bacteria colonize epidermal fissures at lateral root bases and trigger cortical cell death for infection pocket formation. The infection pockets function to facilitate initial intercellular and later intracellular infection thread progression toward the nodule primordium. In this case the common Sym genes were not required for the initial infection pocket formation but are essential for root hair infection and for later nodule organogenesis.13,15 Our observation that the bacteria can colonize the intercellular spaces of both wild-type and mutant rice roots appears to be consistent with that reported in Sesbania rostrata. However, a perplexing question remains: if the infection of rice root hairs resembles infection thread formation in legumes, why common Sym genes are not required for this process? One possibility is that the linear arrangement of bacteria in root hairs is structurally different from infection threads observed in legumes. In contrast to legumes where bacteria are confined to the infection threads until they reached the nodule primordium, in rice roots the whole root hairs were infected, suggesting that the infection thread-like structure observed in rice roots are different with infection threads in legume roots.10 In this case, the rhizobia may enter into the plant tissue through the killing of adjacent epidermal cells or root hairs.22
In summary, rice knockout mutants for DMI3 (CCaMK), CASTOR, and CYCLOPS, which are defective in mycorrhizal symbioses were all successfully colonized by the GFP-tagged Rhizobium leguminosarum bv. trifolii strain R4. Thus, our data suggest that common Sym genes are not required for infection and endophytic colonization of rice roots by nitrogen-fixing rhizobia. This conclusion appears to be supported by the observation that the Sym plasmids are not required for intercellular colonization of rice roots by rhizobia.22 One caveat of our study, however, is that we only used one bacteria strain and it is not clear if there exists strain specificity in the requirement of common Sym genes in endophytic rice-rhizobial associations.
Acknowledgments
The authors thank Barry G. Rolfe, Australian National University, for providing the GFP-tagged Rhizobium leguminosarum bv. trifolii strain R4. This work was supported by the Kentucky Science and Engineering Foundation and by the US. National Science Foundation (grant no. IOS 0640197 to HZ).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/25453
References
- 1.Oldroyd GE. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol. 2013;11:252–63. doi: 10.1038/nrmicro2990. [DOI] [PubMed] [Google Scholar]
- 2.Parniske M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol. 2008;6:763–75. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- 3.Op den Camp R, Streng A, De Mita S, Cao Q, Polone E, Liu W, et al. LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science. 2011;331:909–12. doi: 10.1126/science.1198181. [DOI] [PubMed] [Google Scholar]
- 4.Geurts R, Lillo A, Bisseling T. Exploiting an ancient signalling machinery to enjoy a nitrogen fixing symbiosis. Curr Opin Plant Biol. 2012;15:438–43. doi: 10.1016/j.pbi.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Gobbato E, Marsh JF, Vernié T, Wang E, Maillet F, Kim J, et al. A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr Biol. 2012;22:2236–41. doi: 10.1016/j.cub.2012.09.044. [DOI] [PubMed] [Google Scholar]
- 6.Zhu H, Riely BK, Burns NJ, Ané JM. Tracing nonlegume orthologs of legume genes required for nodulation and arbuscular mycorrhizal symbioses. Genetics. 2005;172:2491–9. doi: 10.1534/genetics.105.051185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charpentier M, Oldroyd G. How close are we to nitrogen-fixing cereals? Curr Opin Plant Biol. 2010;13:556–64. doi: 10.1016/j.pbi.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Prayitno J, Stefaniak J, McIver J, Weinman JJ, Dazzo FB, Ladha JK, et al. Interactions of rice seedlings with bacteria isolated from rice roots. Aust J Plant Physiol. 1999;26:521–35. doi: 10.1071/PP98090. [DOI] [Google Scholar]
- 9.Chi F, Shen S-H, Cheng H-P, Jing Y-X, Yanni YG, Dazzo FB. Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl Environ Microbiol. 2005;71:7271–8. doi: 10.1128/AEM.71.11.7271-7278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perrine-Walker FM, Prayitno J, Rolfe BG, Weinman JJ, Hocart CH. Infection process and the interaction of rice roots with rhizobia. J Exp Bot. 2007;58:3343–50. doi: 10.1093/jxb/erm181. [DOI] [PubMed] [Google Scholar]
- 11.Yanni YG, Rizk RY, Corich V, Squartini A, Ninke K, Philip-Hollingsworth S, et al. Natural endophytic association between Rhizobium leguminosarum bv. trifolii and rice roots and assessments of its potential to promote rice growth. Plant Soil. 1997;194:99–114. doi: 10.1023/A:1004269902246. [DOI] [Google Scholar]
- 12.Bender GL, Nayudu M, Goydych M, Rolfe BG. Early infection events in the nodulation of the non-legume Paraponia andersonii by Bradyrhizobium. Plant Sci. 1987;51:285–93. doi: 10.1016/0168-9452(87)90205-6. [DOI] [Google Scholar]
- 13.Capoen W, Goormachtig S, De Rycke R, Schroeyers K, Holsters M. SrSymRK, a plant receptor essential for symbiosome formation. Proc Natl Acad Sci U S A. 2005;102:10369–74. doi: 10.1073/pnas.0504250102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capoen W, Den Herder J, Rombauts S, De Gussem J, De Keyser A, Holsters M, et al. Comparative transcriptome analysis reveals common and specific tags for root hair and crack-entry invasion in Sesbania rostrata. Plant Physiol. 2007;144:1878–89. doi: 10.1104/pp.107.102178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capoen W, Den Herder J, Sun J, Verplancke C, De Keyser A, De Rycke R, et al. Calcium spiking patterns and the role of the calcium/calmodulin-dependent kinase CCaMK in lateral root base nodulation of Sesbania rostrata. Plant Cell. 2009;21:1526–40. doi: 10.1105/tpc.109.066233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capoen W, Oldroyd G, Goormachtig S, Holsters M. Sesbania rostrata: a case study of natural variation in legume nodulation. New Phytol. 2010;186:340–5. doi: 10.1111/j.1469-8137.2009.03124.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen C, Gao M, Liu J, Zhu H. Fungal symbiosis in rice requires an ortholog of a legume common symbiosis gene encoding a Ca2+/calmodulin-dependent protein kinase. Plant Physiol. 2007;145:1619–28. doi: 10.1104/pp.107.109876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C, Ané JM, Zhu H. OsIPD3, an ortholog of the Medicago truncatula DMI3 interacting protein IPD3, is required for mycorrhizal symbiosis in rice. New Phytol. 2008;180:311–5. doi: 10.1111/j.1469-8137.2008.02612.x. [DOI] [PubMed] [Google Scholar]
- 19.Chen C, Fan C, Gao M, Zhu H. Antiquity and function of CASTOR and POLLUX, the twin ion channel-encoding genes key to the evolution of root symbioses in plants. Plant Physiol. 2009;149:306–17. doi: 10.1104/pp.108.131540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutjahr C, Banba M, Croset V, An K, Miyao A, An G, et al. Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell. 2008;20:2989–3005. doi: 10.1105/tpc.108.062414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yano K, Yoshida S, Müller J, Singh S, Banba M, Vickers K, et al. CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci U S A. 2008;105:20540–5. doi: 10.1073/pnas.0806858105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perrine FM, Prayitno J, Weinman JJ, Dazzo FB, Rolfe BG. Rhizobium plasmids are involved in the inhibition or stimulation of rice growth and development. Aust J Plant Physiol. 2001;28:923–37. [Google Scholar]